A Comparison of Infectious Disease Forecasting Methods across Locations, Diseases, and Time
Abstract
:1. Introduction
2. Results
2.1. Datasets
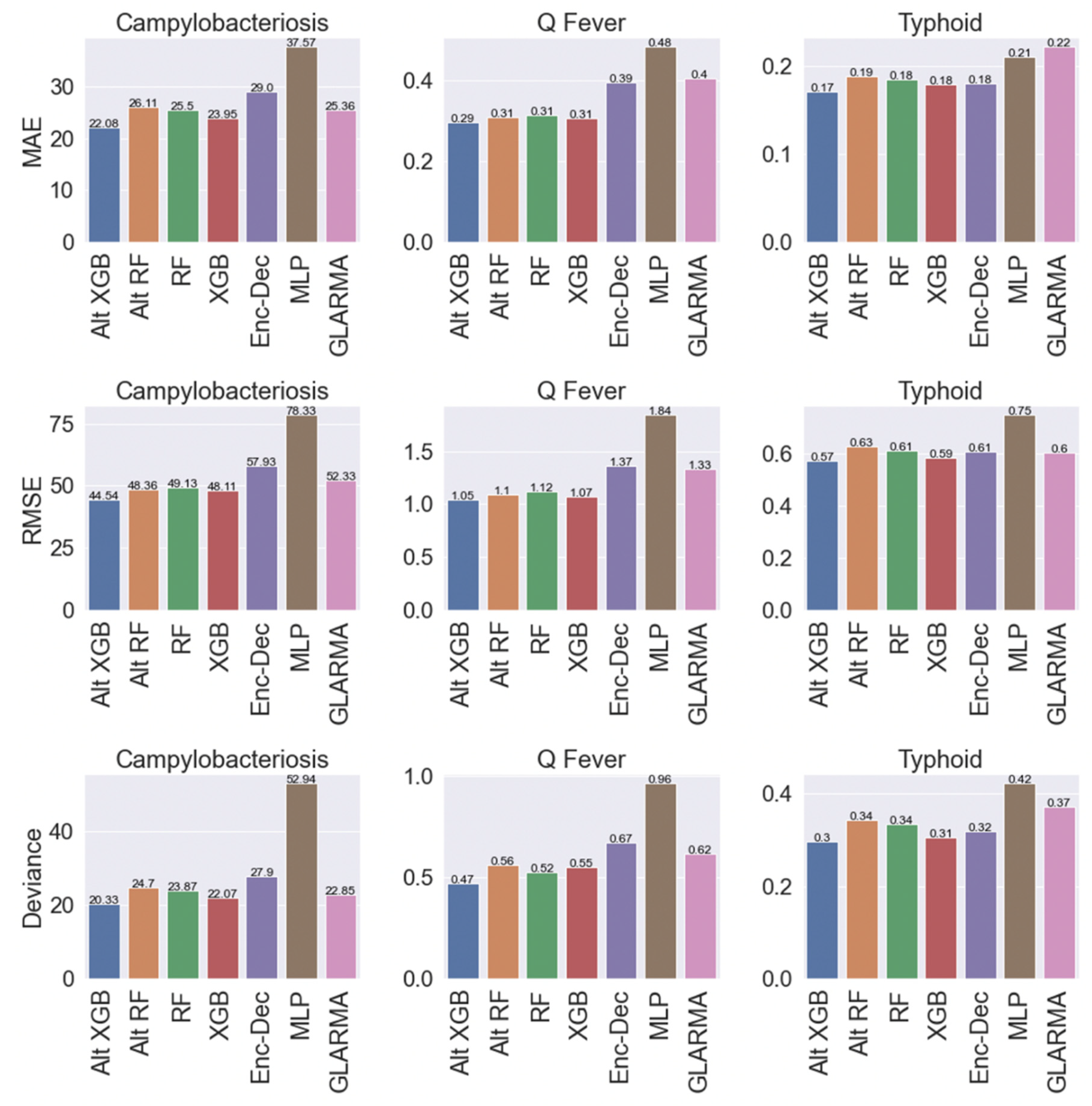
2.2. Overall Model Performance
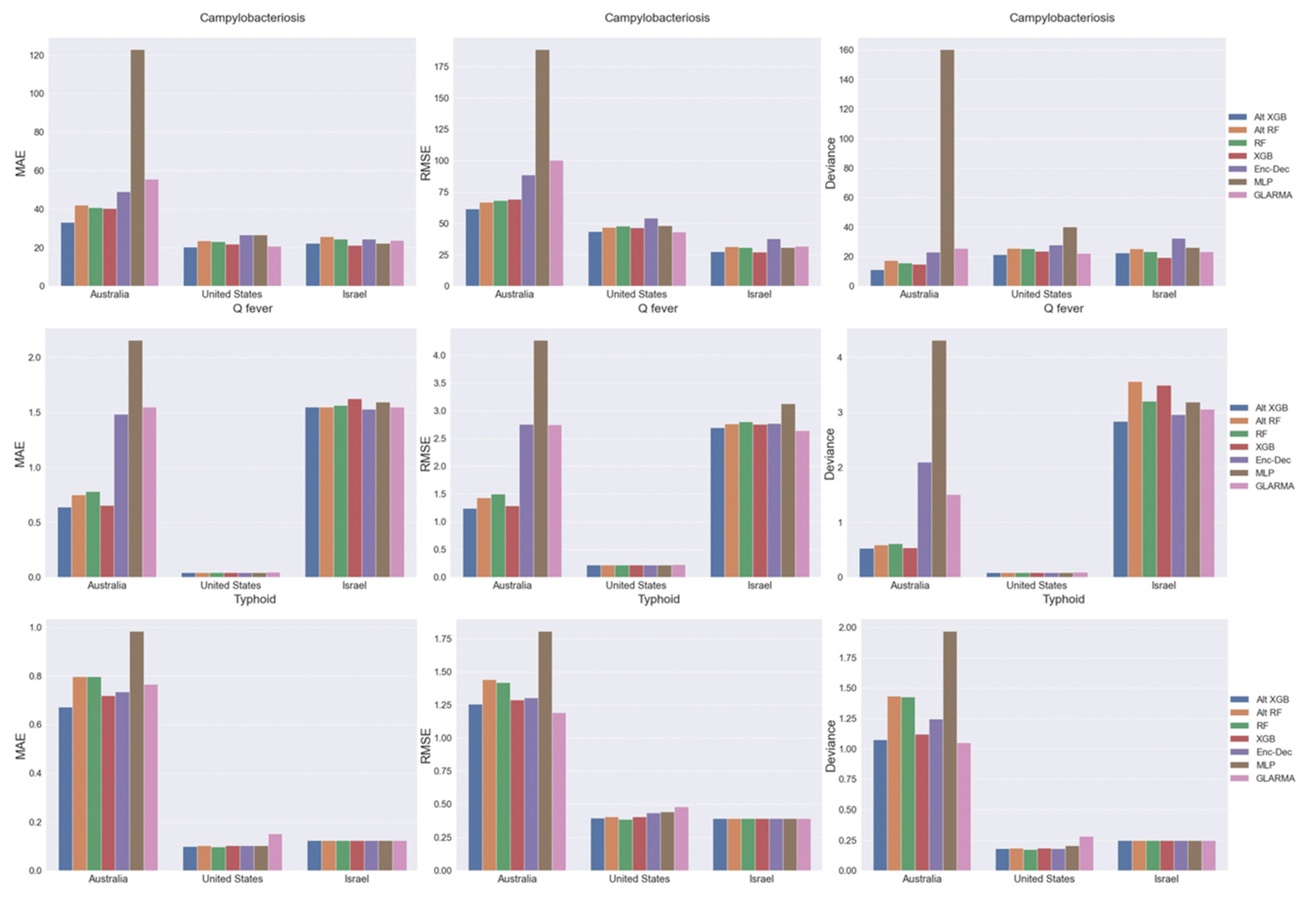
2.3. Model Performance by Country
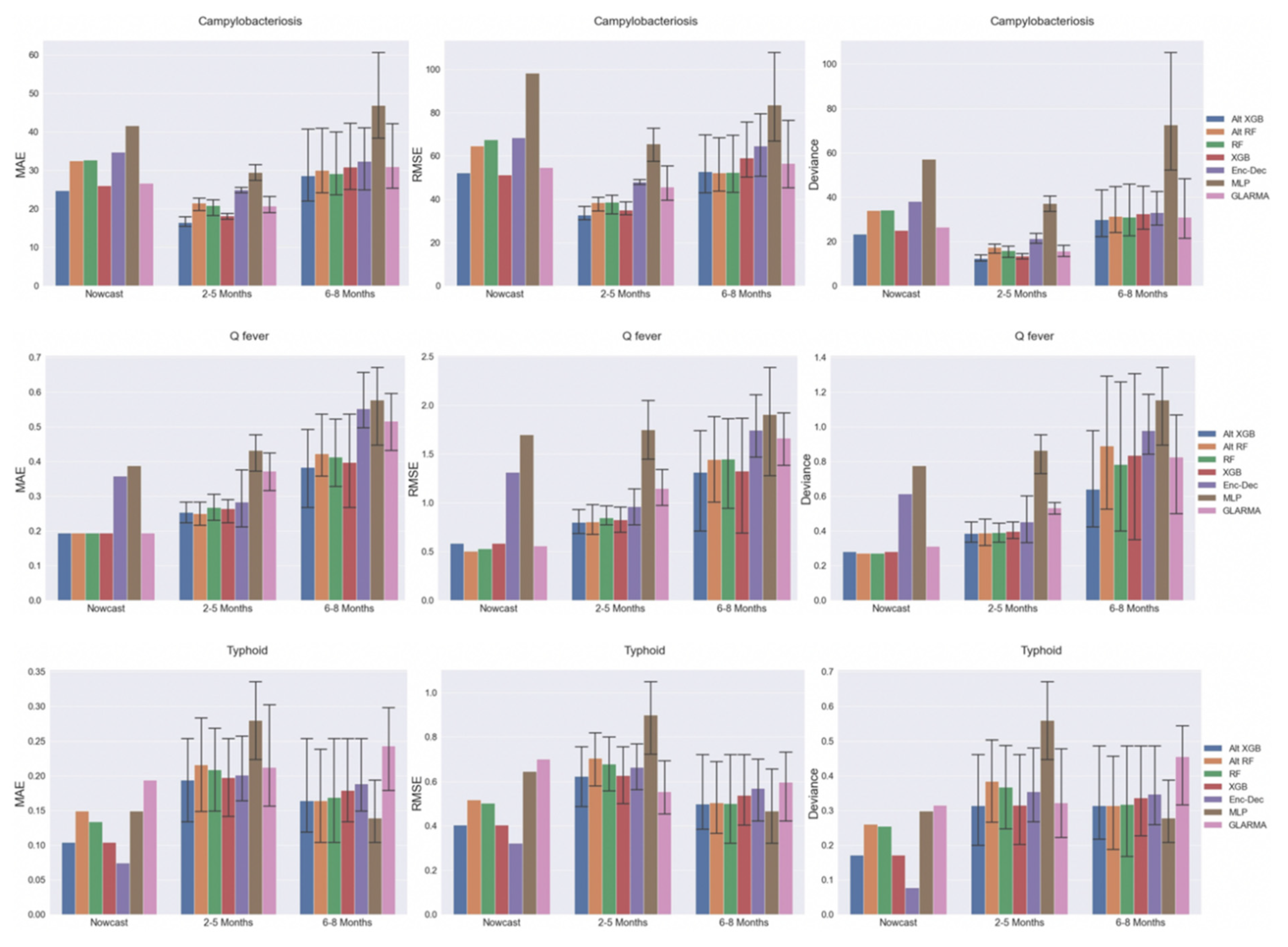
2.4. Model Performance by Time Interval
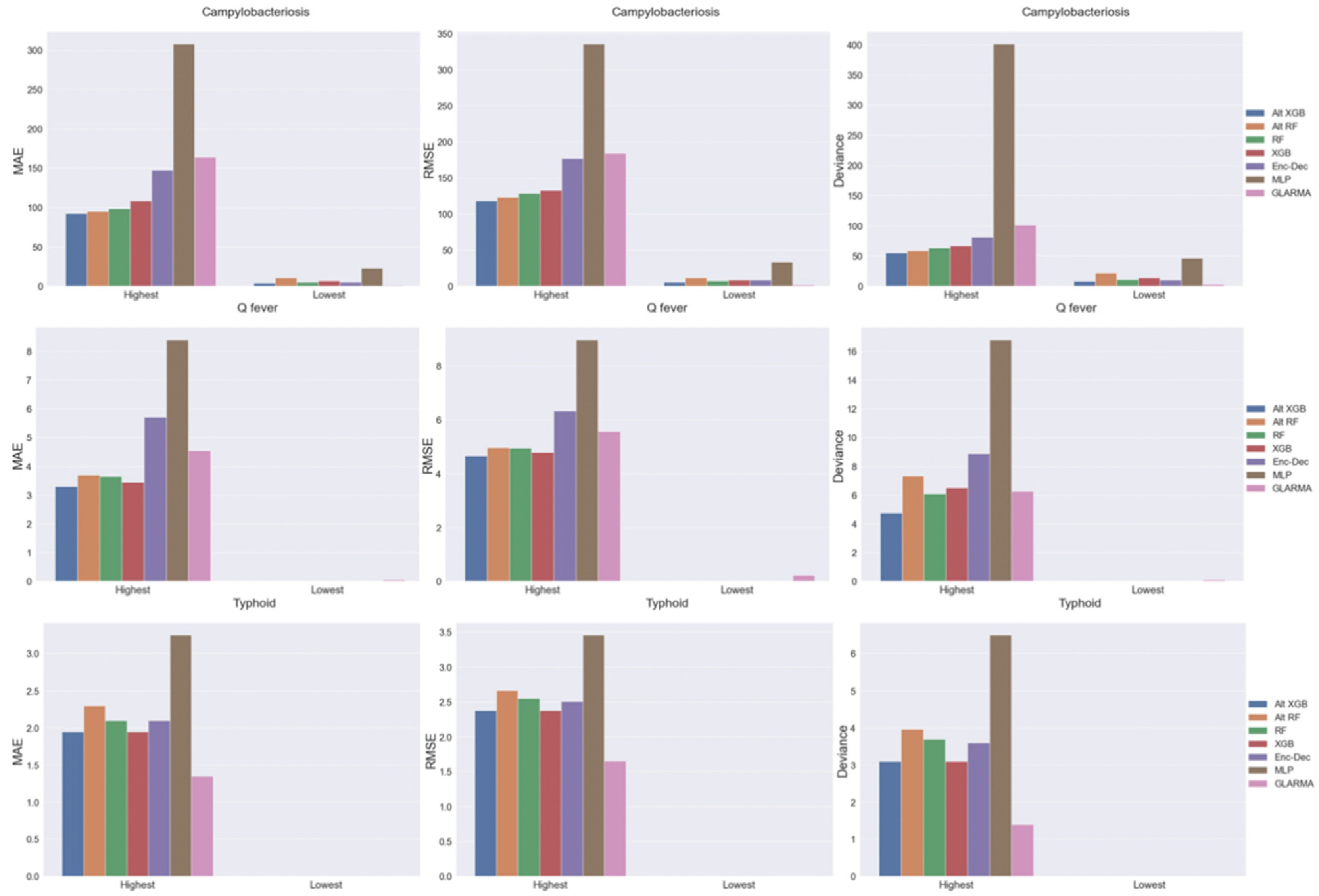
2.5. Model Performance by Disease Incidence
2.6. Summary of Top Model Performance by Disease
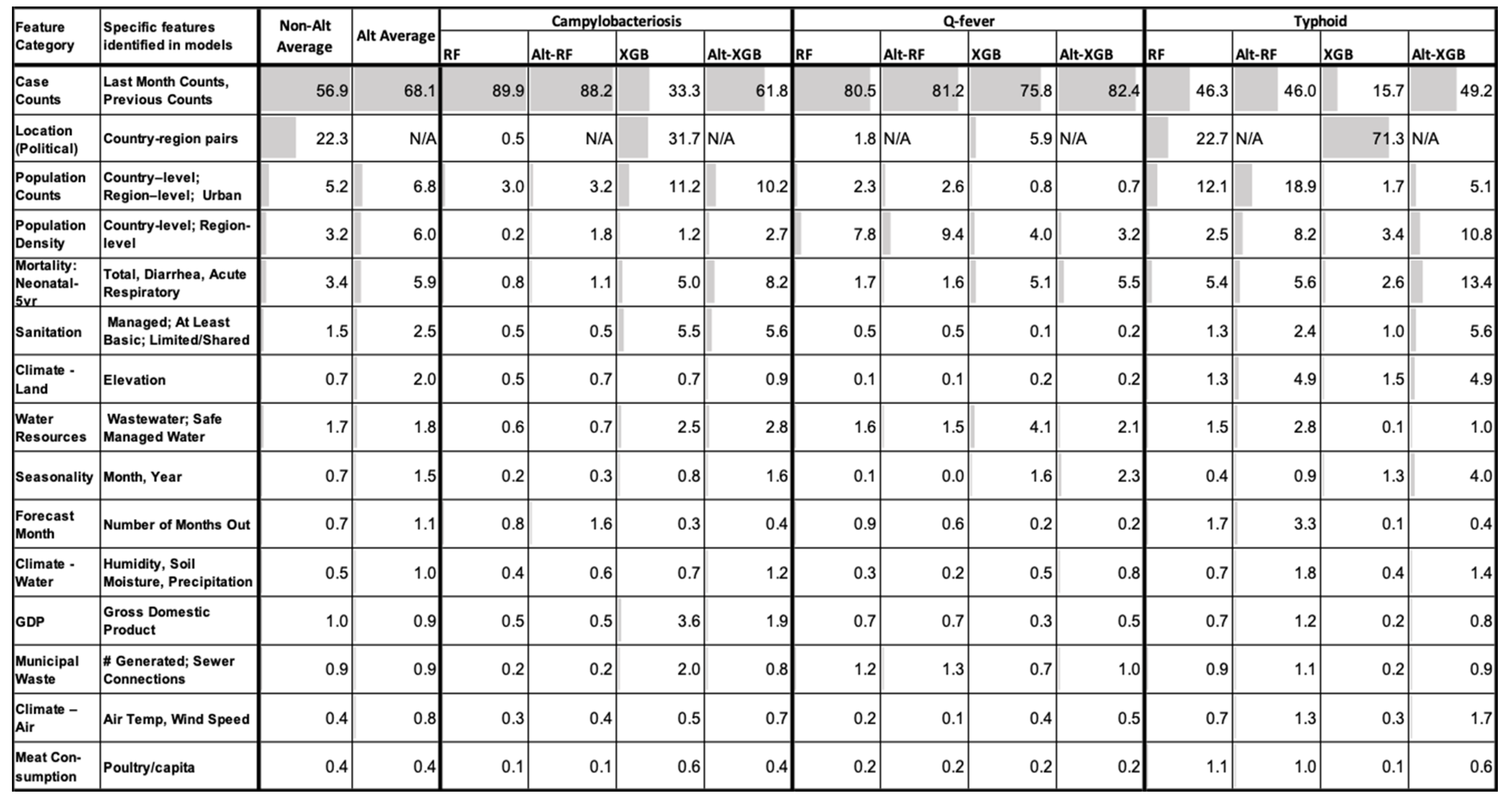
2.7. Feature Importance
3. Discussion
4. Methods
4.1. Data Collection
4.2. Data Engineering
4.3. Data Analysis
4.3.1. Moving Average Models
4.3.2. Tree-Based ML Models
4.3.3. DL Models
4.4. Metrics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| In-Text Abbreviation | Description |
| -Alt | a model trained without country-region as a feature |
| ARIMA | auto-regressive integrated moving average |
| ARIMAX | auto-regressive integrated moving average with exogenous variables |
| DE | encoder-decoder model |
| Deviance | Poisson deviance |
| DL | deep learning |
| DLR | dynamic linear regression |
| GLARMA | generalized linear autoregressive moving averages |
| GRU | gated recurrent unit |
| MAE | mean absolute error |
| ML | machine learning |
| MLP | multi-layer perceptron |
| RF | random forest |
| RMSE | root mean squared error |
| RNN | recurrent neural networks |
| SARIMA | seasonal auto-regressive integrated moving average |
| SIR | susceptible, infectious, and removed |
| SVM | support vector machines |
| XGB | extreme gradient boosted trees |
References
- Bloom, D.E.; Cadarette, D. Infectious Disease Threats in the Twenty-First Century: Strengthening the Global Response. Front. Immunol. 2019, 10, 549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neglected Tropical Diseases. Available online: https://www.who.int/news-room/q-a-detail/neglected-tropical-diseases (accessed on 29 September 2021).
- Hartley, D.M.; Nelson, N.P.; Arthur, R.R.; Barboza, P.; Collier, N.; Lightfoot, N.; Linge, J.P.; van der Goot, E.; Mawudeku, A.; Madoff, L.C.; et al. An overview of internet biosurveillance. Clin. Microbiol. Infect. 2013, 19, 1006–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Shea, J. Digital Disease Detection: A Systematic Review of Event-Based Internet Biosurveillance Systems. Int. J. Med. Inform. 2017, 101, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Bontempi, G.; ben Taieb, S.; le Borgne, Y.A. Machine Learning Strategies for Time Series Forecasting. In European Business Intelligence Summer School; Lecture Notes in Business Information Processing; Springer: Berlin/Heidelberg, Germany, 2012; pp. 62–77. [Google Scholar] [CrossRef]
- Voyant, C.; Notton, G.; Kalogirou, S.; Nivet, M.L.; Paoli, C.; Motte, F.; Fouilloy, A. Machine learning methods for solar radiation forecasting: A review. Renew. Energy 2017, 105, 569–582. [Google Scholar] [CrossRef]
- Makridakis, S.; Spiliotis, E.; Assimakopoulos, V. Statistical and Machine Learning Forecasting Methods: Concerns and Ways Forward. PLoS ONE 2018, 13, e0194889. [Google Scholar] [CrossRef] [Green Version]
- Rashidi, H.H.; Tran, N.K.; Betts, E.V.; Howell, L.P.; Green, R. Artificial Intelligence and Machine Learning in Pathology: The Present Landscape of Supervised Methods. Acad. Pathol. 2019, 6, 2374289519873088. [Google Scholar] [CrossRef]
- Stiglic, G.; Kocbek, P.; Fijacko, N.; Zitnik, M.; Verbert, K.; Cilar, L. Interpretability of Machine Learning-Based Prediction Models in Healthcare. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2020, 10, e1379. [Google Scholar] [CrossRef]
- Fang, X.; Liu, W.; Ai, J.; He, M.; Wu, Y.; Shi, Y.; Shen, W.; Bao, C. Forecasting Incidence of Infectious Diarrhea Using Random Forest in Jiangsu Province, China. BMC Infect. Dis. 2020, 20, 222. [Google Scholar] [CrossRef]
- Kane, M.J.; Price, N.; Scotch, M.; Rabinowitz, P. Comparison of ARIMA and Random Forest Time Series Models for Prediction of Avian Influenza H5N1 Outbreaks. BMC Bioinform. 2014, 15, 276. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Wu, Y.C.; Lin, M.H.; Liu, Y.L.; Tsai, Y.Y.; Wu, J.H.; Pan, K.H.; Ke, C.J.; Chen, C.M.; Liu, D.P.; et al. Applying machine learning models with an ensemble approach for accurate real-time influenza forecasting in Taiwan: Development and validation study. J. Med. Internet Res. 2020, 22, e15394. [Google Scholar] [CrossRef]
- Bagheri, H.; Tapak, L.; Karami, M.; Amiri, B.; Cheraghi, Z. Epidemiological Features of Human Brucellosis in Iran (2011-2018) and Prediction of Brucellosis with Data-Mining Models. J. Res. Health Sci. 2019, 19, e00462. [Google Scholar] [PubMed]
- Alim, M.; Ye, G.H.; Guan, P.; Huang, D.S.; Zhou, B.S.; Wu, W. Comparison of ARIMA Model and XGBoost Model for Prediction of Human Brucellosis in Mainland China: A Time-Series Study. BMJ Open 2020, 10, 39676. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.; Liu, X.; Rajarethinam, J.; Kok, S.Y.; Liang, S.; Tang, C.S.; Cook, A.R.; Ng, L.C.; Yap, G. Mapping Dengue Risk in Singapore Using Random Forest. PLoS Negl. Trop. Dis. 2018, 12, e0006587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Du, S.; Li, T.; Horng, S.J. Time Series Forecasting Using Sequence-to-Sequence Deep Learning Framework. In Proceedings of the 2018 9th International Symposium on Parallel Architectures, Algorithms and Programming (PAAP), Taipei, Taiwan, 26–28 December 2018. [Google Scholar]
- Bouktif, S.; Fiaz, A.; Ouni, A.; Serhani, M.A. Optimal Deep Learning Lstm Model for Electric Load Forecasting Using Feature Selection and Genetic Algorithm: Comparison with Machine Learning Approaches. Energies 2018, 11, 1636. [Google Scholar] [CrossRef] [Green Version]
- Gagne, D.J., II; Haupt, S.E.; Nychka, D.W.; Thompson, G. Interpretable deep learning for spatial analysis of severe hailstorms. Mon. Weather. Rev. 2019, 147, 2827–2845. [Google Scholar] [CrossRef]
- García-Martín, E.; Rodrigues, C.F.; Riley, G.; Grahn, H. Estimation of Energy Consumption in Machine Learning. J. Parallel Distrib. Comput. 2019, 134, 75–88. [Google Scholar] [CrossRef]
- DeFries, R.S.; Chan, J.C.W. Multiple Criteria for Evaluating Machine Learning Algorithms for Land Cover Classification from Satellite Data. Remote Sens. Environ. 2000, 74, 503–515. [Google Scholar] [CrossRef]
- Wanas, N.; Auda, G.; Kamel, M.S.; Karray, F. On the Optimal Number of Hidden Nodes in a Neural Network. Can. Conf. Electr. Comput. Eng. 1998, 2, 918–921. [Google Scholar] [CrossRef]
- Davis, R.; Holan, S.; Lund, R.; Ravishanker, N. Handbook of Discrete-Valued Time Series; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Gasparrini, A.; Scheipl, F.; Armstrong, B.; Kenward, M.G. A Penalized Framework for Distributed Lag Non-Linear Models. Biometrics 2017, 73, 938–948. [Google Scholar] [CrossRef]
- Hase, P.; Chen, C.; Li, O.; Rudin, C. Interpretable Image Recognition with Hierarchical Prototypes. Proc. AAAI Conf. Hum. Comput. Crowdsourcing 2019, 7, 32–40. [Google Scholar]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global Epidemiology of Campylobacter Infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guatteo, R.; Seegers, H.; Taurel, A.F.; Joly, A.; Beaudeau, F. Prevalence of Coxiella Burnetii Infection in Domestic Ruminants: A Critical Review. Vet. Microbiol. 2011, 149, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Keshavamurthy, R.; Singh, B.B.; Kalambhe, D.G.; Aulakh, R.S.; Dhand, N.K. Identification of Risk Factors Associated with Coxiella Burnetii Infection in Cattle and Buffaloes in India. Prev. Vet. Med. 2020, 181, 105081. [Google Scholar] [CrossRef] [PubMed]
- Njeru, J.; Henning, K.; Pletz, M.W.; Heller, R.; Neubauer, H. Q Fever Is an Old and Neglected Zoonotic Disease in Kenya: A Systematic Review. BMC Public Health 2016, 16, 297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, A.K.; DesRoches, C.M.; Kralovec, P.D.; Joshi, M.S. A Progress Report on Electronic Health Records In U.S. Hospitals. Health Aff. 2017, 29, 1951–1957. [Google Scholar] [CrossRef] [Green Version]
- DesRoches, C.M.; Worzala, C.; Joshi, M.S.; Kralovec, P.D.; Jha, A.K. Small, Nonteaching, And Rural Hospitals Continue To Be Slow In Adopting Electronic Health Record Systems. Health Aff. 2017, 31, 1092–1099. [Google Scholar] [CrossRef] [Green Version]
- Desai, A.N.; Kraemer, M.U.; Bhatia, S.; Cori, A.; Nouvellet, P.; Herringer, M.; Cohn, E.L.; Carrion, M.; Brownstein, J.S.; Madoff, L.C.; et al. Real-time epidemic forecasting: Challenges and opportunities. Health Secur. 2019, 17, 268–275. [Google Scholar] [CrossRef]
- Khalsa, H.S.; Cordova, S.R.; Generous, N.; Khalsa, P.S. Epi Archive: Automated Synthesis of Global Notifiable Disease Data. Online J. Public Health Inform. 2019, 11, e280. [Google Scholar] [CrossRef]
- GDAL/OGR Geospatial Data Abstraction Software Library-Google Scholar. Available online: https://scholar.google.com/scholar?hl=en&as_sdt=0%2C48&q=GDAL%2FOGR+geospatial+data+abstraction+software+library&btnG= (accessed on 29 September 2021).
- Little, R.; Rubin, D. Statistical Analysis with Missing Data; John Wiley & Sons: Hoboken, NJ, USA, 2019. [Google Scholar]
- Hancock, J.T.; Khoshgoftaar, T.M. Survey on Categorical Data for Neural Networks. J. Big Data 2020, 7, 28. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar] [CrossRef]
- XGBoost Documentation—Xgboost 1.5.2 Documentation. Available online: https://xgboost.readthedocs.io/en/stable/index.html (accessed on 19 December 2021).
- Pedregosa, F.; Michel, V.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Vanderplas, J.; Cournapeau, D.; Pedregosa, F.; Varoquaux, G.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury Google, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Processing Syst. 2019, 32, 8026–8037. [Google Scholar]
- Willmott, C.; Matsuura, K. Advantages of the Mean Absolute Error (MAE) over the Root Mean Square Error (RMSE) in Assessing Average Model Performance. Clim. Res. 2005, 30, 79–82. [Google Scholar] [CrossRef]
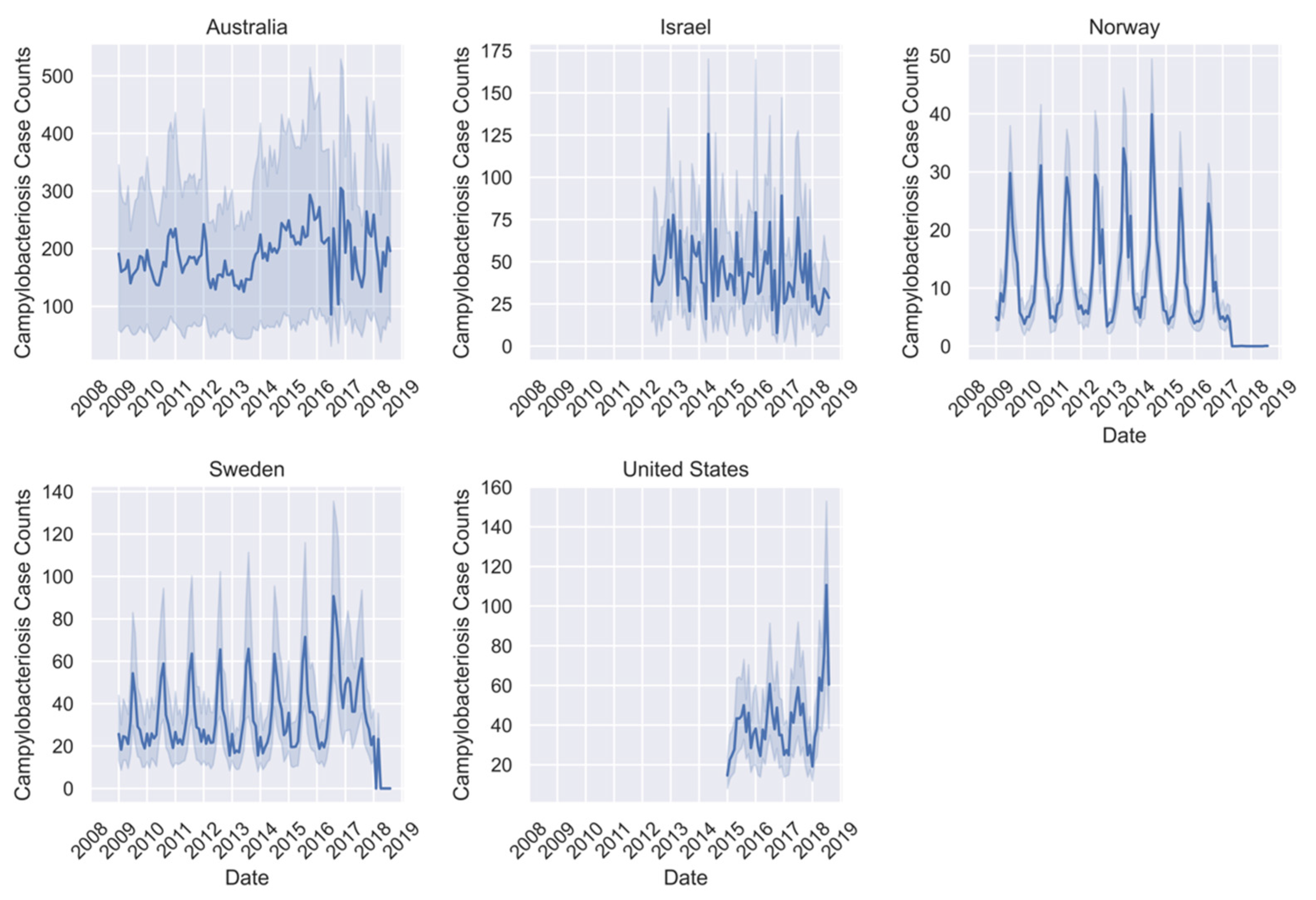
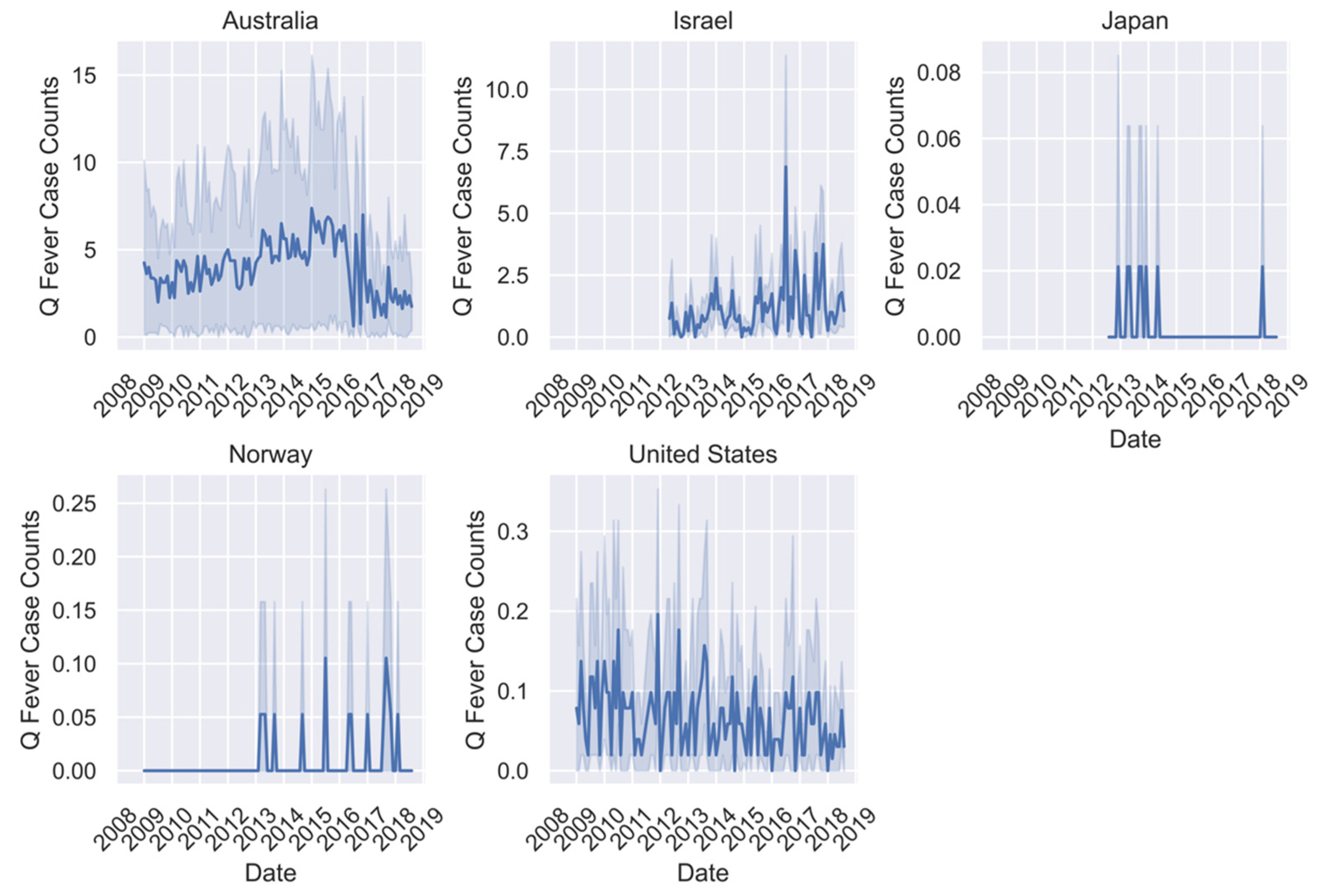
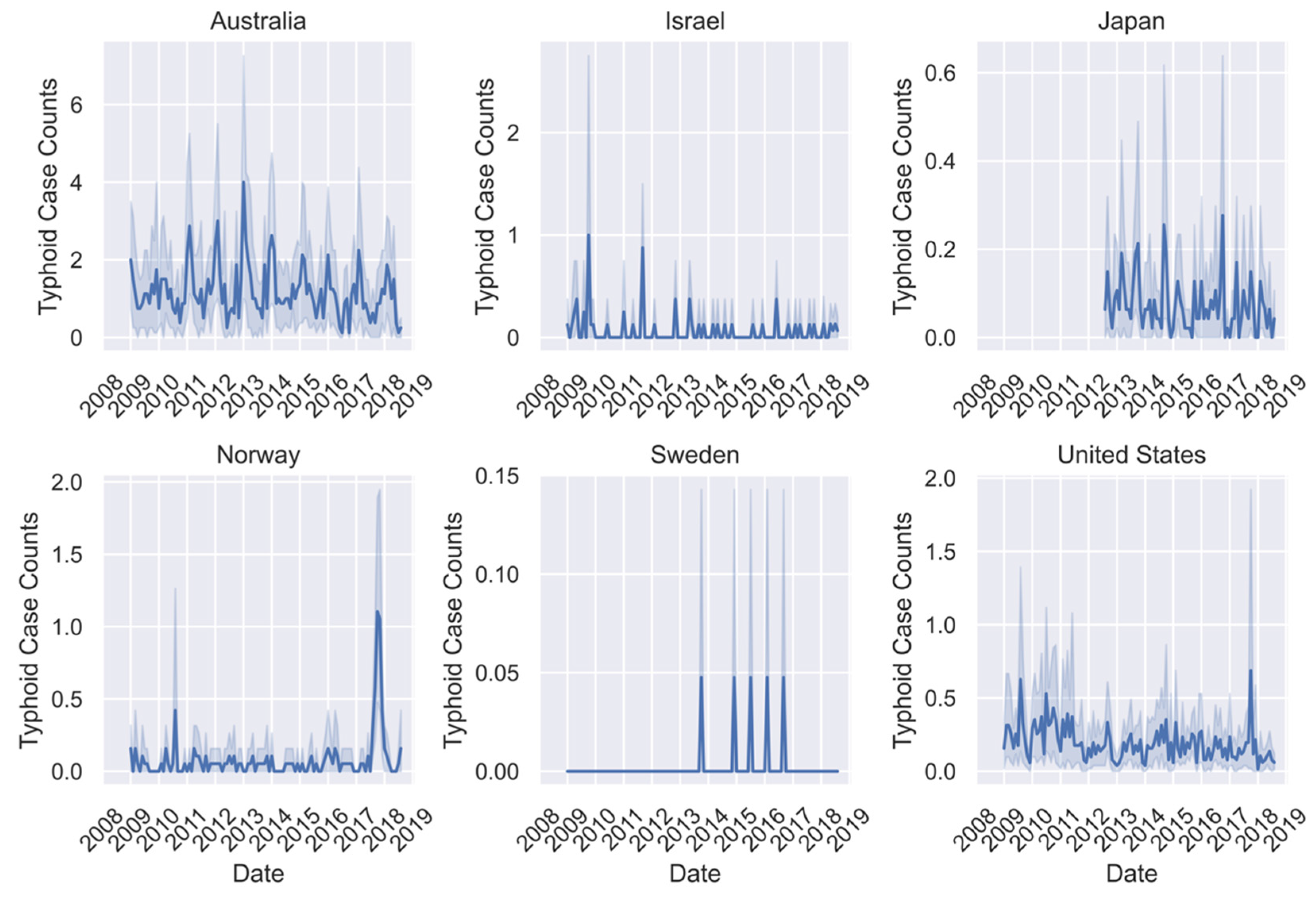
| Campylobacteriosis Case Counts | Q-Fever Case Counts | Typhoid Case Counts | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Min–Max | Total | Mean | Min–Max | Total | Mean | Min–Max | Total | |
| Australia | 190.10 | 0–880 | 182,498 | 3.92 | 0–32 | 3768 | 1.17 | 0–14 | 1122 |
| Israel | 40.64 | 0–361 | 29,429 | 1.12 | 0–21 | 809 | 0.06 | 0–7 | 63 |
| Japan | NA | NA | NA | NA | 0–2 | 10 | 0.07 | 0–7 | 264 |
| Norway | 10.36 | 0–184 | 22,839 | 0.201 | 0–1 | 14 | 0.08 | 0-8 | 169 |
| Sweden | 31.55 | 0–427 | 16,380 | NA | NA | NA | ~0.00 | 0–1 | 5 |
| United States | 42.04 | 0–794 | 98,720 | 0.06 | 0–3 | 423 | 0.19 | 0–29 | 1251 |
| Disease | Location | Nowcast | Short-Term | Long-Term |
|---|---|---|---|---|
| Campylo-bacteriosis | All countries | Alt-XGB | Alt-XGB | Alt-XGB; RF (Both); Alt-XGB |
| Australia | XGB (Both) | Alt-XGB | Alt-XGB | |
| Israel | MLP; MLP; XGB | XGB | XGB; GLARMA; XGB | |
| US | Alt-XGB; GLARMA; Alt-XGB | GLARMA; GLARMA; Alt-XGB | Alt-XGB; Alt-RF; Alt-XGB | |
| Q-Fever | All countries | RF | Alt-RF; Alt-XGB; Alt-XGB | Alt-XGB |
| Australia | GLARMA | Alt-XGB | XGB | |
| Israel | MLP | Enc–Dec; GLARMA; Enc–Dec | Alt-XGB | |
| US | All Models | All Tree-based ML (Alt-XGB *) | All Tree-based ML(Alt-XGB *) | |
| Typhoid | All countries | Enc–Dec | Alt-XGB; GLARMA; XGB (Both) | MLP |
| Australia | GLARMA; XGB(Both), GLARMA; Enc–Dec | GLARMA | MLP; Alt-RF; MLP | |
| Israel | All Models | All Models | All Models | |
| US | MLP | All Tree-based ML | RF; MLP; RF |
| Number of Cases | Country Over All Months | Forecast Time Over All Locations | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Disease | Overall | High Cases | Low Cases (Zero) | Australia | Israel | US | Nowcasting | Short Term | Long Term |
| Campylo- bacteriosis | Alt-XGB | Alt-XGB | GLARMA | Alt-XGB | XGB (Both) | GLARMA, Alt-XGB | XGB | XGB | RF(Both); RF(Both); Alt-XGB |
| Q-fever | Alt-XGB | Alt-XGB | Tree-based, DL | Alt-XGB | GLARMA; Enc–Dec; Alt-XGB | All | RF | Tree-based | Alt-XGB |
| Typhoid | Alt-XGB | GLARMA | All | Alt-XGB; GLARMA; GLARMA | All | RF | Enc–Dec | Alt-XGB; GLARMA; XGB (Both) | MLP |
| Country | Campylobacteriosis | Q-Fever | Typhoid | |||
|---|---|---|---|---|---|---|
| Date Range | # Regions | Date Range | # Regions | Date Range | # Regions | |
| Australia | 2009–2018 | 8 | 2009–2018 | 8 | 2009-2018 | 8 |
| Finland | 2009–2017 | 18 | NA | 0 | NA | 0 |
| Israel | 2012–2018 | 6 | 2012–2018 | 6 | 2009–2018 | 6 |
| Japan | NA | 0 | 2012–2017 | 47 | 2012–2017 | 47 |
| Norway | 2009–2017 | 18 | 2009–2017 | 18 | 2009–2017 | 18 |
| Sweden | 2009–2017 | 21 | NA | 0 | 2009–2017 | 21 |
| United States | 2015–2018 | 51 | 2009–2018 | 51 | 2009–2018 | 51 |
| Data Type | Website | Individual Features | Geographic Location | Geographic Resolution | Time Period | Periodicity |
|---|---|---|---|---|---|---|
| Case Counts | epiarchive.bsvgateway.org accessed on 28 May 2019 | Incidences of select human diseases. | Countries of interest | Region-level | 2009–2018 | Daily |
| Political Borders | gadm.org accessed on 28 May 2019 | Geopolitical borders (country and within country) | Countries of interest | Region-level | 2018 | Single instance |
| Climate | disc.gsfc.nasa.gov; earthdata.nasa.gov accessed on 28 May 2019 | air temperature, humidity, precipitation, soil moisture, and wind speed | Global | Gridded 0.25° × 0.25°, 1° × 1° | 2012–2018 | Monthly |
| Gross Domestic Product | www.ers.usda.gov accessed on 28 May 2019 | Gross Domestic Product | Global | Country-level | Varies | Yearly |
| Elevation | www.diva-gis.org accessed on 28 May 2019 | Digital Elevation Map | Global | 43,200 × 17,200 (30 arc seconds) | NA | NA |
| Mortality | www.who.int accessed on 28 May 2019 | Deaths by country, year, sex, age group, and cause of death. | Global | Country-level | 2009–2018 | Yearly |
| Municipal waste | stats.oecd.org accessed on 28 May 2019 | Municipal waste generation and treatment | Countries of interest | Country-level | 2009–2017 | Yearly |
| Socio-political and Physical data | www.naturalearthdata.com accessed on 28 May 2019 | Country and internal administrative borders; socioeconomic and political attributes | Global | Varies by country; 1: 10 m–110 m | 2019 | Single instance |
| Population | population.un.org accessed on 28 May 2019 | Population by age intervals by location | Global | Country-level | 2009–2015 | Every 5 years |
| Population Density | sedac.ciesin.columbia.edu accessed on 28 May 2019 | Population density | Global | 30 arc-seconds | 2009–2015 | Every 5 years |
| Water Potability and Treatment | stats.oecd.org accessed on 28 May 2019 | Freshwater resources, available water, wastewater treatment plant capacity, surface water | Countries of interest | Country-level | 2009–2017 | Yearly |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dixon, S.; Keshavamurthy, R.; Farber, D.H.; Stevens, A.; Pazdernik, K.T.; Charles, L.E. A Comparison of Infectious Disease Forecasting Methods across Locations, Diseases, and Time. Pathogens 2022, 11, 185. https://doi.org/10.3390/pathogens11020185
Dixon S, Keshavamurthy R, Farber DH, Stevens A, Pazdernik KT, Charles LE. A Comparison of Infectious Disease Forecasting Methods across Locations, Diseases, and Time. Pathogens. 2022; 11(2):185. https://doi.org/10.3390/pathogens11020185
Chicago/Turabian StyleDixon, Samuel, Ravikiran Keshavamurthy, Daniel H. Farber, Andrew Stevens, Karl T. Pazdernik, and Lauren E. Charles. 2022. "A Comparison of Infectious Disease Forecasting Methods across Locations, Diseases, and Time" Pathogens 11, no. 2: 185. https://doi.org/10.3390/pathogens11020185
APA StyleDixon, S., Keshavamurthy, R., Farber, D. H., Stevens, A., Pazdernik, K. T., & Charles, L. E. (2022). A Comparison of Infectious Disease Forecasting Methods across Locations, Diseases, and Time. Pathogens, 11(2), 185. https://doi.org/10.3390/pathogens11020185







