Abstract
Recently described as the sixth base of the DNA macromolecule, the precise role of 5-hydroxymethylcytosine (5hmC) is the subject of debate. Early studies indicate that it is functionally distinct from cytosine DNA methylation (5mC), and there is evidence for 5hmC being a stable derivate of 5mC, rather than just an intermediate of demethylation. Moreover, 5hmC events correlate in time and space with key differentiation steps in mammalian cells. Such events span the three embryonic germ layers and multiple progenitor cell subtypes, suggesting a general mechanism. Because of the growing understanding of the role of progenitor cells in disease origin, we attempted to provide a detailed summary on the currently available literature supporting 5hmC as a key player in adult progenitor cell differentiation. This summary consolidates the emerging role for 5hmC in defining cellular fate.
1. Introduction
Like evidence in a crime scene, cytosine DNA methylation (5mC) can reconstitute diverse information about the origin and context of the host cell [1]. While sex can be easily identified from 5mC data, algorithms designed in recent years use 5mC to also extract information about the major tissue types in which a given cell resides and the age of the donor [2]. Even in complex samples such as blood, DNA methylation can be used to infer the proportion of major cell subpopulations [3], illustrating the tissue- and cell-specificity of 5mC. More recently, several studies showed that, at the level of progenitor cell differentiation, it is the loss of 5mC at particular intragenic regions that precedes a switch in cell identity, as we will show throughout this study. The improved knowledge of how this DNA demethylation occurs is leading to a redefinition of DNA methylation dynamics.
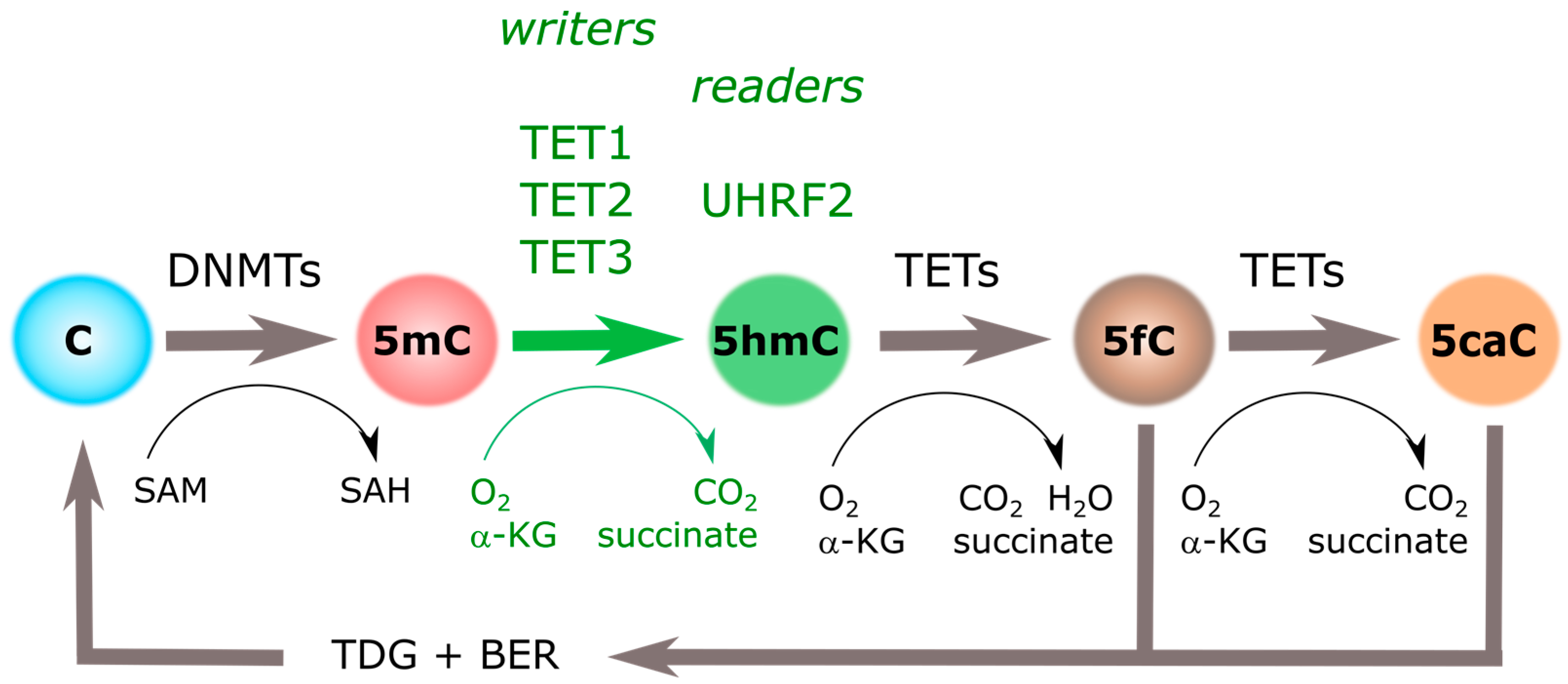
Presence of 5-hydroxymethylcytosine (5hmC) in the genome was suggested in 1972, in rat’s brain and liver DNA [4]. However, it was not until 2009 when 5hmC was identified as an unusual nucleotide when comparing the abundance of 5mC in Purkinje and granule cells [5]. In parallel, Tet Methylcytosine Dioxygenase 1 (TET1) was first described as an enzyme able to convert 5mC to 5hmC by oxidation [6]. Increasing the complexity of this field, additional oxidized derivatives of 5mC (i.e., 5-carboxylcytosine (5caC) and 5-formylcytosine (5fC) [7] and two new members of the TET family (i.e., TET2 and TET3) have been described [8,9]. Although with a slightly distinct chemical construction, all three TET proteins are considered as 5hmC “writers” [8,9], while all three oxidized derivatives of 5mC were initially defined as intermediaries in the active process of DNA demethylation (Figure 1).
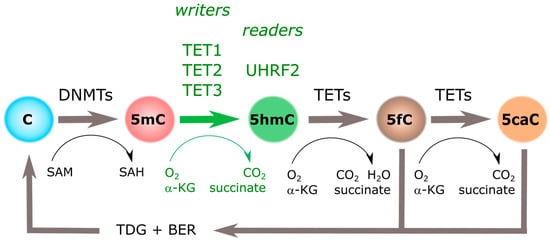
Figure 1.
Cytosine methylation cycle. Cytosine DNA methylation (5mC) groups introduced by DNA Methyl-Transferases (DNMTs) can be iteratively oxidized to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) by the action of Tet Methylcytosine Dioxygenases (TETs). Three “writers” (i.e., TET1, TET2, and TET3), and at least one “reader” (i.e., UHRF2), have been identified for 5hmC. α-KG: α-ketoglutarate; BER: base excision repair; SAH: S-adenosylhomocysteine; SAM: S-adenosylmethionine; TDG: thymine DNA glycosylase.
Ever since the discovery of the “sixth base”, strenuous efforts have been made to characterize the precise role of 5hmC in development. Such roles are becoming more evident as we learn about 5hmC specific genomic localization, its relative stability, and its recognition by other proteins. Based on the available evidence, we illustrate here how 5hmC is consistently found as a dynamic mark during the establishment of cell identity in multiple model systems.
2. 5-Hydroxymethylcytosine: “Localized, Stable and Readable”
Mapping 5hmC location represented a first step in deciphering its function. Early methods such as immunofluorescence (IF) and mass spectrometry (MS) allowed researchers to assess global levels of 5hmC, and high-performance liquid chromatography-mass spectrometry (HPLC-MS) is still the most accurate method for 5hmC global detection [10]. However, the development of locus-specific methods has played a pivotal role in our comprehension of the functional role of 5hmC. In general, affinity-based enrichment methods are widely used and can be easily combined with genome-wide profiling. Affinity-based applications can be divided into antibody and chemical capture-based methods, with both approaches providing highly reproducible results [11], and the vast majority of information on the distribution of 5hmC has been generated by antibody-based methods as 5hmC immunoprecipitation (hMeDIP) [12]. Regarding chemical labelling enrichment technics, those based on β-glycosyltransferase (βGT)-catalyzed 5hmC glycosylation such as glycosylation labelled sequencing (hMe-Seal), glucosylation, periodate oxidation and biotinylation (GLIB), and J-binding protein 1 sequencing (JBP-1) have also been extensively used; however, they are claimed to exhibit relatively low resolution compared to the antibody-based methods [11,13]. More recently, oxidative bisulfite sequencing (oxBS-Seq) [14] and TET assisted bisulfite sequencing (TAB-Seq) [15] were published and, starting now, they will be widely used methods in order to provide single resolution maps of 5hmC. However, both techniques require excessive chemical treatments and therefore can cause DNA damage. Furthermore, oxBS-seq and TAB-Seq require at least 30x sequencing depths to provide reliable, quantitative results.
Globally, 5hmC was shown by IF to be enriched in the euchromatin compartment [16]. Although exposed and accessible to anti-5hmC antibodies in euchromatin, the presence of 5hmC in heterochromatin seems to be hindered by binding proteins under non-denaturizing IF conditions [17]. Novel chemical methods allowed researchers to precisely map the genomic distribution of 5hmC and distinguish it from other cytosine variants. Immunoprecipitation with 5hmC antibodies followed by sequencing (hMeDIP-Seq) provided the first genome-wide map of 5hmC distribution, and confirmed its association with active transcription, CpG islands, 5′ regions of LINE1, CTCF, and pluripotency transcription factor (TF) binding sites and, in general, regions marked by 5mC decrease and active chromatin marks [16]. Upon differentiation from embryonic stem cells (ESCs) into embryonic bodies, the levels of 5hmC decreased. As will be discussed below, with the exception of neurogenesis, this distribution and global decrease of 5hmC is consistent across many models of differentiation. Also in ESCs, 5hmC was described in association with bivalent promoters (i.e., simultaneously occupied by histone marks H3K4me3 and H3K27me3). In this sense, it was recently shown that TET enzymes block aberrant hypermethylation in this type of promoter, ensuring robust lineage-specific transcription upon differentiation [18]. Another type of “bivalency” has been recently described at later stages of differentiation, in which genes simultaneously display promoter 5mC and gene body 5hmC at developmentally important genes [19]. Finally, Ficz et al. also described 5hmC association with non-CpG methylation resulting in strand bias that increased as a response of TET1/2 silencing. Interestingly, whenever 5hmC and 5mC occurred in parallel, gene expression was higher compared to regions in which 5mC occurred alone [16].
Growing evidence confirms the distribution and dynamics of 5hmC during embryonic cell development, outlining that it is functionally distinct from 5mC. However, the relatively low levels of 5hmC in the human genome, as well as the presence of other oxidized cytosine variants (5fC and 5caC), raised fundamental questions regarding the stability of the novel mark. Indirectly, genome-wide acquisition of 5hmC during neurogenesis in the absence of a major 5mC drop suggested that rather than an intermediate of demethylation, 5hmC serves as a stable signal on its own [20,21]. In addition, direct evidence of 5hmC stability in different cell types was obtained from ultrasensitive analytical liquid chromatography-tandem mass spectrometry (LC-MS/MS), showing no change in the levels of 5hmC over cell cycle phases [22]. Moreover, data suggested that 5mC oxidation does not occur on the nascent strand but must be formed in the double-stranded DNA produced immediately after replication [22]. Although several fundamental questions remain to be explained regarding TET binding and interaction affinities, a single, remarkable study described 5hmC-DNA interactions as much more stable and less prone than 5mC-DNA to further oxidation [23]. Favoring the idea that 5hmC is a persistent mark, compared to 5fC and 5caC, 5hmC is a poor substrate for TETs, thus protecting 5hmC from further oxidation [23,24] (reviewed in [25]). In a similar way, 5fC and 5caC are more efficiently recognized by thymine-DNA glycosylase (TDG) for removal and subsequent base excision repair (BER) leading to demethylation [26] (Figure 1). Together, higher affinities of TETs and TDG for 5fC and 5caC ensure a relative abundance and recognition of 5hmC in the genome.
In addition to confined genomic location and biochemical stability, a characteristic of other chromatin marks such as histone modifications is their capacity to be read and interpreted by specific proteins. A computational method that combined 139 published assays (chromatin immunoprecipitation sequencing (ChIP-Seq), MeDIP-Seq and glucosylation, periodate oxidation and biotinylation sequencing (GLIB-Seq), involving 77 chromatin features (3 cytosine modifications, 13 histone marks, and 61 chromatin related proteins), revealed four different co-evolutionary associations of 5hmC with chromatin related proteins: SIN3A/LSD1, TET1/MBD2, MBD2/MLL2, and OGT/TAF1 [27]. The associations of these 5hmC writers/erasers (i.e., TET1, OGT, and LSD1) do not reflect direct physical interactions of the protein pairs but rather their roles in the control of cytosine modifications and gene regulation. Although early studies described several molecules as potential readers of 5hmC, further mass spectrometry (MS) measurements failed to provide direct evidence on binding to 5hmC (reviewed by [28]). A single MS study revealed that the Set and Really Interesting New Gene (RING) finger-associated (SRA) domain of UHRF2 specifically interacts with 5hmC [29]. This discovery was subsequently supported by a structural and biochemical study [30], which demonstrated that the UHRF2-SRA domain, unlike the equivalent domain in UHRF1, does not preferentially bind to hemi-methylated DNA. Instead, it binds to fully hydroxymethylated and hemi-hydroxymethylated DNA 3.2- and 1.5-fold more tightly than it does to hemi-methylated DNA, respectively. In addition, the Chromatin target of Prmt1 (CHTOP)-methylosome complex, which methylates H4R3, has been recently described as a novel molecular interactor of 5hmC in a study conducted in gliomas [31,32].
Interestingly, 5fC and 5caC seem to be associated with numerous molecular readers [28,33,34,35]. Of note, these methylated variations have been shown to cause pause at the transcription rate generated by RNA Polymerase II (Pol II) binding. No similar mechanism has been detected for 5hmC so far, which is in line with the previously described phenomenon that 5hmC is a hallmark of active gene expression.
3. Specific 5-Hydroxymethylcytosine Enrichment Identifies Blood Cell Subtypes
In agreement with its relative stability and readability, recent studies provide ample evidence of the importance of the “sixth base” during differentiation and lineage commitment of several adult cell types. Given the existence of relatively easy approaches to perform in vitro differentiation and selection of well-characterized types of hematopoietic cells, substantial data has been accumulated to favor the role of 5hmC in transcriptional programming of myeloid and lymphoid differentiation (Table 1).

Table 1.
5-hydroxymethylcytosine (5hmC) and blood cell subtypes.
Globally, a drop in 5hmC levels has been described during differentiation from hematopoietic stem cells (HSCs) [36], except for one study showing overall 5hmC increase (without genome context data) [37]. Despite this global reduction, 5hmC remains enriched at particular loci, matching the distribution shown by earlier studies (i.e., promoters and gene bodies). In addition, 5hmC enrichment has been described in enhancers [19,36,38,39,40,41] and lineage-specific TF binding sites [42,43]. In line with this, 5hmC changes are overrepresented in highly transcribed genes. For example, chromatin-accessible-regions are associated with T cell [36,44] and induced natural killers (iNK) T cell development [45], Treg FoxP3 activation [46,47], B cell identity genes [39,41], or Bcl6 binding sites in T follicular helper (Tfh) cells [48] (Figure 2 and Table 1). During mast cell development, it was shown that Tet2 knockout caused differential 5hmC levels in genes enriched with leukocyte differentiation and proliferation, malignant transformation, and mast cell-related processes [40]. Hypo-hydroxymethylated loci overlapped with enhancer regions, suggesting that 5hmC had a role in transcriptional programming [40].
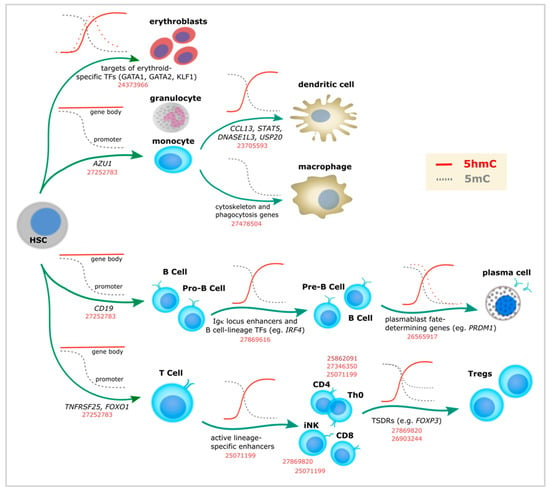
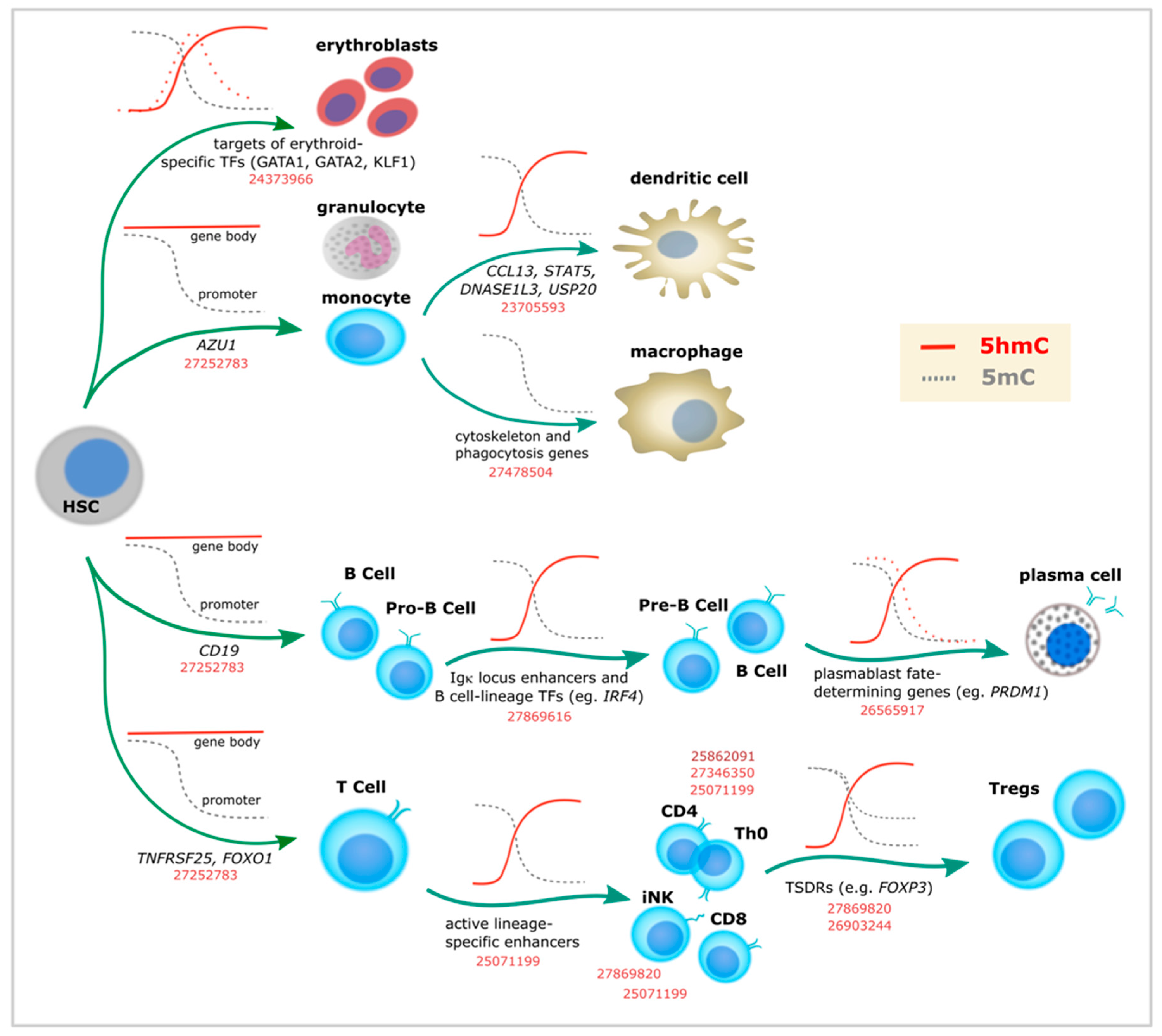
Figure 2.
5-Hydroxymethylcytosine dynamics during blood cell differentiation. Evidence for dynamic changes in 5mC (gray lines) or 5hmC (red lines) at the indicated loci, as associated with immune lineage commitment. Dashed lines represent additional evidence. Corresponding PubMed PMIDs for each study are shown in red. TSDRs: Treg-specific demethylated regions. Th0: naïve T helper cells.
Many of these studies provide mechanistic data supporting the role of TETs in cell differentiation. This is the case for Tet2 (and to a lesser degree Tet3), which is consistently reported in murine models [39,40,43,45,46,47,49] and human cells [37,38,42,50,51,52]. Of note, the role of Tet2 in mast cell differentiation described above [40] is partially independent of 5hmC, as rescuing mast cell developmental changes upon Tet2 knockout was only possible by the re-expression of Tet2 independently of its catalytic activity, which assumes that Tet2 possibly interacts with other complexes (something similar has been shown between TET1 and the SIN3A co-repressor complex [53]).
3.1. Myeloid Lineage
In addition to mast cells, specific redistribution of 5hmC has been described during differentiation of monocytes into macrophages or into dendritic cells [50,51]. Moreover, a unique role of 5hmC in red blood cell development seems well established according to a study that provided a comprehensive view of 5hmC distribution at stem/early progenitors, primitive erythroid progenitors, basophilic, polychromatic, orthochromatic, and reticulocytes [42].
Initially, by mass spectrometry measurements, using both in vitro as well as in vivo models, the authors described a general increase of 5hmC during commitment to the erythroid lineage, followed by remarkable decrease during the subsequent steps of differentiation [42]. In contrast to the dynamic fluctuation of 5hmC, 5mC exhibited modest decrease. Besides negligible levels of TET1 and TET3, TET2 seemed to be involved in the differentiation process, being the highest at the time point of specific erythroid lineage commitment. As for other cell types, in parallel with the overall decrease, elevated levels of 5hmC were detected at particular loci [42]. As erythropoiesis is a replication dependent process, this argues in favor of a stable 5hmC mark at given DNA regions that overlaps with TF binding sites and activating histone marks (i.e., H3K4me1, H3K4me3, H3K9ac, and H3K27ac).
3.2. Lymphoid Lineage
On a pioneer study of 5hmC and T-cell lineage commitment, deep sequencing was performed on purified samples (double positive, CD4 single positive, CD8 single positive, naïve CD4, naïve CD8, and CD4-differentiated Th1 and Th2 T helper cells) enriched by cytosine-5-methylenesulfonate-based immunoprecipitation [36]. In all cell types, 5hmC was found to be highest at promoters and intragenic regions, without enrichment for transcription start sites (TSS). On average, naive T-cells showed higher global 5hmC compared to the previous stage of differentiation, followed by a drop at the later stages (Th1 and Th2). As the reduction of 5hmC was replication-dependent, this initially suggested that 5hmC had been passively diluted in a cell cycle manner. However, in contrast to 5mC, gene body 5hmC exhibited significant positive correlation with gene expression accompanied with Pol II occupancy and enrichment for active histone marks. Moreover, 5hmC was suggested to mark long-range interactions between enhancers and other regulatory regions at the transition points of T-cell development. More recently, using Tet2-Tet3 double knock-out (DKO) mice, the same group showed that TET proteins are essential for maturation of invariant natural killer T cells (iNKT), and its absence generates a T cell antigen receptor (TCR)-mediated expansion of these cells and a lethal lymphocytic infiltrate in lung and liver [45]. Transcriptomic evaluation identified a characteristic profile of natural killer NKT17 cells (expressing RAR-related orphan receptor γ (RORγt) and Interleukin 17) and genes related with malignant transformation.
Of note, studies supporting a role of 5mC/5hmC in immune cell development have been performed in both mouse and human cells (Table 1). For example, analogous DNA methylation dynamics are observed during differentiation of human naive B cells into plasma cells [41]. Most differentially methylated loci identified in this process were demethylated and enriched for enhancers, with demethylation becoming particularly pronounced at terminal stages of development. Initial assessment by selective chemical labeling followed by dot blot showed global loss of 5hmC during differentiation, accompanied by decrease in TETs expression. However, 5hmC was found to be enriched at specific DNA regions, particularly at B-cell identity genes with enhancer chromatin marks (i.e., H3K4me1 and H3K27ac) (Figure 2). As mentioned above, 5hmC at gene bodies correlated with gene expression.
Therefore, 5hmC events take place at all stages of lymphoid cell differentiation, from HSCs to highly specialized cell types (Figure 2). For example, 5hmC seems to define the transition to the lymphoid lineage, the choice between B and T cell types, and between CD4 and CD8 T cells [36]. Furthermore, 5hmC enrichment at specific loci has been described for each of the well specialized fates of naïve T helper (Th0) cells (i.e., Th1, Th2, Th17, Thf, and Treg). Indeed, studies of Treg cell differentiation represent a good example of what has been done in other systems in terms of linking 5hmC to cell identity. Defined as the master TF of Treg programming almost 15 years ago [54], Foxp3 was later shown to be demethylated in particular loci called Treg-specific demethylated regions (TSDRs) (Figure 2). TSDRs were described as the most reliable criterion for natural Treg identification long before the first 5hmC studies became available [55,56]. In addition, TSDRs were suggested as a requirement for lineage stabilization [55,56,57,58] and a way to distinguish Tregs from different origins (i.e., thymic vs. periphery vs. in vitro) [56,57,58,59,60]. While TSDRs were associated with intragenic conserved non-coding sequences (CNS) in the Foxp3 loci displaying a particular chromatin landscape [57], additional TSDRs were found in mice in other immune-related genes as well (i.e., Ctla4, Il2ra, Ikzf4, and Tnfrsf18) [61]. After several additional studies highlighted that CNS2 is specifically demethylated in Tregs [62,63], subsequent work demonstrated that TET proteins were necessary for Foxp3 5hmC enrichment [46,47]. More recently, one additional CNS loci (CNS0) has been described as a FoxP3 super-enhancer and a binding site for the chromatin organizer Satb1 [64].
Overall, mouse and human studies agree on a dynamic redistribution of 5hmC in myeloid and lymphoid differentiation (including both T and B cells). Also, in mice and humans, 5hmC is enriched in gene bodies and enhancers of the corresponding cell identity genes. In addition, in line with its role in blood cell development, TETs mutations are often detected in lymphoid and myeloid malignancies [65]. Indeed, the frequency of frameshift and nonsense mutations in TET2 is estimated to be as high as 45% in patients with chronic myelomonocytic leukemia [66]. Although this places TET mutations as driver events in leukemia, it is worth mentioning that TET2 mutations alone have proven insufficient to cause cancer [65]. Future studies aimed at elucidating a more comprehensive role of TET mutations in the 5mC-5hmC balance may shed light on the dynamic interplay between genetic and epigenetic machinery in disease development, rather than considering mutations as master regulators.
4. 5-Hydroxymethylcytosine Is Redistributed during Terminal Differentiation from All Germ Layers
In addition to most blood cell subtypes, 5hmC has been associated with differentiation into other cell types from mesodermal origin. Moreover, examples also exist for the endodermal and ectodermal germ layers, as described below (Table 2).

Table 2.
5-hydroxymethylcytosine and non-blood cell subtypes.
4.1. Endoderm
Endoderm gives rise to many of the cell types that form the gastrointestinal tract and related organs. The adult liver is relatively rich in 5hmC content, and global redistributions of 5mC [67,68,69] and 5hmC [70] have been described during the fetal to adult liver transition. Specifically, liquid chromatography mass spectrometry (LC-MS) estimations indicate that 5hmC increases from 0.125% to 1% of the total cytosine content. In adult livers, such 5hmC occupancy is overrepresented at genes involved in active catabolic and metabolic processes [71] (Figure 3). While these studies suggest a role of 5hmC in liver physiology, a recent report described a specific 5hmC event during hepatocyte differentiation [72]. TET-dependent 5hmC activation preceded activation of the HNF4A P1 promoter, a master TF of hepatocyte identity (Figure 3). In the same model of hepatocyte differentiation from a bipotent progenitor, we recently profiled 5mC/5hmC using oxidative bisulfite (OxBS) coupled to HumanMethylationEPIC arrays (Illumina, San Diego, CA, USA), and noticed that this process involves 5mC reduction and 5hmC gain partially associated with the acquisition of hepatocyte expression profile (unpublished results). Of note, liver tumor development has been associated with reduced TET activity and 5hmC in human samples [73], as well as in mouse [71] and in vitro models, suggesting 5hmC could be used as a biomarker of hepatocarcinogenesis [74].
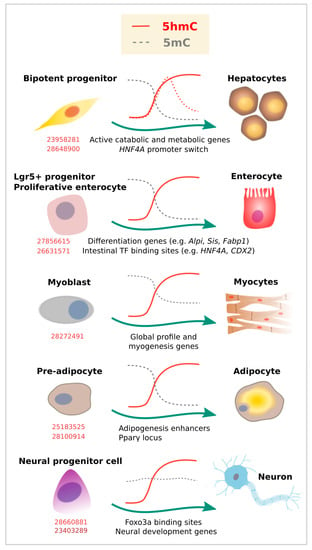
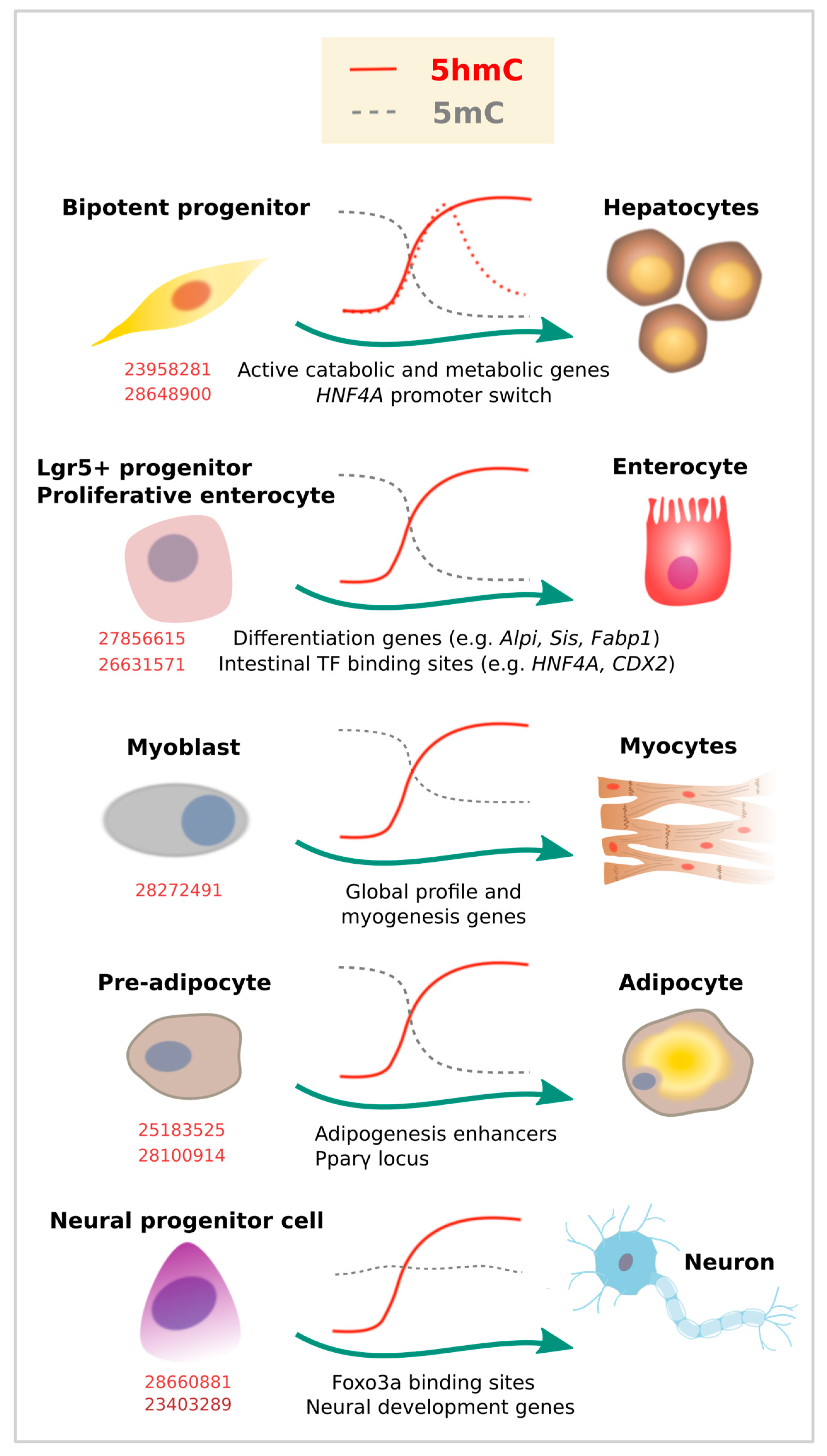
Figure 3.
5-Hydroxymethylcytosine dynamics during cell differentiation. Evidence for dynamic changes in 5mC (gray lines) or 5hmC (red lines) at the indicated loci, as associated with lineage commitment. Dashed lines represent additional evidence. Corresponding PubMed PMIDs for each study are shown in red.
Although 5hmC accumulation at the HNF4A P1 promoter seems to be transient, global higher levels in adult liver suggest stable 5hmC occupancy. In addition, stable accumulation of 5hmC has been described for enterocyte differentiation in well-known TF binding sites and targets involved in this process [75,76]. This contrasts with the pattern shown for blood cell subtypes, at least at earlier stages of blood cell differentiation (i.e., from HSCs) [19], in which there is a more general presence of 5hmC followed by a global drop that respects lineage-related loci, as described above. Interestingly, both in blood cells and endoderm differentiation, 5hmC redistribution was accompanied by a drop in 5mC (compare 5mC kinetics in Figure 2 and Figure 3). Also, in both cases, loss of gene expression in committed cells correlated with a significant decrease in gene body 5hmC levels.
4.2. Mesoderm (Other Than Blood Cells)
Besides blood cells, a more recent base resolution study suggests an important role of 5hmC during terminal chondriocyte differentiation. An increase in global 5hmC IF signal was observed at the first stage of chondrogenic differentiation, followed by decrease at a later stage [77]. Using β-Glucosyltransferase-based chemical labelling and sequencing, the authors benefited from the previously established ATDC5 chondroprogenitor model system and found 5hmC enrichment near TSSs. In a similar way to what was described in immune cell subtypes, the increase in 5hmC was even more evident in gene bodies of key chondrogenic lineage-specific factors (Sox9, 5, and 6, Runx2, and Col2a1) and was associated with transcriptional activation.
Global and localized enrichment of 5hmC has also been described for myocyte [78] and cardiomyocyte development [79,80]. 5hmC enrichment was mainly found at gene bodies and distal regulatory regions, marking active genes enriched for Pol II occupancy, as well as active histone marks. The transition from embryonic into adult heart development was characterized by 5hmC peaks in genes related to heart development, such as actin-filament-based process and regulation of heart contraction. In addition, Tet2 has been implicated in both cardiomyocyte and myocyte development. Tet2 knockout resulted in 5hmC depletion at specific cardiomyocyte gene loci [80]. In a similar way, knockdown of Tet2, but not Tet1, significantly reduced myocyte gene expression (myogenin, Myf6, and myomaker) and impaired myoblast differentiation [78].
At least two studies have shown 5hmC enrichment during adipocyte development, using murine 3T3-L1 preadipocytes as a model (Figure 3). In the first study, the chromatin insulator CTCF was found to interact with TET enzymes to promote 5hmC deposition at adipogenic transcriptional enhancers [81]. In the second study, also TET proteins (and particularly TET2) were shown to be required for adipogenesis by increasing 5hmC at the Pparγ locus and inducing Pparγ gene expression [82].
Finally, a recent study has shown that 5hmC is also redistributed in Sertoli cells, mesodermal cells that are required for spermatogenesis. LC-MS evaluation of primary isolates of rat Sertoli cells from juvenile (7 days) and adult animals showed a puberty-associated reduction of 5mC and increase of 5hmC [83]. 5hmC peaks gained during maturation were related to cell transport, catabolic process, mitochondrion, and endocytosis, which go hand in hand with the fact that adult Sertoli cells have a diverse and highly active metabolism with an abundance of mitochondria.
Similar to blood cell types, these studies relate the differentiation process to an enrichment of 5hmC, suggesting that a general 5hmC shift in genomic distribution could be associated with the upregulation of genes that define cell state.
4.3. Ectoderm
At the time of 5hmC rediscovery, using a wide range of biochemical methods, the newly identified 5-hydroxymethyl-2′-deoxycytidine was shown to constitute 0.6% of total nucleotides in Purkinje cells [5]. Being highly abundant in brain, 5hmC was immediately suggested to have a role in the genomic control of neural functions. Therefore, together with ESCs, neurons of various types, anatomical locations, and levels of differentiation have been some of the best-studied cell types in terms of 5mC/5hmC content, distribution, and function. We refer the readers to specialized reviews in 5hmC and brain function [84,85], and only outline here some of the major findings in the context of dynamic models of neurogenesis.
Similar to other tissue types, 5hmC is enriched in the bodies of actively transcribed genes in postmitotic neurons, and accumulates at the flanking region of enhancers and TF binding sites [15,85,86]. In addition, 5hmC was found to accumulate in adult neurons as synaptogenesis occurs, although its content and distribution varies among different kind of neurons [20]. In line with this, the three TET proteins are expressed in the mammalian brain, and are all involved in neurogenesis and brain physiology [85]. Particularly, neural progenitor cells (NPC) have been reported to be affected by the absence of Tet1 with consequences at the functional level, including in learning and memory [87,88]. Similarly, loss of Tet2 was shown to reduce 5hmC levels (particularly at neurogenic genes) and partially block 5hmC gain upon differentiation [89]. It was also shown that forkhead box O3 (Foxo3a) physically interacts with Tet2 and regulates the expression of genes related to neural stem/progenitor proliferation [89] (Figure 3). Therefore, 5hmC and TETs control the expression of genes and pathways related to neural programs, many of which are developmentally regulated [90]. Specific 5hmC enrichment has been observed at sequences adjacent to p300 sites in genes critical for neuronal differentiation [21], or Notch and Wnt pathway genes in retinal neurogenesis [91].
Another fact that increased the interest for 5hmC was the possibility of methylcytosine binding protein 2 (MeCP2) as a 5hmC reader [53], as this protein was previously associated with neurodevelopmental disorders [92] (reviewed in [93]). Indeed, it was recently shown that distribution of 5hmC in CG and non-CpG dinucleotides is distinct and reflects the binding specificity and genome occupancy of MeCP2 [86]. In addition, loss of MeCP2 led to specific reduction of 5hmC signals in dynamically hydroxymethylated intragenic non-CpGs involved in gene activation [20]. However, while the role of MeCP2 as a reader needs to be further validated [28], other candidate readers have been suggested in the context of neurogenesis and adult brain [29,94].
Therefore, dynamic models of neurogenesis are consistent with an important role of 5hmC, not only during differentiation, but also later in adult neuron life. Contrary to other tissues, 5mC depletion does not necessarily precede 5hmC acquisition [21] (Figure 3). Moreover, the high levels of 5hmC in adult neurons (and capacity to further increase), and the specific distribution across cell types and brain regions, indicate an important functional role in addition to preserving cell identity. Recent studies showing that demethylation occurs rapidly after neuronal or behavioral stimulation [95,96,97], and that 5hmC is dynamically regulated during aging [20], are helping to elucidate such role.
5. Perspectives and Concluding Remarks
Cell identity is acquired through the sequential and combinatorial activity of TFs on lineage-specific enhancers [98]. A global picture is emerging in which, in joint action with TFs and enhancers, TET-catalyzed 5hmC marks identity genes at key differentiation checkpoints. In most cases, global 5hmC seems to be progressively reduced from progenitor to differentiated state, but remains (or is acquired) at relevant lineage-specific loci or at genes related to adult cell function. As illustrated here, examples exist for all embryonic layers, and both epithelial and mesenchymal phenotypes, suggesting a general mechanism. In a time when cell subtypes are being systematically redefined [99], 5hmC promises to be an exquisite marker of cell identity. Moreover, as has been shown for the FoxP3 CNS loci in Treg cells, 5hmC may be able not only to identify a cell subtype, but also its origin, something that gene expression alone cannot do [57,62,63].
Although proliferation status seems to be critical for global 5hmC [22], this does not explain the specificity of 5hmC occupancy during differentiation. Recent studies are shedding light into such mechanisms. For example, interaction between TET proteins and lineage-specific TFs such as the FOXA family have been described [72,89,100]. Although this is better supported at the level of pioneer factors, it is feasible that other TFs involved in lineage commitment are also able to recruit TETs to specific genomic loci. While several TFs are involved in DNA demethylation by interacting with TET proteins (e.g., Nanog during establishment of pluripotency [101], the hematopoiesis-related RUNX1 [102] and EBF1 [103], PU.1 in osteoclast differentiation [104], and PPARγ in adipogenesis [105]), screening systems suggest that many others may share this property [106]. Alternatively, enhancer elements that distinguish cell types may be specifically targeted by TETs based on their chromatin conformation and histone mark profile. Indeed, based on their abundance in gene bodies of active genes and poised enhancers, TETs have been suggested as general transcriptional activators that recruit other proteins to demethylate enhancers [25].
Whether related or not to the preservation of cell identity, this oxidative form of methylated cytosine is emerging as a novel marker in many disorders [107]. Studies assessing 5mC and 5hmC content in different neurodegenerative pathologies, including Alzheimer’s and Huntington’s diseases, haven been reviewed in past years [108]. The levels of 5hmC in brain increase progressively from birth until death [108], and this age-associated increase was significantly higher in an Alzheimer transgenic mice model [109], while there is a disease-associated decrease of 5hmC levels in different brain regions in patients with the same condition [108]. One example that illustrates the importance of 5hmC in maintaining cell phenotype is liver fibrosis. In this pathology, chronic inflammatory injury triggers an adipogenic-to-fibrogenic phenotype switch in hepatic stellate cells, eventually leading to cirrhosis [110]. During different stages of chronic liver injury, 5hmC was found reduced, while hepatic stellate cells became transdifferentiated [111]. Consistent with these data, global 5hmC was shown to be reduced in a rat model of CCl4-induced cirrhosis [112]. It is important to note the recent availability of liver methylome and hydroxymethylome data on Wistar and Sprague-Dawley rats, as well as the genome-wide distribution of enhancer related chromatin marks (i.e., H3K4me1 and H3K27ac), which could represent a baseline reference for future studies of liver pathology [113].
An obvious context in which 5hmC occupancy is expected to be dysregulated is malignant disease (specialized reviews on cancer and TET-dependent 5hmC have been recently published [114,115,116,117,118]). Indeed, while Tet deletions/mutations have been shown to predispose to blood malignancies, cancer was initially associated with a global loss of 5hmC in various tissues [65,66]. Although such reports revealing 5hmC decrease in cancer lacked genome-wide or base-level resolution, studies consistently show that 5hmC profiles are able to distinguish malignant tissues, even under cell culture conditions [119]. If 5hmC does indeed preserve cellular phenotypes, it would be expected to have a protective role against cancer. In line with this, it was suggested that 5hmC is a hallmark from the early intestinal development that prevents neoplastic DNA hypermethylation events [120]. In addition, 5hmC was recently shown to correlate with lower regional CpG > T mutation frequency in cancers from various tissues (i.e., brain, kidney, and blood), also suggesting a protective role [121].
Finally, it has been suggested for years that DNA methylation may represent an interphase between environment and disease. The debate has been centered around which genomic locations are more dynamic or sensitive to external factors, and high interest has been placed on DNA methylation fluctuations at enhancer elements. Nowadays, emerging work suggests an important role of the oxidized forms of 5m—in particular, of 5hmC and the enzymes that establish it. For example, TETs are able to control differentiation into immune cell subtypes in response to specific inflammatory signals and are sensors of diverse metabolic processes. Another interesting example could be the critical role of oxygen concentration (as a co-factor) that influences TET activity and hence regulating cellular differentiation and fate in embryogenesis in which there are graded levels of molecular oxygen [122]. In a similar way, as α-ketoglutarate-dependent dioxygenases, the activity of TETs can be inhibited by 2-hydroxyglutarate in pathological conditions such as aberrant liver progenitor differentiation in presence of IDH1/2 mutations [123], or CD8+ T cell fate under hypoxia [49]. Moreover, TET activity has been shown to be influenced by the availability of vitamins C [46,124,125] and A [125], and hydrogen sulphide [47].
In conclusion, the data sets presented throughout this study lead us to suggest that previously published studies that lack discriminated detection of 5mC and 5hmC (such as the widely used bisulphite conversion) have to be carefully revisited. Furthermore, approaches that are able to distinguish between DNA methylation intermediates should be superior in the future. Evidence points towards 5hmC (rather than 5mC) as a more likely interphase between the environment and dynamic gene expression changes, and most importantly, it seems to regulate the directors of cell fate. Therefore, its distribution in the genome could be used as a “fingerprint” to clearly indicate cell identity. This new potential of the 5hmC not only opens the door to deeper studies of cell differentiation and tissue development, but could also provide a useful tool for early detection and prevention of several conditions associated with the loss of cellular identity such as cancer.
Acknowledgments
We apologize to our colleagues whose works were not directly covered or cited here. S.E. is supported by the French Government, under a grant from the Agence Nationale de la Recherche (Project Investissements d’Avenir UCAJEDI, No. ANR-15-IDEX-01). J.R.R.-A. is the recipient of Fellowships from Consejo Nacional de Ciencia y Tecnología (CONACyT) (CVU 508509) and Programa de Apoyo a los Estudios de Posgrado-Universidad Nacional Autónoma de México (PAEP-UNAM) (No. Cta. 30479367-5). H.H.-V. studies on 5hmC in progenitor and immune cell identity are currently supported by grants from the Agence Française de Recherches sur le VIH/Sida et les Hépatites Virales (ANRS) (grants No. ECTZ47287 and ECTZ50137), and the Institut National du Cancer (INCA) (Grant No. PLBIO17-317, Inserm Plan Cancer No. C14088CS). The authors declare no conflicts of interest.
Author Contributions
S.E., J.R.R.-A. and H.H.-V. wrote jointly this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Penn, N.W.; Suwalski, R.; O’Riley, C.; Bojanowski, K.; Yura, R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972, 126, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Kriaucionis, S.; Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.P.; Yabuuchi, A.; Rao, S.; Huang, Y.; Cunniff, K.; Nardone, J.; Laiho, A.; Tahiliani, M.; Sommer, C.A.; Mostoslavsky, G.; et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011, 8, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chavez, L.; Chang, X.; Wang, X.; Pastor, W.A.; Kang, J.; Zepeda-Martínez, J.A.; Pape, U.J.; Jacobsen, S.E.; Peters, B.; et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Qing, Y.; Tian, Z.; Bi, Y.; Wang, Y.; Long, J.; Song, C.-X.; Diao, J. Quantitation and mapping of the epigenetic marker 5-hydroxymethylcytosine. BioEssays News Rev. Mol. Cell. Dev. Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.P.; Hunter, J.M.; Nestor, C.E.; Dunican, D.S.; Terranova, R.; Moggs, J.G.; Meehan, R.R. Comparative analysis of affinity-based 5-hydroxymethylation enrichment techniques. Nucleic Acids Res. 2013, 41, e206. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-X.; Yi, C.; He, C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat. Biotechnol. 2012, 30, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C.M.; Ren, B. Mapping human epigenomes. Cell 2013, 155, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Booth, M.J.; Branco, M.R.; Ficz, G.; Oxley, D.; Krueger, F.; Reik, W.; Balasubramanian, S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012, 336, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Hon, G.C.; Szulwach, K.E.; Song, C.-X.; Zhang, L.; Kim, A.; Li, X.; Dai, Q.; Shen, Y.; Park, B.; et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012, 149, 1368–1380. [Google Scholar] [CrossRef] [PubMed]
- Ficz, G.; Branco, M.R.; Seisenberger, S.; Santos, F.; Krueger, F.; Hore, T.A.; Marques, C.J.; Andrews, S.; Reik, W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 2011, 473, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, Z.; Jiang, T.; Li, X.; Wang, H. Immunofluorescence imaging strategy for evaluation of the accessibility of DNA 5-hydroxymethylcytosine in chromatins. Anal. Chem. 2017, 89, 5702–5706. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Pan, H.; Doré, L.C.; Shukla, A.; Li, Q.V.; Pelham-Webb, B.; Teijeiro, V.; González, F.; Krivtsov, A.; Chang, C.-J.; et al. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet. 2017. [Google Scholar] [CrossRef]
- Tekpli, X.; Urbanucci, A.; Hashim, A.; Vågbø, C.B.; Lyle, R.; Kringen, M.K.; Staff, A.C.; Dybedal, I.; Mills, I.G.; Klungland, A.; et al. Changes of 5-hydroxymethylcytosine distribution during myeloid and lymphoid differentiation of CD34+ cells. Epigenet. Chromatin 2016, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Szulwach, K.E.; Li, X.; Li, Y.; Song, C.-X.; Wu, H.; Dai, Q.; Irier, H.; Upadhyay, A.K.; Gearing, M.; Levey, A.I.; et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011, 14, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.A.; Qiu, R.; Wu, X.; Li, A.X.; Zhang, H.; Wang, J.; Jui, J.; Jin, S.-G.; Jiang, Y.; Pfeifer, G.P.; et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013, 3, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Bachman, M.; Uribe-Lewis, S.; Yang, X.; Williams, M.; Murrell, A.; Balasubramanian, S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014, 6, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Lu, J.; Cheng, J.; Rao, Q.; Li, Z.; Hou, H.; Lou, Z.; Zhang, L.; Li, W.; Gong, W.; et al. Structural insight into substrate preference for TET-mediated oxidation. Nature 2015, 527, 118–122. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Szyf, M. The elusive role of 5′-hydroxymethylcytosine. Epigenomics 2016, 8, 1539–1551. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Hong, S.; Bhagwat, A.S.; Zhang, X.; Cheng, X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012, 40, 10203–10214. [Google Scholar] [CrossRef] [PubMed]
- Juan, D.; Perner, J.; Carrillo de Santa Pau, E.; Marsili, S.; Ochoa, D.; Chung, H.-R.; Vingron, M.; Rico, D.; Valencia, A. Epigenomic co-localization and co-evolution reveal a key role for 5hmC as a communication hub in the chromatin network of ESCs. Cell Rep. 2016, 14, 1246–1257. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Pfeifer, G.P. Are there specific readers of oxidized 5-methylcytosine bases? BioEssays News Rev. Mol. Cell. Dev. Biol. 2016, 38, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Spruijt, C.G.; Gnerlich, F.; Smits, A.H.; Pfaffeneder, T.; Jansen, P.W.T.C.; Bauer, C.; Münzel, M.; Wagner, M.; Müller, M.; Khan, F.; et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 2013, 152, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Xiong, J.; Wang, M.; Yang, N.; Wong, J.; Zhu, B.; Xu, R.-M. Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol. Cell 2014, 54, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Fanis, P.; Gillemans, N.; Aghajanirefah, A.; Pourfarzad, F.; Demmers, J.; Esteghamat, F.; Vadlamudi, R.K.; Grosveld, F.; Philipsen, S.; van Dijk, T.B. Five friends of methylated chromatin target of protein-arginine-methyltransferase[prmt]-1 (chtop), a complex linking arginine methylation to desumoylation. Mol. Cell. Proteom. MCP 2012, 11, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Takai, H.; Masuda, K.; Sato, T.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Koyama-Nasu, R.; Nasu-Nishimura, Y.; Katou, Y.; Ogawa, H.; et al. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014, 9, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Olanrewaju, Y.O.; Zheng, Y.; Wilson, G.G.; Zhang, X.; Cheng, X. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 2014, 28, 2304–2313. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, Y.; Xu, L.; Xiao, R.; Lu, X.; Chen, L.; Chong, J.; Li, H.; He, C.; Fu, X.-D.; et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature 2015, 523, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.-G.; Zhang, Z.-M.; Dunwell, T.L.; Harter, M.R.; Wu, X.; Johnson, J.; Li, Z.; Liu, J.; Szabó, P.E.; Lu, Q.; et al. Tet3 Reads 5-Carboxylcytosine through Its CXXC Domain and Is a Potential Guardian against Neurodegeneration. Cell Rep. 2016, 14, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Tsagaratou, A.; Äijö, T.; Lio, C.-W.J.; Yue, X.; Huang, Y.; Jacobsen, S.E.; Lähdesmäki, H.; Rao, A. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, E3306–E3315. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Huang, X.; Qiu, H.; Zhao, M.; Liao, W.; Yuan, S.; Xie, Y.; Dai, Y.; Chang, C.; Yoshimura, A.; et al. High salt promotes autoimmunity by TET2-induced DNA demethylation and driving the differentiation of Tfh cells. Sci. Rep. 2016, 6, 28065. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gomez, A.; Li, T.; Kerick, M.; Català-Moll, F.; Comet, N.R.; Rodríguez-Ubreva, J.; de la Rica, L.; Branco, M.R.; Martín, J.; Ballestar, E. TET2- and TDG-mediated changes are required for the acquisition of distinct histone modifications in divergent terminal differentiation of myeloid cells. Nucleic Acids Res. 2017, 45, 10002–10017. [Google Scholar] [CrossRef] [PubMed]
- Lio, C.-W.; Zhang, J.; González-Avalos, E.; Hogan, P.G.; Chang, X.; Rao, A. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife 2016, 5, e18290. [Google Scholar] [CrossRef] [PubMed]
- Montagner, S.; Leoni, C.; Emming, S.; Chiara, G.D.; Balestrieri, C.; Barozzi, I.; Piccolo, V.; Togher, S.; Ko, M.; Rao, A.; et al. TET2 Regulates Mast Cell Differentiation and Proliferation through Catalytic and Non-catalytic Activities. Cell Rep. 2016, 15, 1566–1579. [Google Scholar] [CrossRef] [PubMed]
- Caron, G.; Hussein, M.; Kulis, M.; Delaloy, C.; Chatonnet, F.; Pignarre, A.; Avner, S.; Lemarié, M.; Mahé, E.A.; Verdaguer-Dot, N.; et al. Cell-Cycle-Dependent Reconfiguration of the DNA Methylome during Terminal Differentiation of Human B Cells into Plasma Cells. Cell Rep. 2015, 13, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Madzo, J.; Liu, H.; Rodriguez, A.; Vasanthakumar, A.; Sundaravel, S.; Caces, D.B.D.; Looney, T.J.; Zhang, L.; Lepore, J.B.; Macrae, T.; et al. Hydroxymethylation at Gene Regulatory Regions Directs Stem/Early Progenitor Cell Commitment during Erythropoiesis. Cell Rep. 2014, 6, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Ichiyama, K.; Chen, T.; Wang, X.; Yan, X.; Kim, B.-S.; Tanaka, S.; Ndiaye-Lobry, D.; Deng, Y.; Zou, Y.; Zheng, P.; et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 2015, 42, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Nestor, C.E.; Lentini, A.; Hägg Nilsson, C.; Gawel, D.R.; Gustafsson, M.; Mattson, L.; Wang, H.; Rundquist, O.; Meehan, R.R.; Klocke, B.; et al. 5-Hydroxymethylcytosine Remodeling Precedes Lineage Specification during Differentiation of Human CD4+ T Cells. Cell Rep. 2016, 16, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Tsagaratou, A.; González-Avalos, E.; Rautio, S.; Scott-Browne, J.P.; Togher, S.; Pastor, W.A.; Rothenberg, E.V.; Chavez, L.; Lähdesmäki, H.; Rao, A. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat. Immunol. 2017, 18, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Trifari, S.; Äijö, T.; Tsagaratou, A.; Pastor, W.A.; Zepeda-Martínez, J.A.; Lio, C.-W.J.; Li, X.; Huang, Y.; Vijayanand, P.; et al. Control of Foxp3 stability through modulation of TET activity. J. Exp. Med. 2016, 213, 377–397. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Qu, C.; Zhou, Y.; Konkel, J.; Shi, S.; Liu, Y.; Chen, C.; Liu, S.; Liu, D.; Chen, Y.; et al. Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and Maintain Immune Homeostasis. Immunity 2015, 43, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, H.; Chen, T.; Nallaparaju, K.C.; Yan, X.; Tanaka, S.; Ichiyama, K.; Zhang, X.; Zhang, L.; Wen, X.; et al. Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. 2016, 14, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Tyrakis, P.A.; Palazon, A.; Macias, D.; Lee, K.L.; Phan, A.T.; Veliça, P.; You, J.; Chia, G.S.; Sim, J.; Doedens, A.; et al. The immunometabolite S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 2016, 540, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Wallner, S.; Schröder, C.; Leitão, E.; Berulava, T.; Haak, C.; Beißer, D.; Rahmann, S.; Richter, A.S.; Manke, T.; Bönisch, U.; et al. Epigenetic dynamics of monocyte-to-macrophage differentiation. Epigenet. Chromatin 2016, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Klug, M.; Schmidhofer, S.; Gebhard, C.; Andreesen, R.; Rehli, M. 5-Hydroxymethylcytosine is an essential intermediate of active DNA demethylation processes in primary human monocytes. Genome Biol. 2013, 14, R46. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Wang, Y.; Qu, X.; Li, J.; Hale, J.; Huang, Y.; An, C.; Papoin, J.; Guo, X.; Chen, L.; et al. Distinct roles for TET family proteins in regulating human erythropoiesis. Blood 2017, 129, 2002–2012. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; Christensen, J.; Pedersen, M.T.; Johansen, J.V.; Cloos, P.A.C.; Rappsilber, J.; Helin, K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 2011, 473, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Baron, U.; Floess, S.; Wieczorek, G.; Baumann, K.; Grützkau, A.; Dong, J.; Thiel, A.; Boeld, T.J.; Hoffmann, P.; Edinger, M.; et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3+ conventional T cells. Eur. J. Immunol. 2007, 37, 2378–2389. [Google Scholar] [CrossRef] [PubMed]
- Floess, S.; Freyer, J.; Siewert, C.; Baron, U.; Olek, S.; Polansky, J.; Schlawe, K.; Chang, H.-D.; Bopp, T.; Schmitt, E.; et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007, 5, e38. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Josefowicz, S.; Chaudhry, A.; Peng, X.P.; Forbush, K.; Rudensky, A.Y. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 2010, 463, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Toker, A.; Engelbert, D.; Garg, G.; Polansky, J.K.; Floess, S.; Miyao, T.; Baron, U.; Düber, S.; Geffers, R.; Giehr, P.; et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J. Immunol. 2013, 190, 3180–3188. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-P.; Leonard, W.J. CREB/ATF-dependent T cell receptor-induced Foxp3 gene expression: A role for DNA methylation. J. Exp. Med. 2007, 204, 1543–1551. [Google Scholar] [CrossRef] [PubMed]
- Polansky, J.K.; Kretschmer, K.; Freyer, J.; Floess, S.; Garbe, A.; Baron, U.; Olek, S.; Hamann, A.; von Boehmer, H.; Huehn, J. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 2008, 38, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, N.; Hamaguchi, M.; Morikawa, H.; Sugimura, K.; Tanaka, A.; Ito, Y.; Osaki, M.; Tanaka, Y.; Yamashita, R.; Nakano, N.; et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 2012, 37, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, Y.; LeBlanc, M.; Benner, C.; Zheng, Y. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 2014, 158, 734–748. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Arvey, A.; Chinen, T.; van der Veeken, J.; Gasteiger, G.; Rudensky, A.Y. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 2014, 158, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Ohkura, N.; Kidani, Y.; Vandenbon, A.; Hirota, K.; Kawakami, R.; Yasuda, K.; Motooka, D.; Nakamura, S.; Kondo, M.; et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 2017, 18, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S. Dysregulation of TET2 in hematologic malignancies. Int. J. Hematol. 2017, 105, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Zahid, M.F.; Lasho, T.L.; Finke, C.; Ketterling, R.L.; Gangat, N.; Robertson, K.D.; Hanson, C.A.; Tefferi, A. Number and type of TET2 mutations in chronic myelomonocytic leukemia and their clinical relevance. Blood Cancer J. 2016, 6, e472. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Kasela, S.; Kals, M.; Tamm, R.; Lokk, K.; Barragan, I.; Buurman, W.A.; Deelen, P.; Greve, J.-W.; Ivanov, M.; et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genom. 2014, 15, 860. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Gruppuso, P.A.; Boekelheide, K.; Sanders, J.A. Patterns of gene expression and DNA methylation in human fetal and adult liver. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.A.; Ying, L.; Liesa, M.; Segeritz, C.-P.; Mills, J.A.; Shen, S.S.; Jean, J.; Lonza, G.C.; Liberti, D.C.; Lang, A.H.; et al. Emergence of a stage-dependent human liver disease signature with directed differentiation of alpha-1 antitrypsin-deficient iPS cells. Stem Cell Rep. 2015, 4, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, M.; Kals, M.; Kacevska, M.; Barragan, I.; Kasuga, K.; Rane, A.; Metspalu, A.; Milani, L.; Ingelman-Sundberg, M. Ontogeny, distribution and potential roles of 5-hydroxymethylcytosine in human liver function. Genome Biol. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.P.; Ottaviano, R.; Unterberger, E.B.; Lempiäinen, H.; Muller, A.; Terranova, R.; Illingworth, R.S.; Webb, S.; Kerr, A.R.W.; Lyall, M.J.; et al. Loss of Tet1-associated 5-hydroxymethylcytosine is concomitant with aberrant promoter hypermethylation in liver cancer. Cancer Res. 2016, 76, 3097–3108. [Google Scholar] [CrossRef] [PubMed]
- Ancey, P.-B.; Ecsedi, S.; Lambert, M.-P.; Talukdar, F.R.; Cros, M.-P.; Glaise, D.; Narvaez, D.M.; Chauvet, V.; Herceg, Z.; Corlu, A.; et al. TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors. Stem Cell Rep. 2017, 9, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, Y.; Bai, F.; Zhang, J.-Y.; Ma, S.-H.; Liu, J.; Xu, Z.-D.; Zhu, H.-G.; Ling, Z.-Q.; Ye, D.; et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 2013, 32, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.P.; Hunter, J.M.; Lempiäinen, H.; Müller, A.; Terranova, R.; Moggs, J.G.; Meehan, R.R. Dynamic changes in 5-hydroxymethylation signatures underpin early and late events in drug exposed liver. Nucleic Acids Res. 2013, 41, 5639–5654. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.; Sheaffer, K.L.; Choi, I.; Won, K.-J.; Kaestner, K.H. Epigenetic regulation of intestinal stem cells by Tet1-mediated DNA hydroxymethylation. Genes Dev. 2016, 30, 2433–2442. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.G.; Mariani, C.J.; Wu, F.; Meckel, K.; Butun, F.; Chuang, A.; Madzo, J.; Bissonette, M.B.; Kwon, J.H.; Godley, L.A. TET-catalyzed 5-hydroxymethylcytosine regulates gene expression in differentiating colonocytes and colon cancer. Sci. Rep. 2015, 5, 17568. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.E.B.; Li, Y.H.; Smeriglio, P.; Rath, M.; Wong, W.H.; Bhutani, N. Stable 5-hydroxymethylcytosine (5hmC) acquisition marks gene activation during chondrogenic differentiation. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2016, 31, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Wang, Q.-Q.; Li, J.-W.; Zhang, Y.-M.; An, X.-R.; Hou, J. Ten-eleven translocation-2 (Tet2) is involved in myogenic differentiation of skeletal myoblast cells in vitro. Sci. Rep. 2017, 7, 43539. [Google Scholar] [CrossRef] [PubMed]
- Kranzhöfer, D.K.; Gilsbach, R.; Grüning, B.A.; Backofen, R.; Nührenberg, T.G.; Hein, L. 5′-hydroxymethylcytosine precedes loss of CpG methylation in enhancers and genes undergoing activation in cardiomyocyte maturation. PLoS ONE 2016, 11, e0166575. [Google Scholar] [CrossRef] [PubMed]
- Greco, C.M.; Kunderfranco, P.; Rubino, M.; Larcher, V.; Carullo, P.; Anselmo, A.; Kurz, K.; Carell, T.; Angius, A.; Latronico, M.V.G.; et al. DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Dubois-Chevalier, J.; Oger, F.; Dehondt, H.; Firmin, F.F.; Gheeraert, C.; Staels, B.; Lefebvre, P.; Eeckhoute, J. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res. 2014, 42, 10943–10959. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.; Park, J.H.; Weigel, C.; Liesenfeld, D.B.; Weichenhan, D.; Plass, C.; Seo, D.-G.; Lindroth, A.M.; Park, Y.J. TET-mediated hydroxymethylcytosine at the Pparγ locus is required for initiation of adipogenic differentiation. Int. J. Obes. (2005) 2017, 41, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Landfors, M.; Johansen, J.; Aronsen, J.M.; Vågbø, C.B.; Doré, L.C.; He, C.; Sjaastad, I.; Sætrom, P.; Fedorcsák, P.; Dahl, J.A.; et al. Genome-wide profiling of DNA 5-hydroxymethylcytosine during rat Sertoli cell maturation. Cell Discov. 2017, 3, 17013. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zang, L.; Shu, Q.; Li, X. From development to diseases: the role of 5hmC in brain. Genomics 2014, 104, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Tang, F. Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics 2014, 104, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Mellén, M.; Ayata, P.; Heintz, N. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc. Natl. Acad. Sci. USA 2017, 114, E7812–E7821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-R.; Cui, Q.-Y.; Murai, K.; Lim, Y.C.; Smith, Z.D.; Jin, S.; Ye, P.; Rosa, L.; Lee, Y.K.; Wu, H.-P.; et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 2013, 13, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, A.; Dawlaty, M.M.; Seo, J.; Cheng, A.W.; Meng, J.; Le, T.; Faull, K.F.; Jaenisch, R.; Tsai, L.-H. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 2013, 79, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yao, B.; Chen, L.; Kang, Y.; Li, Y.; Cheng, Y.; Li, L.; Lin, L.; Wang, Z.; Wang, M.; et al. Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat. Commun. 2017, 8, 15903. [Google Scholar] [CrossRef] [PubMed]
- Spiers, H.; Hannon, E.; Schalkwyk, L.C.; Bray, N.J.; Mill, J. 5-hydroxymethylcytosine is highly dynamic across human fetal brain development. BMC Genom. 2017, 18, 738. [Google Scholar] [CrossRef] [PubMed]
- Seritrakul, P.; Gross, J.M. Tet-mediated DNA hydroxymethylation regulates retinal neurogenesis by modulating cell-extrinsic signaling pathways. PLoS Genet. 2017, 13, e1006987. [Google Scholar] [CrossRef] [PubMed]
- Amir, R.E.; Van den Veyver, I.B.; Wan, M.; Tran, C.Q.; Francke, U.; Zoghbi, H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999, 23, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z. Deciphering MECP2-associated disorders: disrupted circuits and the hope for repair. Curr. Opin. Neurobiol. 2017, 48, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Frauer, C.; Hoffmann, T.; Bultmann, S.; Casa, V.; Cardoso, M.C.; Antes, I.; Leonhardt, H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS ONE 2011, 6, e21306. [Google Scholar] [CrossRef] [PubMed]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.K.; Jang, M.-H.; Guo, J.U.; Kitabatake, Y.; Chang, M.-L.; Pow-Anpongkul, N.; Flavell, R.A.; Lu, B.; Ming, G.-L.; Song, H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 2009, 323, 1074–1077. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.G.; Chang, Q.; Lin, Y.; Meissner, A.; West, A.E.; Griffith, E.C.; Jaenisch, R.; Greenberg, M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 2003, 302, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.C.; Fuchs, E. The Yin and Yang of chromatin dynamics in stem cell fate selection. Trends Genet. TIG 2016, 32, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Regev, A.; Teichmann, S.A.; Lander, E.S.; Amit, I.; Benoist, C.; Birney, E.; Bodenmiller, B.; Campbell, P.; Carninci, P.; Clatworthy, M.; et al. Human Cell Atlas Meeting Participants The Human Cell Atlas. eLife 2017, 6. [Google Scholar] [CrossRef]
- Yang, Y.A.; Zhao, J.C.; Fong, K.-W.; Kim, J.; Li, S.; Song, C.; Song, B.; Zheng, B.; He, C.; Yu, J. FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Res. 2016, 44, 8153–8164. [Google Scholar] [CrossRef] [PubMed]
- Costa, Y.; Ding, J.; Theunissen, T.W.; Faiola, F.; Hore, T.A.; Shliaha, P.V.; Fidalgo, M.; Saunders, A.; Lawrence, M.; Dietmann, S.; et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 2013, 495, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Shimizu, Y.; Furuhata, E.; Maeda, S.; Kishima, M.; Nishimura, H.; Enomoto, S.; Hayashizaki, Y.; Suzuki, H. RUNX1 regulates site specificity of DNA demethylation by recruitment of DNA demethylation machineries in hematopoietic cells. Blood Adv. 2017, 1. [Google Scholar] [CrossRef] [PubMed]
- Guilhamon, P.; Eskandarpour, M.; Halai, D.; Wilson, G.A.; Feber, A.; Teschendorff, A.E.; Gomez, V.; Hergovich, A.; Tirabosco, R.; Fernanda, A.M.; et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat. Commun. 2013, 4, 2166. [Google Scholar] [CrossRef] [PubMed]
- de la Rica, L.; Rodríguez-Ubreva, J.; García, M.; Islam, A.B.M.M.K.; Urquiza, J.M.; Hernando, H.; Christensen, J.; Helin, K.; Gómez-Vaquero, C.; Ballestar, E. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013, 14, R99. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, K.; Shinoda, A.; Kano, F.; Sato, R.; Shirahige, K.; Murata, M. PPARγ-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat. Commun. 2013, 4, 2262. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Maeda, S.; Furuhata, E.; Shimizu, Y.; Nishimura, H.; Kishima, M.; Suzuki, H. A screening system to identify transcription factors that induce binding site-directed DNA demethylation. Epigenet. Chromatin 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Yang, F.; Zhao, L.; Bi, C.; Cai, B. Physiological and pathological implications of 5-hydroxymethylcytosine in diseases. Oncotarget 2016, 7, 48813–48831. [Google Scholar] [CrossRef] [PubMed]
- Sherwani, S.I.; Khan, H.A. Role of 5-hydroxymethylcytosine in neurodegeneration. Gene 2015, 570, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Cadena-del-Castillo, C.; Valdes-Quezada, C.; Carmona-Aldana, F.; Arias, C.; Bermúdez-Rattoni, F.; Recillas-Targa, F. Age-dependent increment of hydroxymethylation in the brain cortex in the triple-transgenic mouse model of Alzheimer’s disease. J. Alzheimer’s Dis. JAD 2014, 41, 845–854. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Page, A.; Paoli, P.; Moran Salvador, E.; White, S.; French, J.; Mann, J. Hepatic stellate cell transdifferentiation involves genome-wide remodeling of the DNA methylation landscape. J. Hepatol. 2016, 64, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Aguilera, J.R.; Guerrero-Hernández, C.; Pérez-Molina, R.; Cadena-Del-Castillo, C.E.; Pérez-Cabeza de Vaca, R.; Guerrero-Celis, N.; Domínguez-López, M.; Murillo-de-Ozores, A.R.; Arzate-Mejía, R.; Recillas-Targa, F.; et al. Epigenetic effects of an adenosine derivative in a wistar rat model of liver cirrhosis. J. Cell. Biochem. 2017. [Google Scholar] [CrossRef]
- Thomson, J.P.; Ottaviano, R.; Buesen, R.; Moggs, J.G.; Schwarz, M.; Meehan, R.R. Defining baseline epigenetic landscapes in the rat liver. Epigenomics 2017, 9, 1503–1527. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Rao, A.; Ko, M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med. 2017, 49, e323. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Shi, X.; Guo, L.; Li, Y.; Luo, M.; He, J. Decreased 5-hydroxymethylcytosine levels correlate with cancer progression and poor survival: a systematic review and meta-analysis. Oncotarget 2017, 8, 1944–1952. [Google Scholar] [CrossRef] [PubMed]
- Koivunen, P.; Laukka, T. The TET enzymes. Cell. Mol. Life Sci. 2017. [CrossRef] [PubMed]
- Liang, G.; Weisenberger, D.J. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics 2017, 12, 416–432. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, J.; Collignon, E.; Fuks, F. Portraits of TET-mediated DNA hydroxymethylation in cancer. Curr. Opin. Genet. Dev. 2016, 36, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Foksinski, M.; Zarakowska, E.; Gackowski, D.; Skonieczna, M.; Gajda, K.; Hudy, D.; Szpila, A.; Bialkowski, K.; Starczak, M.; Labejszo, A.; et al. Profiles of a broad spectrum of epigenetic DNA modifications in normal and malignant human cell lines: Proliferation rate is not the major factor responsible for the 5-hydroxymethyl-2′-deoxycytidine level in cultured cancerous cell lines. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Lewis, S.; Stark, R.; Carroll, T.; Dunning, M.J.; Bachman, M.; Ito, Y.; Stojic, L.; Halim, S.; Vowler, S.L.; Lynch, A.G.; et al. 5-hydroxymethylcytosine marks promoters in colon that resist DNA hypermethylation in cancer. Genome Biol. 2015, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Tomkova, M.; McClellan, M.; Kriaucionis, S.; Schuster-Boeckler, B. 5-hydroxymethylcytosine marks regions with reduced mutation frequency in human DNA. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Burr, S.; Caldwell, A.; Chong, M.; Beretta, M.; Metcalf, S.; Hancock, M.; Arno, M.; Balu, S.; Kropf, V.L.; Mistry, R.K.; et al. Oxygen gradients can determine epigenetic asymmetry and cellular differentiation via differential regulation of Tet activity in embryonic stem cells. Nucleic Acids Res. 2017. [CrossRef] [PubMed]
- Saha, S.K.; Parachoniak, C.A.; Ghanta, K.S.; Fitamant, J.; Ross, K.N.; Najem, M.S.; Gurumurthy, S.; Akbay, E.A.; Sia, D.; Cornella, H.; et al. Mutant IDH inhibits HNF-4α to block hepatocyte differentiation and promote biliary cancer. Nature 2014, 513, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan Nair, V.; Song, M.H.; Oh, K.I. Vitamin C Facilitates Demethylation of the Foxp3 Enhancer in a Tet-Dependent Manner. J. Immunol. 2016, 196, 2119–2131. [Google Scholar] [CrossRef] [PubMed]
- Hore, T.A.; von Meyenn, F.; Ravichandran, M.; Bachman, M.; Ficz, G.; Oxley, D.; Santos, F.; Balasubramanian, S.; Jurkowski, T.P.; Reik, W. Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naïve pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. USA 2016, 113, 12202–12207. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).