Beyond the List: Bioagent-Agnostic Signatures Could Enable a More Flexible and Resilient Biodefense Posture Than an Approach Based on Priority Agent Lists Alone
Abstract
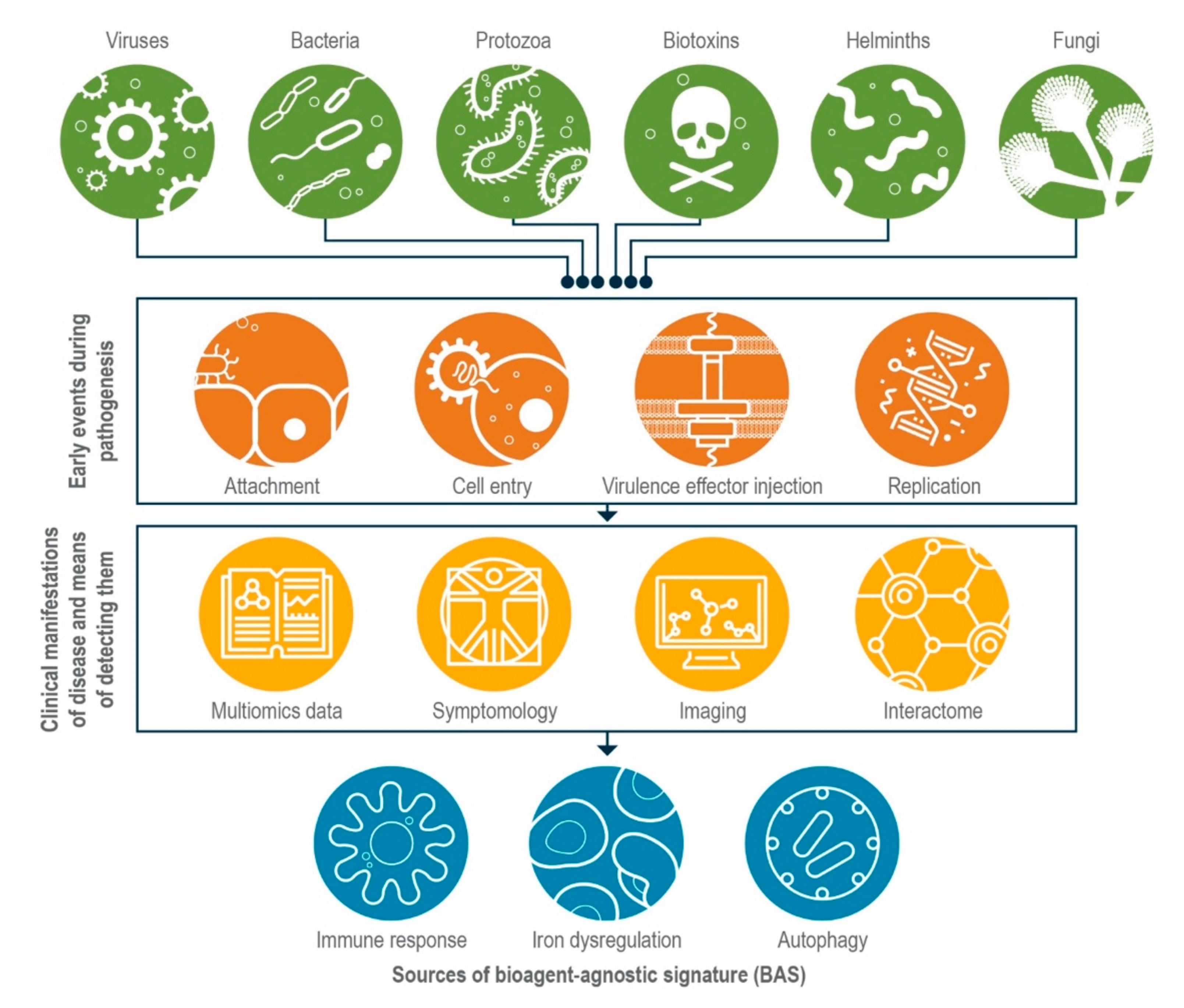
:1. Introduction
2. Common Themes in Host Response Could Form the Basis for Establishing BASs
2.1. Host Innate Immune Response during Infection
2.2. Dysregulation of Iron Homeostasis during Infection
2.3. Autophagy
2.4. Exploiting the Interactome
3. As They Mature, Multi-Omics Approaches May Reveal More BASs
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Select Agents and Toxins List. Available online: https://www.selectagents.gov/sat/list.htm (accessed on 4 August 2021).
- NIAID Emerging Infectious Diseases/Pathogens. Available online: https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens (accessed on 2 November 2021).
- Tacconelli, E.; Magrini, N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 2 November 2021).
- National Academies of Sciences, Engineering, and Medicine. Biodefense in the Age of Synthetic Biology; The National Academies Press: Washington, DC, USA, 2018; p. 188. [Google Scholar]
- Eisfeld, A.J.; Halfmann, P.J.; Wendler, J.P.; Kyle, J.E.; Burnum-Johnson, K.; Peralta, Z.; Maemura, T.; Walters, K.B.; Watanabe, T.; Fukuyama, S.; et al. Multi-platform ’Omics Analysis of Human Ebola Virus Disease Pathogenesis. Cell Host Microbe 2017, 22, 817–829. [Google Scholar] [CrossRef] [Green Version]
- Kyle, J.E.; Burnum-Johnson, K.E.; Wendler, J.P.; Eisfeld, A.J.; Halfmann, P.J.; Watanabe, T.; Sahr, F.; Smith, R.D.; Kawaoka, Y.; Waters, K.M.; et al. Plasma lipidome reveals critical illness and recovery from human Ebola virus disease. Proc. Natl. Acad. Sci. USA 2019, 116, 3919–3928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altman, S.; Bassler, B.L.; Beckwith, J.; Belfort, M.; Berg, H.C.; Bloom, B.; Brenchley, J.E.; Campbell, A.; Collier, R.J.; Connell, N.; et al. An Open Letter to Elias Zerhouni. Science 2005, 307, 1409–1410. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Zerhouni, E.A. NIH Response to Open Letter. Science 2005, 308, 49. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, B.; Qing, X.; Liao, J.; Zhuo, K. Role of Protein Glycosylation in Host-Pathogen Interaction. Cells 2020, 9, 1022. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Nayak, L.; De, R.K. A review on host–pathogen interactions: Classification and prediction. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1581–1599. [Google Scholar] [CrossRef]
- Lopez-Lopez, N.; Gil-Campillo, C.; Diez-Martinez, R.; Garmendia, J. Learning from -omics strategies applied to uncover Haemophilus influenzae host-pathogen inter-actions: Current status and perspectives. Comput. Struct. Biotechnol. J. 2021, 19, 3042–3050. [Google Scholar] [CrossRef]
- Neik, T.X.; Amas, J.; Barbetti, M.; Edwards, D.; Batley, J. Understanding Host–Pathogen Interactions in Brassica napus in the Omics Era. Plants 2020, 9, 1336. [Google Scholar] [CrossRef]
- Turner, A.W.; Wong, D.; Khan, M.D.; Dreisbach, C.N.; Palmore, M.; Miller, C.L. Multi-Omics Approaches to Study Long Non-coding RNA Function in Atherosclerosis. Front. Cardiovasc. Med. 2019, 6. [Google Scholar] [CrossRef]
- Melby, P.C.; Stephens, R.; Dann, S.M. 30—Host Defenses to Protozoa. In Clinical Immunology, 5th ed.; Rich, R.R., Thomas, A.F., William, T.S., Harry, W.S., Jr., Anthony, J.F., Cornelia, M.W., Eds.; Elsevier: London, UK, 2019; pp. 425–435.e1. [Google Scholar]
- Motran, C.C.; Silvane, L.; Chiapello, L.S.; Theumer, M.G.; Ambrosio, L.F.; Volpini, X.; Celias, D.P.; Cervi, L. Helminth Infections: Recognition and Modulation of the Immune Response by Innate Immune Cells. Front. Immunol. 2018, 9, 664. [Google Scholar] [CrossRef] [PubMed]
- Bruno, M.; Dewi, I.M.; Matzaraki, V.; ter Horst, R.; Pekmezovic, M.; Rösler, B.; Groh, L.; Röring, R.J.; Kumar, V.; Li, Y.; et al. Comparative host transcriptome in response to pathogenic fungi identifies common and species-specific transcriptional antifungal host response pathways. Comput. Struct. Biotechnol. J. 2020, 19, 647–663. [Google Scholar] [CrossRef]
- Ali, S.R.; Timmer, A.M.; Bilgrami, S.; Park, E.J.; Eckmann, L.; Nizet, V.; Karin, M. Anthrax Toxin Induces Macrophage Death by p38 MAPK Inhibition but Leads to Inflammasome Activation via ATP Leakage. Immunity 2011, 35, 34–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasper, L.; König, A.; Koenig, P.-A.; Gresnigt, M.S.; Westman, J.; Drummond, R.A.; Lionakis, M.S.; Groß, O.; Ruland, J.; Naglik, J.; et al. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat. Commun. 2018, 9, 4260. [Google Scholar] [CrossRef]
- Pons, B.J.; Pettes-Duler, A.; Naylies, C.; Taieb, F.; Bouchenot, C.; Hashim, S.; Rouimi, P.; Deslande, M.; Lippi, Y.; Mirey, G.; et al. Chronic exposure to Cytolethal Distending Toxin (CDT) promotes a cGAS-dependent type I interferon response. Cell Mol. Life Sci. 2021, 78, 6319–6335. [Google Scholar] [CrossRef] [PubMed]
- Iriti, M.; Faoro, F. Review of innate and specific immunity in plants and animals. Mycopathologia 2007, 164, 57–64. [Google Scholar] [CrossRef]
- Liu, Q.; Rao, Y.; Tian, M.; Zhang, S.; Feng, P. Modulation of Innate Immune Signaling Pathways by Herpesviruses. Viruses 2019, 11, 572. [Google Scholar] [CrossRef] [Green Version]
- Hohl, T.M.; Rivera, A.; Lipuma, L.; Gallegos, A.; Shi, C.; Mack, M.; Pamer, E.G. Inflammatory Monocytes Facilitate Adaptive CD4 T Cell Responses during Respiratory Fungal Infection. Cell Host Microbe 2009, 6, 470–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, M.; Bhatia, S.; Oriss, T.B.; Yarlagadda, M.; Khare, A.; Akira, S.; Saijo, S.; Iwakura, Y.; Junecko, B.A.; Reinhart, T.A.; et al. TNF- from inflammatory dendritic cells (DCs) regulates lung IL-17A/IL-5 levels and neutrophilia versus eosinophilia during persistent fungal infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5360–5365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagundes, C.T.; Amaral, F.A.; Vieira, A.T.; Soares, A.C.; Pinho, V.; Nicoli, J.R.; Vieira, L.Q.; Teixeira, M.M.; Souza DGTransient, T.L.R. Transient TLR Activation Restores Inflammatory Response and Ability To Control Pulmonary Bacterial Infection in Germfree Mice. J. Immunol. 2011, 188, 1411–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.; Gruenbaum, A.; Kanashova, T.; Mertins, P.; Cluzel, P.; Chevrier, N. Pairwise Stimulations of Pathogen-Sensing Pathways Predict Immune Responses to Multi-adjuvant Combinations. Cell Syst. 2020, 11, 495–508. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Persson, E.K.; Uronen-Hansson, H.; Semmrich, M.; Rivollier, A.; Hägerbrand, K.; Marsal, J.; Gudjonsson, S.; Håkansson, U.; Reizis, B.; Kotarsky, K.; et al. IRF4 Transcription-Factor-Dependent CD103+CD11b+ Dendritic Cells Drive Mucosal T Helper 17 Cell Differentiation. Immunity 2013, 38, 958–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlitzer, A.; McGovern, N.; Teo, P.; Zelante, T.; Atarashi, K.; Low, D.; Ho, A.W.; See, P.; Shin, A.; Wasan, P.S.; et al. IRF4 Transcription Factor-Dependent CD11b+ Dendritic Cells in Human and Mouse Control Mucosal IL-17 Cytokine Responses. Immunity 2013, 38, 970–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torchinsky, M.B.; Garaude, J.; Martin, A.P.; Blander, J.M. Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature 2009, 458, 78–82. [Google Scholar] [CrossRef]
- Mashayekhi, M.; Sandau, M.M.; Dunay, I.R.; Frickel, E.M.; Khan, A.; Goldszmid, R.S.; Sher, A.; Ploegh, H.L.; Murphy, T.L.; Sibley, L.D.; et al. CD8α+ Dendritic Cells Are the Critical Source of Interleukin-12 that Controls Acute Infection by Toxoplasma gondii Tachyzoites. Immunity 2011, 35, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelson, B.T.; Bradstreet, T.R.; Hildner, K.; Carrero, J.A.; Frederick, K.E.; Wumesh, K.C.; Belizaire, R.; Aoshi, T.; Schreiber, R.D.; Miller, M.J.; et al. CD8α+ Dendritic Cells Are an Obligate Cellular Entry Point for Productive Infection by Listeria monocytogenes. Immunity 2011, 35, 236–248. [Google Scholar] [CrossRef] [Green Version]
- Thakur, A.; Mikkelsen, H.; Jungersen, G. Intracellular Pathogens: Host Immunity and Microbial Persistence Strategies. J. Immunol. Res. 2019, 2019, 1356540. [Google Scholar] [CrossRef]
- Hieny, S.; Scharton-Kersten, T.; Jankovic, D.; Charest, H.; Germain, R.N.; Sher, A. In Vivo Microbial Stimulation Induces Rapid CD40 Ligand–independent Production of Interleukin 12 by Dendritic Cells and their Redistribution to T Cell Areas. J. Exp. Med. 1997, 186, 1819–1829. [Google Scholar] [CrossRef] [Green Version]
- Dalod, M.; Salazar-Mather, T.P.; Malmgaard, L.; Lewis, C.; Asselin-Paturel, C.; Briere, F.; Trinchieri, G.; Biron, C.A. Interferon alpha/beta and interleukin 12 responses to viral infections: Pathways regulating dendritic cell cytokine expression in vivo. J. Exp. Med. 2002, 195, 517–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Keitany, G.; Li, Y.; Wang, Y.; Ball, H.L.; Goldsmith, E.J.; Orth, K. Yersinia YopJ Acetylates and Inhibits Kinase Activation by Blocking Phosphorylation. Science 2006, 312, 1211–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baruch, K.; Gur-Arie, L.; Nadler, C.; Koby, S.; Yerushalmi, G.; Ben-Neriah, Y.; Yogev, O.; Shaulian, E.; Guttman, C.; Zarivach, R. Metalloprotease type III effectors that specifically cleave JNK and NF-kappa B. Embo J. 2011, 30, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Wang, M.; Liang, S.; Triantafilou, M.; Triantafilou, K. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc. Natl. Acad. Sci. USA 2008, 105, 13532–13537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbibe, L.; Kim, D.W.; Batsche, E.; Pedron, T.; Mateescu, B.; Muchardt, C.; Parsot, C.; Sansonetti, P.J. An injected bacterial effector targets chromatin access for transcription factor NF-kappa B to alter transcription of host genes involved in immune responses. Nat. Immunol. 2007, 8, 47–56. [Google Scholar] [CrossRef]
- Jensen, S.; Thomsen, A.R. Sensing of RNA Viruses: A Review of Innate Immune Receptors Involved in Recognizing RNA Virus Invasion. J. Virol. 2012, 86, 2900–2910. [Google Scholar] [CrossRef] [Green Version]
- Stanifer, M.L.; Guo, C.; Doldan, P.; Boulant, S. Importance of Type I and III Interferons at Respiratory and Intestinal Barrier Surfaces. Front. Immunol. 2020, 11, 608645. [Google Scholar] [CrossRef] [PubMed]
- Negishi, H.; Taniguchi, T.; Yanai, H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb. Perspect. Biol. 2017, 10, a028423. [Google Scholar] [CrossRef]
- Tau, G.; Rothman, P. Biologic functions of the IFN-gamma receptors. Allergy 1999, 54, 1233–1251. [Google Scholar] [CrossRef]
- Holdsworth, S.R.; Gan, P.Y. Cytokines: Names and Numbers You Should Care About. Clin. J. Am. Soc. Nephrol. 2015, 10, 2243–2254. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2013, 14, 36–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upton, J.W.; Chan, F.K. Staying Alive: Cell Death in Antiviral Immunity. Mol. Cell 2014, 54, 273–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brune, W. Inhibition of programmed cell death by cytomegaloviruses. Virus Res. 2011, 157, 144–150. [Google Scholar] [CrossRef]
- Guo, H.; Kaiser, W.J.; Mocarski, E.S. Manipulation of apoptosis and necroptosis signaling by herpesviruses. Z. Hyg. Infekt. 2015, 204, 439–448. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; Liu, M.; Zhao, X.; Jia, R.; et al. Programmed cell death: The battlefield between the host and alpha-herpesviruses and a potential avenue for cancer treatment. Oncotarget 2018, 9, 30704–30719. [Google Scholar] [CrossRef]
- Menachery, V.; Eisfeld, A.J.; Schäfer, A.; Josset, L.; Sims, A.C.; Proll, S.; Fan, S.; Li, C.; Neumann, G.; Tilton, S.C.; et al. Pathogenic Influenza Viruses and Coronaviruses Utilize Similar and Contrasting Approaches to Control Interferon-Stimulated Gene Responses. mBio 2014, 5, e01174-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sims, A.C.; Tilton, S.C.; Menachery, V.; Gralinski, L.E.; Schäfer, A.; Matzke, M.M.; Webb-Robertson, B.-J.; Chang, J.; Luna, M.L.; Long, C.E.; et al. Release of Severe Acute Respiratory Syndrome Coronavirus Nuclear Import Block Enhances Host Transcription in Human Lung Cells. J. Virol. 2013, 87, 3885–3902. [Google Scholar] [CrossRef] [Green Version]
- Cassat, J.E.; Skaar, E.P. Iron in Infection and Immunity. Cell Host Microbe 2013, 13, 509–519. [Google Scholar] [CrossRef] [Green Version]
- Andrews, N.C. Medical progress: Disorders of iron metabolism. N. Engl. J. Med. 1999, 341, 1986–1995. [Google Scholar] [CrossRef]
- Skaar, E.P. The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts. PLoS Pathog. 2010, 6, e1000949. [Google Scholar] [CrossRef]
- Zughaier, S.M.; Cornelis, P. Editorial: Role of Iron in Bacterial Pathogenesis. Front. Cell. Infect. Microbiol. 2018, 8, 344. [Google Scholar] [CrossRef] [Green Version]
- Di Patti, M.C.; Cutone, A.; Polticelli, F.; Rosa, L.; Lepanto, M.S.; Valenti, P.; Musci, G. The ferroportin-ceruloplasmin system and the mammalian iron homeostasis machine: Regulatory pathways and the role of lactoferrin. Biometals 2018, 31, 399–414. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Singh, P.; Srivastava, A. Synthesis, nature and utility of universal iron chelator—Siderophore: A review. Microbiol. Res. 2018, 212–213, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Kramer, J.; Oezkaya, O.; Kuemmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Debarbieux, L.; Wandersman, C. Hemophore-Dependent Heme Acquisition Systems, in Iron Transport in Bacteria; ASM Press: Washington, DC, USA, 2004; pp. 38–47. [Google Scholar]
- Richard, K.L.; Kelley, B.R.; Johnson, J. Heme Uptake and Utilization by Gram-Negative Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2019, 9, 81. [Google Scholar] [CrossRef]
- Ratledge, C.; Dover, L.G. Iron Metabolism in Pathogenic Bacteria. Annu. Rev. Microbiol. 2000, 54, 881–941. [Google Scholar] [CrossRef] [PubMed]
- Goetz, D.H.; Holmes, M.A.; Borregaard, N.; Bluhm, M.E.; Raymond, K.N.; Strong, R.K. The Neutrophil Lipocalin NGAL Is a Bacteriostatic Agent that Interferes with Siderophore-Mediated Iron Acquisition. Mol. Cell 2002, 10, 1033–1043. [Google Scholar] [CrossRef]
- Nairz, M.; Ferring-Appel, D.; Casarrubea, D.; Sonnweber, T.; Viatte, L.; Schroll, A.; Haschka, D.; Fang, F.C.; Hentze, M.W.; Weiss, G.; et al. Iron Regulatory Proteins Mediate Host Resistance to Salmonella Infection. Cell Host Microbe 2015, 18, 254–261. [Google Scholar] [CrossRef] [Green Version]
- Barton, J.C.; Acton, R.T. Hepcidin, iron, and bacterial infection. Vitam. Horm. 2019, 110, 223–242. [Google Scholar]
- Wessling-Resnick, M. Crossing the Iron Gate: Why and How Transferrin Receptors Mediate Viral Entry. Annu. Rev. Nutr. 2018, 38, 431–458. [Google Scholar] [CrossRef] [PubMed]
- Bellmann-Weiler, R.; Lanser, L.; Barket, R.; Rangger, L.; Schapfl, A.; Schaber, M.; Fritsche, G.; Wöll, E.; Weiss, G. Prevalence and Predictive Value of Anemia and Dysregulated Iron Homeostasis in Patients with COVID-19 Infection. J. Clin. Med. 2020, 9, 2429. [Google Scholar] [CrossRef]
- Edeas, M.; Saleh, J.; Peyssonnaux, C. Iron: Innocent bystander or vicious culprit in COVID-19 pathogenesis? Int. J. Infect. Dis. 2020, 97, 303–305. [Google Scholar] [CrossRef]
- Habib, H.M.; Ibrahim, S.; Zaim, A.; Ibrahim, W.H. The role of iron in the pathogenesis of COVID-19 and possible treatment with lactoferrin and other iron chelators. Biomed. Pharmacother. 2021, 136, 111228. [Google Scholar] [CrossRef]
- Sonnweber, T.; Boehm, A.; Sahanic, S.; Pizzini, A.; Aichner, M.; Sonnweber, B.; Kurz, K.; Koppelstätter, S.; Haschka, D.; Petzer, V.; et al. Persisting alterations of iron homeostasis in COVID-19 are associated with non-resolving lung pathologies and poor patients’ performance: A prospective observational cohort study. Respir. Res. 2020, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, M.R.; Lashua, L.P.; Fischer, D.K.; Flitter, B.A.; Eichinger, K.M.; Durbin, J.E.; Sarkar, S.N.; Coyne, C.B.; Empey, K.M.; Bomberger, J.M. Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc. Natl. Acad. Sci. USA 2016, 113, 1642–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hino, K.; Nishina, S.; Sasaki, K.; Hara, Y. Mitochondrial damage and iron metabolic dysregulation in hepatitis C virus infection. Free. Radic. Biol. Med. 2018, 133, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Patton, S.M.; Wang, Q.; Hulgan, T.; Connor, J.R.; Jia, P.; Zhao, Z.; Letendre, S.L.; Ellis, R.J.; Bush, W.S.; Samuels, D.C.; et al. Cerebrospinal fluid (CSF) biomarkers of iron status are associated with CSF viral load, antiretroviral therapy, and demographic factors in HIV-infected adults. Fluids Barriers CNS 2017, 14, 1–16. [Google Scholar] [CrossRef]
- McDermid, J.M.; Jaye, A.; van der Loeff, M.F.S.; Todd, J.; Bates, C.; Austin, S.; Jeffries, D.; Awasana, A.A.; Whittle, H.C.; Prentice, A.M. Elevated Iron Status Strongly Predicts Mortality in West African Adults With HIV Infection. JAIDS J. Acquir. Immune Defic. Syndr. 2007, 46, 498–507. [Google Scholar] [CrossRef]
- Brandtner, A.; Tymoszuk, P.; Nairz, M.; Lehner, G.F.; Fritsche, G.; Vales, A.; Falkner, A.; Schennach, H.; Theurl, I.; Joannidis, M.; et al. Linkage of alterations in systemic iron homeostasis to patients’ outcome in sepsis: A prospective study. J. Intensive Care 2020, 8, 76. [Google Scholar] [CrossRef]
- Keller, M.D.; Torres, V.J.; Cadwell, K. Autophagy and microbial pathogenesis. Cell Death Differ. 2020, 27, 872–886. [Google Scholar] [CrossRef]
- Choy, A.; Dancourt, J.; Mugo, B.; O’Connor, T.J.; Isberg, R.R.; Melia, T.J.; Roy, C.R. The Legionella Effector RavZ Inhibits Host Autophagy Through Irreversible Atg8 Deconjugation. Science 2012, 338, 1072–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Zhou, P.; Cheng, S.; Lu, Q.; Nowak, K.; Hopp, A.-K.; Li, L.; Shi, X.; Zhou, Z.; Gao, W.; et al. A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy. Cell 2019, 178, 552–566. [Google Scholar] [CrossRef]
- Ogawa, M.; Yoshimori, T.; Suzuki, T.; Sagara, H.; Mizushima, N.; Sasakawa, C. Escape of Intracellular Shigella from Autophagy. Science 2005, 307, 727–731. [Google Scholar] [CrossRef]
- Owen, K.A.; Anderson, C.J.; Casanova, J.E. Salmonella Suppresses the TRIF-Dependent Type I Interferon Response in Macrophages. mBio 2016, 7, e02051-15. [Google Scholar] [CrossRef] [Green Version]
- Bravo-Santano, N.; Ellis, J.K.; Mateos, L.M.; Calle, Y.; Keun, H.C.; Behrends, V.; Letek, M. Intracellular Staphylococcus aureus Modulates Host Central Carbon Metabolism To Activate Autophagy. mSphere 2018, 3, e00374-18. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, L.; Mostowy, S.; Sancho-Shimizu, V. Autophagy-Virus Interplay: From Cell Biology to Human Disease. Front. Cell Dev. Biol. 2018, 6, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gannagé, M.; Dormann, D.; Albrecht, R.; Dengjel, J.; Torossi, T.; Rämer, P.C.; Lee, M.; Strowig, T.; Arrey, F.; Conenello, G.; et al. Matrix Protein 2 of Influenza A Virus Blocks Autophagosome Fusion with Lysosomes. Cell Host Microbe 2009, 6, 367–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyei, G.; Dinkins, C.; Davis, A.S.; Roberts, E.; Singh, S.B.; Dong, C.; Wu, L.; Kominami, E.; Ueno, T.; Yamamoto, A.; et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009, 186, 255–268. [Google Scholar] [CrossRef]
- Chiramel, A.I.; Best, S.M. Role of autophagy in Zika virus infection and pathogenesis. Virus Res. 2018, 254, 34–40. [Google Scholar] [CrossRef]
- Killian, M.S. Dual role of autophagy in HIV-1 replication and pathogenesis. AIDS Res. Ther. 2012, 9, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeganeh, A.; Alibhai, F.J.; Tobin, S.W.; Wu, J.; Weisel, R.D.; Li, R.K. High Autophagy Rate in Young Sca-1+Bone Marrow Cells Promotes a Pro-Rejuvenating Phenotype in the Heart via Improving Autophagy. Circulation 2018, 138, A12278. [Google Scholar]
- Lin, Y.; Zhao, Z.; Huang, A.; Lu, M. Interplay between Cellular Autophagy and Hepatitis B Virus Replication: A Systematic Review. Cells 2020, 9, 2101. [Google Scholar] [CrossRef]
- Miller, K.; McGrath, M.E.; Hu, Z.; Ariannejad, S.; Weston, S.; Frieman, M.; Jackson, W.T. Coronavirus interactions with the cellular autophagy machinery. Autophagy 2020, 16, 2131–2139. [Google Scholar] [CrossRef]
- Heaton, N.S.; Randall, G. Dengue Virus-Induced Autophagy Regulates Lipid Metabolism. Cell Host Microbe 2010, 8, 422–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metz, P.; Chiramel, A.; Chatel-Chaix, L.; Alvisi, G.; Bankhead, P.; Mora-Rodríguez, R.; Long, G.; Hamacher-Brady, A.; Brady, N.R.; Bartenschlager, R. Dengue Virus Inhibition of Autophagic Flux and Dependency of Viral Replication on Proteasomal Degradation of the Autophagy Receptor p62. J. Virol. 2015, 89, 8026–8041. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.M.; Ernst, O.; Manes, N.P.; Oyler, B.L.; Fraser, I.D.C.; Goodlett, D.R.; Nita-Lazar, A. Multi-Omics Strategies Uncover Host–Pathogen Interactions. ACS Infect. Dis. 2019, 5, 493–505. [Google Scholar] [CrossRef]
- Messina, F.; Giombini, E.; Montaldo, C.; Sharma, A.A.; Zoccoli, A.; Sekaly, R.P.; Locatelli, F.; Zumla, A.; Maeurer, M.; Capobianchi, M.R.; et al. Looking for pathways related to COVID-19: Confirmation of pathogenic mechanisms by SARS-CoV-2-host interactome. Cell Death Dis. 2021, 12, 788. [Google Scholar] [CrossRef]
- Mitchell, H.D.; Eisfeld, A.J.; Stratton, K.G.; Heller, N.C.; Bramer, L.M.; Wen, J.; McDermott, J.E.; Gralinski, L.E.; Sims, A.C.; Le, M.Q.; et al. The Role of EGFR in Influenza Pathogenicity: Multiple Network-Based Approaches to Identify a Key Regulator of Non-lethal Infections. Front. Cell Dev. Biol. 2019, 7, 200. [Google Scholar] [CrossRef]
- Beerli, C.; Yakimovich, A.; Kilcher, S.; Reynoso, G.V.; Fläschner, G.; Muller, D.J.; Hickman, H.; Mercer, J. Vaccinia virus hijacks EGFR signalling to enhance virus spread through rapid and directed infected cell motility. Nat. Microbiol. 2018, 4, 216–225. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, A.; Riegler, A.N.; Martínez, E.; Beno, S.M.; Ricketts, T.D.; Foxman, E.F.; Orihuela, C.J.; Tettelin, H. An in vivo atlas of host–pathogen transcriptomes during Streptococcus pneumoniae colonization and disease. Proc. Natl. Acad. Sci. USA 2020, 117, 33507–33518. [Google Scholar] [CrossRef] [PubMed]
- Griesenauer, B.; Tran, T.; Fortney, K.R.; Janowicz, D.M.; Johnson, P.; Gao, H.; Barnes, S.; Wilson, L.S.; Liu, Y.; Spinola, S.M. Determination of an Interaction Network between an Extracellular Bacterial Pathogen and the Human Host. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canzler, S.; Schor, J.; Busch, W.; Schubert, K.; Rolle-Kampczyk, U.E.; Seitz, H.; Kamp, H.; Von Bergen, M.; Buesen, R.; Hackermüller, J. Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 2020, 94, 371–388. [Google Scholar] [CrossRef] [Green Version]
- Efremova, M.; Teichmann, S.A. Computational methods for single-cell omics across modalities. Nat. Methods 2020, 17, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; McDermaid, A.; Xu, J.; Chang, Y.; Ma, Q. Integrative Methods and Practical Challenges for Single-Cell Multi-omics. Trends Biotechnol. 2020, 38, 1007–1022. [Google Scholar] [CrossRef]
- Zhu, C.; Preissl, S.; Ren, B. Single-cell multimodal omics: The power of many. Nat. Methods 2020, 17, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Rampler, E.; El Abiead, Y.; Schoeny, H.; Rusz, M.; Hildebrand, F.; Fitz, V.; Koellensperger, G. Recurrent Topics in Mass Spectrometry-Based Metabolomics and Lipidomics-Standardization, Coverage, and Throughput. Anal. Chem. 2021, 93, 519–545. [Google Scholar] [CrossRef]
- De Souza, L.P.; Alseekh, S.; Scossa, F.; Fernie, A.R. Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research. Nat. Methods 2021, 18, 733–746. [Google Scholar] [CrossRef]
- Alfaro, J.A.; Bohländer, P.; Dai, M.; Filius, M.; Howard, C.J.; van Kooten, X.F.; Ohayon, S.; Pomorski, A.; Schmid, S.; Aksimentiev, A.; et al. The emerging landscape of single-molecule protein sequencing technologies. Nat. Methods 2021, 18, 604–617. [Google Scholar] [CrossRef]
- Lam, S.M.; Wang, Z.; Li, B.; Shui, G. High-coverage lipidomics for functional lipid and pathway analyses. Anal. Chim. Acta 2020, 1147, 199–210. [Google Scholar] [CrossRef]
- Orsburn, B. Evaluation of the Sensitivity of Proteomics Methods Using the Absolute Copy Number of Proteins in a Single Cell as a Metric. Proteomes 2021, 9, 34. [Google Scholar] [CrossRef]
- Lopez, R.; Regier, J.; Cole, M.B.; Jordan, M.; Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 2018, 15, 1053–1058. [Google Scholar] [CrossRef]
- Oller-Moreno, S.; Kloiber, K.; Machart, P.; Bonn, S. Algorithmic advances in machine learning for single-cell expression analysis. Curr. Opin. Syst. Biol. 2021, 25, 27–33. [Google Scholar] [CrossRef]
- Ben-David, R. Israel’s MeMed Gets FDA Approval for ‘Breakthrough’ Infection Test. 2021. Available online: https://www.timesofisrael.com/israels-memed-gets-fda-approval-for-breakthrough-infection-test/ (accessed on 2 November 2021).
- Callihan, D.R.; Downing, M.; Meyer, E.; Ochoa, L.A.; Petuch, B.; Tranchell, P.; White, D. Considerations for Laboratory Biosafety and Biosecurity During the Coronavirus Disease 2019 Pandemic: Applying the ISO 35001:2019 Standard and High-Reliability Organizations Principles. Appl. Biosaf. 2021, 26. [Google Scholar] [CrossRef]
- Warsinske, H.; Vashisht, R.; Khatri, P. Host-response-based gene signatures for tuberculosis diagnosis: A systematic comparison of 16 signatures. PLoS Med. 2019, 16, e1002786. [Google Scholar] [CrossRef] [Green Version]
- Ramilo, O.; Mejías, A. Shifting the Paradigm: Host Gene Signatures for Diagnosis of Infectious Diseases. Cell Host Microbe 2009, 6, 199–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gliddon, H.D.; Herberg, J.A.; Levin, M.; Kaforou, M. Genome-wide host RNA signatures of infectious diseases: Discovery and clinical translation. Immunology 2017, 153, 171–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leiser, O.P.; Hobbs, E.C.; Sims, A.C.; Korch, G.W.; Taylor, K.L. Beyond the List: Bioagent-Agnostic Signatures Could Enable a More Flexible and Resilient Biodefense Posture Than an Approach Based on Priority Agent Lists Alone. Pathogens 2021, 10, 1497. https://doi.org/10.3390/pathogens10111497
Leiser OP, Hobbs EC, Sims AC, Korch GW, Taylor KL. Beyond the List: Bioagent-Agnostic Signatures Could Enable a More Flexible and Resilient Biodefense Posture Than an Approach Based on Priority Agent Lists Alone. Pathogens. 2021; 10(11):1497. https://doi.org/10.3390/pathogens10111497
Chicago/Turabian StyleLeiser, Owen P., Errett C. Hobbs, Amy C. Sims, George W. Korch, and Karen L. Taylor. 2021. "Beyond the List: Bioagent-Agnostic Signatures Could Enable a More Flexible and Resilient Biodefense Posture Than an Approach Based on Priority Agent Lists Alone" Pathogens 10, no. 11: 1497. https://doi.org/10.3390/pathogens10111497
APA StyleLeiser, O. P., Hobbs, E. C., Sims, A. C., Korch, G. W., & Taylor, K. L. (2021). Beyond the List: Bioagent-Agnostic Signatures Could Enable a More Flexible and Resilient Biodefense Posture Than an Approach Based on Priority Agent Lists Alone. Pathogens, 10(11), 1497. https://doi.org/10.3390/pathogens10111497





