Abstract
Originally two types of Polycomb Repressive Complexes (PRCs) were described, canonical PRC1 (cPRC1) and PRC2. Recently, a versatile set of complexes were identified and brought up several dilemmas in PRC mediated repression. These new class of complexes were named as non-canonical PRC1s (ncPRC1s). Both cPRC1s and ncPRC1s contain Ring finger protein (RING1, RNF2) and Polycomb group ring finger catalytic (PCGF) core, but in ncPRCs, RING and YY1 binding protein (RYBP), or YY1 associated factor 2 (YAF2), replaces the Chromobox (CBX) and Polyhomeotic (PHC) subunits found in cPRC1s. Additionally, ncPRC1 subunits can associate with versatile accessory proteins, which determine their functional specificity. Homozygous null mutations of the ncPRC members in mice are often lethal or cause infertility, which underlines their essential functions in mammalian development. In this review, we summarize the mouse knockout phenotypes of subunits of the six major ncPRCs. We highlight several aspects of their discovery from fly to mice and emerging role in target recognition, embryogenesis and cell-fate decision making. We gathered data from stem cell mediated in vitro differentiation assays and genetically engineered mouse models. Accumulating evidence suggests that ncPRC1s play profound role in mammalian embryogenesis by regulating gene expression during lineage specification of pluripotent stem cells.
1. Introduction
1.1. Discovery of the Polycomb Repressor System
The concept of epigenetic memory systems brought one of the most important discoveries in the field of developmental biology. The history of the chromatin structure-based repression mechanisms started with the identification of Polycomb (Pc) mutation in Drosophila melanogaster 70 years ago by Pamela Lewis [1,2] (reviewed in [3]). Polycomb gene was named after its peculiar dominant phenotype. Multiple sex combs, a row of thick bristles, appeared on all three pairs of legs of the Pc mutants. This phenotype was striking, since normally sex combs are only found on the first pair of legs of the D. melanogaster males. The first breakthrough of our thinking of epigenetic gene regulation was based on the explanation of the Pc mutant phenotype by Edward B. Lewis. He suggested that the reason for the transformation of the second and third legs towards the appearance of the first leg is that Polycomb gene is needed for the repression of homeotic genes of the Bithorax-complex (BXC) in Drosophila [4].
Eight posterior segments are regulated by the BXC, but it codes only three homeobox genes, Ubx, abd-A and Abd-B [5,6]. The characteristic expression pattern of the three homeotic genes is different in every segment. The segmentally different expression levels of the three homeotic genes are regulated by eight complex specific cis-regulatory regions, which are activated in consecutive segments during embryonic development, in the same order along the head-to-tail body axis as they lined up on the chromosome [7]. Upon activation, cis-regulatory regions remain in an open chromatin conformation according to the “opened for business” hypothesis [8,9]. Due to this sequential activation, all homeotic cis-regulatory regions are in an active chromatin conformation in the most posterior part of the embryo (reviewed in [10]).
In Drosophila, the segment specific expression patterns of homeotic genes are becoming set, by spatially arranged transcription factors, in the first few hours of embryogenesis, which is called the “initiation phase” of BXC regulation. At later stages, in the “maintenance phase”, the transcription factors responsible for this early pattern formation are replaced by two sets of epigenetic regulators (reviewed in [11]). The epigenetic activators belonging to the Trithorax-group keep the active state of the initially activated genes, while Polycomb (PC) protein is one of those epigenetic repressors, which keep the original inactive state of homeotic regulators (described in more details in [12]) (reviewed in [13]). The antagonistic activities of these two types of epigenetic regulators maintain the early expression patterns of the homeotic genes throughout life [14]. In loss of function type heterozygous Polycomb mutants, the amount of PC protein is insufficient; the transmission of the repressed state becomes destabilized. In the course of multiple cell divisions, homeotic genes become active in regions of the body where they should remain silent [15]. The ectopic activation of the homeotic genes in turn, leads to the visible gain-of-function type homeotic transformations, such as the appearance of multiple sex combs. The cause of the lethality of the homozygous Pc null mutant embryos is that all embryonic segments are transformed towards the most posterior segment identity, which is incompatible with life [4].
In the forthcoming years, numerous independent mutants with multiple sex comb phenotypes were identified in Drosophila. These genes all code for epigenetic repressor proteins, similar to PC [15,16,17]. Four independent mutations were recognized with dominant multi sex comb phenotypes: Additional sex combs (Asx), Posterior sex combs (Psc) [18], Polycomblike (Pcl) [19] and Sex comb on midleg (Scm) [20]. Jürgens proposed in 1985 that these factors should be collectively referred as the Polycomb Group of genes (PcG) [21]. Later, six other genes, with similar, but recessive phenotypes were also included in the group: extra sex combs (esc) [22,23], pleiohomeotic (pho) [24], polycombeotic [25], which was later proved to be an allele of the Enhancer of zeste (E(z)) gene [26], super sex combs (sxc) [27], multi sex combs (mxc) [17,28,29] and polyhomeotic (ph) [16,30]. Some members of the group do not have a Polycomb phenotype on their own, but can enhance the phenotype of other genes belonging to the PcG [17], such as Enhancer of Polycomb (E(Pc)) [31], RING1 and YY1 binding protein (Rybp) [32,33] and Suppressor of zeste 2 (Su(z)2), a functional homolog of Psc [34,35,36].
In the next decades, a plethora of additional PcG genes were identified such as Sex combs extra (Sce) [37,38], cramped (crm) [39], Scm related gene containing four mbt domains (Sfmbt) [40] and Suppressor of zeste 12 (Su(z)12) [41], and proved that they all play a role in the transmission of the silent state of homeotic genes. Sixty years after the isolation of the first Pc allele [1], De Ayala et al. published a classical Polycomb screen in Drosophila and described calypso, a non-redundant Drosophila gene showing dominant PcG type homeotic transformations [37]. In D. melanogaster, 17 classical PcG genes have been described so far [42].
PcG genes substantially differ from homeotic genes in many respects. They are ubiquitously expressed, even in the segments where their homeotic target genes are active and cannot themselves convey any positional information [43]. Phenotypes of mutations of PcG genes are the consequence of the ectopic activation of their targets, such as the Antennapedia- and the Bithorax-complex [26,44,45,46,47], segment polarity genes such as engrailed, or cell cycle regulators such as E2F1 [48] or CyclinA [49]. Based on genetic interaction studies, it has long been postulated that PcG proteins interact with each other in Drosophila, form complexes and contribute to a common repressor mechanism. This hypothesis was proven later in mammalian systems as well [50,51,52].
1.2. Evolutionarily Conserved Domains in PcG Proteins
Understanding the molecular mechanism of PcG dependent repression started with the identification of a 37-amino acid (AA) long homologous region the Drosophila PC protein and Heterochromatin associated protein 1 (HP1), called Chromatin Organization Modifier (Chromo) domain [53]. HP1 is a heterochromatin associated chromosomal protein with dosage-dependent effects on Position Effect Variegation (PEV) in Drosophila [54]. PEV is the first recognized epigenetically regulated phenomena, describing a gene expression change depending on the position of the given locus in the genome. The description of PEV is based on the mosaic cell to cell variation of white (w) gene expression [55]. PEV is characteristic to specific chromosomal rearrangements of the w locus, such as wm4 [56]. In flies containing the wm4 rearrangement, the normally euchromatic white gene is displaced into the vicinity of a heterochromatic region. The white gene codes a transporter protein and is responsible for the normal red eye color of D. melanogaster. Consequently, the normal red color disappears from a subpopulation of cells in the eye, indicating the clonal silencing of the locus. The silenced chromatin structure of the tightly packed heterochromatic domain seemed to be able to spread in cis and inactivate the relocated white gene in a subpopulation of cells, which can be detected by white colored clones. The pattern of variegation suggests that the decision of inactivation happens early in development and maintained through multiple cell divisions. Deletion of the Chromo domain abolished the chromosome binding for both HP1 and PC proteins. This suggested that PcG proteins may use analogous mechanisms for the stable transmission of a repressed state, as closed, compacted heterochromatin domains [57]. Finding Chromo domain motifs supported the model that PcG dependent gene repression generates a compact state of the DNA, in which the genes are inaccessible for the transcription machinery [47] such as after HP1 mediated heterochromatin formation [57] (reviewed in [47]). A decade later, this hypothesis was confirmed by Fitzgerald and Bender [58]. According to their experiments, silenced domains of the BXC are more tightly packed and not accessible for DNA polymerase II (POLII) dependent transcription and Flipase (FLP) mediated recombination. The packaging was PcG dependent, the restriction disappeared in PcG mutant backgrounds. Other approaches using super-resolution microscopy came to similar conclusions. Silenced Polycomb domains showed the densest packaging, of all analyzed regions [59]. Intriguingly, longer Pc silenced domains also showed dense packaging. As expected, the packaging states of given regions changed in Embryonic stem (ES) cells during differentiation, accordingly to changes in Pc binding [60]. It was also shown that mutations in the HP1 Chromo domain completely abolished the ability of HP1 to promote silencing. Furthermore, Platero et al. showed that the chimeric HP1-PC protein binds to both heterochromatin and Polycomb binding sites on the chromosome. Domain swapping experiments also proved that the Chromo domain alone does not account for the specific targeting of either PC or HP1, underlining the importance of other protein–protein interactions [61].
Another connection between heterochromatin compaction and PcG dependent repression was suggested by a presence of a conserved C-terminal motif found in the E(Z) protein [26]. This motif was identified not only in a position effect variegation suppressor heterochromatic protein Suppressor of variegation 3-9 SU(VAR)3-9, but, surprisingly, in Trithorax (TRX) protein as well, which is the name giving member of the epigenetic activator Trx-group [62,63] (reviewed in [13]). The conserved motif was later called Su(var)3-9, E(z), Trx (SET) domain, after the names of the three different chromatin regulator factors it was found in (reviewed in [64]). SET domain is not only specific for proteins involved in repression, but it is also necessary for the epigenetic activator function of TRX. Later, many SET domain proteins were found to be histone methyltransferases with different substrate specificity [65,66,67,68]. Finding the SET domain in E(Z) made the first connection between the PcG dependent silencing system and covalent modification of histones.
The second connection between covalent histone modifications and PcG function is linked to Ring finger domain, which is often found in ubiquitinatin ligases. Ring domain was found in three Drosophila PcG proteins PSC, SU(Z)2 and SCE, and the respective mouse homolog proteins Ring finger protein 1 (RING1/RING1A), Ring finger protein 2 (RNF2/RING1B), Polycomb group ring finger catalytic 4 (PCGF4) also known as B Lymphoma Mo-MLV Insertion Region 1 Homolog (BMI1) and Polycomb group ring finger catalytic 2 (PCGF2) also known as MEL18 underlying the evolutionary conservation of the motif [34,69,70]. Some Ring finger proteins were shown to be able to interact with ubiquitin conjugating enzymes [71,72]. SCE and its human homologs RING1/RING1A and RNF2/RING1B are catalytically active ubiquitin ligases [38,73], which were found to be the core catalytic subunits of all mammalian Polycomb Repressive Complex 1s (PRC1s).
The next classical domain of PcG proteins is the Sterile Alpha Motif (SAM), first found in PH and SCM proteins. The SAM domains of the two proteins can mediate both homotypic and heterotypic interactions [74]. SAM domains of PH proteins can form a helical polymer [75]. Mutation of a single amino acid in the SAM domain, which blocks polymerization, ruins the repressor capabilities of PH protein [76]. SAM domain polymerization is essential for the in vivo growth suppressive function of PH too [77]. SAM motif was also found in dSFMBT protein (Table 1). SFMBT protein harbors four consecutive Malignant Brain Tumor (MBT) domains. MBT domains are able to recognize and bind mono- and dimethylated histones [40]. Several studies showed that SAM domain interactions play a crucial role in PcG targeting and clustering, mediate long-range chromatin interactions and affect nuclear organization of repressive centers, called PcG bodies [78] (reviewed in [79]). SAM domain containing subunits of PRCs are prevalent in mammals as well (e.g., SFMBT2, PHC1, PHC2, PHC3, SCMH1 and SCML2), thus polymerization of the SAM domain seems an important feature of PRC mediated silencing.

Table 1.
The composition of mouse non-canonical Polycomb Repressive Complex (ncPRC1) subtypes and alternative names, chromosomal localization, Drosophila orthologs and distinctive protein domains of the encompassing subunits. Core members are found in all ncPRC1 complex subtypes.
Polycomb Group proteins are not related structurally to each other at all, but orthologs of all PcG members identified in Drosophila were found in mammals [80,81,82,83] (reviewed in [84,85]), and even in plants [86,87,88] (reviewed in [89,90,91]). This underpins the importance and evolutionary conserved nature of PcG mediated repression mechanisms. Over the years, it became obvious that the PcG system is not exclusive for homeotic regulation, it acts on thousands of regulatory regions of key developmental genes governing the growth and development of all metazoans (reviewed in [92,93,94,95]).
1.3. Discovery of Polycomb Repressive Complexes
Polycomb and Polyhomeotic [96,97] were identified as components of multimeric protein complexes in D. melanogaster [96]. To clarify the physical interactions that define modules of PcG action, it was necessary to purify the repressor complexes. Purification of the Polycomb repressive complexes opened the way to identify the physically interacting protein subunits and to understand the molecular mechanism of repression.
Saurin et.al purified the first PcG containing protein assembly and called Polycomb Repressive Complex 1 from fruit fly embryonic extracts in 2001. This complex contained PC, PSC, PH and SCE/dRING [94,98] (reviewed in [99]). In the purification scheme another PcG member, SCM, was also identified as a sub-stoichiometric component. A substantial amount of other proteins, which did not belong to the classical PCGs were also identified as different TATA-binding protein associated factors, indicating direct connection and interplay between the epigenetic repressor system and the active transcriptional machinery. The interaction between PC, PSC and RING homologs are largely conserved, PC homolog Chromobox 8 CBX8/HPC3, PSC homolog PCGF4/BMI1 and RING1 were also identified in mammals as interacting partners [100]. Later, similar PRC1 complexes were purified from mammalian cells, all containing the homologs of Drosophila PC, PSC and PH (see Table 1), but lacking most non PcG proteins. Mammalians PRC1 complexes may contain different homologs of the Drosophila PRC1 subunits [51]. The presence of the different paralog PRC1 subunits increases the complexity of the possible repressor mechanisms in mammals. The slightly different complexes with similar stoichiometry likely account for the existence of more dynamic, and tissue specific regulatory systems established in mouse and human (reviewed in [101]). As expected, purified PRC1 complexes were capable to inhibit nucleosome remodeling and transcription in in vitro assays [102,103]. The human PRC1 (hPRC1) complex inhibited nucleosome remodeling, via a mechanism that did not block nuclease accessibility [50] and compact chromatin [104] underlining the functional conservation of the core PRC1 subunits throughout evolution. Surprisingly, in D. melanogaster, PSC alone could also block the nucleosome remodeling activity of the SWI/SNF complex in vitro [102]. The hPRC1 complex were shown to be able to mono-ubiquitylate histone H2A at lysine 119 (K119) [73]. RING subunits are the catalytic E3 ubiquitin ligases in the PRC1 complexes. RING domains are sufficient for H2A ubiquitination, RING1/RING1A and RNF2/RING1B can substitute each other. RING-PCGF interaction enhances the catalytic activity in vitro [105]. The chromatin compaction ability of RNF2/RING1B in ES cells was necessary for proper repression, but by using a catalytically inactive RNF2/RING1B, it was proven that active repressor function of RNF2/RING1B is independent of the ubiquitin ligase activity of the protein [106].The search for different interacting partners of RING proteins lead to the identification of an alternative PRC1 such as complex with similar H2A specific ubiquitin ligase activity, both in mammals and in Drosophila [107,108,109]. The Drosophila complex was called Ring Associated Factors complex (dRAF) [109], while its mammalian counterpart was originally named BCL-6 interacting Corepressor (BCOR) complex [107]. dRAF contained the core components of PRC1, PSC and SCE/dRING, together with a novel subunit, Lysine (K)-specific demethylase 2b (KDM2B) [109], but not included a PC homolog subunit. According to recent terminology it was the first non-canonical PRC1 complex. KDM2B is an F-BOX protein and demethylase specific to H3K36 and required for effective H2A ubiquitination activity of the complex. dRAF complex is able to remove an active histone mark from H3 and at the same time depositing a repressive histone mark to H2A [109].
The next important achievement was to purify an independent multimeric complex, called Polycomb Repressive Complex 2 (PRC2). This complex consists E(Z), SU(Z)12, ESC and Chromatin assembly factor 1 (CAF-1), also called NURF55. PRC2 complex have specific histone methyltransferase activity both in Drosophila [66,67,110] and in human [68]. The human PRC2 (hPRC2) core complex consists the human homologs of the above Drosophila proteins. One of the homologs of E(Z) was found in the purified hPRC2 complex EZH2, together with the only Drosophila ESC homolog Embryonic ectoderm development (EED) and SUZ12. Two CAF-1/NURF55 ortholog proteins were also purified, Retinoblastoma protein associated protein 46 (RBAP46) and Retinoblastoma protein associated protein 48 (RBAP48). The substrate specificity of the hPRC2 complexes was similar. PRC2s methylate histone H3 at the 27th lysine residue (K27). The histone methyltransferase activity of PRC2 is connected to the SET domain of the E(Z) protein, but E(Z) protein is not catalytically active alone [111]. The E(Z)/EZH2 subunits has to be complexed with at least the ESC/EED and SUZ12 homolog subunits to be catalytically active [112,113,114,115,116]. Additional facultative subunits such as PCL [117], Jumonji, AT rich interactive domain 2 (JARID2) [118] and AE binding protein 2 (AEBP2) were shown to target variant PRC2 complexes, and further enhance their catalytic activity [119]. For further details about the subunit diversity of the mammalian PRC2 subcategories, please refer to some recent reports and reviews [120,121,122].
The silencing function of the two types of Polycomb Repressive Complexes is often linked. The Chromo domain in PC and in its mammalian homolog Chromobox proteins (CBXs) are able to specifically recognize and bind the PRC2 made trimethylation marks on on the 27th Lysine (K) of Histone 3 (H3), (H3K27trimet) [123]. On the other hand, the presence of the K119 ubiquitilation on Histone 2A (H2A), produced by Ring protein containing complexes (H2AK119Ub), promotes PRC2 recruitment [124,125]. It was shown that mono-ubiquitination of H2A by PRC1 type complexes creates a binding site for JARID2-AEBP2 containing PRC2 complexes, constituting a positive feedback loop establishing H3K27 trimethylation positive repressed domains [126]. These findings underlined the importance of histone modifications in establishing silenced chromatin domains and revealed the coordinate trans-histone regulation by PcG complexes. The experiments also suggested cooperation between PRC1s and PRC2s, but do not explain how any of the identified PRCs targeted to the regulatory regions of the genes to be repressed. Classical Polycomb target genes in Drosophila contain conserved DNA regions, called Polycomb Response Elements (PREs) [127,128,129,130]. PREs were shown for a long time to mediate binding of PcG proteins and play a crucial role in the organization of repressed chromatin structure. There are 379 sites conserved across the Drosophila species within PcG domains, which can be considered as PREs in the Drosophila genomes [131]. PREs maintain repression through cell divisions, to ensure correct lineage commitment, development and differentiation [132,133]. Neither PRC1, nor PRC2 complexes purified from Drosophila contain any subunit which has PRE specific DNA binding capacity.
The first repressor complex in Drosophila, which contained direct DNA binding subunit, was the Pho-Repressive Complex (PhoRC) [40]. Pho-RC contains two proteins PHO and dSFMBT. PHO is the Drosophila Yin and yang 1 protein (YY1) homolog and has a sequence specific DNA binding domain [134], which was shown to be capable of selectively recognize and bind to a conserved sequence motif identified in PREs [135]. On the other hand, the four MBT domains of dSFMBT are able to selectively bind mono- and dimethylated H3K9 and H4K20, but fail to interact with these residues if they are unmodified or trimethylated [40]. The SAM domain of SFMBT is capable to interact with the SAM domains of both PH and SCM.
Klymenko et al. proposed that the Pho-RC complex is selectively targeted to PRE sequences in the genome. The tethered complex binds the methylated histones in the flanking nucleosomes, which in turn helps to stabilize the repressed chromatin state [40]. Earlier studies detected interaction between PHO and E(Z) by Chromatin immunoprecipitation (ChIP) experiments [136]. The key members of the independent Polycomb repressive complexes, PHO (Pho-RC), E(Z) (PRC2) and PC (PRC1) proteins, were all detected by ChIP experiments at the same position on the most studied Polycomb binding DNA element, the bxd PRE of the UBX gene region in Drosophila in 0–3 h old embryos [137]. The discoveries of the above complexes lead to the first simple and widely accepted model of hierarchical recruitment of Polycomb complexes. According to that, first the DNA binding PhoRC complex lands at the PRE regions, recruiting PRC2 complex, which specifically methylates the neighboring histone H3 at K27 position, creating a mark recognized by the Chromo domain of PC, which in turn leads to PRC1 recruitment [137].
In later experiments neither PRC2 nor PRC1 members were copurified with Pho-RC, nor detected to bind dSFMBT by ChIP. Homozygous null pho mutants can survive to pharate adult stage showing only mild homeotic transformations [24]. Part of the reason for this is redundancy. A second gene coding for a similar YY1 homolog DNA binding protein were also found in Drosophila, called pho-like (phol) [138]. Although the pho/pho; phol/phol double homozygous mutant larvae show strong misexpression of Ubx in wing discs of the third instar larvae, but the localization of the three PRC1 members PC, PSC and PH, as well as PRC2 member E(Z) remained the same for the most identified binding sites on the polytene chromosomes. Only a few exceptional sites were described, where some of the PcG protein are no longer present [138]. This finding would indicate only inferior function of DNA binding YY1 homolog proteins in the targeting of PRC complexes, but pho has a strong maternal contribution in Drosophila, and some of the maternally contributed PHO proteins can persist in third instar larvae, which can be enough for normal targeting function. In the absence of maternal supply of pleiohomeotic, zygotycally mutant pho homozygotes die earlier and exhibit severe homeotic transformations, as well as segmentation defects [24].
Schuettengruber et al. presented an extensive comparative ChIP dataset of five different Drosophila species providing new insights to direct DNA binding factor (PHO, DSP1 and GAGA) dependent Polycomb recruitment. They suggest cooperative interactions between DNA binding recruiters and PRC components instead of the classical hierarchical recruitment models. According to their view, the initial modest sequence specificity for PHO recruitment in PRC binding chromatin domains may be amplified through a PRC1 dependent positive feedback loop. As a consequence of that, in vivo PHO binding landscapes are not necessarily include all of the strongest in vitro predicted PHO binding sites, but reflect a complex function of sequence specific DNA binding factor interactions together with the interactions of PRC components with the DNA binding factors [131].
Later, it was shown that the two SAM domain containing protein dSFMBT and SCM, the subunit of PRC1 can bind each other directly [139] and the crystal structure of the dimer was also determined [140]. The interaction surfaces are mapped to the SAM domains. Both dSFMBT and SCM are essential for PcG dependent repression in Drosophila. PhoRC DNA binding and dSFMBT-SAM SCM-SAM interactions together are capable to target cPRC1 complexes to PRE regions. After cPRC1 recruitment to PREs, SCM-SAM and PH-SAM domain mediated polymerization results in the formation of compacted chromatin domains [140]. Using parallel TAP-tagged purification approaches, it was shown that SCM is the only protein, which is enriched in both PRC1 and PRC2 type complexes. It seems that the SAM domain containing SCM protein can offer the functional link between Polycomb repressive complexes [141].
The fourth Polycomb repressive complex was named Polycomb Repressive Deubiqitinase complex (PR-DUB) in Drosophila. This complex contains two proteins: CALYPSO, a deubiquitinating enzyme belonging to the Ubiquitin C-terminal hydrolase subclass and ASX. PR-DUB complex is capable to bind to the PREs of classical PcG target genes and mutations of its subunits result classical PcG phenotypes. Reconstituted recombinant Drosophila and human PR-DUB complexes were both capable to specifically remove ubiquitin from H2A, but not from H2B [142]. A similar PR-DUB-like complex was identified in mammals and proved to play an important role in the development of chronic myelomonocytic leukemia (CMML) symptoms [143].
2. Mammalian PRC Complexes
2.1. Mammalian PcG Gene Functions: Parallels and Differences between Mammals and Fruit Fly
Mammalian and Drosophila PcG proteins have similar roles, namely the maintenance of homeotic gene expression patterns [144]. In many cases, transgenic constructs expressing mammalian PcG proteins can rescue the Drosophila mutations in their homolog gene, such as in the case of homozygous mutant Sce, which can be rescued by mouse ring finger protein 1 (Ring1) [145]. Similarly, the Pc mutation in Drosophila was rescued by the mouse chromobox 2 (Cbx2/M33) construct [144], and homozygous pho mutants are rescued by the human Yin and yang 1 (Yy1) [146]. Homozygous mutations of mouse PcG genes do not always cause embryonic lethality and can survive to birth, but often show homeotic transformations similarly to their Drosophila counterparts. Axial skeleton alterations were described in many murine PcG mutants, such as in the case of the Pc homolog Cbx2/M33 [147], two Psc homologs Pcgf2/Mel18 [80] and Pcgf4/Bmi1 [148,149,150], both of the two Sce homolog, Ring1/Ring1A [151] and Rnf2/Ring1B [152], the pho homolog Yy1 [153] and in two murine homologs of ph, polyhomeotic homolog 1 (Phc1/Rae28) and polyhomeotic homolog 2 (Phc2) [154,155]. PcG proteins are epigenetic repressors in mammals and have antagonistic functions in homeotic regulation to epigenetic activator proteins belonging to the TrxG, just like in Drosophila [150]. Double mutant combinations of PcG genes in mouse are often found to enhance each other’s phenotype, similarly to Drosophila, such as in the case of Cbx2/M33 and Pcgf4/Bmi1 [156], indicating evolutionary conserved cooperation between different repressor complexes in mammalians.
Some of these differences are easier to comprehend, bearing in mind that in several cases Drosophila PcG genes have more than one orthologs in mammalians. There are at least 37 PcG genes identified in mammals compared to the 17 classical PcG members in Drosophila (see Table 1 for non-canonical members) The partial compensation between orthologs often results in less severe phenotypes of mouse PcG mutants than in their Drosophila counterparts. Moreover, expansion and diversification of PcG genes contributed significantly to the complexity of epigenetic gene repression mechanisms in mammalians [157]. Different orthologs can have more complex tissue and developmental stage specific functions in mammals then in fruit fly. Many PcG null mutations of one of the orthologs in mice can survive to later embryonic stages; some of them are even viable, showing very mild phenotypes [94] (see Table 2, Table 3, Table 4 and Table 5). Furthermore, analyzing the mouse phenotypes of late lethal PcG mutations, it became obvious that although these factors play profound role in homeotic regulation [158,159], their epigenetic repressor functions are also important in hematopoiesis [160,161,162,163] (reviewed in [164] and [165]), neuronal [154,166] (reviewed in [167]) cardiac [168,169,170] and skeletal muscle differentiation [171], as well as in the maintenance of germ cell fate [172,173,174,175].

Table 2.
Phenotypes of ncPRC core subunit knockouts. Phenotypes of homozygous knockout mouse mutants and ES cells are listed.

Table 3.
Phenotypes of ncPRC1.1 subunit knockouts. Phenotypes of homozygous knockout mouse mutants and ES cells are listed.

Table 4.
Phenotypes of ncPRC1.2 and ncPRC1.4 subunit knockouts. Phenotypes of homozygous knockout mouse mutants and ES cells are listed.

Table 5.
Phenotypes of ncPRC1.3 and ncPRC1.5 subunit knockouts. Phenotypes of homozygous knockout mouse mutants and ES cells are listed.
Homozygous mutations in PRC subunits can often lead to very early, post-implantation lethality in mouse, before the development of any segmented tissues such as in the case of Rybp−/− [166], or result gastrulation arrest, as in the homozygous Eed mutants [176]. The early lethal phase of many PcG gene mutations makes it impossible to examine the role of the coded protein in the regulation of the majority of presumptive target genes, which would only be activated in later developmental stages. After the identification of hundreds of PcG target genes, including transcription factors and signaling components for key developmental pathways both in mouse and human [92,177] it become important to seek for additional model system to study the late activated target genes of the PcG repressors. This led to the application of embryonic stem (ES) cell-based systems to study Polycomb function (see Section 4).
2.2. Classification of Mammalian PRC Complexes
PcG proteins form two major types of complexes: PRC1 and PRC2. Both complexes contain core subunits present in all the sub complexes of the family. Additionally, accessory proteins may interact with the core subunits, which have regulatory functions in targeting to specific chromatin domains or to modulating the catalytic activity of the core complex. PRC1 complexes are further divided into canonical PRC1s (cPRC1s) and non-canonical PRC1s (ncPRC1s). The cPRC1 is the functional homolog of Drosophila PRC1s. The ncPRC1s more resemble to dRAF complexes and do not harbor a PC homolog subunit, but contain RYBP/YAF2 subunits instead. PRC1s are subdivided based on the type PCGFs (PCGF1, 2, 3, 4, 5 and 6) they encompass. Different PCGFs have defined functions, altered binding specificity towards specific accessory subunits [178]. PCGF subunits play critical role in determining the functions of versatile PRC1s. The recently used nomenclature of the different PRC1 complexes (PRC1.1, PRC1.2, PRC1.3, PRC1.4, PRC1.5 and PRC1.6) is based on which PCGF subunit they contain [52]. Accordingly, the PCGF1 containing complex is PRC1.1, the PCGF2 protein containing complex is PRC1.2 and so on. It is worth keeping in mind that PCGF2/MEL18 and PCGF4/BMI1 were originally identified only in CBX protein containing cPRC1 complexes [52], whilst cPRC1.2 and ncPRC1.4 type complexes were not found. Later, systematic mapping of the human complexome (using a robust affinity purification mass spectrometry approach), revealed detectable interactions between PCGF2 and PCGF4 with RYBP or YAF2 [179]. As PCGF2 and PCGF4 do not interact with any additional subunits besides RING1/RING2 and RYBP/YAF2 it is possible, that ncPRC1.2 and ncPRC1.4 correspond to several transient intermedier products of PRC1 holo-complex assemblies. Hierarchical clustering of the interactions revealed four groups of complexes defined by the six PCGF proteins: (a) PRC1.1; (b) PRC1.2-PRC1.4; (c) PRC1.3–PRC1.5; and (d) PRC1.6 [179]. Not only the similar interaction network, but also the regulated gene sets and distinctive function indicate closer connection between the indicated pairs of complexes. For example, PRC1.3–PRC1.5 mainly function as a transcriptional activator in ES cells, promoting the expression of many genes involved in mesoderm differentiation via interaction of TEX10 transcription factor [180]. Only ncPRC1.6 complex provides link to heterochromatin control system, with its HP1 homolog subunits CBX1 and CBX3. In germ cell differentiation and meiosis regulation ncPRC1.6 has a specific role [173,181,182].
3. Canonical PRC1 Complexes
The composition of mammalian PRC1s are more diverse then their Drosophila counterparts [51,52,183]. Canonical PRC1 complexes consist of one of the five of PC homolog proteins in mammals (CBX2, CBX4, CBX6, CBX7 and CBX8), one of the three PH-like subunits (PHC1, PHC2 and PHC3), any of the six identified PSC homologs (PCGF1–6) together with the two paralogs of the core catalytic ubiquitin ligase subunit, SCE (RING1/RING1A, RNF2/RING1B) (Table 1) [94]. By definition, all PRC1s consist of homologs of these four core members we call “canonical” (cPRC1). The composition, targeting and activity of the various cPRC1s changes dynamically during embryonic development, cell differentiation or in cancer (reviewed in [184,185,186,187]). This is in accordance with the notion that PcG complexes are involved in repressing several thousand genes in mammalian genomes, and the pool of regulatory target genes are not simply tissue specific, but it is changing spatiotemporally as differentiation proceeds (reviewed in [188]). CBX proteins play a crucial role in this process by in fine-tuning lineage commitment and differentiation events. This was first shown in ES cells, where maintenance of pluripotency primarily depends on PRC1s encompassing CBX7, while after lineage commitment, CBX2 and CBX4 have become more abundant in the complexes [189].
The question, how specific subunit exchange in PRC1 complexes happens is still under debate. However, some aspects of the subunit swap can be explained by the presence of a “Polycomb regulatory loop”, which was discovered in ES cells. Morey at al. have shown that the promoters of Cbx2, Cbx4, Cbx8, Phc2 and Pcgf4/Bmi1 are occupied by repressor proteins RING1 and SUZ12 and decorated mainly with the H3K27 repressive marks in pluripotent stem cells. On the other hand, the promoters of Cbx7, Phc1 and Pcgf2/Mel18 contain only active histone marks. This correlates with the high expression of Cbx7, Phc1 and Pcgf2/Mel18 in pluripotent ES cells. When ES cells differentiate and form embryoid bodies (EB), the protein levels of CBX2/MEL18 and CBX4/BMI1 increases and CBX7 decreases [189]. According to these data, the subunit exchange in similar PRC1 complexes can be simply the consequence of the changes of expression level of different paralog genes and the collateral difference of paralog protein concentration. The probability of the more abundant paralogs associating with the repressive complexes is higher and they make the more prevalent subunits. The switch was later explained by microRNA mediated regulation of Cbx7 when ES cells differentiate [190].
Recent studies revealed similar switching mechanism in mouse hematopoietic stem cells (HSCs). Like in ES cells, CBX7 is also required for self-renewal while CBX2, CBX4 and CBX8 for differentiation of HSCs [191].
Recent data suggest that long non-coding RNAs (lncRNAs) may have functions in fine-tuning repression in a cell-lineage specific manner. They initiate or stabilize PRC1 recruitment to a given gene loci [192]. However, only a few specific lncRNAs that guide PRC1 activity have been characterized [193,194,195].
4. Studying Mammalian PcG Functions with Embryonic Stem Cell and Mouse Models
In this review, we rely mostly on data derived from in vitro stem cell-based differentiation experiments and in vivo knockout mouse models (see Table 2, Table 3, Table 4 and Table 5). The two model systems have several advantages.
4.1. Advantages of Mouse Models in Studying Gene Function during Development
Genetically engineered mouse models are powerful tools of studying loss (knockout) of gene function and gain (transgenic) of function in vivo. Using mice has numerous advantages:
- Mice are experimentally tractable mammalian model systems.
- Mice have short reproducing time, easy to breed and maintain.
- Mice have relatively short gestation (20 days) and big litter size (5 to 15 pups), brief time for sexual maturity and rapid generation time, which makes an ideal model for studying embryonic development.
- Mice have small size and are easy to handle.
- Mice have close similarities with human development and disease.
- The genome size, number of genes and genomic organization of mice are similar to humans.
- Mice are suitable for derivation of stem cells, such as ES cells, which can be re-introduced to the mouse germline.
4.2. Advantages of ES Cells in Studying Gene Function during Development
ES cells are pluripotent cells, which are capable of unlimited self-renewal and differentiating to all cell types of the body [196]. They are ideal model systems for studying developmental processes and have multiple advantages over other systems, such as in vivo mouse models:
- There is no need of sacrificing animals since mutations can be introduced to a broad variety of well characterized ES cell lines on versatile genetic background [197,198].
- If required, ES cells lines can be newly established from mice or other species (e.g., bovine, pig) by isolating morula or blastocyst stage embryos [198].
- There are reproducible experimental conditions and a more serum/animal free environment (small molecules, inhibitors and proteins).
- Well scalable thus high throughput experiments can be executed.
- There is no limitation of starting material due to unlimited self-renewal.
- ES cells can be differentiated to all cell types of the body.
- Differentiation conditions can be tightly controlled, which is highly desired for industrial applications.
- Culture conditions for maintaining ES cells and for differentiating them could be internationally standardized conferring high reproducibility to the experimental systems.
- Finally, studying human development by the utilization of existing and approved human ES cell lines [199] circumvents the ethical barriers, as it does not require destruction of preimplantation human embryos.
With the usage of these peculiar cells, it is possible to study different phases of embryonic development in vitro and analyze the functions of different PcG genes during ES cell based differentiation.
ES cell lines were generated from versatile PcG mutations, which enabled the investigation of different PcG functions in this system. Accumulating evidence suggests that some PcG proteins are essential for ES cell differentiation (reviewed in [200]) and genome architecture [201]. Over thousands of developmental genes, driving cell fate decisions are repressed in a PcG protein-dependent manner in ES cells (reviewed in [202,203]).
Due to many advantages of the ES cell based in vitro model systems, nowadays, ES cells are extensively used for classical protein purification studies, mass spectrometry-based schemes [108] and for ChIP experiments followed by massive parallel sequencing of targets [204,205,206]. Taken together, the ES cell based in vitro differentiation system increases feasibility for getting sufficient amount of wild type and mutant cells from reproducible differentiation schemes.
In ES cells a specific bivalent marking, overlapping repressive (H3K27 trimethylation) and activating (H3K4 trimethylation) is found on the nucleosomes of the promoters of more than 2000 genes. These bivalent markings suggest that certain genes are poised for both activation and repression in stem cells [204,207]. Serine 2 phosphorylation of RNA polymerase II is present in the coding regions of the active genes, but it is absent from paused and poised complexes. The phosphorylation of Serine 5, on the bivalent promoters indicates that these genes are ready for transcription [208]. At the same time, PcG protein binding regions of developmental regulator genes are found in close proximity to bivalent markings [209]. These bivalent domains are segregated to two categories; some of them are occupied by both PRC1 and PRC2, while in the other category only PRC2 member proteins are detected. It seems that the presence of PRC1 complex can block RNA polymerase II at the elongation phase [210]. The promoter regions of overwhelming majority of key developmental genes are bound to both PRC1 PRC2 and efficiently retain the H3K27 mark during differentiation and remain stably repressed [204,207]. Computational analysis of the dataset from different ChIP-seq experiments also showed that genome-wide locations of PRC1 and PRC2 complexes can be successfully predicted with the use of mapping locations sizes of CpG islands in parallel with different activator motifs. The main conclusion was that large CpG islands depleted of activating motifs are bound by both PRC complexes in pluripotent stem cells [204,207]. In more recent studies bivalent domains were connected to different nucleosome remodeling complexes too (reviewed in [211]). There are numerous genetic variants of ES cell lines have been made in the past decades. Early knockout models were generated from substrains of inbred 129 mice and not on C57BL/6 background, which is the most frequently used inbred strain. Alterations in genetic background may subtly affect experimental conditions and phenotype. To circumvent this disadvantage, several commercially available recent efforts have been made to utilize common sources of ES cells, such as C57BL6/6NTAC. As of August 2016, the IMPC reported 22,277 mouse lines derived from C57BL6/6NTAC ES cells.
5. General Description of Mammalian ncPRC1s
RING-PCGF dimers form the catalytic core of all PRC1 complexes. In ncPRCs, RYBP or its close homolog YAF2 protein interacts directly with the RING homologs [52,183,189,190,212]. In cPRC1s, the same interaction surface is occupied by the PC or PH homolog subunits [107,213,214]. NcPRC1s were previously called atypical, variant or PRC1-like [52,215], but by now all of these complexes are generally referred to as non-canonical PRC1s.
A structural study proved that RING-CBX and RING-RYBP/YAF2 binding is mutually exclusive. The interacting Cbox domain of CBX homolog proteins and the C-terminal domain of RYBP bind to the same interaction pocket with comparable affinities of the C-terminal ubiquitin fold region of RING1B. This C-terminal region of RING proteins was also identified in PCGF homolog proteins and later called the Ring-finger And WD40 associated Ubiquitin-Like (RAWUL) domain [216]. RAWUL is one of the most important protein–protein interactive motifs in PRC1 proteins. Although RAWUL domains are conserved between RING and PCGF orthologs, different RAWULs have highly specific affinities toward protein partners.
There is a sufficient sequence similarity between the Cbox region of different CBX proteins, but Cbox region shares little sequence similarity of the RAWUL binding regions of RYBP and YAF2. Moreover, both Cbox region and RYBP protein are intrinsically disorganized and only fold upon binding to the RAWUL domain of RNF2/RING1B. The resolved structure of RING1B-RYBP C-terminal domain complex shows that RYBP interacting with RING1B in a very similar way to CBX7 despite the significant sequence differences. Both proteins form a nearly identical intermolecular β-sheet, flanked with a loop structures which in turn are completely different in the two proteins [217]. Both β-sheets and loop structures are required for stable binding to RNF2/RING1B and needed for repression [217]. A specific point mutation in the RAWUL domain of SCE/dRING protein in Drosophila dRING1Y370A, which disrupts the binding capacity of the protein to both PC and RYBP homologs, behaves as a dominant negative allele. The Tyr370 position in dRING1 is equivalent to TYR 262 of human RING1B, a key residue required for stable complex formation with both PC and RYBP. This specific mutation in dRING prevents chromatin association and lead to compromised Polycomb repression [217].
The discovery of ncPRC1s came as a surprise for two reasons. According to the widely accepted model of hierarchical recruitment of Polycomb repressive complexes (see Section 1.3) at a time, PRC1 complexes thought to target chromatin by the recognition of the repressive H3K27 trimethylation mark with the Chromo domain of PC and its homolog CBXs [137]. The lack of PC homologs in ncPRC1s lead to the conclusion that these complexes are targeted differently than their canonical counterparts. The simple model of hierarchical recruitment hypothesis was later exceeded even in the case of cPRC1 recruitment. In Drosophila, 30% of SCE/dRING1 binding sites on the giant chromosome are not bound by PC and PH [145]. The independent targeting of ncPRC1s was supported by the previous finding that, in mouse ES cells, 20% of the RING1B binding sites has no H3K27 trimethylation marking at all (244 gene promoters H3K27 negative out of 1219 total RING1B binding sites) [218]. There is a ncPRC1 type repressor complex in Drosophila, which does not contain PC or PH protein, but has SCE/dRING subunit and capable to H2A specific monoubiquitilation, called dRAF. This complex can also specifically target chromatin without a Chromo and SAM domain containing subunits. dRAF is responsible for the bulk H2A monoubiqitilation in Drosophila [109]. Finding of these ncPRC1 complexes in different model organisms brought up alternative targeting models of PRCs besides the classical, hierarchical recruitment. It seems that the order of recruitment can be reversed. In some cases, H3K27 trimethylation independent targeting of ncPRC1 complexes can precede that of PRC2 complexes [219]. According to the more recent extension of this model RYBP containing complexes are targeted first and the effective H2A mono-ubiqitilation drives PRC2 recruitment and consequent Polycomb domain formation in mammals [124]. In Su(z)12 mutant background, H3K27 methylation is nearly completely abolished, but PRC1 targeting remains intact in Drosophila, which is in accordance with the newest model [219].
Certain ncPRCs are recruited to CpG islands by KDM2B, independently of H3K27 trimethylation by PRC2 [212,220,221,222], but KDM2B subunit is not found in all variant ncPRC1 complexes. It is also worth to keep in mind that KDM2B does not recognize specific DNA sequences, similar to many other transcription factors. It binds to CpG islands, consequently KDM2B cannot be responsible alone for sequence specific recruiting of the complex. It remains another open question, which subunits target the ncPRC1 complexes when more than one potential DNA binding protein is present. For example, in the case of ncPRC1.6, there are four subunits identified with direct DNA binding capacity (Myc Associated Factor X (MAX), Max gene associated (MGA), E2F dimerization partner 1 (TFDP1) and E2F transcription factor 6 (E2F6)). Theoretically, any of these sequence specific DNA binding subunits can target the whole repressor complex to the regulatory regions of multiple genes. Further studies need to clarify the exact role and cooperation of sequence specific DNA binding proteins in the recognition of the in vivo targets during development.
On the other hand, the lack of SAM domain containing PH subunits, in ncPRC1s indicated that the targeting and chromatin compaction mechanism of ncPRC1 complexes must be different too. SAM domain mediates essential interaction between independent Polycomb repressive complexes. In Drosophila, the SAM domain of dSFMBT is tethered to the PRE regions by the specific DNA binding protein PHO. Then dSFMBT/SAM interaction with SAM domain containing proteins of cPRC1 complexes nucleates the recruitment, while SCM-SAM PH-SAM mediated polymerization between separate cPRC1s results the compaction of the repressed domain [140]. The question remains unanswered: if HPH is missing from non-canonical complexes, which other SAM domain containing subunit, or different chromatin compaction mechanisms can substitute the function of HPHs SAM domain?
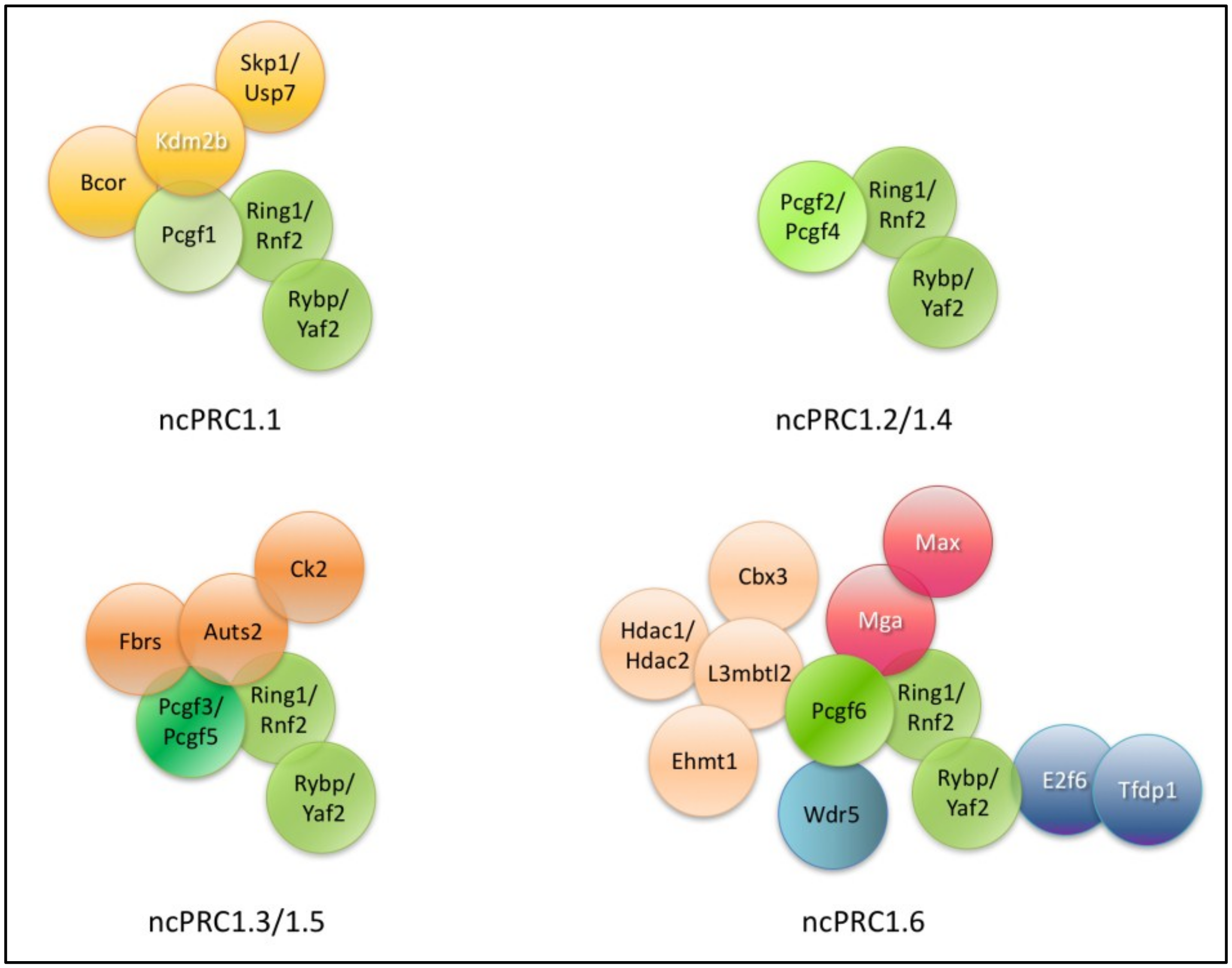
In this review, we focus on the mouse and human ncPRC1 complexes (Figure 1). All ncPRC1 complexes are catalytically active H2AK119 ubiquitin ligases. In fact, ncPRC1 complexes are more effective ubiquitin ligases, than their canonical counterparts [52]. NcPRC1s can be recruited and ubiquitylate their targets independently of PRC2 mediated H3K27 trimethylation [212,220,221,222].
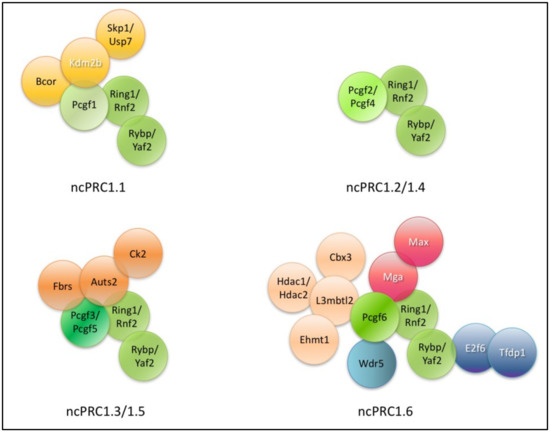
Figure 1.
Encompassing subunits of different ncPRC1 subtypes. Core members as the catalytic subunits Really interesting new protein 1/E3 ubiquitin-protein ligase (RING1), Ring finger protein 2 (RNF2), the Polycomb group ring finger (PCGF) variants and the non-canonical PRC1 definitive RYBP homolog subunits are indicated by different shades of green (RING1/RNF2, PCGF1–6 and RING1 and YY1 binding protein (RYBP) and its close homolog YY1 associated factor 2 (YAF2). The names of specific DNA binding proteins are labeled with white letters (Lysine specific demethylase 2b (KDM2B), Max gene associated (MGA), Myc Associated Factor X (MAX), E2F dimerization partner 1 (TFDP1) and E2F transcription factor 6 (E2F6). Proteins acting as heterodimers (RING1/RYBP, MGA/MAX and E2F6-TFDP1) are indicated with identical coloring.
The molecular diversity of ncPRC1 complexes are mainly explained by diversity of PSC homolog PCGF subunits. All PCGF paralog subunits are able to directly associate RING1A/B proteins, by their RING domain, but they are bound to paralog specific partners due to the minor alterations of their RAWUL domain, which changes their binding specificity. Therefore, the type of the PCGF subunit has profound role in defining the PRC1 complex stoichiometry. As different PCGF paralog proteins can interact with profoundly different partner proteins, subunit composition, targeting mechanism and repressor activity of the differently numbered complexes can highly differ (these aspects are discussed in Section 8.3).
Non-canonical PRC1 complexes often have different biological functions in mouse ES cells than classical PRC1s. Genes regulated by RYBP containing complexes rarely completely silenced, often highly transcribed, and primarily involved in the regulation of metabolism and cell cycle progression. CBX7 containing complexes are typically found on promoters of completely silenced genes, which are involved in early lineage commitment of ES cells [223]. The binding site of CBX7- and RYBP-containing PRC1 complexes overlaps in certain genes, but on other targets their binding are mutually exclusive. These findings indicate that different PRC1 subtypes help to establish a complex pattern of gene regulation and regulate both common and non-overlapping aspects of ES cell pluripotency and differentiation. The targeting mechanism and subunits of different ncPRC1 complexes can be altered profoundly; consequently, different complexes have altered binding specificity and different target specificity.
6. Core Subunits of ncPRCs
6.1. Core Members of ncPRCs
The ncPRC core members are found in all ncPRC1 complexes (Figure 1). These are one of the two catalytic RING subunits one of the six PCGF paralogs and RYBP or YAF2 (Table 2).
RING1 was identified as a protein with a zinc-binding motif belonging to the zinc finger type of Ring domains [224]. The Ring1 null mutant mice are viable however, they exhibit anterior homeotic transformations of the axial skeleton with subtle changes in Hox gene expression. Surprisingly, Ring1 overexpression results similar homeotic transformations caused by the depletion of the gene expression [151]. The relatively mild phenotype of the Ring1 null mutant mice can be partially explained by the functional redundancy with a ubiquitously expressed and highly related Rnf2, which may compensate for the lack of Ring1 during development.
RNF2 was first identified as an interactor of the Polycomb homolog protein M33/CBX2 [225]. Rnf2 null mutant mice are embryonic lethal at gastrulation due to growth arrest of the embryonic and extraembryonic tissues [226]. Mice bearing a hypomorphic Rnf2 allele showed posterior homeotic transformations of the axial skeleton [152]. This phenotype is complementary to the anterior homeotic transformation phenotype of the Ring1 null mutant mice, which suggests that Ring1/Rnf2 together participates in the specification of the antero-posterior axis during development.
By crossing conditional Rnf2flox/flox mouse line with a RERTert mouse deleter line (Cre-ERT2::Polr2a), Calés et al. found that inactivation of Rnf2 increased the proliferative rate of myeloid progenitors but does not cause major alterations in hematopoietic differentiation [227].
Rnf2 homozygous null ES cells have no visible morphological alterations in comparison to the wild type and can be cultured for more than 20 passages [228,229]. Others described depletion of H2A ubiquitination in Rnf2 null mutant ES cells [228]. Further in vitro studies revealed that loss of Rnf2 in ES cells caused downregulation of some PcG proteins (MPH1, PHC1/2, CBX2, PCGF2 and RYBP) and upregulation of developmental control genes (Cdx2, Eomes, Hand1, Foxa2, Hnf4, Hoxa1 and Nestin) [229,230]. Other ES cell studies found that members of TGF-β-signaling pathway and Bmp signaling transcriptionally upregulated upon loss of Rnf2 [231]. Inactivation of Rnf2 resulted in premature neural differentiation of embryonic neural stem cells (NSCs) in conditional Rnf2 mouse mutant [232], the same mouse line that Calés et al. used [227]. Further ES cell studies revealed that RNF2 target genes repress Wnt signaling [223]. One of the latest in vitro studies showed that knockdown of Rnf2 promotes cell-cycle arrest and apoptosis in prostate cancer cells [233] and gastric cancer cells [234].
Further in vitro studies revealed that Ring1/Rnf2 double mutant showed proliferation arrest, ES cells lost its cell morphology, several genes involved in differentiation were upregulated while several PRC1 components downregulated. Ring1, Rnf2 and Oct3/4 appear to be functionally linked together in mediating stem cell identity [235]. Others also found that failure in S-phase entry, proliferative arrest and p21 upregulation in Ring1/Rnf2 double mutant cells [236]. In vivo studies found that Bmp signaling is altered in the mandibular molar of Ring1−/−; Rnf2cko/cko mice [237].
Notably, there is a compensatory mechanism between Ring1 and Rnf2 in their function, which is also reflected by the increased expression of RING1 protein observed in the Rnf2 null ES cells. Both Ring1 and Rnf2 are repressing developmental genes in ES cells and together they are required for the maintenance of ES cell identity. When both Ring1 and Rnf2 genes are ablated, ES cells tend to differentiate and lose pluripotency. This is also linked to pluripotency marker Oct4 expression, indicating a potential functional link between Oct4, Ring1 and Rnf2 [235].
RYBP and its paralog, YAF2 are intrinsically disordered proteins with a RanBP2-type zinc finger motif at their N-terminal end [238]. RYBP was first isolated as an interacting partner of the PcG protein RING1/RNF2, YY1 and CBX2/M33 [239]. RYBP/YAF2 has no known DNA binding consensus sequences however RYBP is able to bind DNA nonspecifically [238]. Both RYBP/YAF2 and CBX proteins, despite their dissimilar conformation, contact the same region of RNF2 [217]. In vivo mouse studies showed that Rybp is essential for embryonic development as the homozygous Rybp null mutant mice dies at implantation (Table 2). Furthermore, the portion of the heterozygotes exhibited forebrain overgrowth, disrupted neural tube closure, exencephaly, lack of cerebellum [166], retinal coloboma, malformed lenses, incomplete closure of the optic fissure (Table 2) [240]. This suggests that the effect of Rybp depletion is dosage dependent and Rybp belongs to the dosage sensitive genes. RYBP target genes repress Wnt signaling [223] and others found that in undifferentiated myoblasts, TGFβ-Smad3 signaling enhances the recruitment of a complex, which contains YY1 and RYBP [241] confirming a possible tight functional link between RYBP and YY1. RYBP and YY1 also mediates the expression of miR-125a, which is a downstream mediator of Notch signaling [242], a signaling pathway important for epithelial–mesenchymal transition and numerous differentiation steps during lineage commitment of progenitor cells. Due to the early embryonic lethality of the Rybp homozygous null mice, the role of Rybp in later stages of embryonic development and lineage commitment was further studied in conditional gene ablations systems. The role of Rybp in hematopoiesis was further studied when RYBP function was conditionally ablated during adult hematopoiesis by the Mx1-Cre reporter. As a result, Rybp deletion caused an increase in the number of B-1 progenitors and the loss of B-2 progenitors [243] revealing its role in progenitor fate decisions. This study was the first to reveal a function of Rybp in cell fate decision making and lineage commitment. Parallel in vitro studies also showed that Rybp has important role in differentiation of stem cells. Whilst the Rybp−/− ES cells are viable, can proliferate and can be maintained for an unlimited time, they do not form contractile cardiomyocytes (CMCs) (Table 2) [244], and form less matured neurons, astrocytes and oligodendrocytes in vitro (Table 2) [245]. These results are in agreement with previous in vivo studies suggesting the role of Rybp in differentiation rather than self-renewal [166,240]. Collectively, these data suggest that Rybp is dispensable for self-renewal, required for proper differentiation of mouse ES cells in vitro and important for differentiation and certain early cell fate decision steps in vivo.
The human YAF2 was isolated as an interacting partner for YY1 [246]. Later, it was found that YAF2 also interacts with MYCN [247], MYC [248] and RING1/RNF2 [249]. YAF2 is paralog of RYBP [239] and together they constitute a functionally different cofactor family for YY1 and E4TF1/hGABP transcription factors [250]. YAF2 has promyogenic regulatory role, since YAF2 overexpression stimulates myogenic promoter activity in C2 myoblasts [246]. It was also found that Yaf2 enhances MycN dependent transcriptional activation in human neuroblastoma cell lines [247], however inhibits MYC mediated transactivation and transformation [248] suggesting its potential role in tumorigenic processes. Importantly, YAF2 serves as a mediator to bridge between YY1 and PRC complex proteins, such as EZH2 or RING1/RNF2 [251] and this interaction is essential for PRC recruitment in mammals [252]. No Yaf2 homozygous null mice have been reported yet, which keep their function in a shadow (Table 2).
6.2. The Function of Yy1
YY1 is a PcG protein with DNA binding ability [253] (Table 1). The protein has never been purified in cPRC or ncPRC complexes, but it can interact with ncPRC1 core subunits, such as RYBP [239]. YY1 was isolated simultaneously by two independent research teams, based on its binding ability to adenovirus P5 promoter [254], immunoglobulin κ3′ enhancer and the immunoglobulin heavy-chain µE1 site [255]. YY1 acted as a repressor on all of these targets [254,255]. Yy1 homozygous null mutant mice die at the time of implantation and heterozygotes exhibit neurulation defects [256] similarly to the Rybp mutants [166]. This may suggest that the two genes and their products are functioning in common developmental pathways or in close connection (e.g., as heterodimers or binding partners). Ectopic expression of YY1 inhibits, while knockdown of endogenous YY1 enhances TGFβ- and BMP-induced cell differentiation [257]. Yy1 heterozygous mice show anterior homeotic transformations of the axial skeleton [153] and impaired peripheral myelination [258]. Hoxb1 gene expression is sensitive to Yy1 dosage: in Yy1+/− ES cells, the Hoxb1 expression is upregulated [153]. Conditional gene ablation studies of the normal Yy1 function have proven multiple and complex role of Yy1 in embryonic development and endoderm specification. Epiblast specific deletion of Yy1 results a defect in epithelial to mesenchymal transition (EMT) due to impaired E-Cadherin expression of the streak descendants [259]. The same study has also shown that the expression of Nodal, which is a critical component of gastrulation, is sensitive for the YY1 dosage: ablation of YY1 resulted multiple impairments in definitive endoderm patterning and coincided with misexpression of genes required for definitive endoderm development of the embryo proper (Foxa2, Shh, Fgf4, Fgf8 and Lefty2) [259]. The latest studies found that YY1 interacts with SMAD7 and the interaction is attenuated by TGF-β signaling, and SMAD7 and YY1 together inhibit TGF-β-induced transcription in the nucleus [260]. RYBP and YY1 mediates the expression of miR-125a, which is a downstream mediator of Notch signaling [242] and YY1 is required for posttranscriptional stability of pluripotency factors, OCT4 and SOX2 [261].
Studies on hypomorphic Yy1 murine model (Yy1flox/flox) revealed that reduction of YY1 levels impairs embryonic growth and viability and pinpoint dose-dependent function of YY1 [262]. Mesp1-cre mediated knockout of Yy1 (Mesp1-YY1) at early developmental stage (at E6.5) resulted in abolished cardiac lineage formation and YY1 together with GATA4 transcriptionally activates early cardiac enhancer, Nkx2.5 [263]. A latest study found that MHC-cre mediated Yy1 null mice (MHC-YY1) displayed congenital heart defects with defective cardiomyocyte proliferation and increased apoptosis, while Nkx2.5-cre mediated Yy1 null mouse embryos (Nkx2.5-YY1) showed hypoplastic endocardial cushions in atrioventricular canal and outflow tract and died around E13.5 [170]. They also identified downstream targets of YY1 that are cardiac morphogenesis regulators (Mesp1, Cited1, eHAND and Dll) [170].
6.3. Detailed Description of the Composition and Function of Different ncPRC1 Type Complexes
6.3.1. ncPRC1.1
The ncPRC1.1 complex consists of PCGF1, KDM2B also known as F-Box and Leucine-rich repeat protein 10 (FBXL10), BCOR and S-Phase Kinase associated Protein 1a SKP1/USP7 (Figure 1) (Table 3) [107,108].
The PCGF1 containing complex was originally called BCOR. PRC1.1 is similar to dRAF complex of Drosophila. The complex is abundant in the nervous system [264], has effective H2A ubiquitin ligase and histone demethylase activity, and targets the CpG islands by KDM2B [265,266] (reviewed in [267]). Resolving the structure of PCGF1-BCOR subcomplex revealed the basis of the binding selectivity of different PCGFs [178].
PCGF1 (also known as Nervous System Polycomb 1 (NSPC1)) was first found in the developing peripheral nervous system as a protein with an N-terminal RING finger domain (Table 1), and as a protein mediating ubiquitination and protein–protein interaction [268]. PCGF1 directly interacts with BCOR, RNF2 and RYBP [52,107,269]. Knockdown of Pcgf1 in HeLa and embryonal carcinoma cell line NT2/D1 results in significant reduction of H2A ubiquitination and DNA demethylation [270]. Yan et al. have recently shown that PCGF1 promotes ectodermal and mesodermal fate during ES cell lineage specification via EB formation [264]. They also found that Pcgf1 deletion in ES cells, established by CRISPR/Cas9 technology, does not interfere with self-renewal but impairs differentiation, likely by preserving high level expression of key pluripotency factors (Oct4 and Nanog) [264].
BCL-6 interacting Corepressor (BCOR) is named for its function as a corepressor, which can potentiate B-cell Lymphoma 6 protein (BCL6) repression [271] (Table 3). BCOR protein is also a member of the BCOR-complex [107], which is named after it. There is a protein–protein interaction motif identified in BCOR called PCGF ubiquitin fold discriminator (PFUD) (Table 1), which selectively binds the RAWUL domain of PCGF1 and can discriminate between different PCGF RAWUL domains [178]. Wamstad et al. found several functions of BCOR both in vivo and in vitro: (1) in bcorneo mutant mice, which exhibit a parent-of-origin effect (with paternally imprinted X-inactivation), it indicates that Bcor is indispensable in extra-embryonic tissue; (2) loss of Bcor results in delayed repression of pluripotency factor Oct3/4 and delayed activation of genes responsible for ectodermal and mesodermal lineage specification during in vitro differentiation of ES cells; and (3) BCOR is also required for formation of primitive erythrocytes [272] and, in Bcor mutants, there is a failure of heart looping [273]. Mutations of Bcor has been found in acute myeloid leukemia [274], in retinoblastoma [275], in medulloblastoma [276] and in kidney sarcoma [277,278], suggesting its role in tumorigenic processes. Bcor ablation resulted in higher rate of myeloid cell proliferation and differentiation of CRE induced conditional Bcor knockout (Bcor−/Y) mouse bone marrow cells [279].
KDM2B is an F-BOX containing histone demethylase (Table 1), a key member of the BCOR-complex as it is essential for recruitment of the ncPRC1 complex [107] and also an interactor of RING1/RNF2 [108]. The CpG binding activity of the protein targets ncPRC1.1 complexes specifically to CpGIs [265,266] (reviewed in [267]). Absence of Kdm2b is perinatally lethal in mice due to incomplete neural tube closure, exencephaly, altered cell-cycle processes in neural precursors and reduced number of spermatozoa [280]. Hemizygous loss of the CxxC motif, which is responsible for DNA binding of KDM2B results in a homeotic transformation in mice and loss of ncPRC1 genomic occupancy, supporting its role in the recruitment of ncPRC1.1 to target locus [124]. Knockdown of Kdm2b in primary mouse embryonic fibroblasts inhibits cell proliferation and induces cellular senescence in a pRb- and p53 pathway dependent manner [281]. Knockdown of Kdm2b in mouse ES cells induces early differentiation processes and upregulates mesoderm and endoderm specific genes (Gata4, Gata6, Sox7, Sox17, Eomes and Foxa2) [282]. Knockdown KDM2B in different types of human breast cancer cell lines (MDAMB-231, MCF7 and T47D) upregulates WNT1, but inhibits the Wnt/β-catenin signaling [283,284]. Ablation of KDM2B in ES cells causes DNA hypermethylation [285]. One of the latest studies found that Kdm2b is required for hematopoietic cell development and, during this process, KDM2B regulates cell lineage commitment in cooperation with PcG and TrxG complexes [162].
S-Phase Kinase Associated Protein 1a (SKP1) was first identified in a complex with human CyclinA–Cyclin-Dependent Kinase 2 (CDK2) in conjunction with SKP2 [286] and later SKP1 was found promoting ubiquitination and degradation of various cell cycle regulators [287,288]. Inactivation of Skp1 (in Cul-1 deletion mutant (Cul1-N252), where Skp1 is inactivated) results in hypoplasia and reduced proliferation in the lymphoid organs, however, after a latent period, the Cul1 mutant mice develop T-cell lymphomas with high penetrance (Table 1) [289]. SKP1 is significantly reduced in sporadic Parkinson’s disease and RNAi-mediated silencing of SKP1A in neuronal cells increases susceptibility to cell death, whereas intranigral stereotaxic injection of a lentiviral vector targeting SKP1A induces pathological and behavioral deficits in mice [290].
Ubiquitin Specific Peptidase 7 (USP7), also known as Herpesvirus-Associated Ubiquitin-Specific Protease (Hausp) was originally identified as an interaction partner of Infected Cell Protein no. 0 (ICP0), also named Vmw110, a gene with a role in the initiation of the viral lytic life cycle in herpes simplex virus 1 [291]. Further studies have shown that USP7 can bind Transformed Mouse 3T3 cell Double Minute 2 (MDM2), as well as regulate its stability and functions through deubiquitination [292], and other results showed the importance of this interaction: lack of USP7 resulted in the stabilization of p53 due to the loss of MDM2 [293]. Usp7 knockout mice die during early embryonic development between embryonic days E6.5 and E7.5. Usp7 knockout embryos showed p53 activation, reduction in proliferation, but no apparent increase in apoptosis (Table 3) [294]. Deletion of Usp7 in neural cells resulted in neonatal lethality, brains from these mice (lacking Usp7 specifically in the brain) displayed hypoplasia and deficiencies in development, which were mainly caused by p53-mediated apoptosis; and neural cell survival and brain development of Usp7-mutant mice can largely be restored in the p53 null background (Table 3) [295]. USP7 knockdown by short hairpin RNA (shRNA) promotes neuronal differentiation and disrupts self-renewal in human neural progenitor cells [296]. A latest study identified de novo heterozygous loss-of-function mutations of USP7 in individuals with a neurodevelopmental disorder, featuring intellectual disability and autism spectrum disorder (Table 1) [297]. Knockdown of USP7 in human adipose-derived stem cells (hASCs) with shRNAs compromises osteogenic differentiation both in vitro and in vivo [298].
6.3.2. ncPRC1.2 and ncPRC1.4
The ncPRC1.2 and ncPRC1.4 complexes consist of RING1/RNF2, PCGF2 or PCGF4, respectively, and RYBP/YAF2 subunits only (Figure 1) (Table 4). No additional subunits were purified, thus determining the tissue specific subunit composition of these kind of ncPRCs remains to be further elucidated. In the first large scale purification of different PRC1 type complexes PCGF2 and PCGF4 was found only in cPRC1s [52]. Pcgf2/Mel18 and Pcgf4/Bmi1 functions are partially redundant. Knockout mutants of both proteins are viable, but display paralog specific phenotypic alterations. The Pcgf2/Mel18 and Pcgf4/Bmi1 double mutant embryos exhibit severe growth retardation, accelerated apoptosis and defects in the maintenance of stable Hox cluster gene expression [299].
Pcgf2 was isolated as a complementary DNA from B16 mouse melanoma cells [300]. Mice lacking Pcgf2 exhibit strong growth retardation, posterior transformations of the axial skeleton and die around 3–6 weeks after birth due to obstruction of the lower intestine (Table 4) [80]. Absence of Pcgf2 also causes defects in T and B lymphocyte development and hypertrophy of intestinal smooth muscle (Table 4) [301]. Others found that, in pcgf2 homozygous null mutant [301] T progenitors Hes1, one of the target genes of the Notch signaling pathway, is downregulated [302]. PCGF2 is repressing negative regulators of the Bmp pathway [303]. Blocking of Pcgf2 up-regulates the expression of the WNT/TCF target Jagged1, a Notch ligand, and consequently activates the Notch pathway [304]. PCGF2 represses pluripotency genes, lineage specification genes, late cardiac differentiation genes, and negative regulators of the Bmp pathway but positively regulates expression of key mesoderm transcription factors (Tbx5, Tbx20, Nkx2.5, Mixl1 and Myl7) during cardiac differentiation due to direct activation of several mesoderm specific genes (Gata4, Hand1, Lhx1 and Six2) [305]. Pcgf2/Mel18 depletion by two specific shRNAs suggested that the protein is not necessary to the self-renewal of ES cells and required for the stability of PHC1 and CBX7 proteins [305]. Genome wide ChIP coupled massive parallel sequencing analysis of PCGF2/MEL18 binding sites in during cardiac differentiation of ES cells revealed that PCGF2 has two antagonistic functions. Mechanistically PCGF2/MEL18 has classical repressive function during early cardiac differentiation. It is a negative regulator of pluripotency genes and members of the BMP pathway. However, at the same time, PCGF2/MEL18 is a positive regulator of key mesodermal transcription factors. The protein is required to specify PRC1 function in a developmental stage and context specific manner [305].
Pcgf4/Bmi1 is an oncogene and has essential functions during normal development and adult life. Homozygous null Pcgf4/Bmi1 mouse mutant phenotypes are attributed mainly to the depletion of the classical cPRC1 complexes (Table 4). Pcgf4/Bmi1 functions required for postnatal stem cell maintenance of multiple tissues including hematopoietic [306,307] and neural [308,309]. Pcgf4/Bmi1 null mutant mice are viable but display progressive postnatal growth retardation, defect in hematopoiesis and neurological abnormalities manifested by seizures [148]. Furthermore, Pcgf4/Bmi1 is required for the self-renewal of NSCs in the peripheral and central nervous systems but not for their proliferation or differentiation (Table 4) [310]. Ablation of Pcgf4/Bmi1 function in mice led to decrease in the number of bone marrow progenitor cells. Pcgf4 is also involved in hematopoiesis because a loss-of-function allele causes a profound decrease in bone marrow progenitor cells.
6.3.3. ncPRC1.3 and ncPRC1.5
Polycomb group ring finger 3 (PCGF3) or Polycomb group ring finger 5 (PCGF5 ) paralogs are found in ncPRC1.3 and ncPRC1.5 complexes, which harbor the catalytic RING subunit, RYBP/YAF2 and Fibrosin (FBRS) (Figure 1) (Table 5). Two accessory subunits of the complex are Casein kinase 2 alpha prime polypeptide (CSNK2A2), also known as Casein kinase 2 (CK2) and Autism susceptibility candidate 2 (AUTS2), which were identified in the central nervous system (CNS). The closest relative of PCGF3 is PCGF5. The similar functions of the two paralogs (PCGF3 and PCGF5) in mouse embryonic development and in response to Xist RNA regulation has recently been established [311]. Almeida et al. generated Pcgf3−/− and Pcgf5−/− single and double mutant ES cell lines, where Xist expression could be induced by doxycycline. In the double mutant cells, Xist dependent H2AK119Ub and H3K27trimet deposition was strongly reduced. This study has also shown that both ncPRC1.3 and ncPRC1.5 mediates ubiquitylation of histone H2A signals and initiates the recruitment of both PRC1 and of PRC2 in response to Xist RNA expression. When mutations were introduced to the mouse germline, single mutants of Pcgf3−/− and Pcgf5−/− were viable with no apparent phenotype. However, Pcgf3−/− and Pcgf5−/− double mutant mice exhibited female-specific embryo lethality at mid-gestation (E9.5–E10.5). Male embryos were grossly normal looking but female embryos were resorbed and their placenta was abnormally formed (lack of throphoblast and labyrinth cell layers). Finally, in the double homozygous mutants, Xist mediated gene silencing was compromised. These suggest that Pcgf3 and Pcgf5 has important role in both embryonic and extraembryonic development and that their functions are at least partially redundant.
Pcgf3 homozygous null mutant mice are viable with no apparent phenotype. Increasing body of evidence suggest that the functions of PCGF3 can be fully substituted by its paralog PCGF5.
The effect of Pcgf5 gene ablation in mouse embryonic development and hematopoietic lineages was extensively studied ([312] and see above at Pcgf3). Pcgf5 null mutant mice are viable without any obvious phenotype. Similarly, wild type and Pcgf5 null hematopoietic stem and progenitor cells (HSPCs) exhibited morphologically identical phenotype. This was unexpected considering that Pcgf5 is broadly expressed in HPSCs. A possible explanation for this apparent controversy is that Pcgf5 either has no specific function in HSCSc maintenance, proliferation and differentiation or other PCGFs (e.g., PCGF3) may compensate for its function. Notably, at molecular level, the H2AK119ub1 levels of the Pcgf5 null mutants were reduced in comparison to the control but this reduction had no effect on global gene expression. Genome-wide studies suggested that PRC1.5 complexes encompassing CK2 and AUTS2 are recruited to active genes, uncovering a new function of the PRC1.5 in activating transcription ([313] and see below).
CK2 belongs to the serine/threonine-selective protein kinase family. CK2 is consisted of two α (α and α’) and two β (β and β’) subunits, foremost of which α is responsible for the catalytic and β for the regulatory activity. Since the mammalian CK2 interacts with over hundreds of target genes, the phenotypes of different subunit mutations are versatile. In mice, the Ck2β gene activity is essential for early embryonic development; homozygous null embryos unable to develop beyond the blastocyst stage during implantation due to their diminished proliferation capacity [314], CK2β is required for stem cell maintenance: investigators were unable to generate Ck2β−/− ES cells or mouse embryonic fibroblasts (MEFs). Due to the critical role of CK2β in stem cell maintenance and embryonic development, the tissue specific role of Ck2β was studied in conditional gene ablation experiments. CK2β is able to bind a plethora of key signaling components and transcription factors and therefore this kinase has important role in multiple aspects of mammalian development (reviewed in [315,316]). Among these, CK2β binds to OLIG2, an essential transcription factor for oligodendrocyte lineage specification of neural progenitors. CK2β activates OLIG2 by phosphorylation. This was first shown by a stem cell mediated in vitro differentiation system, where a conditional mutation was introduced to the Ck2β genomic locus in ES cells. When this mutation was introduced to mice and the mice were crossed with nestin-cre reporter breeding pairs resulting offspring with defect in oligodendrogenesis in their telencephalon. This experiment showed that CK2β is required for normal oligodendrocyte lineage specification and that CK2β is a positive regulator of oligodendrogenesis [317].
The minor CK2α’ catalytic subunit has critical role in germ cell development at all stages of spermatogenesis and homozygous null Ck2α−/− male mice are infertile [318,319]. The phenotype resembles human globozoospermia. Mice lacking the more abundant, major CK2α subunit die at mid-gestation (E9.5–E10.5) due to neural tube defects, improper branchial arch and heart development. The latest one is likely the causative of the mid-gestational lethality [320]. Multiple studies indicate the importance of normal CK2 function is the specification of muscle, neuronal, adipose, osteogenic, hematopoietic, or germ lineage differentiation and stem cells self-renewal process. These studies clearly indicate that the subunits of CK2 cannot substitute each other, indicating their unique roles in nearly all developmental and differentiation processes. Organisms and organs cannot develop without CK2.
Finally, due to the broad substrate specificity of CK2 subunits, it is beyond the scope of this review to list all of the mouse CK2 mutants generated so far. There are numerous review articles and book chapters describing mouse mutants and the role of CK2 in embryonic and adult development (see review in [321]).
AUTS2 protein has been isolated as a component of ncPRC1.5 in the CNS. ncPRC1.5 containing AUTS2 is also frequently called PRC1-AUTS2 or ncPRC1.5-AUTS2, which is consisted of PCGF5, CK2B, RING1/RNF2 and RYBP/YAF2. This complex has some unique properties among PRCs: it can activate the target genes, which is in contrast to the repressive function of the cPRC1 members. Activation by this complex is achieved by CK2B phosphorylation of RING1/RNF2 and by the AUTS2-mediated recruitment of P300. RING1/RNF2 phosphorylation leads to neutralization of its activity and recruitment of P300 responsible for gene activation.
There is no homozygous null Auts2 mutant has been made so far. However, the Auts2 gene is deleted conditionally in neuronal progenitor cells by the Cre transgenic Nestin promoter [313]. The mutation was named as Auts2tm1.1Dare or Tg(Nes-Cre)1Kln/0 where exon 7 of the Auts2 gene is deleted by the Cre transgenic Nestin promoter. The Auts2 mutation led to developmental defects of the CNS in mice, which resembled the AUTS2 disruptions in human [322,323]. In the mouse model, conditional homozygous Auts2 mutants were viable and the heterozygotes showed a broad range of intermediate phenotypes such as impairments in basic motor skills, or cognition characteristics of the AUTS2 human disease symptoms. Auts2 has a long (coded by 19 exons) and short isoforms (coded by 11 exons). In ES cells the long isoform is expressed exclusively and when ES cells committed to neural lineages the short Auts2 isoform also starts to be expressed. Auts2+/− ES cells have premature neuronal differentiation during in vitro corticogenesis resulting accelerated cell death of the Auts2+/− corticoids. Mesodermal gene expression is also upregulated during this process [324]. These data suggest that Auts2 has a role in shifting ES cell proliferation to differentiation perhaps via selective interactions with the ncPRC1.3/1.5.
Fibrosin (FBRS) is a lymphokine, which is secreted by activated lymphocytes that induces fibroblast proliferation [325]. Gao et al. have isolated FBRS by tandem affinity purification (TAP) as an interacting partner of PCGF3 and PCGF5 [52]. FBRS stimulates expression of alpha SMA-producing cells in culture and plays an important role in wound healing, scaring and fibrotic diseases by up regulating myofibroblasts expression. There is no available homozygous null mouse model of Fbrs till now and the role of FBRS cell maintenance and differentiation is uncharacterized. FBRS, as part of the ncPRC1, most likely connects PRCs to the process of wound healing and regeneration but the details of this biological process are not yet known.
6.3.4. ncPRC1.6
All ncPRC1.6 complexes are characterized by the presence of Polycomb group ring finger 6 (PCGF6), also known as Mel18 and Bmi1-Like RING finger protein (MBLR) subunit. Numerous ncPRC1.6-type complexes were purified during the years. These complexes typically contain RING1/RNF2, PCGF6, either RYBP, or YAF2, Lethal(3)Malignant Brain Tumor-Like protein 2 (L3MBTL2), E2F6, EHMT2/G9A, EHMT1/GLP and either CBX1 or CBX3 (Figure 1) (Table 6) [52,179,206,213,215]. The similar complex (PRC1L4) purified by using L3MBTL3 as a bate was devoid of detectable amount of EHMT2/G9A, but contained two histone deacetylases, Histone Deacetylase 1 (HDAC1) and Histone Deacetylase 2 (HDAC2), and a histone demethylase lysine (K)-specific Demethylase 5C (KDM5C/JARID1C) and Retinoblastoma Binding Protein 4, chromatin remodeling factor 4/7 (RBBP4/7), also known as CAF1 subunits, identified in PRC2 complexes [214].

Table 6.
Phenotypes of ncPRC1.6 subunit knockouts. Phenotypes of homozygous knockout mouse mutants and ES cells are listed.
The first non-canonical PRC1-type repressor complex ever purified belongs to the ncPRC1.6 type complexes. The complex was found in HeLa cells fifteen years ago and originally named E2F6.com-1, because a double tagged E2F6 protein was used in the purification scheme as a bate [213]. It was well known at a time that E2F proteins are generally responsible for the activation of genes required for proliferation, cell cycle progression and DNA replication [326,327,328]. The isolated E2F6.com-1 complex seemed to be peculiar because it had nothing to do with promoting S phase, or apoptosis, as it was expected from E2F containing complexes, it acted in senescent cells in the G0 phase of the cell cycle [213]. E2F6 protein differs from other E2Fs, because although it has E2F type DNA binding and the dimerization domain, it is “truncated” it does not have a C-terminal transactivation domain; it cannot interact with pRB, thus cannot trans-activate the target genes. Besides E2F6, three additional proteins with different sequence specific DNA binding abilities were identified in the E2F6.com-1: (a) the transcription Factor TFDP1/DP1, which has been previously identified as direct binding partner of E2F proteins [329,330,331]; and a pair of DNA binding proteins: (b) MAX; and (c) MGA, which were known to be involved in transcriptional control and recruitment of coactivator and corepressor complexes [332]. MAX and MGA heterodimers (MAX/MGA) have a different binding specificity towards both E-box (MYC binding sites) and T-box (Brachyury sites) sequences [333]. This suggested that ncPRC1.6 can be recruited to a variety of target genes by its different DNA binding subunits.
The E2F6.com-1 contained two paralog SET domain containing H3K9 specific histone methyltransferase proteins, EHMT1 and EHMT2 (also known as NG36/G9a/KMT1C) [65,334]. The complex has a CBX3/HP1γ subunit, which can recognize H3K9 methylated histones and was implicated in silencing of euchromatic genes upon recognition of H3K9 specific methylation mark [335]. The presence of HP1 homolog subunits provides a link to heterochromatic control system. In other words, the complex contained a specific H3K9 histone marking module and a subunit which is able to recognize it. Unexpectedly, RYBP was not isolated as a member of the E2F6.com-1 complex however it was previously shown that RYBP can interact with E2F6 [336,337]. NcPRC1.6 type complexes are the most abundant ncPRC1s found in mouse ES cells and have a key role in maintaining pluripotency. During neuronal and hematopoietic differentiation, their amount declines [206,338].
PCGF6 protein is a master regulator of stem cell identity and has important role in differentiation. The effect of Pcgf6 gene ablation was studied only in stem cell based in vitro model systems. No knockout mice were generated and studied. It was found that Pcgf6 expresses at high level in ES cells and acts as a positive regulator of transcription. Pcgf6 expression declines upon differentiation. Zhao et al. found that Pcgf6−/− ES cells were indistinguishable from wild type, based on gross morphology and pluripotency gene expression. However, Pcgf6−/− ES cells had decreased proliferation (doubling time 13–22 h), which was rescued by re-introduction Pcgf6 to the cells. At molecular level, Pcgf6 null ES cells showed overexpression of genes involved in differentiation, especially in spermatogenesis and tend to differentiate towards endoderm when they formed EBs in vitro. Importantly, Pcgf6 deletion in MEF did not have any effect on proliferation, suggesting that PCGF6 is important for stem cell-renewal but its function is dispensable in differentiated cells [339]. The genomic binding profile of PCGF6 is similar to TRXG activators and coincides to active chromatin marks. Knock down of Pcgf6 reduced the expression of core pluripotency regulators Oct4, Sox2 and Nanog [338,340]. Conversely Pcgf6 overexpression prevented downregulation of these factors, impaired differentiation and enhanced reprograming [340]. Consistent with a role in pluripotency PCGF6 could replace SOX2 in the generation of germline competent induced pluripotent stem cells (iPS). Zdzieblo et al. found that PCGF6 represses genes involved in spermatogenesis and mesodermal differentiation [338]. PCGF6 is expressed in meiotic and post-meiotic male germ cells in the testis. It could negatively regulate the proliferation of immortalized mouse spermatogenic cells (GC-2 spd) [341].
L3mbtl2 is an oncogene, which is also essential for mouse development. The gene is widely expressed. Homozygous l3mbtl2 mutant mice seemed to develop normally until day 6.5, but one day later they show growth retardation and failed to gastrulate further. Development of the extraembryonic tissues was also disturbed. At day 6.5 l3mbtl2−/− mutants did not show pro-amniotic cavity, chorion, or amnion [215]. L3mbtl2 regulates proliferation of ES cells, but not required for maintenance of ES cell identity. L3mntl2 loss in ES cells affects the expression of over 900 genes. Mutant ES cells are not able to form cavitating EBs. Both the Zn finger domain and the four methylated histone recognition module MBT domains of the proteins are essential for the protein function of the protein. This is one of the central module of PRC1.6 complexes are capable to interact with NuRd complex components, PRC2 component and transcription factors at the same time [214]. L3MBTL2 protein can be SUMOylated. SUMOylation facilitate its repressor ability especially on targets bound with low efficiency, such as pro-implamatory genes [342].
Max gene is also essential in mouse. Loss of Max function resulted generalized developmental arrest both in embryonic and extraembryonic tissues at early post-implantation, after the dilution of maternal Max stores [343]. MAX is a repressor of germ cell related genes [181]. Complete ablation of Max expression in ES cells results in special cytological changes reminiscent of meiosis [182]. MAX bound to MGA in ncPRC1.6 complex. MGA, the dual specificity-DNA binding protein contains T-box and basic-helix-loop-helix-zipper domains too. MGA depletion in mouse leads to the death of ICM cells in vivo and ES cells in vitro [344].
Depletion of E2f6 in mouse cause axial transformations [345] and derepression of germ cell specific genes [346].
The loss of the binding partner of E2F6, TFDP1 leads death in utero in mouse and compromises the development of the trophectoderm derived tissues [347].
In summary, ncPRC1.6 complex members have a profound role in controlling pluripotency and cell proliferation, in the development of extraembryonic tissues and germ cell lineage commitment, as well as in the regulation of the entry to meiosis (reviewed in [348]).
7. Targeting of Different ncPRC1 Complexes
Different PcG complexes act redundantly [349]. The majority of our knowledge about PcG repression is based on research done on the two classical complexes, the H3K27 specific histone methyltransferase PRC2 and cPRC1. As cPRC1 complexes all contain a Chromo domain protein PC-CBX homolog, which can recognize the specific methylation mark, the mechanism of PcG-dependent repression seemed simple. First, the H3k27 trimethylation mark is made by PRC2, and then it is recognized by PRC1. However, everything is turned upside down by the identification of a new set of repressive complexes, the variant or ncPRC1s [52,212,215].
ncPRC1 complexes lack the CBX and PHC subunits. The discovery of these ncPRC1 complexes directly challenged the accepted classical hierarchical model of Polycomb recruitment described above. As ncPRC1 complexes have no Pc homolog subunit, they are not able to recognize and bind H3K27 tri-methylated histones. Since these complexes do not recognize the PRC2 made histone mark, they must be targeted differently than it was supposed for their canonical counterparts.
First the role of two CpG island binding proteins was recognized. It is proven that the H3K36 specific demethylases KDM2B and FBX10 can target ncPRC1.1 complexes to CpG islands. The subsequent effective H2AK119 monoubiquitilation facilitates the subsequent recruitment of PRC2 and the initiation of a repressed Polycomb domain [124,266]. The discovery of this new targeting mechanism not only explained the targeting of PRC1.1 type complexes, but at the same time challenged the classical hierarchical recruitment model widely accepted for more than a decade [124]. The strongest argument against the prevalence of classical PRC2 H3K27 methylation dependent targeting mechanism was presented by Tavares et al. [212]. They found that RYBP-RING containing variant PRC1 complexes targeted normally in the EED mutant, despite the almost complete loss of PRC2 dependent H3K27 trimethylation. Similarly, in Drosophila, in Su(z)12 mutant background, H3K27 methylation is nearly completely abolished, but PRC1 targeting remains intact [219]. At the same time, these experiments underlined the collateral importance of additional targeting subunits in effective ubiquitination activity of ncPRC1 complexes. FBX10 depletion not only caused a decrease in RNF2 binding to target genes, but at the same time resulted to major loss of H2Ak119 ubiquitilation and serious defect in ES cell differentiation ([266], reviewed in [350]). CpG targeted ncPRC1.1 complexes are also active during pre-implantation development and play important role in establishing cellular plasticity of the early embryo [351]. Different ncPRC complexes, such as ncPRC1.6, can use independent targeting mechanism, and have a different, but just as important role in ES cell fate regulation [339]. The ncPRC1.6 complex does not harbor KDM2B or FBX10 subunits. The target genes of ncPRC1.1 and ncPRC1.6 are highly different [173]. It seems PRC1.6 complex is mainly targeted by its DNA binding subunits. MAX/MGA heterodimers can independently associate with both PCGF6 and L3MBT2 subunits and target the PRC1.6 complex. The H3K9 specific methyltransferase subunits of the complex, in turn, repress germ cell specific genes in ES cells [173].
ncPRC1.3–ncPRC1.5 mainly function as a transcriptional activator in ES cells and targeted differently than the above-mentioned complexes. In recent studies, it was shown that the association of the complexes with the TEX10 transcription factor plays important role in targeting. TEX10 and p300 connection with ncPRC1.3–ncPRC1.5 necessary for promoting the expression of many genes involved in mesoderm differentiation [180].
The role of another DNA binding protein YY1 in targeting ncPRC1 complexes is still in debate. Although this protein has never been purified with any described ncPRC1 complexes, YY1 can strongly associates with both RYBP [239] and YAF2, and functionally important to PcG targeting both in Drosophila [251,252] and in human, as illustrated with the example of the human HOX cluster [352]. Nowadays, a possibility of similar DNA binding protein mediated targeting is established in the case of cPRC1 complexes too. The role of some core binding transcription factors, such as RUNX1/CBFβ, are well established through their association with PCGF4/BMI1 [353]. Now, it seems that different non-canonical and canonical PRC1 complexes possess many alternative targeting possibilities to reach their distinctive set of target genes.
8. Conclusions and Future Questions
8.1. There Are Profound Differences in PRC Function between Fly and Mice
The classical epigenetic function of PcG proteins is to maintain the repressed state of the target genes through cell divisions. Homologs of all Drosophila PcG proteins were found in mammals suggesting the existence of a conserved epigenetic repression mechanism by the Polycomb system. However, the activating histone H3 Lys 4 trimethylation (H3K4me3) marks and the repressive H3 Lys 27 trimethylation (H3K27me3) marks are generally exclusive in the fly genome; no real bivalent domains were found. [14]. Bivalent domains are characteristics for the mammalian genome, especially in ES cells. These bivalent domains make possible a timely activation of target genes while maintaining repression in the absence of differentiation signals. Detailed examination of the PcG function revealed other surprising differences. In Drosophila, the gene expression pattern of homeotic PcG target genes is initialized in early embryogenesis and remains the same afterwards. However, in mammalian cells, dynamic interactions of regulators initialize the expression pattern of Polycomb target genes for more prolonged period during development. PRCs, both canonical and non-canonical, are “anchored” to the promoters of target genes in the ES cell genomes and “waiting for differentiation signals” to act then and initiate the next step of differentiation. Different PRC complexes act during different developmental stages; some of the repressive complexes are assembled tissue specific, which is a profound characteristic of the mammalian development. In mammals, ncPRCs have distinctive role in cell lineage commitment and differentiation, which makes ncPRCs a unique regulator of mammalian development. NcPRCs seem to be one of the major safeguards of the development of the embryo proper in mammals.
8.2. The Function of ncPRC1 Subunits Are often Essential for Mammalian Development
Although, the majority of mPRC/hPRC subunits are comprehensively characterized at the protein level, still little is known about their interplay and dynamics during differentiation. To reveal the special importance of the function of different PRC1 complexes, many of the identified subunits were knocked out in mouse and mutated in ES cells. The majority of these mutations were generated decades ago [148], before the discovery of the canonical and non-canonical PRC1 complexes. Gene ablation of the non-canonical RYBP subunit results peri-implantational lethality in mice [166] and ES cells have compromised differentiation abilities [244,245]. Gene ablation of two ncPRC1.1 members, Usp7 [294] and Kdm2b [280], or the ncPRC1.6 member l3mbtl2, is also embryonic lethal [215], which highlights their essential function in embryogenesis. Double mutant combinations of PcG genes in mouse are often found to enhance each other’s phenotype, similar to Drosophila, e.g., in the case of Cbx2/M33 and Pcgf4/Bmi1 [156], indicating evolutionarily conserved cooperation between different repressor complexes in mammalians. However, in most cases, we still do not know whether the given phenotype is a consequence of the loss of canonical or non-canonical Polycomb repressor function. To clarify this important question, research laboratories will need to circumvent several obstacles. One obstacle is that, unfortunately, the catalytic core unit of both cPRC1s and ncPRC1 complexes (RING-PCGF) is the same, making their phenotypes and biological functions indistinguishable. On the other hand, many of the specific accessory subunits of different PRC1 complex subtypes are also identified in other complexes with versatile activities, making phenotypic analyses equally difficult.
In some cases, no observed phenotypic alteration was found in the loss of function mutants of ncPRC1 subunit coding genes due to the functional redundancy among paralogs. In these cases, the survival of the single knockouts can be explained by functional compensation between the paralogs. For example, Pcgf3−/− and Pcgf5−/− single mutants are viable with no apparent phenotype [311]. However, Pcgf3−/− and Pcgf5−/− double mutant mice exhibit female-specific embryo lethality at mid-gestation, pinpointing the common functions of these two subunits in X chromosome inactivation [311]. Since PCGF3 and PCGF5 are members of the ncPRC1s and not of the cPRCs, it is likely that ncPRCs have important functions in recruiting PcGs during X chromosome inactivation. Future studies will need to establish the nature of interaction between ncPRC1.3–ncPRC1.5 and corresponding region of Xist [311].
8.3. The Dosage and Interactions of ncPRC1 Subunits Is Critical for Mammalian Development
The classification of ncPRCs is based on their PCGF subunits. In different cell types, the importance and amount of different ncPRC1 variants can differ significantly. The different PCGF subunits recruit diverse set of associated proteins. A plethora (83) of new ncPRC1.1 interacting proteins were identified linking non-canonical Polycomb repression to cell cycle regulation and pluripotency [269]. Consequently, different ncPRC1 complexes have altered gene targets and profoundly different function. The expression and abundance of different PCGF paralogs is changing during differentiation. For example, PCGF6 occupies 60% of RING1/RNF2 complexes in ES cells. During neuronal differentiation of ES cells to neuronal progenitor cells PCGF6 exchanged to PCGF4, which become a predominant RING1/RNF2 interactor [206]. The Gene Balance Hypothesis states that the stoichiometry of members of multi-subunit complexes affects the function of the whole due to the kinetics and mode of assembly [354]. Many PcG genes have dominant phenotypes and different PcG genes generally show strong genetic interaction. PcG dependent repression is dosage dependent. These support the Gene Balance Hypothesis. The differentiation induced gene expression changes alter the abundance of different subunits and consequently affect the probability of the different sub-complex assemblies. Changes in the concentrations of different proteins and versatile subunit composition of different ncPRC1 allow high degree of flexibility, both for repression function and for alternative interactions during lineage specification.
We are just coming to recognize another dimension of complexity. Highly sensitive affinity purification mass spectrometry (MS) approaches revealed a whole new set of interactor proteins both from Drosophila embryos [355] and from mammalian cells [51,179,269]. The identification of the new players further increases the diversity of the targeting and interaction possibilities of the Polycomb system. It turned out that not only cell type specific expression pattern of paralogs determines the assembly of different ncPRC1 complexes, but differentiation stage specific accessory proteins can also play an important role in the process. The question remains open: how do the exact subunit compositions of the different complexes change during development and what is the driving source of tissue specific regulatory mechanisms?
8.4. The Assembly of ncPRC1 Subunits Is still under Debate
Research laboratories made significant progress to understand the catalytic and regulatory functions of subunits of different Polycomb repressive complexes. Numerous co-crystallized protein structures are determined, helping to understand the assembly and spatial interactions of the identified domains and conserved motifs of the variant complexes (reviewed in [356]). It appears that the huge megaDalton sized protein assemblies build up in a well-defined, step by step process. First, different sub-complexes are formed, as described in the case of ncPRC1.1 [357]. It was found that KDM2B/SKP1 heterodimer can only bind to PCGF1 RAWUL domain after the formation of a PCGF1/BCORL1 heterodimer. The BCORL1 PUFD domain preceding the RAWUL domain produces the extended interface only after heterodimer formation for interaction of the preformed KDM2B/SKP1 sub-complex [357]. Subsequently, the complex grows even larger after contacting the RING1 (or RNF2) and RYBP/YAF2 module. The coexistence of differently assembled modules is reflected in the wide range of estimated size of PRC1 complexes [52]. With the usage of cross linking coupled MS purification, a refined map of interacting regions has been determined in numerous cases [206,358], but there are still many open questions, such as: how do different ncPRCs assemble and react for diverse differentiation signals and transcription factors?
8.5. The Activator Function of ncPRC1s Opens New Perspectives in Gene Regulation
One of the variant ncPRCs, PRC1-AUTS, is not a repressor. The CK2 subunit of the complex neutralizes PRC1 repressing activity and subsequent recruitment of P300 by AUTS2 leads to neural gene activation [313]. Similar activating function of PRC1s was described in spermatogenesis. RNF2 was shown to activate transcription of Sall4 [359].
PRC1 complexes might mitigate the expression of these transcriptionally active genes. It turned out that there are two kinds of targets of PRC1s: repressed genes associated with unproductive RNAPII, and expressed genes bound to active, elongating RNAPII [360]. Both types of genes are marked by large regions of H3K4me3 and smaller regions of H3K27me3. These bivalent marking in ES cells is present on basic, transcription factor coding genes expressed at low levels and silenced developmental genes keeping them poised for activation [209].
Recently, Kar et al. combined single-cell RNA-sequencing data and genome-wide analyses of RNAPII and PRC occupancy in mES cells. They have shown that PRC-active genes are clustered on chromosomes and interacting with active enhancers to promote a stabilization of gene expression noise [361]. The fact that there are actively transcribed and repressed promoters, which are both occupied by PRCs in different single ES cells, confers a dynamic state to these cells. A key feature of PcG complexes in both flies and mammals is that, although they are present in all cells, their binding to a specific target gene depends on the antecedent activity and the chromatin state of that gene.
8.6. More Emphasis Is Placed on Integrative Approaches to Analyze ncPRC1 Functions in Differentiation
By utilizing ES cells and different in vitro differentiation assays, researchers accessed reproducible amount of starting material for their high throughput assays. Combining the results of independent high throughput sequencing coupled ChIP experiments, the binding sites and targets of PRCs are more or less known. However, independent, systematic, quantitative and integrative studies are necessary to understand the dynamics, functions and interactions of the different repressive complexes during differentiation [206]. To achieve this, it is necessary to perform MS based proteomic studies with parallel profiling of the genome wide binding sites of PRC1 and PRC2 type complexes and covalent histone modifications during differentiation. Because of the technical difficulties these types of integrative studies are still sparse. Using this type of combined approach, it was possible to identify stem cell specific and neural specific interactors in ES cells. Kloet et al. followed Polycomb dynamics. It turned out that a global downregulation of the amount of PRC2 occurs during differentiation, which is not reflected in the relatively steady global H3K27 trimethylation levels. A specific switch occurred between ncPRC1 and cPRC1 complexes during differentiation, which was accompanied by a drastic change in genome wide RNF2 binding. PRC1 binding was observed in at active promoters and enhancers without concomitant presence of PR2 [206].
8.7. NcPRCs Have a Profound Role in Extraembryonic Lineage Commitment
Relatively few studies deal with the role of ncPRC1 subunits in extraembryonic development. Stem cells can differentiate to all somatic tissues of the body but not to extraembryonic tissues. This might be among the reasons why only limited amount of studies examine the role of ncPRC constituents in placental development. In addition, there are only several research laboratories specialized on extraembryonic development and skilled scientist required for analyzing the extraembryonic defects of ncPRCs, which are often problematic. Extraembryonic tissues start to form at the time of implantation and hereafter when the growing blastocysts invading the uterus and the differentiation and expansion of trophectoderm has been initiated. There are numerous ncPRC components required for normal extraembryonic development, such as the catalytic subunit RNF2; two of the characteristic ncPRC1.1 subunits BCOR and USP7 and three of the ncPRC1.6 subunits MAX, L3MBTL2 and TFDP1. Furthermore, ncPRC1.3 and ncPRC1.5 complexes also play a part. Double homozygous Pcgf3/Pcgf5 null mutant mice also exhibit extraembryonic developmental defects, underlying the general role of ncPRCs in extraembryonic lineage development.
8.8. The Role of ccPRCs in Initiating and Maintaining Naïve vs. Primed Pluripotent States Are Not Established
Finally, future studies need to clarify the exact role of ncPRCs in initiating and maintaining the different pluripotent states. Our knowledge regarding these intriguing states of pluripotency is still limited. Establishing regulatory loops leading to these two states of pluripotency is a basic question of contemporary developmental biology. To answer this question is urged by the fact that only naïve state ES cells can be transmitted to the germline, mouse epiblast stem (EpiES) cells cannot. We know already that PRC2 is required only for maintaining pluripotency for primed and not naïve ES cells [362], but we do not understand the function of any of the PRCs in this basic developmental process. The question arises, is only PRC2 required for maintaining pluripotency in primed ES cells, or cPRC1 and ncPRC1 are also required for its maintenance PRC2 also specifies ectoderm lineages in primed but not in naïve ES cells indicating that PRCs have critical, cell fate decision making roles at the early stages of development. It is worth to keeping in mind that there are marked differences among mammals, e.g., mouse vs. human ES cells, even in the expression of key pluripotency factors, suggesting multiple role of PRCs in maintaining stemness among species [363].
9. Concluding Remarks
The past decades of ES cell mediated gene targeting experiments have allowed us to make significant strides towards understanding how ncPRC1 complexes regulate multiple aspects of development and lineage commitment. However, many aspects and connections are yet to be clarified. The interactions and interdependence of canonical and non-canonical Polycomb repressive complexes still hold many secrets. Deeper understanding of ncPRCs regulation will lead to fundamental knowledge of development and lineage commitment and hold promises to design new approaches to control differentiation.
Acknowledgments
We thank Eva Monostori and Gabriella Tick as well as members of the Pirity laboratory, for stimulating discussions and critical reading of the manuscript. This work was supported by National Research, Development and Innovation Office GINOP-2.3.2-15-2016-00001 and GINOP-2.3.2-15-2016-00039.
Author Contributions
Izabella Bajusz, Gergő Kovács and Melinda K. Pirity wrote jointly this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lewis, P.H. New mutants. Drosoph. Inf. Serv. 1947, 21, 69. [Google Scholar]
- Lewis, P.H. Pc: Polycomb. Drosoph. Inf. Serv. 1949, 21, 69. [Google Scholar]
- Schuettengruber, B.; Bourbon, H.-M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Herrero, E.; Vernós, I.; Marco, R.; Morata, G. Genetic organization of Drosophila bithorax complex. Nature 1985, 313, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Tiong, S.; Bone, L.M.; Whittle, J.R. Recessive lethal mutations within the bithorax-complex in Drosophila. Mol. Gen. Genet. 1985, 200, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Karch, F.; Weiffenbach, B.; Peifer, M.; Bender, W.; Duncan, I.; Celniker, S.; Crosby, M.; Lewis, E.B. The abdominal region of the bithorax complex. Cell 1985, 43, 81–96. [Google Scholar] [CrossRef]
- Peifer, M.; Karch, F.; Bender, W. The Bithorax complex: Control of segmental identity. Genes Dev. 1987, 1, 891–898. [Google Scholar] [CrossRef]
- Bowman, S.K.; Deaton, A.M.; Domingues, H.; Wang, P.I.; Sadreyev, R.I.; Kingston, R.E.; Bender, W. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife 2014, 3, e02833. [Google Scholar] [CrossRef] [PubMed]
- Maeda, R.K.; Karch, F. The open for business model of the bithorax complex in Drosophila. Chromosoma 2015, 124, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Maeda, R.K.; Karch, F. The ABC of the BX-C: The bithorax complex explained. Development 2006, 133, 1413–1422. [Google Scholar] [CrossRef] [PubMed]
- Bajusz, I.; Sipos, L.; Pirity, M.K. Nucleotide substitutions revealing specific functions of Polycomb group genes. Mol. Genet. Metab. 2015, 114, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.J.; Paro, R. Trithorax and Polycomb group-dependent regulation: A tale of opposing activities. Development 2015, 142, 2876–2887. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Ganapathi, M.; Leblanc, B.; Portoso, M.; Jaschek, R.; Tolhuis, B.; van Lohuizen, M.; Tanay, A.; Cavalli, G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009, 7, e13. [Google Scholar] [CrossRef] [PubMed]
- Denell, R.E.; Frederick, R.D. Homoeosis in Drosophila: A description of the Polycomb lethal syndrome. Dev. Biol. 1983, 97, 34–47. [Google Scholar] [CrossRef]
- Simon, J.; Chiang, A.; Bender, W. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 1992, 114, 493–505. [Google Scholar] [PubMed]
- Landecker, H.L.; Sinclair, D.A.R.; Brock, H.W. Screen for enhancers of Polycomb and Polycomblike in Drosophila melanogaster. Dev. Genet. 1994, 15, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Nüsslein-Volhard, C.; Wieschaus, E.; Kluding, H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster—I. Zygotic loci on the second chromosome. Wilhelm Roux’s Arch. Dev. Biol. 1984, 193, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Duncan, I.M. Polycomblike: A gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics 1982, 102, 49–70. [Google Scholar] [PubMed]
- Kennison, J.A.; Tamkun, J.W. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. USA 1988, 85, 8136–8140. [Google Scholar] [CrossRef] [PubMed]
- Jürgens, G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 1985, 316, 153–155. [Google Scholar] [CrossRef]
- Struhl, G. A gene product required for correct initiation of segmental determination in Drosophila. Nature 1981, 293, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Struhl, G. Role of the esc+ gene product in ensuring the selective expression of segment-specific homeotic genes in Drosophila. J. Embryol. Exp. Morphol. 1983, 76, 297–331. [Google Scholar] [PubMed]
- Gehring, W. A recessive lethal [l(4)29] with a homeotic effect in D. melanogaster. Drosoph. Inf. Serv. 1970, 45, 5. [Google Scholar]
- Phillips, M.D.; Shearn, A. Mutations in polycombeotic, a Drosophila polycomb-group gene, cause a wide range of maternal and zygotic phenotypes. Genetics 1990, 125, 91–101. [Google Scholar] [PubMed]
- Jones, R.S.; Gelbart, W.M. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 1990, 126, 185–199. [Google Scholar] [PubMed]
- Ingham, P.W. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell 1984, 37, 815–823. [Google Scholar] [CrossRef]
- Santamaría, P.; Randsholt, N.B. Characterization of a region of the X chromosome of Drosophila including multi sex combs (mxc), a Polycomb group gene which also functions as a tumour suppressor. Mol. Gen. Genet. 1995, 246, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Campbell, R.B.; Sinclair, D.A.; Couling, M.; Brock, H.W. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol. Gen. Genet. 1995, 246, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Dura, J.M.; Brock, H.W.; Santamaria, P. Polyhomeotic: A gene of Drosophila melanogaster required for correct expression of segmental identity. Mol. Gen. Genet. 1985, 198, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Russell, M.A.; Denell, R.E. Homoeosis in Drosophila: A new enhancer of polycomb and related homoeotic mutations. Genetics 1983, 105, 357–370. [Google Scholar] [PubMed]
- Bejarano, F.; González, I.; Vidal, M.; Busturia, A. The Drosophila RYBP gene functions as a Polycomb-dependent transcriptional repressor. Mech. Dev. 2005, 122, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, I.; Aparicio, R.; Busturia, A. Functional Characterization of the dRYBP Gene in Drosophila. Genetics 2008, 179, 1373–1388. [Google Scholar] [CrossRef] [PubMed]
- Brunk, B.P.; Martin, E.C.; Adler, P.N. Drosophila genes Posterior Sex Combs and Suppressor two of zeste encode proteins with homology to the murine bmi-1 oncogene. Nature 1991, 353, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.C.; Chou, T.B.; Bender, W. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 1995, 140, 231–243. [Google Scholar] [PubMed]
- Beuchle, D.; Struhl, G.; Müller, J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 2001, 128, 993–1004. [Google Scholar] [PubMed]
- de Ayala Alonso, A.G.; Gutierrez, L.; Fritsch, C.; Papp, B.; Beuchle, D.; Muller, J. A Genetic Screen Identifies Novel Polycomb Group Genes in Drosophila. Genetics 2007, 176, 2099–2108. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, L.; Oktaba, K.; Scheuermann, J.C.; Gambetta, M.C.; Ly-Hartig, N.; Müller, J. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development 2012, 139, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Girard, F.; Bello, B.; Affolter, M.; Gehring, W.J. The cramped gene of Drosophila is a member of the Polycomb-group, and interacts with mus209, the gene encoding Proliferating Cell Nuclear Antigen. Development 1997, 124, 3385–3394. [Google Scholar] [PubMed]
- Klymenko, T.; Papp, B.; Fischle, W.; Köcher, T.; Schelder, M.; Fritsch, C.; Wild, B.; Wilm, M.; Müller, J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006, 20, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Birve, A.; Sengupta, A.K.; Beuchle, D.; Larsson, J.; Kennison, J.A.; Rasmuson-Lestander, A.; Müller, J. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 2001, 128, 3371–3379. [Google Scholar] [PubMed]
- Schwartz, Y.B.; Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Paro, R. Mechanisms of heritable gene repression during development of Drosophila. Curr. Opin. Cell Biol. 1993, 5, 999–1005. [Google Scholar] [CrossRef]
- Struhl, G.; Akam, M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 1985, 4, 3259–3264. [Google Scholar] [PubMed]
- Wedeen, C.; Harding, K.; Levine, M. Spatial regulation of Antennapedia and bithorax gene expression by the Polycomb locus in Drosophila. Cell 1986, 44, 739–748. [Google Scholar] [CrossRef]
- Dura, J.M.; Ingham, P. Tissue- and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos by the polyhomeotic gene. Development 1988, 103, 733–741. [Google Scholar] [PubMed]
- Paro, R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990, 6, 416–421. [Google Scholar] [CrossRef]
- Ji, J.-Y.; Miles, W.O.; Korenjak, M.; Zheng, Y.; Dyson, N.J. In Vivo Regulation of E2F1 by Polycomb Group Genes in Drosophila. G3 Genes|Genomes|Genetics 2012, 2, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.-M.; Colomb, S.; Déjardin, J.; Bantignies, F.; Cavalli, G. Polycomb group-dependent Cyclin A repression in Drosophila. Genes Dev. 2006, 20, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Levine, S.S.; Weiss, A.; Erdjument-Bromage, H.; Shao, Z.; Tempst, P.; Kingston, R.E. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 2002, 22, 6070–6078. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, J.; Völkel, P.; Rosnoblet, C.; Le Faou, P.; Angrand, P.-O. Interaction Proteomics Analysis of Polycomb Proteins Defines Distinct PRC1 Complexes in Mammalian Cells. Mol. Cell. Proteom. 2011, 10, M110.002642. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF Homologs, CBX Proteins, and RYBP Define Functionally Distinct PRC1 Family Complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Paro, R.; Hogness, D.S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. USA 1991, 88, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Festenstein, R.; Sharghi-Namini, S.; Fox, M.; Roderick, K.; Tolaini, M.; Norton, T.; Saveliev, A.; Kioussis, D.; Singh, P. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat. Genet. 1999, 23, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. Types of visible variations induced by X-rays in Drosophila. J. Genet. 1930, 22, 299–334. [Google Scholar] [CrossRef]
- Gowen, J.W.; Gay, E.H. Eversporting as a Function of the Y-Chromosome in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1933, 19, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Messmer, S.; Franke, A.; Paro, R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992, 6, 1241–1254. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fitzgerald, D.P.; Bender, W. Polycomb group repression reduces DNA accessibility. Mol. Cell. Biol. 2001, 21, 6585–6597. [Google Scholar] [CrossRef] [PubMed]
- Boettiger, A.N.; Bintu, B.; Moffitt, J.R.; Wang, S.; Beliveau, B.J.; Fudenberg, G.; Imakaev, M.; Mirny, L.A.; Wu, C.; Zhuang, X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016, 529, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Ji, F.; Sunwoo, H.; Jain, G.; Lee, J.T.; Sadreyev, R.I.; Dekker, J.; Kingston, R.E. Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol. Cell 2017, 65, 432–446.e5. [Google Scholar] [CrossRef] [PubMed]
- Platero, J.S.; Hartnett, T.; Eissenberg, J.C. Functional analysis of the chromo domain of HP1. EMBO J. 1995, 14, 3977–3986. [Google Scholar] [PubMed]
- Tschiersch, B.; Hofmann, A.; Krauss, V.; Dorn, R.; Korge, G.; Reuter, G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994, 13, 3822–3831. [Google Scholar] [PubMed]
- Orlando, V. Polycomb, epigenomes, and control of cell identity. Cell 2003, 112, 599–606. [Google Scholar] [CrossRef]
- Alvarez-Venegas, R.; Avramova, Z. SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene 2002, 285, 25–37. [Google Scholar] [CrossRef]
- Tachibana, M.; Sugimoto, K.; Fukushima, T.; Shinkai, Y. SET Domain-containing Protein, G9a, Is a Novel Lysine-preferring Mammalian Histone Methyltransferase with Hyperactivity and Specific Selectivity to Lysines 9 and 27 of Histone H3. J. Biol. Chem. 2001, 276, 25309–25317. [Google Scholar] [CrossRef] [PubMed]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef] [PubMed]
- van Lohuizen, M.; Frasch, M.; Wientjens, E.; Berns, A. Sequence similarity between the mammalian bmi-1 proto-oncogene and the Drosophila regulatory genes Psc and Su(z)2. Nature 1991, 353, 353–355. [Google Scholar] [CrossRef] [PubMed]
- Asano, H.; Ishida, A.; Hasegawa, M.; Ono, T.; Yoshida, M.C.; Taniguchi, M.; Kanno, M. The mouse Mel-18 “RING-finger” gene: Genomic organization, promoter analysis and chromosomal assignment. DNA Seq. 1993, 3, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Noel, G.; Niedenthal, R.; Tamura, T.; Harbers, K. A family of structurally related RING finger proteins interacts specifically with the ubiquitin-conjugating enzyme UbcM4. FEBS Lett. 1999, 454, 257–261. [Google Scholar] [PubMed]
- Lorick, K.L.; Jensen, J.P.; Fang, S.; Ong, A.M.; Hatakeyama, S.; Weissman, A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 1999, 96, 11364–11369. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.J.; Kyba, M.; Bornemann, D.; Morgan, K.; Brock, H.W.; Simon, J. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 1997, 17, 6683–6692. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.A.; Gingery, M.; Pilpa, R.M.; Bowie, J.U. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 2002, 9, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, M.C.; Müller, J. O-GlcNAcylation prevents aggregation of the Polycomb group repressor polyhomeotic. Dev. Cell 2014, 31, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.K.; Leal, B.Z.; Chadwell, L.V.; Wang, R.; Ilangovan, U.; Kaur, Y.; Junco, S.E.; Schirf, V.; Osmulski, P.A.; Gaczynska, M.; et al. The Growth-Suppressive Function of the Polycomb Group Protein Polyhomeotic Is Mediated by Polymerization of Its Sterile Alpha Motif (SAM) Domain. J. Biol. Chem. 2012, 287, 8702–8713. [Google Scholar] [CrossRef] [PubMed]
- Wani, A.H.; Boettiger, A.N.; Schorderet, P.; Ergun, A.; Münger, C.; Sadreyev, R.I.; Zhuang, X.; Kingston, R.E.; Francis, N.J. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat. Commun. 2016, 7, 10291. [Google Scholar] [CrossRef] [PubMed]
- Qiao, F.; Bowie, J.U. The many faces of SAM. Sci. STKE 2005, 2005, re7. [Google Scholar] [CrossRef] [PubMed]
- Akasaka, T.; Kanno, M.; Balling, R.; Mieza, M.A.; Taniguchi, M.; Koseki, H. A role for mel-18, a Polycomb group-related vertebrate gene, during theanteroposterior specification of the axial skeleton. Development 1996, 122, 1513–1522. [Google Scholar] [PubMed]
- Gould, A. Functions of mammalian Polycomb group and trithorax group related genes. Curr. Opin. Genet. Dev. 1997, 7, 488–494. [Google Scholar] [CrossRef]
- Gunster, M.J.; Satijn, D.P.; Hamer, K.M.; den Blaauwen, J.L.; de Bruijn, D.; Alkema, M.J.; van Lohuizen, M.; van Driel, R.; Otte, A.P. Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol. Cell. Biol. 1997, 17, 2326–2335. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.L.; Lee, I.; Bloyer, S.; Bozza, S.; Chevalier, J.; Dahl, A.; Bodner, C.; Helgason, C.D.; Hess, J.L.; Humphries, R.K.; et al. Additional sex combs-like 1 belongs to the enhancer of trithorax and polycomb group and genetically interacts with Cbx2 in mice. Dev. Biol. 2010, 337, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Lanzuolo, C.; Orlando, V. Memories from the Polycomb Group Proteins. Annu. Rev. Genet. 2012, 46, 561–589. [Google Scholar] [CrossRef] [PubMed]
- Grossniklaus, U.; Paro, R. Transcriptional Silencing by Polycomb-Group Proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019331. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.; Puangsomlee, P.; Martin, M.; Long, D.; Meyerowitz, E.M.; Coupland, G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 1997, 386, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.M.; Danilevskaya, O.N.; Hermon, P.; Helentjaris, T.G.; Phillips, R.L.; Kaeppler, H.F.; Kaeppler, S.M. Sequence Relationships, Conserved Domains, and Expression Patterns for Maize Homologs of the Polycomb Group Genes E(z), esc, and E(Pc). PLANT Physiol. 2002, 128, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.; Makarevich, G. Epigenetic mechanisms governing seed development in plants. EMBO Rep. 2006, 7, 1223–1227. [Google Scholar] [CrossRef] [PubMed]
- Derkacheva, M.; Hennig, L. Variations on a theme: Polycomb group proteins in plants. J. Exp. Bot. 2014, 65, 2769–2784. [Google Scholar] [CrossRef] [PubMed]
- Calonje, M. PRC1 Marks the Difference in Plant PcG Repression. Mol. Plant 2014, 7, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Pu, L.; Sung, Z.R. PcG and trxG in plants—Friends or foes. Trends Genet. 2015, 31, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.; Villar, C.B.R. Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 2008, 18, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Roure, V.; Bantignies, F. Polycomb group-mediated gene silencing mechanisms: Stability versus flexibility. Epigenomics 2009, 1, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 2010, 35, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Trask, M.C.; Mager, J. Complexity of polycomb group function: Diverse mechanisms of target specificity. J. Cell. Physiol. 2011, 226, 1719–1721. [Google Scholar] [CrossRef] [PubMed]
- Franke, A.; DeCamillis, M.; Zink, D.; Cheng, N.; Brock, H.W.; Paro, R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992, 11, 2941–2950. [Google Scholar] [PubMed]
- Tie, F.; Furuyama, T.; Harte, P.J. The Drosophila Polycomb Group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development 1998, 125, 3483–3496. [Google Scholar] [PubMed]
- Saurin, A.J.; Shao, Z.; Erdjument-Bromage, H.; Tempst, P.; Kingston, R.E. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 2001, 412, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Bárdos, J.I.; Saurin, A.J.; Tissot, C.; Duprez, E.; Freemont, P.S. HPC3 Is a New Human Polycomb Orthologue That Interacts and Associates with RING1 and Bmi1 and Has Transcriptional Repression Properties. J. Biol. Chem. 2000, 275, 28785–28792. [Google Scholar] [CrossRef] [PubMed]
- Satijn, D.P.; Otte, A.P. Polycomb group protein complexes: Do different complexes regulate distinct target genes? Biochim. Biophys. Acta 1999, 1447, 1–16. [Google Scholar] [CrossRef]
- Francis, N.J.; Saurin, A.J.; Shao, Z.; Kingston, R.E. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 2001, 8, 545–556. [Google Scholar] [CrossRef]
- King, I.F.G.; Emmons, R.B.; Francis, N.J.; Wild, B.; Müller, J.; Kingston, R.E.; Wu, C.-T. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol. 2005, 25, 6578–6591. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Francis, N.J.; Kingston, R.E.; Woodcock, C.L. Chromatin Compaction by a Polycomb Group Protein Complex. Science 2004, 306, 1574–1577. [Google Scholar] [CrossRef] [PubMed]
- Buchwald, G.; van der Stoop, P.; Weichenrieder, O.; Perrakis, A.; van Lohuizen, M.; Sixma, T.K. Structure and E3-ligase activity of the Ring–Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 2006, 25, 2465–2474. [Google Scholar] [CrossRef] [PubMed]
- Eskeland, R.; Leeb, M.; Grimes, G.R.; Kress, C.; Boyle, S.; Sproul, D.; Gilbert, N.; Fan, Y.; Skoultchi, A.I.; Wutz, A.; et al. Ring1B Compacts Chromatin Structure and Represses Gene Expression Independent of Histone Ubiquitination. Mol. Cell 2010, 38, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Gearhart, M.D.; Corcoran, C.M.; Wamstad, J.A.; Bardwell, V.J. Polycomb Group and SCF Ubiquitin Ligases Are Found in a Novel BCOR Complex That Is Recruited to BCL6 Targets. Mol. Cell. Biol. 2006, 26, 6880–6889. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, C.; Sánchez, I.; Demmers, J.A.A.; Rodriguez, P.; Strouboulis, J.; Vidal, M. Proteomics Analysis of Ring1B/Rnf2 Interactors Identifies a Novel Complex with the Fbxl10/Jhdm1B Histone Demethylase and the Bcl6 Interacting Corepressor. Mol. Cell. Proteom. 2007, 6, 820–834. [Google Scholar] [CrossRef] [PubMed]
- Lagarou, A.; Mohd-Sarip, A.; Moshkin, Y.M.; Chalkley, G.E.; Bezstarosti, K.; Demmers, J.A.A.; Verrijzer, C.P. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008, 22, 2799–2810. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science. 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.-W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, Y. SUZ12 Is Required for Both the Histone Methyltransferase Activity and the Silencing Function of the EED-EZH2 Complex. Mol. Cell 2004, 15, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Lazzerini Denchi, E.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. [Google Scholar] [CrossRef] [PubMed]
- Ketel, C.S.; Andersen, E.F.; Vargas, M.L.; Suh, J.; Strome, S.; Simon, J.A. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol. Cell. Biol. 2005, 25, 6857–6868. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nekrasov, M.; Wild, B.; Müller, J. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 2005, 6, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, N.D.; Yee, D.; Chen, A.; Kalantry, S.; Chamberlain, S.J.; Otte, A.P.; Magnuson, T. The Murine Polycomb Group Protein Eed Is Required for Global Histone H3 Lysine-27 Methylation. Curr. Biol. 2005, 15, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, M.; Klymenko, T.; Fraterman, S.; Papp, B.; Oktaba, K.; Köcher, T.; Cohen, A.; Stunnenberg, H.G.; Wilm, M.; Müller, J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007, 26, 4078–4088. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Margueron, R.; Ku, M.; Chambon, P.; Bernstein, B.E.; Reinberg, D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010, 24, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, K.; Kim, J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009, 37, 2940–2950. [Google Scholar] [CrossRef] [PubMed]
- Aldiri, I.; Vetter, M.L. PRC2 during vertebrate organogenesis: A complex in transition. Dev. Biol. 2012, 367, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Veneti, Z.; Gkouskou, K.; Eliopoulos, A. Polycomb Repressor Complex 2 in Genomic Instability and Cancer. Int. J. Mol. Sci. 2017, 18, 1657. [Google Scholar] [CrossRef] [PubMed]
- Moritz, L.E.; Trievel, R.C. Structure, mechanism, and regulation of polycomb repressive complex 2. J. Biol. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Fischle, W.; Wang, Y.; Jacobs, S.A.; Kim, Y.; Allis, C.D.; Khorasanizadeh, S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003, 17, 1870–1881. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L. P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 Complex-Dependent H2A Ubiquitylation Drives PRC2 Recruitment and Polycomb Domain Formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Dienstbier, M.; Hassan, R.; Schermelleh, L.; Sharif, J.; Blackledge, N.P.; De Marco, V.; Elderkin, S.; Koseki, H.; Klose, R.; et al. Targeting Polycomb to Pericentric Heterochromatin in Embryonic Stem Cells Reveals a Role for H2AK119u1 in PRC2 Recruitment. Cell Rep. 2014, 7, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.T.C.; Müller, C.W.; Vermeulen, M.; Müller, J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Bienz, M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991, 10, 3147–3155. [Google Scholar] [PubMed]
- Simon, J.; Chiang, A.; Bender, W.; Shimell, M.J.; O’Connor, M. Elements of the Drosophila Bithorax Complex That Mediate Repression by Polycomb Group Products. Dev. Biol. 1993, 158, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.S.; Rastelli, L.; Pirrotta, V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994, 13, 2553–2564. [Google Scholar] [PubMed]
- Chiang, A.; O’Connor, M.B.; Paro, R.; Simon, J.; Bender, W. Discrete Polycomb-binding sites in each parasegmental domain of the bithorax complex. Development 1995, 121, 1681–1689. [Google Scholar] [PubMed]
- Schuettengruber, B.; Oded Elkayam, N.; Sexton, T.; Entrevan, M.; Stern, S.; Thomas, A.; Yaffe, E.; Parrinello, H.; Tanay, A.; Cavalli, G. Cooperativity, Specificity, and Evolutionary Stability of Polycomb Targeting in Drosophila. Cell Rep. 2014, 9, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Ringrose, L.; Rehmsmeier, M.; Dura, J.-M.; Paro, R. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev. Cell 2003, 5, 759–771. [Google Scholar] [CrossRef]
- Müller, J.; Kassis, J.A. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 2006, 16, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, C.; Brown, J.L.; Kassis, J.A.; Müller, J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 1999, 126, 3905–3913. [Google Scholar] [PubMed]
- Mihaly, J.; Mishra, R.K.; Karch, F. A conserved sequence motif in Polycomb-response elements. Mol. Cell 1998, 1, 1065–1066. [Google Scholar] [CrossRef]
- Poux, S.; Melfi, R.; Pirrotta, V. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 2001, 15, 2509–2514. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Brown, J.L.; Cao, R.; Zhang, Y.; Kassis, J.A.; Jones, R.S. Hierarchical Recruitment of Polycomb Group Silencing Complexes. Mol. Cell 2004, 14, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; Fritsch, C.; Mueller, J.; Kassis, J.A. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 2003, 130, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Grimm, C.; Matos, R.; Ly-Hartig, N.; Steuerwald, U.; Lindner, D.; Rybin, V.; Müller, J.; Müller, C.W. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 2009, 28, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Frey, F.; Sheahan, T.; Finkl, K.; Stoehr, G.; Mann, M.; Benda, C.; Müller, J. Molecular basis of PRC1 targeting to Polycomb response elements by PhoRC. Genes Dev. 2016, 30, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; McElroy, K.A.; Jung, Y.L.; Alekseyenko, A.A.; Zee, B.M.; Park, P.J.; Kuroda, M.I. Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila. Genes Dev. 2015, 29, 1136–1150. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, J.C.; de Ayala Alonso, A.G.; Oktaba, K.; Ly-Hartig, N.; McGinty, R.K.; Fraterman, S.; Wilm, M.; Muir, T.W.; Müller, J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 2010, 465, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Seshasayee, D.; Noubade, R.; French, D.M.; Liu, J.; Chaurushiya, M.S.; Kirkpatrick, D.S.; Pham, V.C.; Lill, J.R.; Bakalarski, C.E.; et al. Loss of the Tumor Suppressor BAP1 Causes Myeloid Transformation. Science 2012, 337, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Gaunt, S.; Lawrence, P.A. Function of the Polycomb protein is conserved in mice and flies. Development 1995, 121, 2847–2852. [Google Scholar] [PubMed]
- Gorfinkiel, N.; Fanti, L.; Melgar, T.; Garcı́a, E.; Pimpinelli, S.; Guerrero, I.; Vidal, M. The Drosophila Polycomb group gene Sex combs extra encodes the ortholog of mammalian Ring1 proteins. Mech. Dev. 2004, 121, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Atchison, L.; Ghias, A.; Wilkinson, F.; Bonini, N.; Atchison, M.L. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003, 22, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Coré, N.; Bel, S.; Gaunt, S.J.; Aurrand-Lions, M.; Pearce, J.; Fisher, A.; Djabali, M. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development 1997, 124, 721–729. [Google Scholar] [PubMed]
- van der Lugt, N.M.; Domen, J.; Linders, K.; van Roon, M.; Robanus-Maandag, E.; te Riele, H.; van der Valk, M.; Deschamps, J.; Sofroniew, M.; van Lohuizen, M. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994, 8, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Alkema, M.J.; van der Lugt, N.M.T.; Bobeldijk, R.C.; Berns, A.; van Lohuizen, M. Transformation of axial skeleton due to overexpression of bmi-1 in transgenic mice. Nature 1995, 374, 724–727. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.D.; Hess, J.L.; Yu, B.D.; Ernst, P.; van Lohuizen, M.; Berns, A.; van der Lugt, N.M.; Shashikant, C.S.; Ruddle, F.H.; Seto, M.; et al. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. USA 1999, 96, 14372–14377. [Google Scholar] [CrossRef] [PubMed]
- del Mar Lorente, M.; Marcos-Gutiérrez, C.; Pérez, C.; Schoorlemmer, J.; Ramírez, A.; Magin, T.; Vidal, M. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 2000, 127, 5093–5100. [Google Scholar] [PubMed]
- Suzuki, M.; Mizutani-Koseki, Y.; Fujimura, Y.-I.; Miyagishima, H.; Kaneko, T.; Takada, Y.; Akasaka, T.; Tanzawa, H.; Takihara, Y.; Nakano, M.; Masumoto, H.; Vidal, M.; Isono, K.-I.; Koseki, H. Involvement of the Polycomb-group gene Ring1B in the specification of the anterior-posterior axis in mice. Development 2002, 129, 4171–4183. [Google Scholar] [PubMed]
- Lorente, M.; Pérez, C.; Sánchez, C.; Donohoe, M.; Shi, Y.; Vidal, M. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mech. Dev. 2006, 123, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Takihara, Y.; Tomotsune, D.; Shirai, M.; Katoh-Fukui, Y.; Nishii, K.; Motaleb, M.A.; Nomura, M.; Tsuchiya, R.; Fujita, Y.; Shibata, Y.; et al. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development 1997, 124, 3673–3682. [Google Scholar] [PubMed]
- Isono, K.-I.; Fujimura, Y.-I.; Shinga, J.; Yamaki, M.; O-Wang, J.; Takihara, Y.; Murahashi, Y.; Takada, Y.; Mizutani-Koseki, Y.; Koseki, H. Mammalian Polyhomeotic Homologues Phc2 and Phc1 Act in Synergy To Mediate Polycomb Repression of Hox Genes. Mol. Cell. Biol. 2005, 25, 6694–6706. [Google Scholar] [CrossRef] [PubMed]
- Bel, S.; Coré, N.; Djabali, M.; Kieboom, K.; Van der Lugt, N.; Alkema, M.J.; Van Lohuizen, M. Genetic interactions and dosage effects of Polycomb group genes in mice. Development 1998, 125, 3543–3551. [Google Scholar] [PubMed]
- Whitcomb, S.J.; Basu, A.; Allis, C.D.; Bernstein, E. Polycomb Group proteins: An evolutionary perspective. Trends Genet. 2007, 23, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.; Magnuson, T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997, 13, 167–170. [Google Scholar] [CrossRef]
- Prezioso, C.; Orlando, V. Polycomb proteins in mammalian cell differentiation and plasticity. FEBS Lett. 2011, 585, 2067–2077. [Google Scholar] [CrossRef] [PubMed]
- Ohta, H.; Sawada, A.; Kim, J.Y.; Tokimasa, S.; Nishiguchi, S.; Humphries, R.K.; Hara, J.; Takihara, Y. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J. Exp. Med. 2002, 195, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Raaphorst, F.M. Self-renewal of hematopoietic and leukemic stem cells: A central role for the Polycomb-group gene Bmi-1. Trends Immunol. 2003, 24, 522–524. [Google Scholar] [CrossRef]
- Andricovich, J.; Kai, Y.; Peng, W.; Foudi, A.; Tzatsos, A. Histone demethylase KDM2B regulates lineage commitment in normal and malignant hematopoiesis. J. Clin. Investig. 2016, 126, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Searle, N.E.; Pillus, L. Critical genomic regulation mediated by Enhancer of Polycomb. Curr. Genet. 2017. [Google Scholar] [CrossRef] [PubMed]
- Takihara, Y.; Hara, J. Polycomb-group genes and hematopoiesis. Int. J. Hematol. 2000, 72, 165–172. [Google Scholar] [PubMed]
- Vidal, M.; Starowicz, K. Polycomb complexes PRC1 and their function in hematopoiesis. Exp. Hematol. 2017, 48, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Pirity, M.K.; Locker, J.; Schreiber-Agus, N. Rybp/DEDAF Is Required for Early Postimplantation and for Central Nervous System Development. Mol. Cell. Biol. 2005, 25, 7193–7202. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.; Kroll, K.L. The roles and regulation of Polycomb complexes in neural development. Cell Tissue Res. 2015, 359, 65–85. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-G.; Chen, J.; Sadoshima, J.; Lee, Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol. Cell. Biol. 2004, 24, 10151–10160. [Google Scholar] [CrossRef] [PubMed]
- He, A.; Ma, Q.; Cao, J.; von Gise, A.; Zhou, P.; Xie, H.; Zhang, B.; Hsing, M.; Christodoulou, D.C.; Cahan, P.; et al. Polycomb Repressive Complex 2 Regulates Normal Development of the Mouse Heart. Circ. Res. 2012, 110, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Beketaev, I.; Zhang, Y.; Kim, E.Y.; Yu, W.; Qian, L.; Wang, J. Critical role of YY1 in cardiac morphogenesis. Dev. Dyn. 2015, 244, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Kee, H.J.; Kim, J.-R.; Nam, K.-I.; Park, H.Y.; Shin, S.; Kim, J.C.; Shimono, Y.; Takahashi, M.; Jeong, M.H.; Kim, N.; et al. Enhancer of Polycomb1, a Novel Homeodomain Only Protein-binding Partner, Induces Skeletal Muscle Differentiation. J. Biol. Chem. 2007, 282, 7700–7709. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Starmer, J.; Fedoriw, A.M.; Yee, D.; Magnuson, T. Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 2014, 28, 2056–2069. [Google Scholar] [CrossRef] [PubMed]
- Endoh, M.; Endo, T.A.; Shinga, J.; Hayashi, K.; Farcas, A.; Ma, K.-W.; Ito, S.; Sharif, J.; Endoh, T.; Onaga, N.; et al. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife 2017, 6. [Google Scholar] [CrossRef]
- Eun, S.H.; Feng, L.; Cedeno-Rosario, L.; Gan, Q.; Wei, G.; Cui, K.; Zhao, K.; Chen, X. Polycomb Group Gene E(z) Is Required for Spermatogonial Dedifferentiation in Drosophila Adult Testis. J. Mol. Biol. 2017, 429, 2030–2041. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Shi, Z.; Chen, X. Enhancer of polycomb coordinates multiple signaling pathways to promote both cyst and germline stem cell differentiation in the Drosophila adult testis. PLOS Genet. 2017, 13, e1006571. [Google Scholar] [CrossRef] [PubMed]
- Faust, C.; Schumacher, A.; Holdener, B.; Magnuson, T. The eed mutation disrupts anterior mesoderm production in mice. Development 1995, 121, 273–285. [Google Scholar] [PubMed]
- Bracken, A.P.; Dietrich, N.; Pasini, D.; Hansen, K.H.; Helin, K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006, 20, 1123–1136. [Google Scholar] [CrossRef] [PubMed]
- Junco, S.E.; Wang, R.; Gaipa, J.C.; Taylor, A.B.; Schirf, V.; Gearhart, M.D.; Bardwell, V.J.; Demeler, B.; Hart, P.J.; Kim, C.A. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure 2013, 21, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Huang, Y.; Zhang, J.; Liu, M.; Ji, H.; Wang, C.; Cao, N.; Li, C.; Xia, Y.; Jiang, Q.; et al. Polycomb group RING finger protein 3/5 activate transcription via an interaction with the pluripotency factor Tex10 in embryonic stem cells. J. Biol. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Maeda, I.; Okamura, D.; Tokitake, Y.; Ikeda, M.; Kawaguchi, H.; Mise, N.; Abe, K.; Noce, T.; Okuda, A.; Matsui, Y. Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells. Nat. Commun. 2013, 4, 1754. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Hirasaki, M.; Hishida, T.; Wu, J.; Okamura, D.; Ueda, A.; Nishimoto, M.; Nakachi, Y.; Mizuno, Y.; Okazaki, Y.; et al. Loss of MAX results in meiotic entry in mouse embryonic and germline stem cells. Nat. Commun. 2016, 7, 11056. [Google Scholar] [CrossRef] [PubMed]
- Luis, N.M.; Morey, L.; Di Croce, L.; Benitah, S.A. Polycomb in Stem Cells: PRC1 Branches Out. Cell Stem Cell 2012, 11, 16–21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schuettengruber, B.; Cavalli, G. Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 2009, 136, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- Richly, H.; Aloia, L.; Di Croce, L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011, 2, e204. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Zhang, Y.; Sun, T.; Cheng, B. Epigenetic regulation by polycomb group complexes: Focus on roles of CBX proteins. J. Zhejiang Univ. Sci. B 2014, 15, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; O’Loghlen, A. PRC1 complex diversity: Where is it taking us? Trends Cell Biol. 2014, 24, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol. Cell 2013, 49, 808–824. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Pascual, G.; Cozzuto, L.; Roma, G.; Wutz, A.; Benitah, S.A.; Di Croce, L. Nonoverlapping Functions of the Polycomb Group Cbx Family of Proteins in Embryonic Stem Cells. Cell Stem Cell 2012, 10, 47–62. [Google Scholar] [CrossRef] [PubMed]
- O’Loghlen, A.; Muñoz-Cabello, A.M.; Gaspar-Maia, A.; Wu, H.-A.; Banito, A.; Kunowska, N.; Racek, T.; Pemberton, H.N.; Beolchi, P.; Lavial, F.; et al. MicroRNA Regulation of Cbx7 Mediates a Switch of Polycomb Orthologs during ESC Differentiation. Cell Stem Cell 2012, 10, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Klauke, K.; Radulović, V.; Broekhuis, M.; Weersing, E.; Zwart, E.; Olthof, S.; Ritsema, M.; Bruggeman, S.; Wu, X.; Helin, K.; et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat. Cell Biol. 2013, 15, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N. Noncoding RNA and Polycomb recruitment. RNA 2013, 19, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.-M. Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, C.; Liu, W.; Zhang, J.; Ohgi, K.A.; Grinstein, J.D.; Dorrestein, P.C.; Rosenfeld, M.G. ncRNA- and Pc2 Methylation-Dependent Gene Relocation between Nuclear Structures Mediates Gene Activation Programs. Cell 2011, 147, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Feng, Y.; Zhang, D.; Zhao, S.D.; Hu, Z.; Greshock, J.; Zhang, Y.; Yang, L.; Zhong, X.; Wang, L.-P.; et al. A Functional Genomic Approach Identifies FAL1 as an Oncogenic Long Noncoding RNA that Associates with BMI1 and Represses p21 Expression in Cancer. Cancer Cell 2014, 26, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Boiani, M.; Schöler, H.R. Developmental cell biology: Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005, 6, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Joyner, A.L. Gene Targeting: A Practical Approach; Oxford University Press: Oxford, UK, 2000; ISBN 9780199637928. [Google Scholar]
- Brook, F.A.; Gardner, R.L. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA 1997, 94, 5709–5712. [Google Scholar] [CrossRef] [PubMed]
- Sivarajah, S.; Raj, G.S.; Mathews, A.J.V.; Sahib, N.B.E.; Hwang, W.S.; Crook, J.M. The generation of GLP-grade human embryonic stem cell banks from four clinical-grade cell lines for preclinical research. Vitr. Cell. Dev. Biol. Anim. 2010, 46, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Aloia, L.; Di Stefano, B.; Di Croce, L. Polycomb complexes in stem cells and embryonic development. Development 2013, 140, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Mas, G.; Di Croce, L. The role of Polycomb in stem cell genome architecture. Curr. Opin. Cell Biol. 2016, 43, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Sauvageau, G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010, 7, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Surface, L.E.; Thornton, S.R.; Boyer, L.A. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 2010, 7, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Ku, M.; Koche, R.P.; Rheinbay, E.; Mendenhall, E.M.; Endoh, M.; Mikkelsen, T.S.; Presser, A.; Nusbaum, C.; Xie, X.; Chi, A.S.; et al. Genomewide Analysis of PRC1 and PRC2 Occupancy Identifies Two Classes of Bivalent Domains. PLoS Genet. 2008, 4, e1000242. [Google Scholar] [CrossRef] [PubMed]
- Creppe, C.; Palau, A.; Malinverni, R.; Valero, V.; Buschbeck, M. A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation. PLoS Genet. 2014, 10, e1004851. [Google Scholar] [CrossRef] [PubMed]
- Kloet, S.L.; Makowski, M.M.; Baymaz, H.I.; van Voorthuijsen, L.; Karemaker, I.D.; Santanach, A.; Jansen, P.W.T.C.; Di Croce, L.; Vermeulen, M. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 2016, 23, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Mantsoki, A.; Devailly, G.; Joshi, A. CpG island erosion, polycomb occupancy and sequence motif enrichment at bivalent promoters in mammalian embryonic stem cells. Sci. Rep. 2015, 5, 16791. [Google Scholar] [CrossRef] [PubMed]
- Stock, J.K.; Giadrossi, S.; Casanova, M.; Brookes, E.; Vidal, M.; Koseki, H.; Brockdorff, N.; Fisher, A.G.; Pombo, A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007, 9, 1428–1435. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Brookes, E.; Pombo, A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep. 2009, 10, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Harikumar, A.; Meshorer, E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015, 16, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Ishiguro, K.-I.; Gaubatz, S.; Livingston, D.M.; Nakatani, Y. A Complex with Chromatin Modifiers That Occupies E2F- and Myc-Responsive Genes in G0 Cells. Science 2002, 296, 1132–1136. [Google Scholar] [CrossRef] [PubMed]
- Trojer, P.; Cao, A.R.; Gao, Z.; Li, Y.; Zhang, J.; Xu, X.; Li, G.; Losson, R.; Erdjument-Bromage, H.; Tempst, P.; et al. L3MBTL2 Protein Acts in Concert with PcG Protein-Mediated Monoubiquitination of H2A to Establish a Repressive Chromatin Structure. Mol. Cell 2011, 42, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Whyte, W.A.; Anderssen, E.; Apostolou, E.; Chen, H.-H.; Akbarian, S.; Bronson, R.T.; Hochedlinger, K.; Ramaswamy, S.; Young, R.A.; et al. The Polycomb Group Protein L3mbtl2 Assembles an Atypical PRC1-Family Complex that Is Essential in Pluripotent Stem Cells and Early Development. Cell Stem Cell 2012, 11, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pulido, L.; Devos, D.; Sung, Z.R.; Calonje, M. RAWUL: A new ubiquitin-like domain in PRC1 Ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genom. 2008, 9, 308. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Taylor, A.B.; Leal, B.Z.; Chadwell, L.V.; Ilangovan, U.; Robinson, A.K.; Schirf, V.; Hart, P.J.; Lafer, E.M.; Demeler, B.; et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 2010, 18, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K.; et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Kahn, T.G.; Dorafshan, E.; Schultheis, D.; Zare, A.; Stenberg, P.; Reim, I.; Pirrotta, V.; Schwartz, Y.B. Interdependence of PRC1 and PRC2 for recruitment to Polycomb Response Elements. Nucleic Acids Res. 2016, 44, 10132–10149. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Sengupta, A.K.; Kubicek, S.; Mechtler, K.; Spahn, L.; Koseki, H.; Jenuwein, T.; Wutz, A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006, 25, 3110–3122. [Google Scholar] [CrossRef] [PubMed]
- Puschendorf, M.; Terranova, R.; Boutsma, E.; Mao, X.; Isono, K.; Brykczynska, U.; Kolb, C.; Otte, A.P.; Koseki, H.; Orkin, S.H.; et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 2008, 40, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Richly, H.; Rocha-Viegas, L.; Ribeiro, J.D.; Demajo, S.; Gundem, G.; Lopez-Bigas, N.; Nakagawa, T.; Rospert, S.; Ito, T.; Di Croce, L. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 2010, 468, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Aloia, L.; Cozzuto, L.; Benitah, S.A.; Di Croce, L. RYBP and Cbx7 Define Specific Biological Functions of Polycomb Complexes in Mouse Embryonic Stem Cells. Cell Rep. 2013, 3, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Lovering, R.; Hanson, I.M.; Borden, K.L.; Martin, S.; O’Reilly, N.J.; Evan, G.I.; Rahman, D.; Pappin, D.J.; Trowsdale, J.; Freemont, P.S. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc. Natl. Acad. Sci. USA 1993, 90, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Schoorlemmer, J.; Marcos-Gutiérrez, C.; Were, F.; Martínez, R.; García, E.; Satijn, D.P.E.; Otte, A.P.; Vidal, M. Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 1997, 16, 5930–5942. [Google Scholar] [CrossRef] [PubMed]
- Voncken, J.W.; Roelen, B.A.J.; Roefs, M.; de Vries, S.; Verhoeven, E.; Marino, S.; Deschamps, J.; van Lohuizen, M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA 2003, 100, 2468–2473. [Google Scholar] [CrossRef] [PubMed]
- Cales, C.; Roman-Trufero, M.; Pavon, L.; Serrano, I.; Melgar, T.; Endoh, M.; Perez, C.; Koseki, H.; Vidal, M. Inactivation of the Polycomb Group Protein Ring1B Unveils an Antiproliferative Role in Hematopoietic Cell Expansion and Cooperation with Tumorigenesis Associated with Ink4a Deletion. Mol. Cell. Biol. 2008, 28, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- de Napoles, M.; Mermoud, J.E.; Wakao, R.; Tang, Y.A.; Endoh, M.; Appanah, R.; Nesterova, T.B.; Silva, J.; Otte, A.P.; Vidal, M.; et al. Polycomb Group Proteins Ring1A/B Link Ubiquitylation of Histone H2A to Heritable Gene Silencing and X Inactivation. Dev. Cell 2004, 7, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, Y.-I.; Isono, K.-I.; Vidal, M.; Endoh, M.; Kajita, H.; Mizutani-Koseki, Y.; Takihara, Y.; van Lohuizen, M.; Otte, A.; Jenuwein, T.; et al. Distinct roles of Polycomb group gene products in transcriptionally repressed and active domains of Hoxb8. Development 2006, 133, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- Leeb, M.; Wutz, A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J. Cell Biol. 2007, 178, 219–229. [Google Scholar] [CrossRef] [PubMed]
- van der Stoop, P.; Boutsma, E.A.; Hulsman, D.; Noback, S.; Heimerikx, M.; Kerkhoven, R.M.; Voncken, J.W.; Wessels, L.F.A.; van Lohuizen, M. Ubiquitin E3 Ligase Ring1b/Rnf2 of Polycomb Repressive Complex 1 Contributes to Stable Maintenance of Mouse Embryonic Stem Cells. PLoS ONE 2008, 3, e2235. [Google Scholar] [CrossRef] [PubMed]
- Román-Trufero, M.; Méndez-Gómez, H.R.; Pérez, C.; Hijikata, A.; Fujimura, Y.; Endo, T.; Koseki, H.; Vicario-Abejón, C.; Vidal, M. Maintenance of Undifferentiated State and Self-Renewal of Embryonic Neural Stem Cells by Polycomb Protein Ring1B. Stem Cells 2009, 27, 1559–1570. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Jiao, D.; Han, D.; Wu, J.; Wei, F.; Zheng, G.; Guo, Z.; Xi, W.; Yang, F.; Xie, P.; et al. Knockdown of RNF2 induces cell cycle arrest and apoptosis in prostate cancer cells through the upregulation of TXNIP. Oncotarget 2017, 8, 5323–5338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, Z.; Han, Y.; Yao, R.; Yue, L.; Xu, Y.; Zhang, J. Rnf2 knockdown reduces cell viability and promotes cell cycle arrest in gastric cancer cells. Oncol. Lett. 2017, 13, 3817–3822. [Google Scholar] [CrossRef] [PubMed]
- Endoh, M.; Endo, T.A.; Endoh, T.; Fujimura, Y. I.; Ohara, O.; Toyoda, T.; Otte, A.P.; Okano, M.; Brockdorff, N.; Vidal, M.; et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 2008, 135, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
- Bravo, M.; Nicolini, F.; Starowicz, K.; Barroso, S.; Cales, C.; Aguilera, A.; Vidal, M. Polycomb RING1A- and RING1B-dependent histone H2A monoubiquitylation at pericentromeric regions promotes S-phase progression. J. Cell Sci. 2015, 128, 3660–3671. [Google Scholar] [CrossRef] [PubMed]
- Lapthanasupkul, P.; Feng, J.; Mantesso, A.; Takada-Horisawa, Y.; Vidal, M.; Koseki, H.; Wang, L.; An, Z.; Miletich, I.; Sharpe, P.T. Ring1a/b polycomb proteins regulate the mesenchymal stem cell niche in continuously growing incisors. Dev. Biol. 2012, 367, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Neira, J.L.; Román-Trufero, M.; Contreras, L.M.; Prieto, J.; Singh, G.; Barrera, F.N.; Renart, M.L.; Vidal, M. The Transcriptional Repressor RYBP Is a Natively Unfolded Protein Which Folds upon Binding to DNA †. Biochemistry 2009, 48, 1348–1360. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.; Marcos-Gutiérrez, C.; del Mar Lorente, M.; Moreno, J.C.; Vidal, M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999, 18, 3404–3418. [Google Scholar] [CrossRef] [PubMed]
- Pirity, M.K.; Wang, W.-L.; Wolf, L.V.; Tamm, E.R.; Schreiber-Agus, N.; Cvekl, A. Rybp, a polycomb complex-associated protein, is required for mouse eye development. BMC Dev. Biol. 2007, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; He, H.-B.; Zhang, W.-Y.; Zhang, H.-X.; Bai, J.-B.; Liu, H.-Z.; Cao, J.-H.; Chang, K.-C.; Li, X.-Y.; Zhao, S.-H. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013, 4, e668. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-L.; Huang, F.; He, F.; Gao, C.-C.; Liang, S.-Q.; Ma, P.-F.; Dong, G.-Y.; Han, H.; Qin, H.-Y. Forced Activation of Notch in Macrophages Represses Tumor Growth by Upregulating miR-125a and Disabling Tumor-Associated Macrophages. Cancer Res. 2016, 76, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Calés, C.; Pavón, L.; Starowicz, K.; Pérez, C.; Bravo, M.; Ikawa, T.; Koseki, H.; Vidal, M. Role of Polycomb RYBP in Maintaining the B-1-to-B-2 B-Cell Lineage Switch in Adult Hematopoiesis. Mol. Cell. Biol. 2016, 36, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Ujhelly, O.; Szabo, V.; Kovacs, G.; Vajda, F.; Mallok, S.; Prorok, J.; Acsai, K.; Hegedus, Z.; Krebs, S.; Dinnyes, A.; et al. Lack of Rybp in Mouse Embryonic Stem Cells Impairs Cardiac Differentiation. Stem Cells Dev. 2015, 24, 2193–2205. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.; Szabo, V.; Pirity, M.K. Absence of Rybp Compromises Neural Differentiation of Embryonic Stem Cells. Stem Cells Int. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kalenik, J.L.; Chen, D.; Bradley, M.E.; Chen, S.J.; Lee, T.C. Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Res. 1997, 25, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Bannasch, D.; Mädge, B.; Schwab, M. Functional interaction of Yaf2 with the central region of MycN. Oncogene 2001, 20, 5913–5919. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mädge, B.; Geisen, C.; Möröy, T.; Schwab, M. Yaf2 inhibits Myc biological function. Cancer Lett. 2003, 193, 171–176. [Google Scholar] [CrossRef]
- Kaneko, T.; Miyagishima, H.; Hasegawa, T.; Mizutani-Koseki, Y.; Isono, K.; Koseki, H. The mouse YAF2 gene generates two distinct transcripts and is expressed in pre-and postimplantation embryos. Gene 2003, 315, 183–192. [Google Scholar] [CrossRef]
- Sawa, C.; Yoshikawa, T.; Matsuda-Suzuki, F.; Deléhouzée, S.; Goto, M.; Watanabe, H.; Sawada, J.; Kataoka, K.; Handa, H. YEAF1/RYBP and YAF-2 are functionally distinct members of a cofactor family for the YY1 and E4TF1/hGABP transcription factors. J. Biol. Chem. 2002, 277, 22484–22490. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, F.; Pratt, H.; Atchison, M.L. PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J. Cell. Biochem. 2009, 109. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Wilkinson, F.H.; Colavita, K.; Fennelly, C.; Atchison, M.L. YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment. Nucleic Acids Res. 2014, 42, 2208–2223. [Google Scholar] [CrossRef] [PubMed]
- Yant, S.R.; Zhu, W.; Millinoff, D.; Slightom, J.L.; Goodman, M.; Gumucio, D.L. High affinity YY1 binding motifs: Identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 1995, 23, 4353–4362. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Seto, E.; Chang, L.S.; Shenk, T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell 1991, 67, 377–388. [Google Scholar] [CrossRef]
- Park, K.; Atchison, M.L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3’ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc. Natl. Acad. Sci. USA 1991, 88, 9804–9808. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, M.E.; Zhang, X.; McGinnis, L.; Biggers, J.; Li, E.; Shi, Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 1999, 19, 7237–7244. [Google Scholar] [CrossRef] [PubMed]
- Kurisaki, K.; Kurisaki, A.; Valcourt, U.; Terentiev, A.A.; Pardali, K.; Ten Dijke, P.; Heldin, C.-H.; Ericsson, J.; Moustakas, A. Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 2003, 23, 4494–4510. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Kim, J.Y.; Dupree, J.; Tewari, A.; Melendez-Vasquez, C.; Svaren, J.; Casaccia, P. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat. Neurosci. 2010, 13, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Trask, M.C.; Tremblay, K.D.; Mager, J. Yin-Yang1 is required for epithelial-to-mesenchymal transition and regulation of Nodal signaling during mammalian gastrulation. Dev. Biol. 2012, 368, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Pan, J.; Xiong, W.; Cheng, M.; Sun, Y.; Zhang, S.; Chen, Y. Yin Yang 1 (YY1) synergizes with Smad7 to inhibit TGF-β signaling in the nucleus. Sci. China Life Sci. 2014, 57, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Wallingford, M.C.; Hiller, J.; Zhang, K.; Mager, J. YY1 Is Required for Posttranscriptional Stability of SOX2 and OCT4 Proteins. Cell. Reprogram. 2017, 19, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Affar, E.B.; Gay, F.; Shi, Y.; Liu, H.; Huarte, M.; Wu, S.; Collins, T.; Li, E.; Shi, Y. Essential Dosage-Dependent Functions of the Transcription Factor Yin Yang 1 in Late Embryonic Development and Cell Cycle Progression. Mol. Cell. Biol. 2006, 26, 3565–3581. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, S.; Karra, R.; Passer, D.; Deutsch, M.-A.; Krane, M.; Feistritzer, R.; Sturzu, A.; Domian, I.; Saga, Y.; Wu, S.M. Essential and Unexpected Role of Yin Yang 1 to Promote Mesodermal Cardiac Differentiation. Circ. Res. 2013, 112, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhao, W.; Huang, Y.; Tong, H.; Xia, Y.; Jiang, Q.; Qin, J. Loss of Polycomb Group Protein Pcgf1 Severely Compromises Proper Differentiation of Embryonic Stem Cells. Sci. Rep. 2017, 7, 46276. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. Elife 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Johansen, J.V.; Helin, K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell 2013, 49, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.A.; Bracken, A.P. A “complex” issue: Deciphering the role of variant PRC1 in ESCs. Cell Stem Cell 2013, 12, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.; Blanc, I.; Maes, J.; Fellous, M.; Robert, B.; McElreavey, K. NSPc1, a novel mammalian Polycomb gene, is expressed in neural crest-derived structures of the peripheral nervous system. Mech. Dev. 2001, 102, 219–222. [Google Scholar] [CrossRef]
- Oliviero, G.; Munawar, N.; Watson, A.; Streubel, G.; Manning, G.; Bardwell, V.; Bracken, A.P.; Cagney, G. The variant Polycomb Repressor Complex 1 component PCGF1 interacts with a pluripotency sub-network that includes DPPA4, a regulator of embryogenesis. Sci. Rep. 2015, 5, 18388. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Gong, Y.; Yue, J.; Qiang, B.; Yuan, J.; Peng, X. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008, 36, 3590–3599. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.D.; Fischle, W.; Verdin, E.; Bardwell, V.J. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000, 14, 1810–1823. [Google Scholar] [PubMed]
- Wamstad, J.A.; Corcoran, C.M.; Keating, A.M.; Bardwell, V.J. Role of the Transcriptional Corepressor Bcor in Embryonic Stem Cell Differentiation and Early Embryonic Development. PLoS ONE 2008, 3, e2814. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.J.; Vollmer, M.; Tamplin, O.; Lu, M.; Biechele, S.; Gertsenstein, M.; van Campenhout, C.; Floss, T.; Kuhn, R.; Wurst, W.; et al. Phenotypic annotation of the mouse X chromosome. Genome Res. 2010, 20, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, V.; Tiacci, E.; Holmes, A.B.; Kohlmann, A.; Martelli, M.P.; Kern, W.; Spanhol-Rosseto, A.; Klein, H.-U.; Dugas, M.; Schindela, S.; et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 2011, 118, 6153–6163. [Google Scholar] [CrossRef] [PubMed]
- Temming, P.; Corson, T.W.; Lohmann, D.R. Retinoblastoma tumorigenesis: Genetic and epigenetic changes walk hand in hand. Future Oncol. 2012, 8, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Pugh, T.J.; Weeraratne, S.D.; Archer, T.C.; Pomeranz Krummel, D.A.; Auclair, D.; Bochicchio, J.; Carneiro, M.O.; Carter, S.L.; Cibulskis, K.; Erlich, R.L.; et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012, 488, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Kenny, C.; Bausenwein, S.; Lazaro, A.; Furtwängler, R.; Gooskens, S.L.; van den Heuvel Eibrink, M.; Vokuhl, C.; Leuschner, I.; Graf, N.; Gessler, M.; et al. Mutually exclusive BCOR internal tandem duplications and YWHAE-NUTM2 fusions in clear cell sarcoma of kidney: Not the full story. J. Pathol. 2016, 238, 617–620. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.-C.; Sung, Y.-S.; Zhang, L.; Huang, S.-C.; Argani, P.; Chung, C.T.; Graf, N.S.; Wright, D.C.; Kellie, S.J.; Agaram, N.P.; et al. Recurrent BCOR Internal Tandem Duplication and YWHAE-NUTM2B Fusions in Soft Tissue Undifferentiated Round Cell Sarcoma of Infancy. Am. J. Surg. Pathol. 2016, 40, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Gearhart, M.D.; Gery, S.; Shojaee, S.; Yang, H.; Sun, H.; Lin, D.; Bai, J.; Mead, M.; Zhao, Z.; et al. BCOR regulates myeloid cell proliferation and differentiation. Leukemia 2016, 30, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Tokunaga, A.; Sakamoto, R.; Yoshida, N. Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol. Cell. Neurosci. 2011, 46, 614–624. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Kallin, E.M.; Tsukada, Y.; Zhang, Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15Ink4b. Nat. Struct. Mol. Biol. 2008, 15, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Shen, L.; Wan, M.; Taranova, O.; Wu, H.; Zhang, Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 2013, 15, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Kottakis, F.; Foltopoulou, P.; Sanidas, I.; Keller, P.; Wronski, A.; Dake, B.T.; Ezell, S.A.; Shen, Z.; Naber, S.P.; Hinds, P.W.; et al. NDY1/KDM2B Functions as a Master Regulator of Polycomb Complexes and Controls Self-Renewal of Breast Cancer Stem Cells. Cancer Res. 2014, 74, 3935–3946. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Gao, Y.; Zhang, Z.; Cao, Q.; Zhang, X.; Zou, J.; Cao, Y. Kdm2a/b Lysine Demethylases Regulate Canonical Wnt Signaling by Modulating the Stability of Nuclear β-Catenin. Dev. Cell 2015, 33, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Boulard, M.; Edwards, J.R.; Bestor, T.H. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat. Genet. 2015, 47, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kobayashi, R.; Galaktionov, K.; Beach, D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell 1995, 82, 915–925. [Google Scholar] [CrossRef]
- Bai, C.; Sen, P.; Hofmann, K.; Ma, L.; Goebl, M.; Harper, J.W.; Elledge, S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996, 86, 263–274. [Google Scholar] [CrossRef]
- Connelly, C.; Hieter, P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 1996, 86, 275–285. [Google Scholar] [CrossRef]
- Piva, R.; Liu, J.; Chiarle, R.; Podda, A.; Pagano, M.; Inghirami, G. In vivo interference with Skp1 function leads to genetic instability and neoplastic transformation. Mol. Cell. Biol. 2002, 22, 8375–8387. [Google Scholar] [CrossRef] [PubMed]
- Mandel, S.A.; Fishman-Jacob, T.; Youdim, M.B.H. Modeling sporadic Parkinson’s disease by silencing the ubiquitin E3 ligase component, SKP1A. Parkinsonism Relat. Disord. 2009, 15, S148–S151. [Google Scholar] [CrossRef]
- Everett, R.D.; Meredith, M.; Orr, A.; Cross, A.; Kathoria, M.; Parkinson, J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997, 16, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Cummins, J.M.; Rago, C.; Kohli, M.; Kinzler, K.W.; Lengauer, C.; Vogelstein, B. Tumour suppression: Disruption of HAUSP gene stabilizes p53. Nature 2004, 428. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Brooks, C.L.; Kon, N.; Gu, W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 2004, 13, 879–886. [Google Scholar] [CrossRef]
- Kon, N.; Kobayashi, Y.; Li, M.; Brooks, C.L.; Ludwig, T.; Gu, W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene 2010, 29, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Kon, N.; Zhong, J.; Kobayashi, Y.; Li, M.; Szabolcs, M.; Ludwig, T.; Canoll, P.D.; Gu, W. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ. 2011, 18, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wu, Q.; Guryanova, O.A.; Cheng, L.; Shou, W.; Rich, J.N.; Bao, S. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat. Cell Biol. 2011, 13, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.-H.; Fountain, M.D.; Fon Tacer, K.; Xia, F.; Bi, W.; Kang, S.-H.L.; Patel, A.; Rosenfeld, J.A.; Le Caignec, C.; et al. USP7 Acts as a Molecular Rheostat to Promote WASH-Dependent Endosomal Protein Recycling and Is Mutated in a Human Neurodevelopmental Disorder. Mol. Cell 2015, 59, 956–969. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Lv, L.; Li, W.; Zhang, X.; Jiang, Y.; Ge, W.; Zhou, Y. Protein deubiquitinase USP7 is required for osteogenic differentiation of human adipose-derived stem cells. Stem Cell Res. Ther. 2017, 8, 186. [Google Scholar] [CrossRef] [PubMed]
- Akasaka, T.; van Lohuizen, M.; van der Lugt, N.; Mizutani-Koseki, Y.; Kanno, M.; Taniguchi, M.; Vidal, M.; Alkema, M.; Berns, A.; Koseki, H. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 2001, 128, 1587–1597. [Google Scholar] [PubMed]
- Tagawa, M.; Sakamoto, T.; Shigemoto, K.; Matsubara, H.; Tamura, Y.; Ito, T.; Nakamura, I.; Okitsu, A.; Imai, K.; Taniguchi, M. Expression of novel DNA-binding protein with zinc finger structure in various tumor cells. J. Biol. Chem. 1990, 265, 20021–20026. [Google Scholar] [PubMed]
- Akasaka, T.; Tsuji, K.; Kawahira, H.; Kanno, M.; Harigaya, K.; Hu, L.; Ebihara, Y.; Nakahata, T.; Tetsu, O.; Taniguchi, M.; et al. The role of mel-18, a mammalian Polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity 1997, 7, 135–146. [Google Scholar] [CrossRef]
- Miyazaki, M.; Kawamoto, H.; Kato, Y.; Itoi, M.; Miyazaki, K.; Masuda, K.; Tashiro, S.; Ishihara, H.; Igarashi, K.; Amagai, T.; et al. Polycomb group gene mel-18 regulates early T progenitor expansion by maintaining the expression of Hes-1, a target of the Notch pathway. J. Immunol. 2005, 174, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Hod-Dvorai, R.; Jacob, E.; Boyko, Y.; Avni, O. The binding activity of Mel-18 at the Il17a promoter is regulated by the integrated signals of the TCR and polarizing cytokines. Eur. J. Immunol. 2011, 41, 2424–2435. [Google Scholar] [CrossRef] [PubMed]
- Won, H.-Y.; Lee, J.-Y.; Shin, D.-H.; Park, J.-H.; Nam, J.-S.; Kim, H.-C.; Kong, G. Loss of Mel-18 enhances breast cancer stem cell activity and tumorigenicity through activating Notch signaling mediated by the Wnt/TCF pathway. FASEB J. 2012, 26, 5002–5013. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Santanach, A.; Blanco, E.; Aloia, L.; Nora, E.P.; Bruneau, B.G.; Di Croce, L. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell 2015, 17, 300–315. [Google Scholar] [CrossRef] [PubMed]
- Oguro, H.; Yuan, J.; Ichikawa, H.; Ikawa, T.; Yamazaki, S.; Kawamoto, H.; Nakauchi, H.; Iwama, A. Poised Lineage Specification in Multipotential Hematopoietic Stem and Progenitor Cells by the Polycomb Protein Bmi1. Cell Stem Cell 2010, 6, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.-L.; Yeung, J.; Zeisig, B.B.; Popov, N.; Huijbers, I.; Barnes, J.; Wilson, A.J.; Taskesen, E.; Delwel, R.; Gil, J.; et al. Functional Crosstalk between Bmi1 and MLL/Hoxa9 Axis in Establishment of Normal Hematopoietic and Leukemic Stem Cells. Cell Stem Cell 2011, 8, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, S.W.M.; Valk-Lingbeek, M.E.; van der Stoop, P.P.M.; Jacobs, J.J.L.; Kieboom, K.; Tanger, E.; Hulsman, D.; Leung, C.; Arsenijevic, Y.; Marino, S.; et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005, 19, 1438–1443. [Google Scholar] [CrossRef] [PubMed]
- Zencak, D.; Lingbeek, M.; Kostic, C.; Tekaya, M.; Tanger, E.; Hornfeld, D.; Jaquet, M.; Munier, F.L.; Schorderet, D.F.; van Lohuizen, M.; et al. Bmi1 Loss Produces an Increase in Astroglial Cells and a Decrease in Neural Stem Cell Population and Proliferation. J. Neurosci. 2005, 25, 5774–5783. [Google Scholar] [CrossRef] [PubMed]
- Molofsky, A.V.; Pardal, R.; Iwashita, T.; Park, I.-K.; Clarke, M.F.; Morrison, S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 2003, 425, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.; Pintacuda, G.; Masui, O.; Koseki, Y.; Gdula, M.; Cerase, A.; Brown, D.; Mould, A.; Innocent, C.; Nakayama, M.; et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 2017, 356, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Si, S.; Nakajima-Takagi, Y.; Aoyama, K.; Oshima, M.; Saraya, A.; Sugishita, H.; Nakayama, M.; Ishikura, T.; Koseki, H.; Iwama, A. Loss of Pcgf5 Affects Global H2A Monoubiquitination but Not the Function of Hematopoietic Stem and Progenitor Cells. PLoS ONE 2016, 11, e0154561. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Lee, P.; Stafford, J.M.; von Schimmelmann, M.; Schaefer, A.; Reinberg, D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 2014, 516, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Buchou, T.; Vernet, M.; Blond, O.; Jensen, H.H.; Pointu, H.; Olsen, B.B.; Cochet, C.; Issinger, O.-G.; Boldyreff, B. Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol. Cell. Biol. 2003, 23, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G.; Meggio, F.; Moro, S. The dark side of protein kinase CK2 inhibition. Curr. Med. Chem. 2011, 18, 2867–2884. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G.; Pinna, L.A.; Moro, S. Kinase CK2 inhibition: An update. Curr. Med. Chem. 2013, 20, 671–693. [Google Scholar] [CrossRef] [PubMed]
- Huillard, E.; Ziercher, L.; Blond, O.; Wong, M.; Deloulme, J.-C.; Souchelnytskyi, S.; Baudier, J.; Cochet, C.; Buchou, T. Disruption of CK2beta in embryonic neural stem cells compromises proliferation and oligodendrogenesis in the mouse telencephalon. Mol. Cell. Biol. 2010, 30, 2737–2749. [Google Scholar] [CrossRef] [PubMed]
- Seldin, D.C.; Xu, X.; Toselli, P.A.; Russell, L.D. Globozoospermia in mice lacking the casein kinase II alpha’ catalytic subunit. Nat. Genet. 1999, 23, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Escalier, D.; Silvius, D.; Xu, X. Spermatogenesis of mice lacking CK2alpha’: Failure of germ cell survival and characteristic modifications of the spermatid nucleus. Mol. Reprod. Dev. 2003, 66, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.Y.; Dominguez, I.; Toselli, P.; Landesman-Bollag, E.; O’Brien, C.; Seldin, D.C. The Alpha Catalytic Subunit of Protein Kinase CK2 Is Required for Mouse Embryonic Development. Mol. Cell. Biol. 2008, 28, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.M.; Revuelta-Cervantes, J.; Dominguez, I. CK2 in Embryonic Development. In Protein Kinase CK2; John Wiley & Sons, Inc.: Oxford, UK, 2013; pp. 129–168. ISBN 9781118482490. [Google Scholar]
- Sultana, R.; Yu, C.-E.; Yu, J.; Munson, J.; Chen, D.; Hua, W.; Estes, A.; Cortes, F.; de la Barra, F.; Yu, D.; et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics 2002, 80, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Oksenberg, N.; Ahituv, N. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet. 2013, 29, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Rothkoff, G.; Reisman, M.; Tal, N.; Shani, O.; Nissim-Rafinia, M.; Meshorer, E.; Shifman, S. Isoform Dependent Functions of AUTS2 During Neuronal Differentiation. Eur. Neuropsychopharmacol. 2017, 27, S372–S373. [Google Scholar] [CrossRef]
- Prakash, S.; Robbins, P.W. Cloning and Analysis of the cDNA for Human Fibrosin, a Novel Fibrogenic Lymphokine. DNA Cell Biol. 1998, 17, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Trimarchi, J.M.; Lees, J.A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002, 3, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Cam, H.; Dynlacht, B.D. Emerging roles for E2F: Beyond the G1/S transition and DNA replication. Cancer Cell 2003, 3, 311–316. [Google Scholar] [CrossRef]
- Polager, S.; Ginsberg, D. E2F—at the crossroads of life and death. Trends Cell Biol. 2008, 18, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Bandara, L.R.; Buck, V.M.; Zamanian, M.; Johnston, L.H.; La Thangue, N.B. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 1993, 12, 4317–4324. [Google Scholar] [PubMed]
- Helin, K.; Wu, C.L.; Fattaey, A.R.; Lees, J.A.; Dynlacht, B.D.; Ngwu, C.; Harlow, E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 1993, 7, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Krek, W.; Livingston, D.M.; Shirodkar, S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science 1993, 262, 1557–1560. [Google Scholar] [CrossRef] [PubMed]
- Grandori, C.; Cowley, S.M.; James, L.P.; Eisenman, R.N. The Myc/Max/Mad Network and the Transcriptional Control of Cell Behavior. Annu. Rev. Cell Dev. Biol. 2000, 16, 653–699. [Google Scholar] [CrossRef] [PubMed]
- Hurlin, P.J.; Steingrìmsson, E.; Copeland, N.G.; Jenkins, N.A.; Eisenman, R.N. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999, 18, 7019–7028. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.E.; Campbell, R.D.; Sanderson, C.M. Novel NG36/G9a gene products encoded within the human and mouse MHC class III regions. Mamm. Genome 2001, 12, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.O.; Mattei, M.G.; Horsley, D.; Cowell, I.G.; Singh, P.B. The gene and pseudogenes of Cbx3/mHP1 gamma. DNA Seq. 2001, 12, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Trimarchi, J.M.; Fairchild, B.; Wen, J.; Lees, J.A. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 2001, 98, 1519–1524. [Google Scholar] [CrossRef] [PubMed]
- Schlisio, S.; Halperin, T.; Vidal, M.; Nevins, J.R. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002, 21, 5775–5786. [Google Scholar] [CrossRef] [PubMed]
- Zdzieblo, D.; Li, X.; Lin, Q.; Zenke, M.; Illich, D.J.; Becker, M.; Müller, A.M. Pcgf6, a Polycomb Group Protein, Regulates Mesodermal Lineage Differentiation in Murine ESCs and Functions in iPS Reprogramming. Stem Cells 2014, 32, 3112–3125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Tong, H.; Huang, Y.; Yan, Y.; Teng, H.; Xia, Y.; Jiang, Q.; Qin, J. Essential Role for Polycomb Group Protein Pcgf6 in Embryonic Stem Cell Maintenance and a Noncanonical Polycomb Repressive Complex 1 (PRC1) Integrity. J. Biol. Chem. 2017, 292, 2773–2784. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-S.; Chang, K.-Y.; Dang, J.; Rana, T.M. Polycomb Group Protein Pcgf6 Acts as a Master Regulator to Maintain Embryonic Stem Cell Identity. Sci. Rep. 2016, 6, 26899. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, J.; He, L.; Lin, Y.; Wu, J. Knockdown of polycomb-group RING finger 6 modulates mouse male germ cell differentiation in vitro. Cell. Physiol. Biochem. 2015, 35, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Stielow, C.; Stielow, B.; Finkernagel, F.; Scharfe, M.; Jarek, M.; Suske, G. SUMOylation of the polycomb group protein L3MBTL2 facilitates repression of its target genes. Nucleic Acids Res. 2014, 42, 3044–3058. [Google Scholar] [CrossRef] [PubMed]
- Shen-Li, H.; O’Hagan, R.C.; Hou, H.; Horner, J.W.; Lee, H.W.; DePinho, R.A. Essential role for Max in early embryonic growth and development. Genes Dev. 2000, 14, 17–22. [Google Scholar] [PubMed]
- Washkowitz, A.J.; Schall, C.; Zhang, K.; Wurst, W.; Floss, T.; Mager, J.; Papaioannou, V.E. Mga is essential for the survival of pluripotent cells during peri-implantation development. Development 2015, 142, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Storre, J.; Elsässer, H.-P.; Fuchs, M.; Ullmann, D.; Livingston, D.M.; Gaubatz, S. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 2002, 3, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Pohlers, M.; Truss, M.; Frede, U.; Scholz, A.; Strehle, M.; Kuban, R.-J.; Hoffmann, B.; Morkel, M.; Birchmeier, C.; Hagemeier, C. A Role for E2F6 in the Restriction of Male-Germ-Cell-Specific Gene Expression. Curr. Biol. 2005, 15, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Kohn, M.J.; Bronson, R.T.; Harlow, E.; Dyson, N.J.; Yamasaki, L. Dp1 is required for extra-embryonic development. Development 2003, 130, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Hirasaki, M.; Okuda, A. Does MAX open up a new avenue for meiotic research? Dev. Growth Differ. 2017, 59, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Leeb, M.; Pasini, D.; Novatchkova, M.; Jaritz, M.; Helin, K.; Wutz, A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010, 24, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Rose, N.R.; Klose, R.J. Targeting Polycomb systems to regulate gene expression: Modifications to a complex story. Nat. Rev. Mol. Cell Biol. 2015, 16, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Torres-Padilla, M.-E. Characterization of non-canonical Polycomb Repressive Complex 1 subunits during early mouse embryogenesis. Epigenetics 2016, 11, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.J.; Kharchenko, P.V.; Daheron, L.; Park, P.J.; Kingston, R.E. A Region of the Human HOXD Cluster that Confers Polycomb-Group Responsiveness. Cell 2010, 140, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Mazor, T.; Huang, H.; Huang, H.-T.; Kathrein, K.L.; Woo, A.J.; Chouinard, C.R.; Labadorf, A.; Akie, T.E.; Moran, T.B.; et al. Direct Recruitment of Polycomb Repressive Complex 1 to Chromatin by Core Binding Transcription Factors. Mol. Cell 2012, 45, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Veitia, R.A. The gene balance hypothesis: Implications for gene regulation, quantitative traits and evolution. New Phytol. 2010, 186, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Strübbe, G.; Popp, C.; Schmidt, A.; Pauli, A.; Ringrose, L.; Beisel, C.; Paro, R. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc. Natl. Acad. Sci. USA 2011, 108, 5572–5577. [Google Scholar] [CrossRef] [PubMed]
- Chittock, E.C.; Latwiel, S.; Miller, T.C.R.; Müller, C.W. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 2017, 45, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.J.; Gearhart, M.D.; Taylor, A.B.; Nanyes, D.R.; Ha, D.J.; Robinson, A.K.; Artigas, J.A.; Lee, O.J.; Demeler, B.; Hart, P.J.; et al. KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1. Structure 2016, 24, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.R.; King, H.W.; Blackledge, N.P.; Fursova, N.A.; Ember, K.J.; Fischer, R.; Kessler, B.M.; Klose, R.J. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Maezawa, S.; Hasegawa, K.; Yukawa, M.; Sakashita, A.; Alavattam, K.G.; Andreassen, P.R.; Vidal, M.; Koseki, H.; Barski, A.; Namekawa, S.H. Polycomb directs timely activation of germline genes in spermatogenesis. Genes Dev. 2017, 31, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Brookes, E.; de Santiago, I.; Hebenstreit, D.; Morris, K.J.; Carroll, T.; Xie, S.Q.; Stock, J.K.; Heidemann, M.; Eick, D.; Nozaki, N.; et al. Polycomb Associates Genome-wide with a Specific RNA Polymerase II Variant, and Regulates Metabolic Genes in ESCs. Cell Stem Cell 2012, 10, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Kar, G.; Kim, J.K.; Kolodziejczyk, A.A.; Natarajan, K.N.; Torlai Triglia, E.; Mifsud, B.; Elderkin, S.; Marioni, J.C.; Pombo, A.; Teichmann, S.A. Flipping between Polycomb repressed and active transcriptional states introduces noise in gene expression. Nat. Commun. 2017, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Liang, Z.; Xing, Q.; Zhang, T.; Wang, B.; Tian, S.; Huang, W.; Zhang, Y.; Yao, J.; Zhu, Y.; et al. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naïve ESCs. Nat. Commun. 2017, 8, 672. [Google Scholar] [CrossRef] [PubMed]
- Blakeley, P.; Fogarty, N.M.E.; del Valle, I.; Wamaitha, S.E.; Hu, T.X.; Elder, K.; Snell, P.; Christie, L.; Robson, P.; Niakan, K.K. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 2015, 142, 3151–3165. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).