Drosophila DNA-Binding Proteins in Polycomb Repression
Abstract
1. Introduction
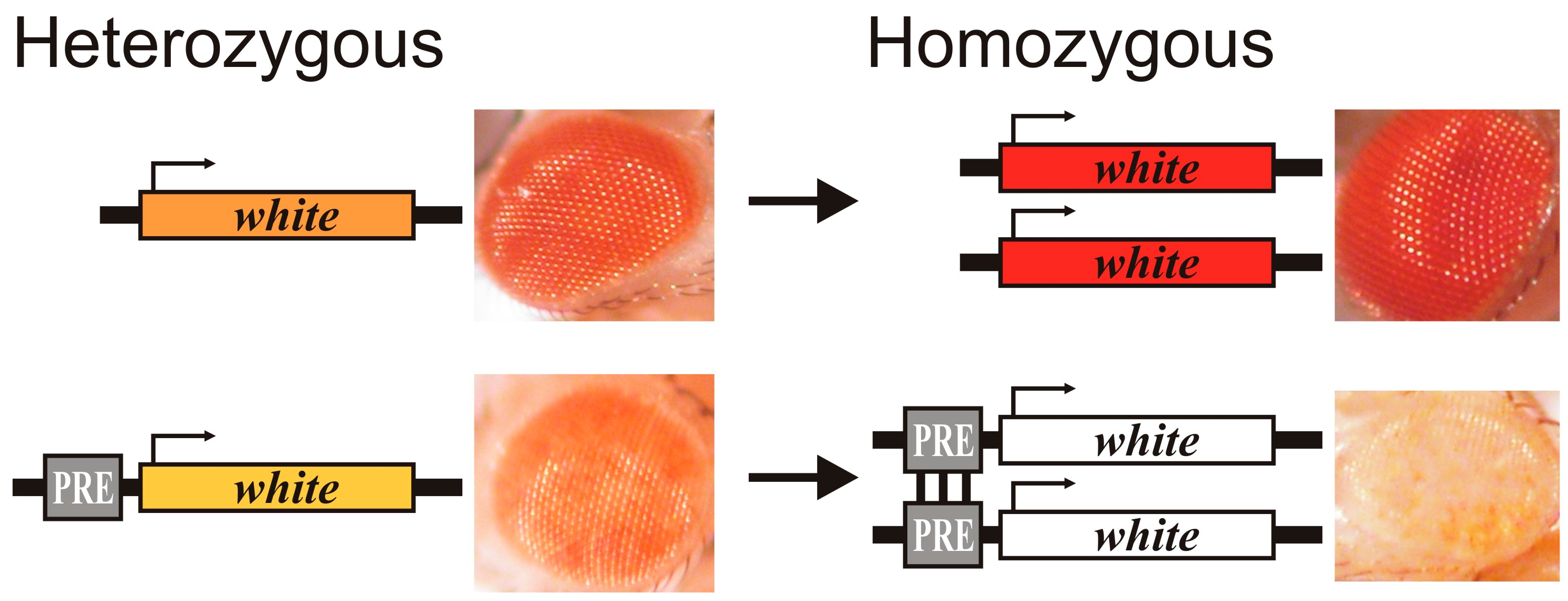
2. Properties of Polycomb Response Elements
3. Role of PRE DNA-Binding Proteins in Repression
4. Role of Protein Binding Sites in Repression
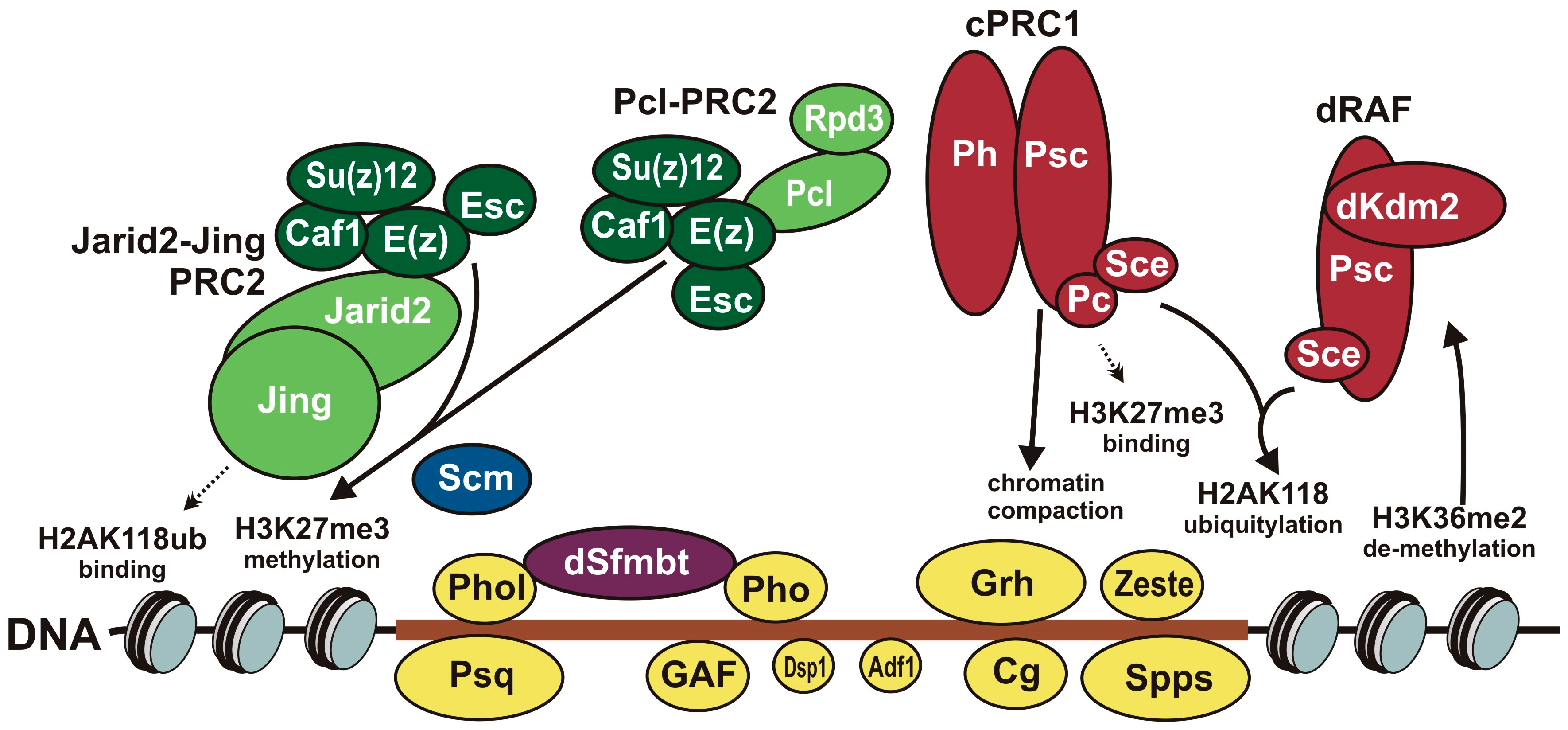
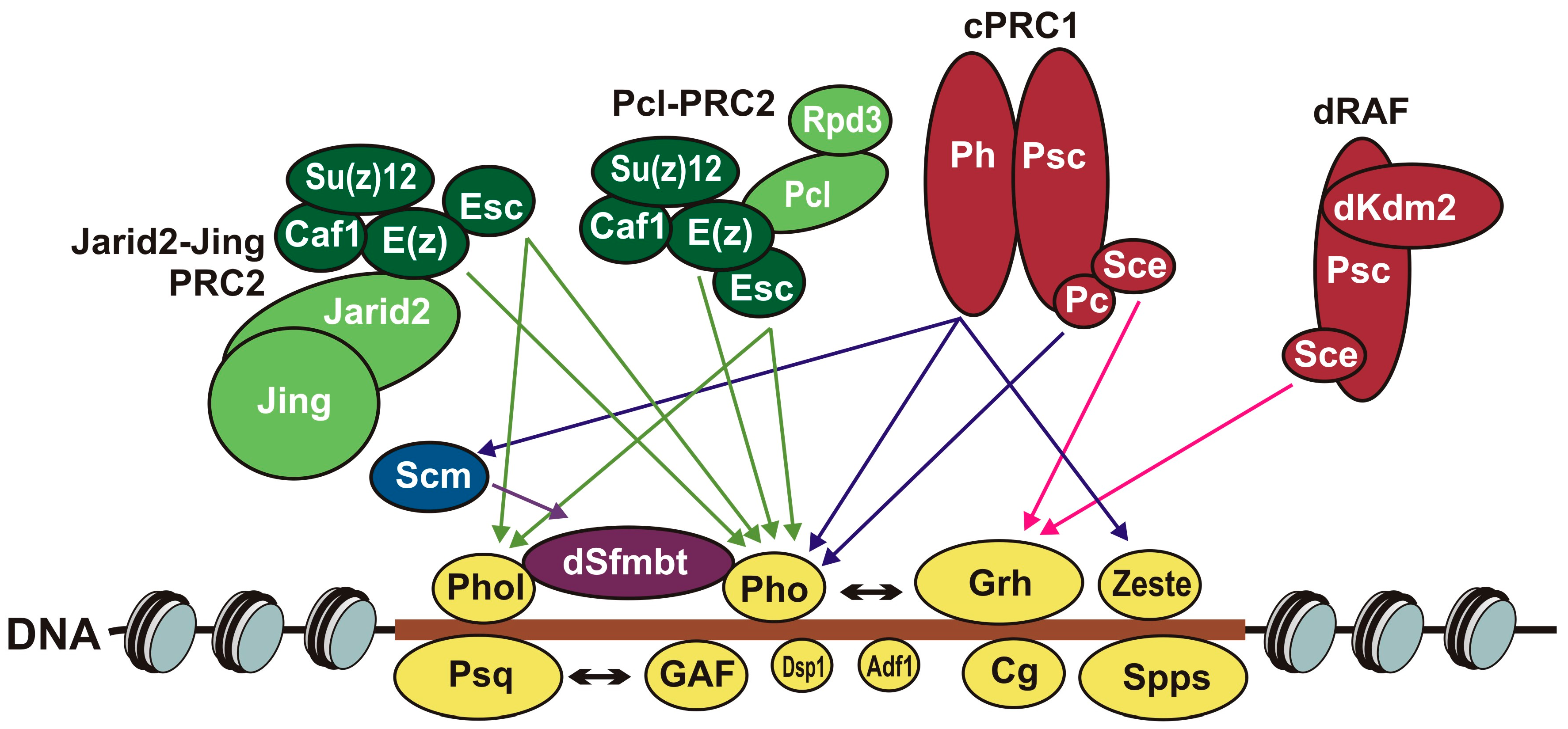
5. Role of DNA-Binding Proteins and Their Binding Sites in Recruitment of Polycomb Group Complexes
6. Multiple Interactions between Proteins on PRE
6.1. Contacts between DNA-Binding Proteins
6.2. Contacts between DNA-Binding Proteins and Subunits of PcG Complexes
6.3. Contacts between Different PcG Complexes
7. Consequences of Multiple Contacts Formed by PRE DNA-Binding Proteins
7.1. Indirect Contacts with PRE
7.2. Redundant Activity of DNA-Binding Proteins
7.3. Assistance between Proteins
8. Much More DNA-Binding Proteins Waits to Be Identified
9. Model of Combinatorial Recruitment
10. Conclusions and Future Perspective
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chetverina, D.A.; Elizar’ev, P.V.; Lomaev, D.V.; Georgiev, P.G.; Erokhin, M.M. Control of the gene activity by polycomb and trithorax group proteins in Drosophila. Russ. J. Genet. 2017, 53, 157–177. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Paro, R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019331. [Google Scholar] [CrossRef] [PubMed]
- Kassis, J.A.; Kennison, J.A.; Tamkun, J.W. Polycomb and trithorax group genes in Drosophila. Genetics 2017, 206, 1699–1725. [Google Scholar] [CrossRef] [PubMed]
- Kingston, R.E.; Tamkun, J.W. Transcriptional regulation by trithorax-group proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019349. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Breen, T.R.; Duncan, I.M. Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev. Biol. 1986, 118, 442–456. [Google Scholar] [CrossRef]
- Jürgens, G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 1985, 316, 153–155. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Struhl, G. A gene product required for correct initiation of segmental determination in Drosophila. Nature 1981, 293, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Kennison, J.A.; Tamkun, J.W. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. USA 1988, 85, 8136–8140. [Google Scholar] [CrossRef] [PubMed]
- Kwong, C.; Adryan, B.; Bell, I.; Meadows, L.; Russell, S.; Manak, J.R.; White, R. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 2008, 4, e1000178. [Google Scholar] [CrossRef] [PubMed]
- Negre, N.; Hennetin, J.; Sun, L.V.; Lavrov, S.; Bellis, M.; White, K.P.; Cavalli, G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006, 4, e170. [Google Scholar] [CrossRef] [PubMed]
- Oktaba, K.; Gutierrez, L.; Gagneur, J.; Girardot, C.; Sengupta, A.K.; Furlong, E.E.; Muller, J. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 2008, 15, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Ganapathi, M.; Leblanc, B.; Portoso, M.; Jaschek, R.; Tolhuis, B.; van Lohuizen, M.; Tanay, A.; Cavalli, G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009, 7, e13. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Y.B.; Kahn, T.G.; Nix, D.A.; Li, X.Y.; Bourgon, R.; Biggin, M.; Pirrotta, V. Genome-wide analysis of polycomb targets in Drosophila melanogaster. Nat. Genet. 2006, 38, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Tolhuis, B.; de Wit, E.; Muijrers, I.; Teunissen, H.; Talhout, W.; van Steensel, B.; van Lohuizen, M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006, 38, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Aloia, L.; Di Stefano, B.; Di Croce, L. Polycomb complexes in stem cells and embryonic development. Development 2013, 140, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.; Kroll, K.L. The roles and regulation of polycomb complexes in neural development. Cell Tissue Res. 2015, 359, 65–85. [Google Scholar] [CrossRef] [PubMed]
- Dressler, G.R.; Patel, S.R. Epigenetics in kidney development and renal disease. Transl. Res. 2015, 165, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Laugesen, A.; Helin, K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 2014, 14, 735–751. [Google Scholar] [CrossRef] [PubMed]
- Mozgova, I.; Hennig, L. The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 2015, 66, 269–296. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Y.B.; Pirrotta, V. A new world of polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013, 14, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Sowpati, D.T.; Ramamoorthy, S.; Mishra, R.K. Expansion of the polycomb system and evolution of complexity. Mech. Dev. 2015, 138 Pt 2, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.Z.; Yan, Y.; Wang, X.X.; Jiang, Y.; Xu, H.E. EZH2: Biology, disease, and structure-based drug discovery. Acta Pharmacol. Sin. 2014, 35, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Tiffen, J.; Gallagher, S.J.; Hersey, P. Ezh2: An emerging role in melanoma biology and strategies for targeted therapy. Pigment Cell Melanoma Res. 2015, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Pasini, D.; Di Croce, L. Emerging roles for polycomb proteins in cancer. Curr. Opin. Genet. Dev. 2016, 36, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Morey, L.; Santanach, A.; Di Croce, L. Pluripotency and epigenetic factors in mouse embryonic stem cell fate regulation. Mol. Cell. Biol. 2015, 35, 2716–2728. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N. Chromosome silencing mechanisms in X-chromosome inactivation: Unknown unknowns. Development 2011, 138, 5057–5065. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N.; Turner, B.M. Dosage compensation in mammals. Cold Spring Harb. Perspect. Biol. 2015, 7, a019406. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.R.; Bartolomei, M.S. Chromatin regulators of genomic imprinting. Biochim. Biophys. Acta 2014, 1839, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Kheradmand Kia, S.; Solaimani Kartalaei, P.; Farahbakhshian, E.; Pourfarzad, F.; von Lindern, M.; Verrijzer, C.P. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenet. Chromatin 2009, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Wakeling, L.A.; Ions, L.J.; Escolme, S.M.; Cockell, S.J.; Su, T.; Dey, M.; Hampton, E.V.; Jenkins, G.; Wainwright, L.J.; McKay, J.A.; et al. SIRT1 affects DNA methylation of polycomb group protein target genes, a hotspot of the epigenetic shift observed in ageing. Hum. Genom. 2015, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.; Di Foggia, V. Polycomb group genes in the regeneration of the healthy and pathological skeletal muscle. Neuropathol. Appl. Neurobiol. 2015, 42, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Hamada, Y.; Bando, T.; Nakamura, T.; Ishimaru, Y.; Mito, T.; Noji, S.; Tomioka, K.; Ohuchi, H. Leg regeneration is epigenetically regulated by histone H3K27 methylation in the cricket Gryllus bimaculatus. Development 2015, 142, 2916–2927. [Google Scholar] [CrossRef] [PubMed]
- Cheedipudi, S.; Puri, D.; Saleh, A.; Gala, H.P.; Rumman, M.; Pillai, M.S.; Sreenivas, P.; Arora, R.; Sellathurai, J.; Schroder, H.D.; et al. A fine balance: Epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic Acids Res. 2015, 43, 6236–6256. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.H.; Chiu, L.; Yu, Y.L.; Shyu, W.C. The potential roles of EZH2 in regenerative medicine. Cell Transpl. 2015, 24, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Mallen-St Clair, J.; Soydaner-Azeloglu, R.; Lee, K.E.; Taylor, L.; Livanos, A.; Pylayeva-Gupta, Y.; Miller, G.; Margueron, R.; Reinberg, D.; Bar-Sagi, D. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev. 2012, 26, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef]
- Muller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef] [PubMed]
- Pengelly, A.R.; Copur, O.; Jackle, H.; Herzig, A.; Muller, J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor polycomb. Science 2013, 339, 698–699. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.; Schotta, G.; Lein, S.; Kubicek, S.; Krauss, V.; Jenuwein, T.; Reuter, G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004, 18, 2973–2983. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.S.; Gelbart, W.M. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 1990, 126, 185–199. [Google Scholar] [PubMed]
- Simon, J.; Chiang, A.; Bender, W. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 1992, 114, 493–505. [Google Scholar] [PubMed]
- Ketel, C.S.; Andersen, E.F.; Vargas, M.L.; Suh, J.; Strome, S.; Simon, J.A. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol. Cell. Biol. 2005, 25, 6857–6868. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Birve, A.; Sengupta, A.K.; Beuchle, D.; Larsson, J.; Kennison, J.A.; Rasmuson-Lestander, A.; Muller, J. Su(z)12, a novel Drosophila polycomb group gene that is conserved in vertebrates and plants. Development 2001, 128, 3371–3379. [Google Scholar] [PubMed]
- Nekrasov, M.; Klymenko, T.; Fraterman, S.; Papp, B.; Oktaba, K.; Kocher, T.; Cohen, A.; Stunnenberg, H.G.; Wilm, M.; Muller, J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007, 26, 4078–4088. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Prasad-Sinha, J.; Birve, A.; Rasmuson-Lestander, A.; Harte, P.J. A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol. Cell. Biol. 2003, 23, 3352–3362. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Banerjee, R.; Stratton, C.A.; Prasad-Sinha, J.; Stepanik, V.; Zlobin, A.; Diaz, M.O.; Scacheri, P.C.; Harte, P.J. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009, 136, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Herz, H.M.; Mohan, M.; Garrett, A.S.; Miller, C.; Casto, D.; Zhang, Y.; Seidel, C.; Haug, J.S.; Florens, L.; Washburn, M.P.; et al. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol. Cell. Biol. 2012, 32, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Lagarou, A.; Mohd-Sarip, A.; Moshkin, Y.M.; Chalkley, G.E.; Bezstarosti, K.; Demmers, J.A.; Verrijzer, C.P. dKMD2 couples histone H2A ubiquitylation to histone H3 demethylation during polycomb group silencing. Genes Dev. 2008, 22, 2799–2810. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.; Muller, C.W.; Vermeulen, M.; Muller, J. Histone H2A monoubiquitination promotes histone H3 methylation in polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Pengelly, A.R.; Kalb, R.; Finkl, K.; Muller, J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015, 29, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Francis, N.J.; Saurin, A.J.; Shao, Z.; Kingston, R.E. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 2001, 8, 545–556. [Google Scholar] [CrossRef]
- Saurin, A.J.; Shao, Z.; Erdjument-Bromage, H.; Tempst, P.; Kingston, R.E. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 2001, 412, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.T.; Bender, W.; Kingston, R.E. Stabilization of chromatin structure by PRC1, a polycomb complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef]
- Francis, N.J.; Kingston, R.E.; Woodcock, C.L. Chromatin compaction by a polycomb group protein complex. Science 2004, 306, 1574–1577. [Google Scholar] [CrossRef] [PubMed]
- King, I.F.; Emmons, R.B.; Francis, N.J.; Wild, B.; Muller, J.; Kingston, R.E.; Wu, C.T. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol. 2005, 25, 6578–6591. [Google Scholar] [CrossRef] [PubMed][Green Version]
- King, I.F.; Francis, N.J.; Kingston, R.E. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol. Cell. Biol. 2002, 22, 7919–7928. [Google Scholar] [CrossRef] [PubMed]
- Fischle, W.; Wang, Y.; Jacobs, S.A.; Kim, Y.; Allis, C.D.; Khorasanizadeh, S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by polycomb and HP1 chromodomains. Genes Dev. 2003, 17, 1870–1881. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Zhang, Y.; Xu, R.M. Structural basis for specific binding of polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003, 17, 1823–1828. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Trupke, J.; Ringrose, L. The quest for mammalian Polycomb response elements: Are we there yet? Chromosoma 2016, 125, 471–496. [Google Scholar] [CrossRef] [PubMed]
- Di Croce, L.; Helin, K. Transcriptional regulation by polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Kassis, J.A.; Brown, J.L. Polycomb group response elements in Drosophila and vertebrates. Adv. Genet. 2013, 81, 83–118. [Google Scholar] [PubMed]
- Steffen, P.A.; Ringrose, L. What are memories made of? How polycomb and trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 2014, 15, 340–356. [Google Scholar] [CrossRef] [PubMed]
- van Kruijsbergen, I.; Hontelez, S.; Veenstra, G.J. Recruiting polycomb to chromatin. Int. J. Biochem. Cell Biol. 2015, 67, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.; Scholer, A.; Pachkov, M.; Balwierz, P.J.; Jorgensen, H.; Stadler, M.B.; van Nimwegen, E.; Schubeler, D. Modeling of epigenome dynamics identifies transcription factors that mediate polycomb targeting. Genome Res. 2013, 23, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, N.; Lerdrup, M.; Landt, E.; Agrawal-Singh, S.; Bak, M.; Tommerup, N.; Rappsilber, J.; Sodersten, E.; Hansen, K. Rest-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012, 8, e1002494. [Google Scholar] [CrossRef] [PubMed]
- Schorderet, P.; Lonfat, N.; Darbellay, F.; Tschopp, P.; Gitto, S.; Soshnikova, N.; Duboule, D. A genetic approach to the recruitment of PRC2 at the HoxD locus. PLoS Genet. 2013, 9, e1003951. [Google Scholar] [CrossRef] [PubMed]
- Sing, A.; Pannell, D.; Karaiskakis, A.; Sturgeon, K.; Djabali, M.; Ellis, J.; Lipshitz, H.D.; Cordes, S.P. A vertebrate polycomb response element governs segmentation of the posterior hindbrain. Cell 2009, 138, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.J.; Kharchenko, P.V.; Daheron, L.; Park, P.J.; Kingston, R.E. A region of the human HoxD cluster that confers polycomb-group responsiveness. Cell 2010, 140, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.S.; Rastelli, L.; Pirrotta, V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994, 13, 2553–2564. [Google Scholar] [PubMed]
- Chiang, A.; O‘Connor, M.B.; Paro, R.; Simon, J.; Bender, W. Discrete polycomb-binding sites in each parasegmental domain of the bithorax complex. Development 1995, 121, 1681–1689. [Google Scholar] [PubMed]
- Simon, J.; Chiang, A.; Bender, W.; Shimell, M.J.; O‘Connor, M. Elements of the Drosophila bithorax complex that mediate repression by polycomb group products. Dev. Biol. 1993, 158, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Gindhart, J.G., Jr.; Kaufman, T.C. Identification of polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics 1995, 139, 797–814. [Google Scholar] [PubMed]
- Ray, P.; De, S.; Mitra, A.; Bezstarosti, K.; Demmers, J.A.; Pfeifer, K.; Kassis, J.A. Combgap contributes to recruitment of Polycomb group proteins in Drosophila. Proc. Natl. Acad. Sci. USA 2016, 113, 3826–3831. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Y.B.; Kahn, T.G.; Stenberg, P.; Ohno, K.; Bourgon, R.; Pirrotta, V. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 2010, 6, e1000805. [Google Scholar] [CrossRef] [PubMed]
- Zink, B.; Engstrom, Y.; Gehring, W.J.; Paro, R. Direct interaction of the Polycomb protein with Antennapedia regulatory sequences in polytene chromosomes of Drosophila melanogaster. EMBO J. 1991, 10, 153–162. [Google Scholar] [PubMed]
- Brown, J.L.; Kassis, J.A. Architectural and functional diversity of polycomb group response elements in Drosophila. Genetics 2013, 195, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Dejardin, J.; Rappailles, A.; Cuvier, O.; Grimaud, C.; Decoville, M.; Locker, D.; Cavalli, G. Recruitment of Drosophila polycomb group proteins to chromatin by DSP1. Nature 2005, 434, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Kassis, J.A. Unusual properties of regulatory DNA from the Drosophila engrailed gene: Three “pairing-sensitive” sites within a 1.6-kb region. Genetics 1994, 136, 1025–1038. [Google Scholar] [PubMed]
- Kassis, J.A. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 2002, 46, 421–438. [Google Scholar] [PubMed]
- Cavalli, G.; Paro, R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 1998, 93, 505–518. [Google Scholar] [CrossRef]
- Cavalli, G.; Paro, R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 1999, 286, 955–958. [Google Scholar] [CrossRef] [PubMed]
- Chetverina, D.A.; Mikhailova, A.V.; Georgiev, P.G.; Erokhin, M.M. PRE/TRE elements act as transcription activators in Drosophila S2 cells. Dokl. Biochem. Biophys. 2017, 472, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Erceg, J.; Pakozdi, T.; Marco-Ferreres, R.; Ghavi-Helm, Y.; Girardot, C.; Bracken, A.P.; Furlong, E.E. Dual functionality of cis-regulatory elements as developmental enhancers and Polycomb response elements. Genes Dev. 2017, 31, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.; Barrio, L.; Cano, D.; Fiuza, U.M.; Muzzopappa, M.; Milan, M. Enhancer-pre communication contributes to the expansion of gene expression domains in proliferating primordia. Development 2011, 138, 3125–3134. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Martinez, A.M.; Iovino, N.; Cavalli, G. Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011, 12, 799–814. [Google Scholar] [CrossRef] [PubMed]
- Beisel, C.; Buness, A.; Roustan-Espinosa, I.M.; Koch, B.; Schmitt, S.; Haas, S.A.; Hild, M.; Katsuyama, T.; Paro, R. Comparing active and repressed expression states of genes controlled by the polycomb/trithorax group proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 16615–16620. [Google Scholar] [CrossRef] [PubMed]
- Erokhin, M.; Elizar’ev, P.; Parshikov, A.; Schedl, P.; Georgiev, P.; Chetverina, D. Transcriptional read-through is not sufficient to induce an epigenetic switch in the silencing activity of polycomb response elements. Proc. Natl. Acad. Sci. USA 2015, 112, 14930–14935. [Google Scholar] [CrossRef] [PubMed]
- Langlais, K.K.; Brown, J.L.; Kassis, J.A. Polycomb group proteins bind an engrailed pre in both the “on” and “off” transcriptional states of engrailed. PLoS ONE 2012, 7, e48765. [Google Scholar] [CrossRef] [PubMed]
- Americo, J.; Whiteley, M.; Brown, J.L.; Fujioka, M.; Jaynes, J.B.; Kassis, J.A. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a polycomb group response element from the Drosophila engrailed gene. Genetics 2002, 160, 1561–1571. [Google Scholar] [PubMed]
- Maurange, C.; Paro, R. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 2002, 16, 2672–2683. [Google Scholar] [CrossRef] [PubMed]
- Poux, S.; Kostic, C.; Pirrotta, V. Hunchback-independent silencing of late Ubx enhancers by a Polycomb group response element. EMBO J. 1996, 15, 4713–4722. [Google Scholar] [PubMed]
- Rank, G.; Prestel, M.; Paro, R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 2002, 22, 8026–8034. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Y.B. Chapter 6—Cooperative recruitment of polycomb complexes by polycomb response elements. In Polycomb Group Proteins; Pirrotta, V., Ed.; Academic Press: New York, NY, USA, 2017; pp. 111–129. [Google Scholar]
- Brown, J.L.; Mucci, D.; Whiteley, M.; Dirksen, M.L.; Kassis, J.A. The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1998, 1, 1057–1064. [Google Scholar] [CrossRef]
- Fritsch, C.; Brown, J.L.; Kassis, J.A.; Muller, J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 1999, 126, 3905–3913. [Google Scholar] [PubMed]
- Brown, J.L.; Fritsch, C.; Mueller, J.; Kassis, J.A. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 2003, 130, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Hagstrom, K.; Muller, M.; Schedl, P. A polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 1997, 146, 1365–1380. [Google Scholar] [PubMed]
- Strutt, H.; Cavalli, G.; Paro, R. Co-localization of polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997, 16, 3621–3632. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, J.W.; Argiropoulos, B.; Brock, H.W. Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol. Cell. Biol. 2001, 21, 4528–4543. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.H.; Chang, Y.L.; Yang, C.C.; Pan, I.C.; King, B. Pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Mol. Cell. Biol. 2002, 22, 6261–6271. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; Kassis, J.A. Spps, a Drosophila Sp1/KLF family member, binds to PRE and is required for PRE activity late in development. Development 2010, 137, 2597–2602. [Google Scholar] [CrossRef] [PubMed]
- Blastyak, A.; Mishra, R.K.; Karch, F.; Gyurkovics, H. Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between Grainyhead and Pleiohomeotic. Mol. Cell. Biol. 2006, 26, 1434–1444. [Google Scholar] [CrossRef] [PubMed]
- Hur, M.W.; Laney, J.D.; Jeon, S.H.; Ali, J.; Biggin, M.D. Zeste maintains repression of Ubx transgenes: Support for a new model of Polycomb repression. Development 2002, 129, 1339–1343. [Google Scholar] [PubMed]
- Orsi, G.A.; Kasinathan, S.; Hughes, K.T.; Saminadin-Peter, S.; Henikoff, S.; Ahmad, K. High-resolution mapping defines the cooperative architecture of polycomb response elements. Genome Res. 2014, 24, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J. A recessive lethal [l(4)29] with a homeotic effect in D. melanogaster. Drosoph. Inform. Serv. 1970, 45, 103. [Google Scholar]
- Kwon, S.H.; Kim, S.H.; Chung, H.M.; Girton, J.R.; Jeon, S.H. The Drosophila pleiohomeotic mutation enhances the polycomblike and polycomb mutant phenotypes during embryogenesis and in the adult. Int. J. Dev. Biol. 2003, 47, 389–395. [Google Scholar] [PubMed]
- Busturia, A.; Lloyd, A.; Bejarano, F.; Zavortink, M.; Xin, H.; Sakonju, S. The MCP silencer of the Drosophila Abd-B gene requires both pleiohomeotic and GAGA factor for the maintenance of repression. Development 2001, 128, 2163–2173. [Google Scholar] [PubMed]
- Kahn, T.G.; Stenberg, P.; Pirrotta, V.; Schwartz, Y.B. Combinatorial interactions are required for the efficient recruitment of pho repressive complex (PhoRC) to polycomb response elements. PLoS Genet. 2014, 10, e1004495. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D.; Faulk, C.; Kim, J. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007, 35, 3442–3452. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.; Zhang, X.; Olashaw, N.; Seto, E. Molecular cloning and functional characterization of the transcription factor YY2. J. Biol. Chem. 2004, 279, 25927–25934. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Brown, J.L.; Cao, R.; Zhang, Y.; Kassis, J.A.; Jones, R.S. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 2004, 14, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Farkas, G.; Gausz, J.; Galloni, M.; Reuter, G.; Gyurkovics, H.; Karch, F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 1994, 371, 806–808. [Google Scholar] [CrossRef] [PubMed]
- van Steensel, B.; Delrow, J.; Bussemaker, H.J. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl. Acad. Sci. USA 2003, 100, 2580–2585. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, R.C.; Lis, J.T. GAGA factor binding to DNA via a single trinucleotide sequence element. Nucleic Acids Res. 1998, 26, 2672–2678. [Google Scholar] [CrossRef] [PubMed]
- Matharu, N.K.; Hussain, T.; Sankaranarayanan, R.; Mishra, R.K. Vertebrate homologue of Drosophila GAGA factor. J. Mol. Biol. 2010, 400, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Kumar, A.S.; Mishra, R.K. Vertebrate gaf/thpok: Emerging functions in chromatin architecture and transcriptional regulation. Cell. Mol. Life Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Siegel, V.; Jongens, T.A.; Jan, L.Y.; Jan, Y.N. Pipsqueak, an early acting member of the posterior group of genes, affects vasa level and germ cell-somatic cell interaction in the developing egg chamber. Development 1993, 119, 1187–1202. [Google Scholar] [PubMed]
- Lehmann, M.; Siegmund, T.; Lintermann, K.G.; Korge, G. The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem. 1998, 273, 28504–28509. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; Grau, D.J.; DeVido, S.K.; Kassis, J.A. An Sp1/KLF binding site is important for the activity of a Polycomb group response element from the Drosophila engrailed gene. Nucleic Acids Res. 2005, 33, 5181–5189. [Google Scholar] [CrossRef] [PubMed]
- Kaczynski, J.; Cook, T.; Urrutia, R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003, 4, 206. [Google Scholar] [CrossRef] [PubMed]
- Decoville, M.; Giacomello, E.; Leng, M.; Locker, D. DSP1, an HMG-like protein, is involved in the regulation of homeotic genes. Genetics 2001, 157, 237–244. [Google Scholar] [PubMed]
- Malarkey, C.S.; Churchill, M.E. The high mobility group box: The ultimate utility player of a cell. Trends Biochem. Sci. 2012, 37, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Stros, M. HMGB proteins: Interactions with DNA and chromatin. Biochim. Biophys. Acta 2010, 1799, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J.; Kafatos, F.C. Developmental function of Elf-1: An essential transcription factor during embryogenesis in Drosophila. Genes Dev. 1991, 5, 1672–1683. [Google Scholar] [CrossRef] [PubMed]
- Dynlacht, B.D.; Attardi, L.D.; Admon, A.; Freeman, M.; Tjian, R. Functional analysis of NTF-1, a developmentally regulated Drosophila transcription factor that binds neuronal cis elements. Genes Dev. 1989, 3, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Wang, S.; Westholm, J.O.; Dai, Q.; Matsuda, R.; Hosono, C.; Bray, S.; Lai, E.C.; Samakovlis, C. Genome-wide identification of Grainy head targets in Drosophila reveals regulatory interactions with the POU domain transcription factor Vvl. Development 2017, 144, 3145–3155. [Google Scholar] [CrossRef] [PubMed]
- Tuckfield, A.; Clouston, D.R.; Wilanowski, T.M.; Zhao, L.L.; Cunningham, J.M.; Jane, S.M. Binding of the RING polycomb proteins to specific target genes in complex with the grainyhead-like family of developmental transcription factors. Mol. Cell. Biol. 2002, 22, 1936–1946. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.L.; Colvin, R.A.; Mellin, A.F. The Drosophila zeste locus is nonessential. Genetics 1989, 123, 145–155. [Google Scholar] [PubMed]
- Biggin, M.D.; Bickel, S.; Benson, M.; Pirrotta, V.; Tjian, R. Zeste encodes a sequence-specific transcription factor that activates the ultrabithorax promoter in vitro. Cell 1988, 53, 713–722. [Google Scholar] [CrossRef]
- Moses, A.M.; Pollard, D.A.; Nix, D.A.; Iyer, V.N.; Li, X.Y.; Biggin, M.D.; Eisen, M.B. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2006, 2, e130. [Google Scholar] [CrossRef] [PubMed]
- DeZazzo, J.; Sandstrom, D.; de Belle, S.; Velinzon, K.; Smith, P.; Grady, L.; DelVecchio, M.; Ramaswami, M.; Tully, T. nalyot, a mutation of the Drosophila Myb-related Adf1 transcription factor, disrupts synapse formation and olfactory memory. Neuron 2000, 27, 145–158. [Google Scholar] [CrossRef]
- Timmerman, C.; Suppiah, S.; Gurudatta, B.V.; Yang, J.; Banerjee, C.; Sandstrom, D.J.; Corces, V.G.; Sanyal, S. The Drosophila transcription factor Adf-1 (nalyot) regulates dendrite growth by controlling FasII and staufen expression downstream of CaMKII and neural activity. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 11916–11931. [Google Scholar] [CrossRef] [PubMed]
- Klymenko, T.; Papp, B.; Fischle, W.; Kocher, T.; Schelder, M.; Fritsch, C.; Wild, B.; Wilm, M.; Muller, J. A polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006, 20, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Mihaly, J.; Barges, S.; Spierer, A.; Karch, F.; Hagstrom, K.; Schweinsberg, S.E.; Schedl, P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 2001, 21, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, M.; Yusibova, G.L.; Zhou, J.; Jaynes, J.B. The DNA-binding polycomb-group protein pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development 2008, 135, 4131–4139. [Google Scholar] [CrossRef] [PubMed]
- Shimell, M.J.; Peterson, A.J.; Burr, J.; Simon, J.A.; O’Connor, M.B. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev. Biol. 2000, 218, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Horard, B.; Tatout, C.; Poux, S.; Pirrotta, V. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 2000, 20, 3187–3197. [Google Scholar] [CrossRef] [PubMed]
- Biggin, M.D.; Tjian, R. Transcription factors that activate the ultrabithorax promoter in developmentally staged extracts. Cell 1988, 53, 699–711. [Google Scholar] [CrossRef]
- Cutler, G.; Perry, K.M.; Tjian, R. Adf-1 is a nonmodular transcription factor that contains a TAF-binding myb-like motif. Mol. Cell. Biol. 1998, 18, 2252–2261. [Google Scholar] [CrossRef] [PubMed]
- Laney, J.D.; Biggin, M.D. Redundant control of Ultrabithorax by zeste involves functional levels of zeste protein binding at the Ultrabithorax promoter. Development 1996, 122, 2303–2311. [Google Scholar] [PubMed]
- Loubiere, V.; Delest, A.; Thomas, A.; Bonev, B.; Schuettengruber, B.; Sati, S.; Martinez, A.M.; Cavalli, G. Coordinate redeployment of PRC1 proteins suppresses tumor formation during Drosophila development. Nat. Genet. 2016, 48, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Han, Z.; Chen, H.; Yang, B.; Yang, X.; Xia, Y.; Pan, C.; Fu, L.; Zhang, S.; Han, H.; et al. A positive role for polycomb in transcriptional regulation via H4K20me1. Cell Res. 2016, 26, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Pherson, M.; Misulovin, Z.; Gause, M.; Mihindukulasuriya, K.; Swain, A.; Dorsett, D. Polycomb repressive complex 1 modifies transcription of active genes. Sci. Adv. 2017, 3, e1700944. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, C.A.; Misulovin, Z.; Gause, M.; Koenig, A.; Gohara, D.W.; Watson, A.; Dorsett, D. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013, 9, e1003560. [Google Scholar] [CrossRef] [PubMed]
- Schwendemann, A.; Lehmann, M. Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proc. Natl. Acad. Sci. USA 2002, 99, 12883–12888. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.D.; Brown, J.L.; Kassis, J.A. Characterization of the polycomb group response elements of the Drosophila melanogaster invected locus. Mol. Cell. Biol. 2010, 30, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Ringrose, L.; Paro, R. Polycomb/trithorax response elements and epigenetic memory of cell identity. Development 2007, 134, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Shearn, A. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics 1989, 121, 517–525. [Google Scholar] [PubMed]
- Kang, H.; McElroy, K.A.; Jung, Y.L.; Alekseyenko, A.A.; Zee, B.M.; Park, P.J.; Kuroda, M.I. Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila. Genes Dev. 2015, 29, 1136–1150. [Google Scholar] [CrossRef] [PubMed]
- Strubbe, G.; Popp, C.; Schmidt, A.; Pauli, A.; Ringrose, L.; Beisel, C.; Paro, R. Polycomb purification by in vivo biotinylation tagging reveals cohesin and trithorax group proteins as interaction partners. Proc. Natl. Acad. Sci. USA 2011, 108, 5572–5577. [Google Scholar] [CrossRef] [PubMed]
- Okulski, H.; Druck, B.; Bhalerao, S.; Ringrose, L. Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenet. Chromatin 2011, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jahren, N.; Miller, E.L.; Ketel, C.S.; Mallin, D.R.; Simon, J.A. Comparative analysis of chromatin binding by Sex Comb on Midleg (Scm) and other polycomb group repressors at a Drosophila Hox gene. Mol. Cell. Biol. 2010, 30, 2584–2593. [Google Scholar] [CrossRef] [PubMed]
- Kahn, T.G.; Dorafshan, E.; Schultheis, D.; Zare, A.; Stenberg, P.; Reim, I.; Pirrotta, V.; Schwartz, Y.B. Interdependence of PRC1 and PRC2 for recruitment to Polycomb response elements. Nucleic Acids Res. 2016, 44, 10132–10149. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Oded Elkayam, N.; Sexton, T.; Entrevan, M.; Stern, S.; Thomas, A.; Yaffe, E.; Parrinello, H.; Tanay, A.; Cavalli, G. Cooperativity, specificity, and evolutionary stability of polycomb targeting in Drosophila. Cell Rep. 2014, 9, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Frey, F.; Sheahan, T.; Finkl, K.; Stoehr, G.; Mann, M.; Benda, C.; Muller, J. Molecular basis of PRC1 targeting to polycomb response elements by PhoRC. Genes Dev. 2016, 30, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Bonchuk, A.; Denisov, S.; Georgiev, P.; Maksimenko, O. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J. Mol. Biol. 2011, 412, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Lomaev, D.; Mikhailova, A.; Erokhin, M.; Shaposhnikov, A.V.; Moresco, J.J.; Blokhina, T.; Wolle, D.; Aoki, T.; Ryabykh, V.; Yates, J.R., 3rd; et al. The GAGA factor regulatory network: Identification of GAGA factor associated proteins. PLoS ONE 2017, 12, e0173602. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.H.; Chang, Y.L. Isolation and characterization of CHRASCH, a polycomb-containing silencing complex. Methods Enzymol. 2004, 377, 267–282. [Google Scholar] [PubMed]
- Poux, S.; Melfi, R.; Pirrotta, V. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 2001, 15, 2509–2514. [Google Scholar] [CrossRef] [PubMed]
- Mohd-Sarip, A.; Venturini, F.; Chalkley, G.E.; Verrijzer, C.P. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 2002, 22, 7473–7483. [Google Scholar] [CrossRef] [PubMed]
- Grimm, C.; Matos, R.; Ly-Hartig, N.; Steuerwald, U.; Lindner, D.; Rybin, V.; Muller, J.; Muller, C.W. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 2009, 28, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Grau, D.J.; Antao, J.M.; Kingston, R.E. Functional dissection of polycomb repressive complex 1 reveals the importance of a charged domain. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.A.; Sawaya, M.R.; Cascio, D.; Kim, W.; Bowie, J.U. Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J. Biol. Chem. 2005, 280, 27769–27775. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.J.; Kyba, M.; Bornemann, D.; Morgan, K.; Brock, H.W.; Simon, J. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 1997, 17, 6683–6692. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, T.; Zuijderduijn, L.M.; Mohd-Sarip, A.; Verrijzer, C.P. GAGA facilitates binding of pleiohomeotic to a chromatinized polycomb response element. Nucleic Acids Res. 2003, 31, 4147–4156. [Google Scholar] [CrossRef] [PubMed]
- Mohd-Sarip, A.; Cleard, F.; Mishra, R.K.; Karch, F.; Verrijzer, C.P. Synergistic recognition of an epigenetic DNA element by pleiohomeotic and a polycomb core complex. Genes Dev. 2005, 19, 1755–1760. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, T.; Rehmsmeier, M. JPREdictor: A versatile tool for the prediction of cis-regulatory elements. Nucleic Acids Res. 2006, 34, W546–W550. [Google Scholar] [CrossRef] [PubMed]
- Hauenschild, A.; Ringrose, L.; Altmutter, C.; Paro, R.; Rehmsmeier, M. Evolutionary plasticity of polycomb/trithorax response elements in Drosophila species. PLoS Biol. 2008, 6, e261. [Google Scholar] [CrossRef] [PubMed]
- Ringrose, L.; Rehmsmeier, M.; Dura, J.M.; Paro, R. Genome-wide prediction of polycomb/trithorax response elements in Drosophila melanogaster. Dev. Cell 2003, 5, 759–771. [Google Scholar] [CrossRef]
- Zeng, J.; Kirk, B.D.; Gou, Y.; Wang, Q.; Ma, J. Genome-wide polycomb target gene prediction in Drosophila melanogaster. Nucleic Acids Res. 2012, 40, 5848–5863. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, K.; Kim, J. AEBP2 as a potential targeting protein for polycomb repression complex PRC2. Nucleic Acids Res. 2009, 37, 2940–2950. [Google Scholar] [CrossRef] [PubMed]
- Kugler, S.J.; Nagel, A.C. A novel Pzg-NURF complex regulates Notch target gene activity. Mol. Biol. Cell 2010, 21, 3443–3448. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.A.; Bajpe, P.K.; Bassett, A.; Moshkin, Y.M.; Kozhevnikova, E.; Bezstarosti, K.; Demmers, J.A.; Travers, A.A.; Verrijzer, C.P. Drosophila transcription factor Tramtrack69 binds MEP1 to recruit the chromatin remodeler NuRD. Mol. Cell. Biol. 2010, 30, 5234–5244. [Google Scholar] [CrossRef] [PubMed]
- Alfieri, C.; Gambetta, M.C.; Matos, R.; Glatt, S.; Sehr, P.; Fraterman, S.; Wilm, M.; Muller, J.; Muller, C.W. Structural basis for targeting the chromatin repressor Sfmbt to Polycomb response elements. Genes Dev. 2013, 27, 2367–2379. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.R.; Liu, J.L. CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genom. 2014, 41, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Wilkinson, F.H.; Colavita, K.; Fennelly, C.; Atchison, M.L. YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment. Nucleic Acids Res. 2014, 42, 2208–2223. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 2004, 15, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Ciferri, C.; Lander, G.C.; Maiolica, A.; Herzog, F.; Aebersold, R.; Nogales, E. Molecular architecture of human polycomb repressive complex 2. eLife 2012, 1, e00005. [Google Scholar] [CrossRef] [PubMed]
- Landeira, D.; Sauer, S.; Poot, R.; Dvorkina, M.; Mazzarella, L.; Jorgensen, H.F.; Pereira, C.F.; Leleu, M.; Piccolo, F.M.; Spivakov, M.; et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA polymerase II to developmental regulators. Nat. Cell Biol. 2010, 12, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Margueron, R.; Ku, M.; Chambon, P.; Bernstein, B.E.; Reinberg, D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010, 24, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Pasini, D.; Cloos, P.A.; Walfridsson, J.; Olsson, L.; Bukowski, J.P.; Johansen, J.V.; Bak, M.; Tommerup, N.; Rappsilber, J.; Helin, K. Jarid2 regulates binding of the polycomb repressive complex 2 to target genes in ES cells. Nature 2010, 464, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Valouev, A.; Swigut, T.; Zhang, J.; Zhao, Y.; Sidow, A.; Wysocka, J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 2009, 139, 1290–1302. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Mazor, T.; Huang, H.; Huang, H.T.; Kathrein, K.L.; Woo, A.J.; Chouinard, C.R.; Labadorf, A.; Akie, T.E.; Moran, T.B.; et al. Direct recruitment of polycomb repressive complex 1 (PRC1) to chromatin by core binding transcription factors. Mol. Cell 2012, 45, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Herranz, N.; Pasini, D.; Diaz, V.M.; Franci, C.; Gutierrez, A.; Dave, N.; Escriva, M.; Hernandez-Munoz, I.; Di Croce, L.; Helin, K.; et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 2008, 28, 4772–4781. [Google Scholar] [CrossRef] [PubMed]
- Jermann, P.; Hoerner, L.; Burger, L.; Schubeler, D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc. Natl. Acad. Sci. USA 2014, 111, E3415–E3421. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.D.; Smith, A.J.; De Gobbi, M.; Flenley, M.; Hughes, J.R.; Vernimmen, D.; Ayyub, H.; Sharpe, J.A.; Sloane-Stanley, J.A.; Sutherland, L.; et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012, 31, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Mendenhall, E.M.; Koche, R.P.; Truong, T.; Zhou, V.W.; Issac, B.; Chi, A.S.; Ku, M.; Bernstein, B.E. GC-rich sequence elements recruit PRC in mammalian ES cells. PLoS Genet. 2010, 6, e1001244. [Google Scholar] [CrossRef] [PubMed]
- Ballare, C.; Lange, M.; Lapinaite, A.; Martin, G.M.; Morey, L.; Pascual, G.; Liefke, R.; Simon, B.; Shi, Y.; Gozani, O.; et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 2012, 19, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Brien, G.L.; Gambero, G.; O’Connell, D.J.; Jerman, E.; Turner, S.A.; Egan, C.M.; Dunne, E.J.; Jurgens, M.C.; Wynne, K.; Piao, L.; et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 2012, 19, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, C.; Cech, T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 2015, 21, 2007–2022. [Google Scholar] [CrossRef] [PubMed]
- Ringrose, L. Noncoding RNAs in polycomb and trithorax regulation: A quantitative perspective. Annu. Rev. Genet. 2017, 51, 385–411. [Google Scholar] [CrossRef] [PubMed]

| Protein | Viability/Phenotype of Homozygous Mutants | Affect Homeotic Phenotype of Other Mutations | Protein Domains | Sequence Specificity | Human Homologs |
|---|---|---|---|---|---|
| Pho | Die as pharate adults, PcG homeotic phenotype [6,111] | Enhance Pc [112] | Four C2H2-type zinc fingers | GCCAT core [101,113]; genome-wide consensus [11,13,14,114]. | YY1/YY2 [100,102,115,116] |
| Phol | Viable, but females are sterile, no homeotic phenotype [102], | Enhance pho [102] | Four C2H2-type zinc fingers | GCCAT core [102]; genome-wide consensus [14,114]. Pho and Phol show different genome-binding profile in vivo [14,114,117]. | YY1/YY2 [100,102,115,116] |
| GAF | Die at third instar larvae stage, no homeotic phenotype [118] | Enhance Pc, Ph [104,105,113]; Ubx [118] | One C2H2-type zinc finger | d(GA)n motif genome-wide [119], GAG minimal [120]. | c-Krox/Th-POK [121,122] |
| Psq | Viable, no homeotic phenotype [123] | Enhance Ph, Pc, Scm [105,106] | Four HTH domains | d(GA)n motif; prefers longer GA stretch than GAF [124]. | Not reported |
| Cg | Die at pupal stage, not a typical PcG phenotype [79] | Pho (enhance lethality) [79] | 11 C2H2-type zinc fingers | GTGT motif genome-wide [79] | Not reported |
| Spps | Die as pharate adults, no homeotic phenotype [107] | Pho (enhance lethality) [107] | Three C2H2-type zinc finger motifs | (G/A)(G/A)GG(C/T)G [125]. | Sp1/KLF proteins [126] |
| Dsp1 | Die prematurely at adult stage, very low fertility, PcG/TrxG homeotic phenotype [127] | Enhance trx, Ubx; enhance and suppress Pc [127] | Two HMG box domains | Binds GAAAA in Fab7PRE [83], relatively weak correlation with genome-wide binding [14]. | HMGB1/2 [128,129] |
| Grh | Die at embryo stage, no homeotic phenotype [130] | Enhance Sce [108] | Grainy head DNA binding domain | (T/C)NAAC(C/T)GGT(T/C)(T/C)TGCGG [131]. Genome-wide consensus [132]. | CP2 [133] |
| Z | Viable and fertile, no homeotic phenotype [134] | - | Myb/SANT-like DNA binding domain | (с/T)GAG(C/T)G [135]; genome-wide consensus [136]. | Not reported |
| Adf1 | Die at late embryo—early larvae stage, no homeotic phenotype [137] | Enhance pho [110] | MADF | СGGC(A/G)G [110,138]. | Not reported |
| Required for Recruitment of the: | ||||
|---|---|---|---|---|
| Protein(s) | PhoRC | Scm | PRC1 | PRC2 |
| Pho (PhoRC) | - | NO, at the bxdPRE, in Drosophila cells [158]. | PARTIAL: YES, at the bxdPRE in Drosophila cells [117,158]; NO, at the bxdPRE in larvae [102,117]. | PARTIAL: YES, at the bxdPRE in Drosophila cells [117,158]. NO, at the bxdPRE in larvae [117]. |
| Pho and Phol (PhoRC) | - | PARTIAL: YES, at few sites on larval polytene chromosomes [102,159]. | PARTIAL: YES, at the bxdPRE [117], at few sites on larval polytene chromosomes [102]. | PARTIAL: YES, at the bxdPRE in larvae [117]. NO, on larval polytene chromosomes (X, 3R) [102]. |
| Scm | NO, at the bxdPRE in Drosophila cells and larvae [158]. | - | PARTIAL: YES, at the bxdPRE in Drosophila cells and larvae [158]. NO, on polytene chromosomes in larvae [155]. | PARTIAL: YES, at the bxdPRE in Drosophila cells and larvae [158].PARTIAL in larvae (only strongly stained bands are affected) [155]. |
| Pc (PRC1) | NO, at the bxdPRE in Drosophila cells and larvae [117,158]. | NO, at the bxdPRE in Drosophila cells and larvae [158], on larval polytene chromosomes [155]. | - | PARTIAL: NO, at the bxdPRE in Drosophila cells and larvae [117,158]. YES, at few sites on larval polytene chromosomes [155]. |
| Ph (PRC1) | PARTIAL: in Drosophila embryos [160] | Not reported. | - | Not reported. |
| Psc and its homologue Suppressor of zeste 2 (Su(z)2) (PRC1/dRAF) | PARTIAL: in Drosophila cells [114,159] | Not reported. | - | PARTIAL: in Drosophila cells [159] |
| E(z) (PRC2) | NO, at the bxdPRE in Drosophila cells and larvae [117,158]. | NO, at the bxdPRE in Drosophila cells [158], on larval polytene chromosomes [155]. | PARTIAL: YES, at the bxdPRE in Drosophila cells and larvae [117,158]. NO, on larval polytene chromosomes [155] | - |
| Su(z)12 (PRC2) | NO, in Drosophila cells [159]. | Not reported. | PARTIAL: in Drosophila cells [159] | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erokhin, M.; Georgiev, P.; Chetverina, D. Drosophila DNA-Binding Proteins in Polycomb Repression. Epigenomes 2018, 2, 1. https://doi.org/10.3390/epigenomes2010001
Erokhin M, Georgiev P, Chetverina D. Drosophila DNA-Binding Proteins in Polycomb Repression. Epigenomes. 2018; 2(1):1. https://doi.org/10.3390/epigenomes2010001
Chicago/Turabian StyleErokhin, Maksim, Pavel Georgiev, and Darya Chetverina. 2018. "Drosophila DNA-Binding Proteins in Polycomb Repression" Epigenomes 2, no. 1: 1. https://doi.org/10.3390/epigenomes2010001
APA StyleErokhin, M., Georgiev, P., & Chetverina, D. (2018). Drosophila DNA-Binding Proteins in Polycomb Repression. Epigenomes, 2(1), 1. https://doi.org/10.3390/epigenomes2010001





