Improving Coronary Artery Disease Diagnosis in Cardiac MRI with Self-Supervised Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Supervised Pretext and Unsupervised Pretext Algorithms
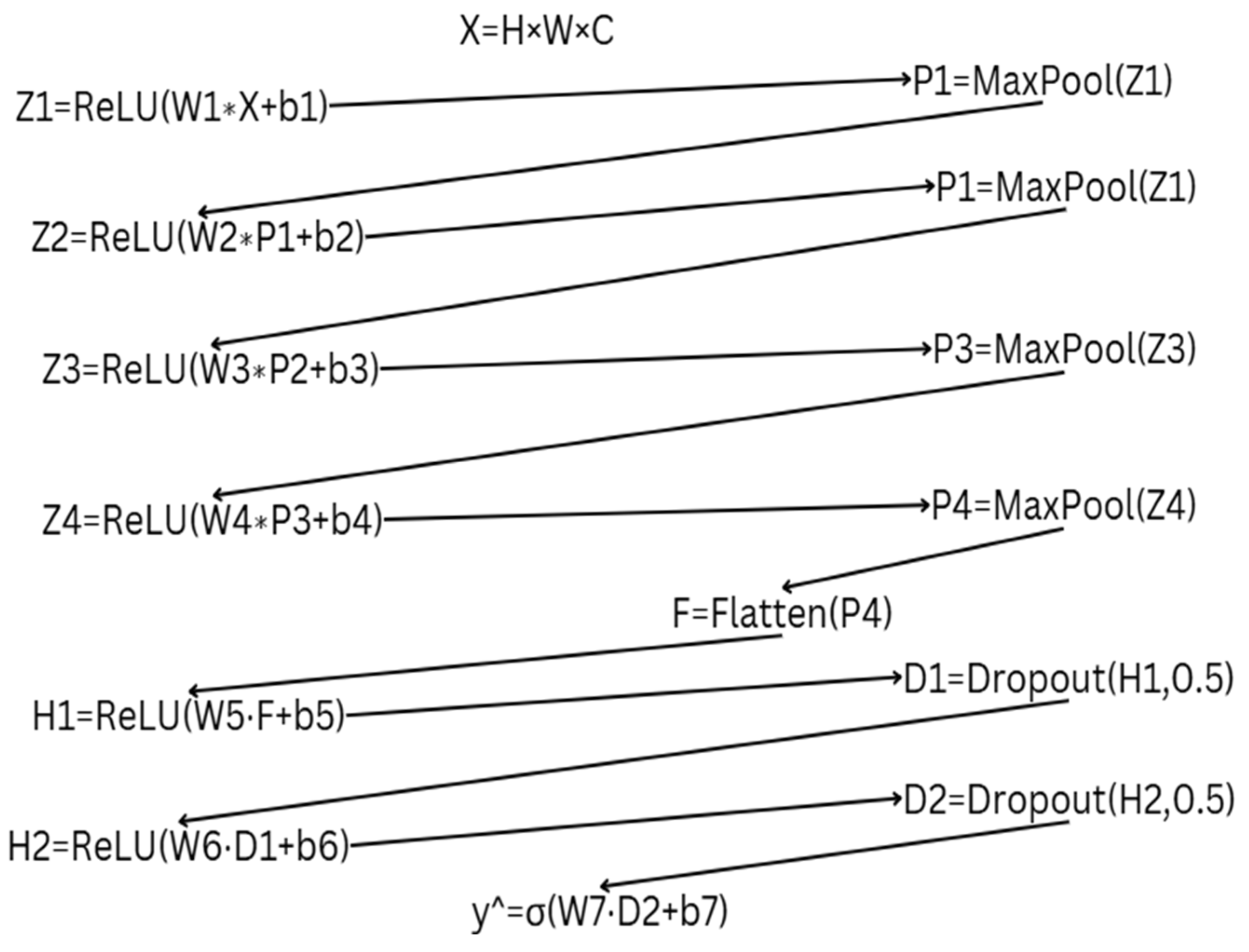
2.2. Mathematical Representation of Two Approaches
2.3. Dataset and Pre-Processing
2.4. Pretext Algorithms
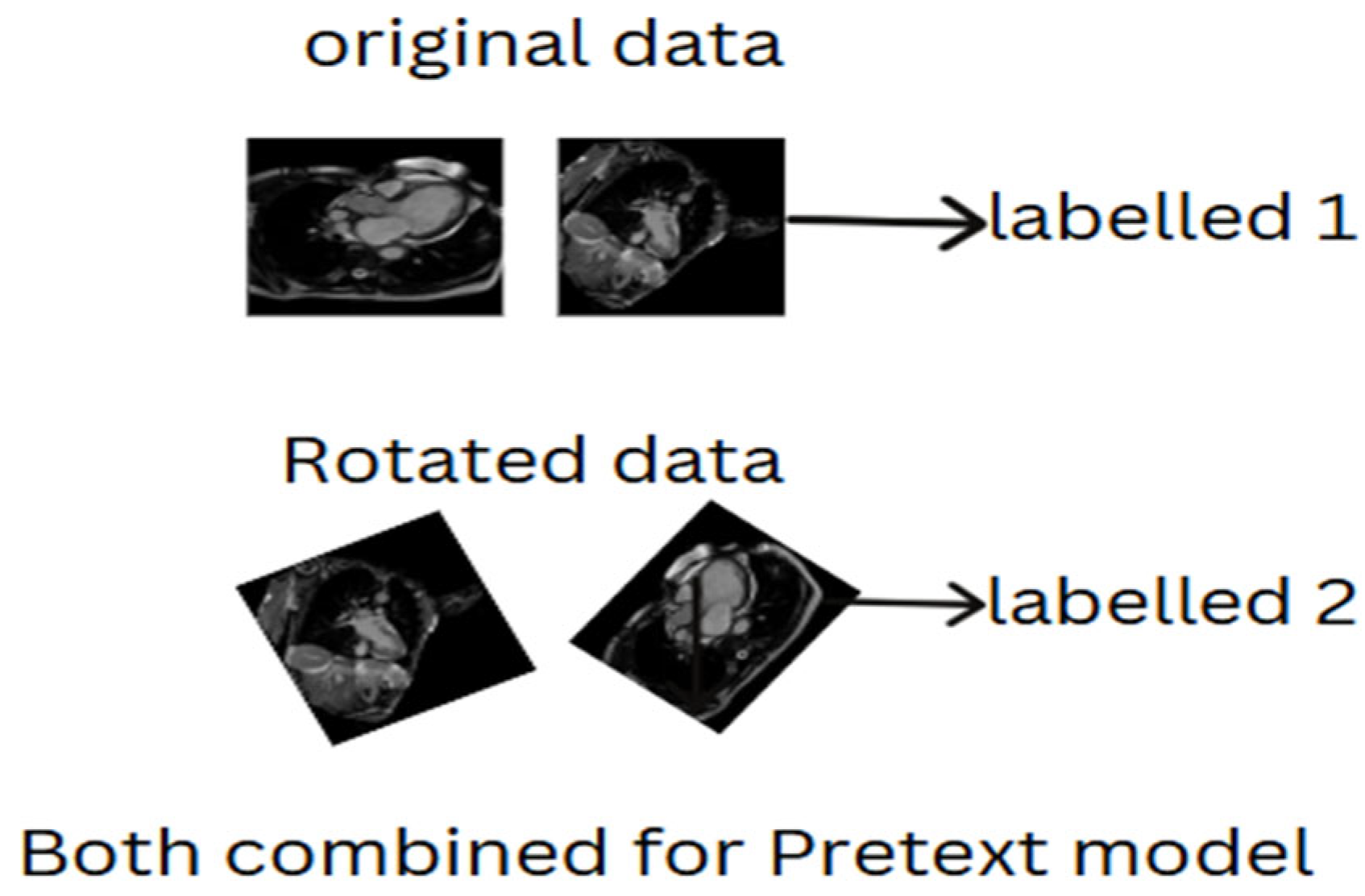
2.4.1. Selection of Pretext Tasks
2.4.2. Self-Predictive Pretext
- 1.
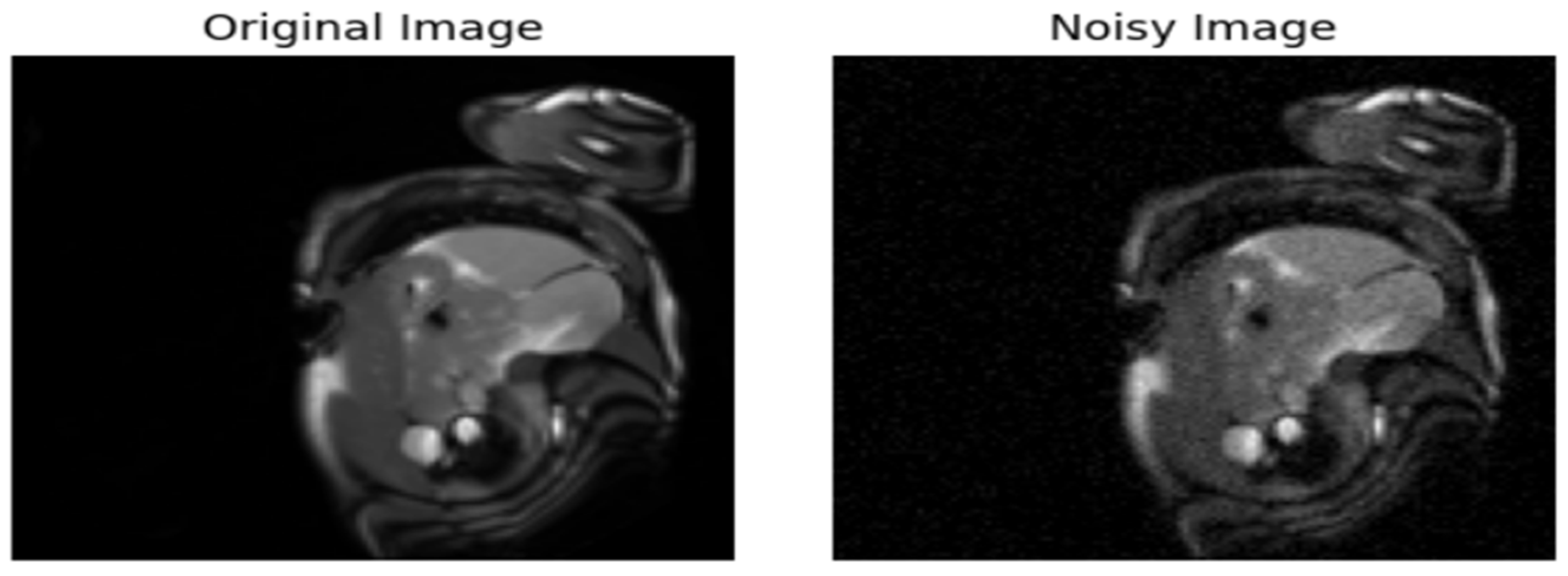
- Introduction of Gaussian Noise to the Original Data
- 2.
- Formulation
- 3.
- Augmenting the original images
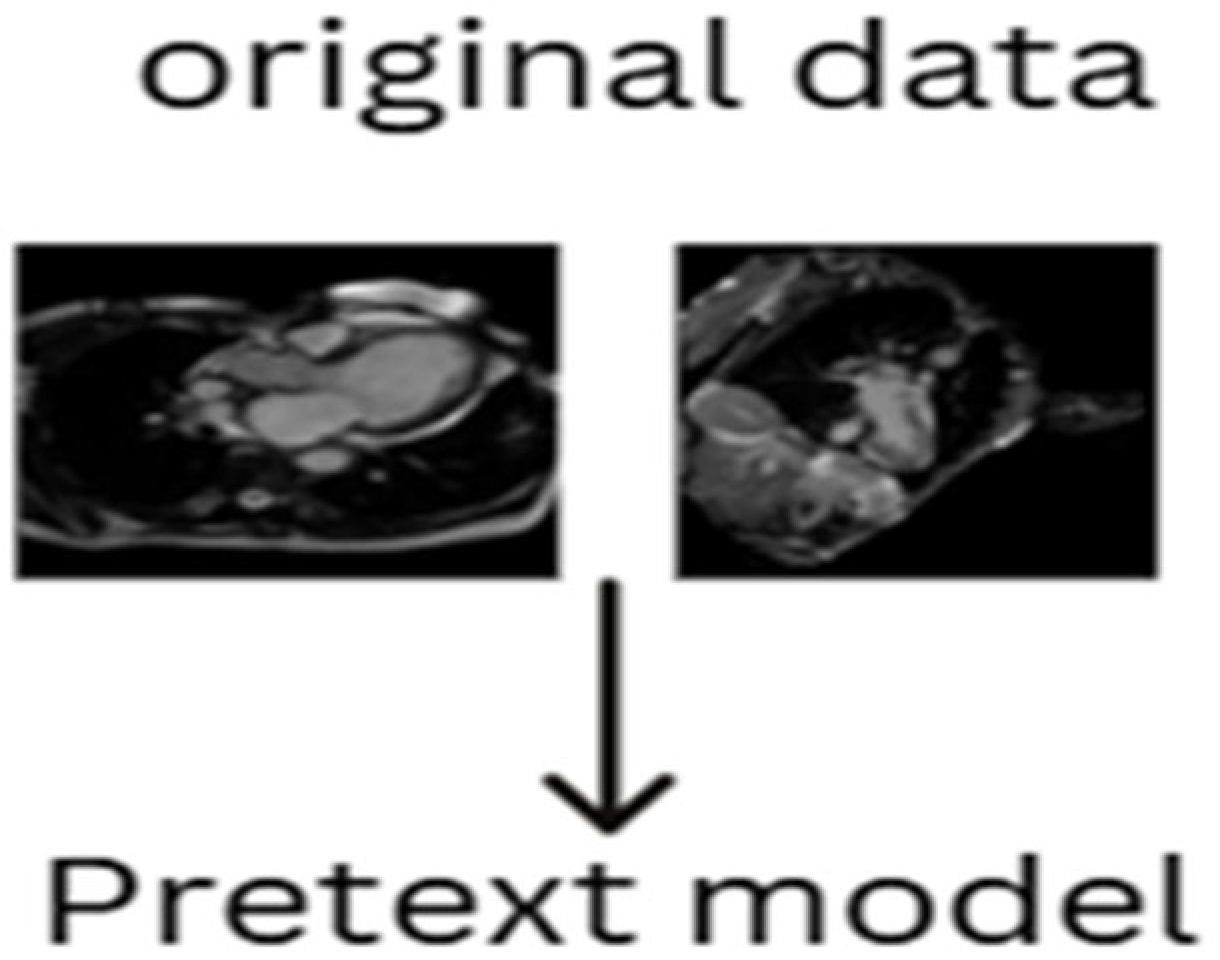
2.4.3. Generative Pretext Model
2.5. Downstream Task
3. Results
4. Discussion
4.1. Quantitative Analysis
4.2. Statistical Analysis of the Results
4.3. Limitations of the Study and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SSL | Self-Supervised Learning |
| OOD | Out-of-Distribution |
| PDG | Projected Gradient Descent |
| FGSM | Fast Gradient Sign Method |
| CAD | Coronary Artery Disease |
References
- Bastanlar, Y.; Orhan, S.; Bastanlar, Y.; Orhan, S. Self-Supervised Contrastive Representation Learning in Computer Vision. In Artificial Intelligence Annual Volume; IntechOpen: London, UK, 2022; pp. 1–14. [Google Scholar] [CrossRef]
- Wu, Z.; Xiong, Y.; Yu, S.X.; Lin, D. Unsupervised Feature Learning via Non-Parametric Instance Discrimination. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3733–3742. [Google Scholar] [CrossRef]
- Vyas, A.; Jammalamadaka, N.; Zhu, X.; Das, D.; Kaul, B.; Willke, T.L. Out-of-Distribution Detection Using an Ensemble of Self Supervised Leave-out Classifiers. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 550–564. [Google Scholar] [CrossRef]
- Rafiee, N.; Gholamipoor, R.; Adaloglou, N.; Jaxy, S.; Ramakers, J.; Kollmann, M. Self-supervised Anomaly Detection by Self-distillation and Negative Sampling. In International Conference on Artificial Neural Networks; Springer Nature: Cham, Switzerland, 2022; pp. 459–470. [Google Scholar] [CrossRef]
- Mohseni, S.; Pitale, M.; Yadawa, J.B.S.; Wang, Z. Self-Supervised Learning for Generalizable Out-of-Distribution Detection. Proc. AAAI Conf. Artif. Intell. 2020, 34, 5216–5223. [Google Scholar] [CrossRef]
- Kim, M.; Tack, J.; Hwang, S.J. Adversarial Self-Supervised Contrastive Learning. Adv. Neural Inf. Process. Syst. 2020, 33, 2983–2994. [Google Scholar]
- Hendrycks, D.; Mazeika, M.; Kadavath, S.; Song, D. Using Self-Supervised Learning Can Improve Model Robustness and Uncertainty. Adv. Neural Inf. Process. Syst. 2019, 1403, 15663–15674. [Google Scholar]
- Krishnan, R.; Rajpurkar, P.; Topol, E.J. Self-supervised learning in medicine and healthcare. Nat. Biomed. Eng. 2022, 6, 1346–1352. [Google Scholar] [CrossRef]
- Cheung, W.K.; Bell, R.; Nair, A.; Menezes, L.J.; Patel, R.; Wan, S.; Chou, K.; Chen, J.; Torii, R.; Davies, R.H.; et al. A Computationally Efficient Approach to Segmentation of the Aorta and Coronary Arteries Using Deep Learning. IEEE Access 2021, 9, 108873–108888. [Google Scholar] [CrossRef]
- Cheung, W.K.; Kalindjian, J.; Bell, R.; Nair, A.; Menezes, L.J.; Patel, R.; Wan, S.; Chou, K.; Chen, J.; Torii, R.; et al. A 3D deep learning classifier and its explainability when assessing coronary artery disease. arXiv 2023, arXiv:2308.00009. [Google Scholar] [CrossRef]
- Lauer, M.S.; Kiley, J.P.; Mockrin, S.C.; Mensah, G.A.; Hoots, W.K.; Patel, Y.; Cook, N.L.; Patterson, A.P.; Gibbons, G.H. National Heart, Lung, and Blood Institute (NHLBI) Strategic Visioning. J. Am. Coll. Cardiol. 2015, 65, 1130. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Jaiswal, A.; Babu, A.R.; Zadeh, M.Z.; Banerjee, D.; Makedon, F. A Survey on Contrastive Self-Supervised Learning. Technologies 2020, 9, 2. [Google Scholar] [CrossRef]
- Khalid, U.; Kaya, B.; Kaya, M. Contrastive Self-Supervised Learning for Cocoa Disease Classification. In Proceedings of the 10th International Conference on Smart Computing and Communication (ICSCC), Bali, Indonesia, 25–27 July 2024; pp. 413–416. [Google Scholar] [CrossRef]
- Yu, J.; Yin, H.; Xia, X.; Chen, T.; Li, J.; Huang, Z. Self-Supervised Learning for Recommender Systems: A Survey. IEEE Trans. Knowl. Data Eng. 2024, 36, 335–355. [Google Scholar] [CrossRef]
- Noroozi, M.; Favaro, P. Unsupervised learning of visual representations by solving jigsaw puzzles. In European Conference on Computer Vision; Springer International Publishing: Cham, Switzerland, 2016; pp. 69–84. [Google Scholar] [CrossRef]
- Gidaris, S.; Singh, P.; Komodakis, N. Unsupervised Representation Learning by Predicting Image Rotations. In Proceedings of the 6th International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar] [CrossRef]
- Caron, M.; Bojanowski, P.; Joulin, A.; Douze, M. Deep Clustering for Unsupervised Learning of Visual Features. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 132–149. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Y.; Fu, H. AE2-Nets: Autoencoder in Autoencoder Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2577–2585. [Google Scholar] [CrossRef]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. In Proceedings of the International Conference on Machine Learning, Proceedings of the PmLR, Vienna, Austria, 13–18 July 2020; pp. 1597–1607. [Google Scholar]
- Chen, X.; Fan, H.; Girshick, R.; He, K. Improved Baselines with Momentum Contrastive Learning. arXiv 2003, arXiv:2003.04297. [Google Scholar] [CrossRef]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked Autoencoders Are Scalable Vision Learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2022; pp. 16000–16009. [Google Scholar] [CrossRef]
- Grill, J.B.; Strub, F.; Altché, F.; Tallec, C.; Richemond, P.H.; Buchatskaya, E.; Doersch, C.; Pires, B.A.; Guo, Z.; Azar, M.G.; et al. Bootstrap Your Own Latent-A New Approach to Self-Supervised Learning. Adv. Neural Inf. Process. Syst. 2020, 33, 21271–21284. [Google Scholar]
- Khozeimeh, F.; Sharifrazi, D.; Izadi, N.H.; Joloudari, J.H.; Shoeibi, A.; Alizadehsani, R.; Tartibi, M.; Hussain, S.; Sani, Z.A.; Khodatars, M.; et al. RF-CNN-F: Random forest with convolutional neural network features for coronary artery disease diagnosis based on cardiac magnetic resonance. Sci. Rep. 2022, 12, 11178. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wu, H.; Zhao, B.; Wang, G.; Simonetti, O. Open-Access Cardiac MRI (OCMR) Dataset; Ohio State Cardiac MRI Raw Data Website. Available online: https://www.ocmr.info (accessed on 3 October 2025).
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. Adv. Neural Inf. Process. Syst. 2014, 27. [Google Scholar]
- Khan, A.; Jin, W.; Haider, A.; Rahman, M.; Wang, D. Adversarial Gaussian Denoiser for Multiple-Level Image Denoising. Sensors 2021, 21, 2998. [Google Scholar] [CrossRef]
- Huang, S.C.; Pareek, A.; Jensen, M.; Lungren, M.P.; Yeung, S.; Chaudhari, A.S. Self-supervised learning for medical image classification: A systematic review and implementation guidelines. NPJ Digit. Med. 2023, 6, 74. [Google Scholar] [CrossRef]
- Tomar, D.; Bozorgtabar, B.; Lortkipanidze, M.; Vray, G.; Rad, M.S.; Thiran, J.P. Self-Supervised Generative Style Transfer for One-Shot Medical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 1998–2008. [Google Scholar] [CrossRef]
- Wei, C.; Xie, S.M.; Ma, T. Why Do Pretrained Language Models Help in Downstream Tasks? An Analysis of Head and Prompt Tuning. Adv. Neural Inf. Process. Syst. 2021, 34, 16158–16170. [Google Scholar]
- Dong, H.; Cheng, Z.; He, X.; Zhou, M.; Zhou, A.; Zhou, F.; Liu, A.; Han, S.; Zhang, D. Table Pre-training: A Survey on Model Architectures, Pre-training Objectives, and Downstream Tasks. IJCAI Int. Jt. Conf. Artif. Intell. 2022, 5426–5435. [Google Scholar] [CrossRef]
- Ucan, M.; Kaya, B.; Kaya, M. Generating Medical Reports With a Novel Deep Learning Architecture. Int. J. Imaging Syst. Technol. 2025, 35, e70062. [Google Scholar] [CrossRef]
- Laganà, F.; Faccì, A.R. Parametric optimisation of a pulmonary ventilator using the Taguchi method. J. Electr. Eng. 2025, 76, 265–274. [Google Scholar] [CrossRef]
- Pratticò,, D.; Laganà,, F.; Oliva, G.; Fiorillo, A.S.; Pullano, S.A.; Calcagno, S.; De Carlo, D.; La Foresta, F. Sensors and Integrated Electronic Circuits for Monitoring Machinery on Wastewater Treatment: Artificial Intelligence Approach. IEEE Sensors Appl. Symp. SAS 2024, 1–6. [CrossRef]
- Laganà, F.; Pellicanò, D.; Arruzzo, M.; Pratticò, D.; Pullano, S.A.; Fiorillo, A.S. FEM-Based Modelling and AI-Enhanced Monitoring System for Upper Limb Rehabilitation. Electronics 2025, 14, 2268. [Google Scholar] [CrossRef]





| Literature | Description | Generation of Pseudo-Label |
|---|---|---|
| [16] | Arrangement of image segments and tasking the pretext model to predict the correct order | Pseudo-labels for the correct puzzles |
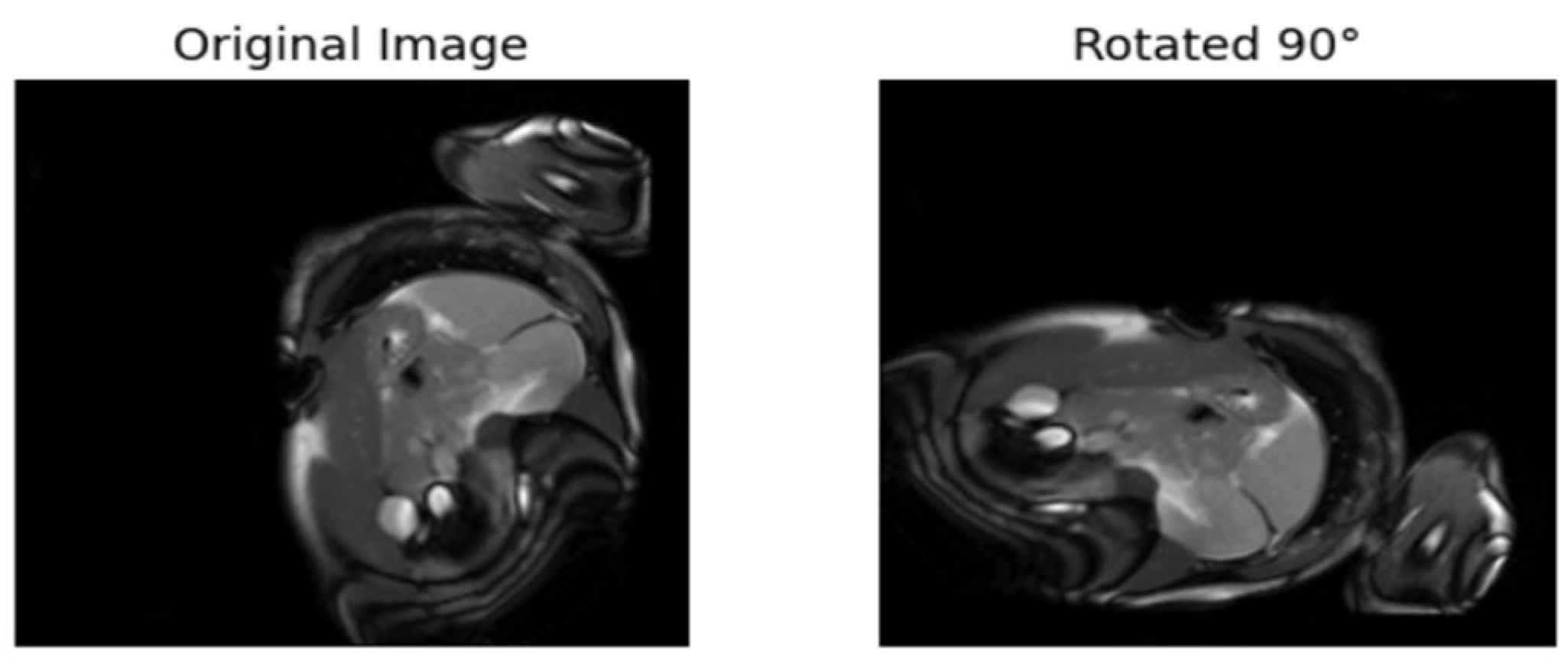
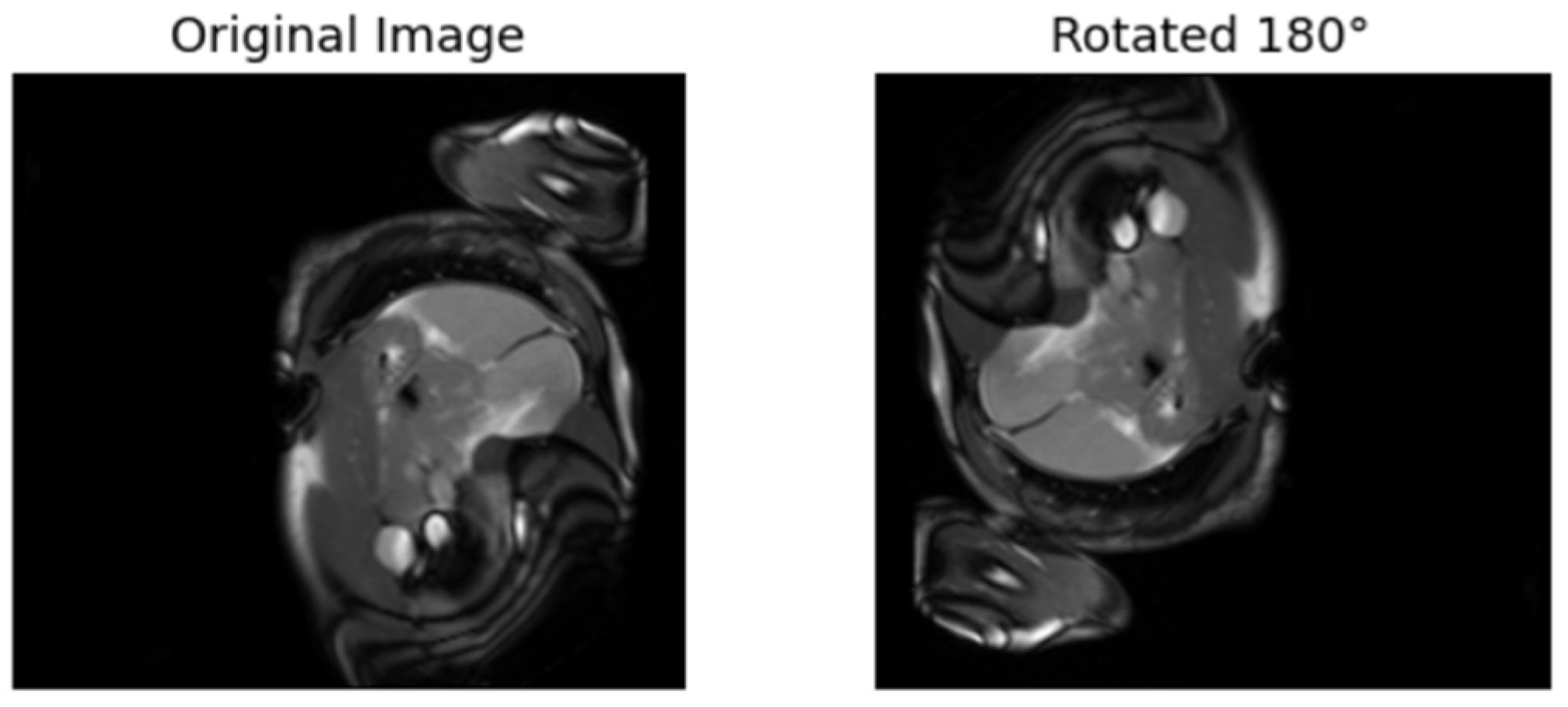
| [17] | Rotates the images 90°, 180°, etc. The model is trained to predict the rotation. | Labels are from the rotated angles |
| [14] | Uses image augmentation. The pretext is to differentiate between the original image and the augmented images | Label from the original images and that of the augmented images |
| [18] | This approach uses clustering, and the pretext model is trained with a classification-based objective. | Pseudo-labels are obtained from the clustering |
| Literature | Description | Generation of Label or Not |
|---|---|---|
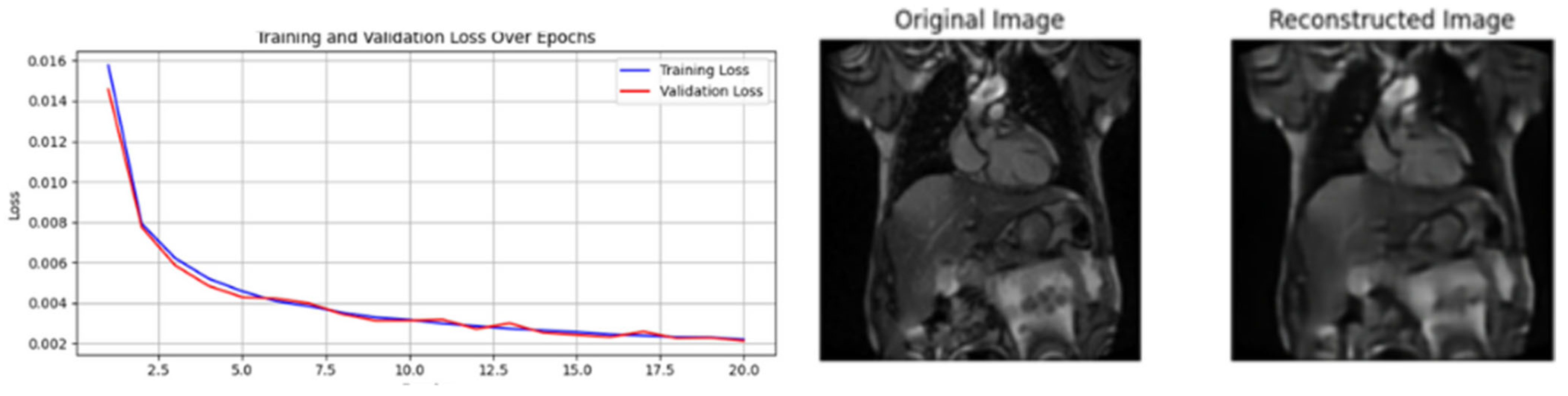
| [19] | Reconstruction of images | Direct from the original images no label needed |
| [20,21] | Similarities between images with Moco having a memory bank for negative samples | Based on contrastive learning and contrastive loss, respectively |
| [22] | Predicting missing part | Reconstruction of missing pixels |
| [23] | Learns features without negative sample only based on self-distillation | No pseudo-labels needed |
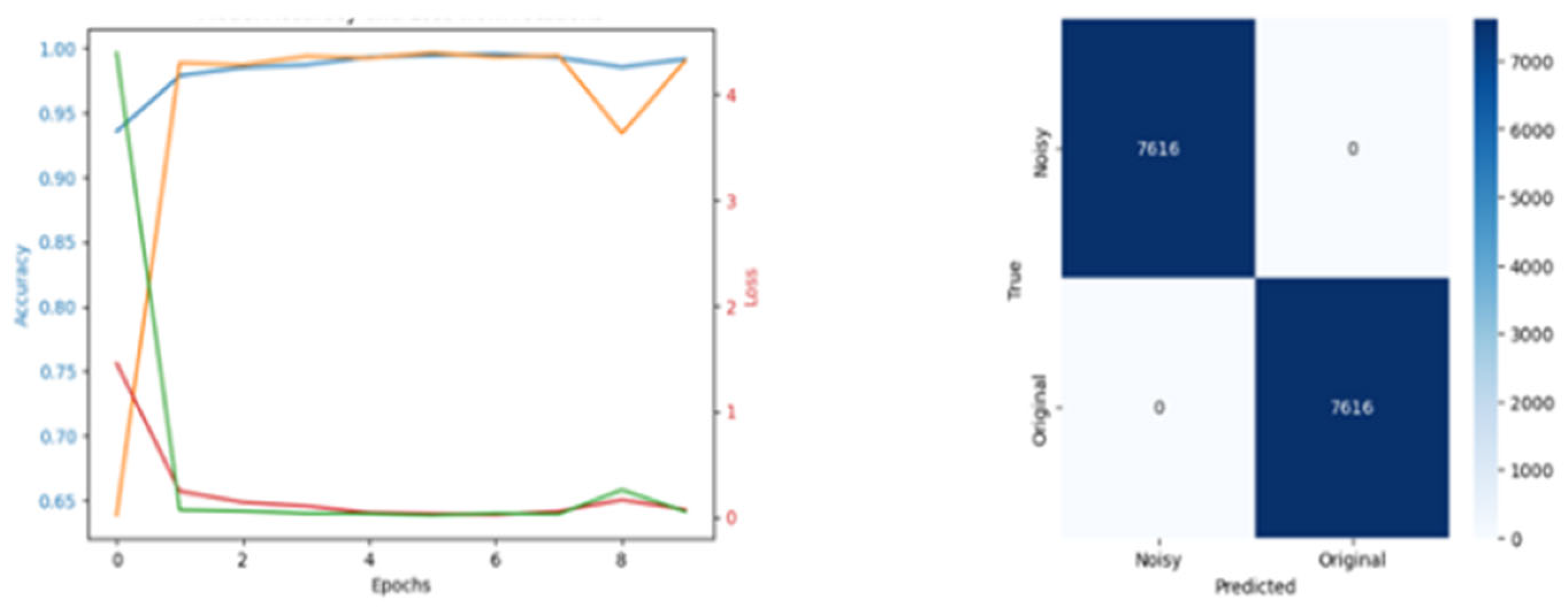
| % Reduction | Training Accuracy | Val Accuracy | Training Loss | Val Loss | PGD AND FGSM Attack | OOD |
|---|---|---|---|---|---|---|
| None | 0.989 | 0.988 | 0.0019 | 0.0014 | No effect | 0.001 |
| 20 | 0.990 | 0.998 | 0,07 | 0.04 | No effect | 0.005 |
| 50 | 0.999 | 0.999 | 0.08 | 0.014 | No effect | 0.04 |
| 70 | 0.999 | 0.999 | 0.05 | 0.47 | Less effect | 0.4 |
| Percentage | Training Accuracy | Val Accuracy | Training Loss | Val Loss | PGD Attack and FGSM Attack | OOD |
|---|---|---|---|---|---|---|
| None | 0.99 | 0.99 | 0.02 | 0.015 | No effect | 0.01 |
| 20 | 0.97 | 0.98 | 0.04 | 0.098 | No effect | 0.1 |
| 50 | 0.84 | 0.80 | 0.5 | 0.6 | There is an effect | 0.34 |
| 70 | 0.70 | 0.72 | 0.6 | 0.7 | There is an effect | 0.49 |
| Percentage | Training Accuracy | Val Accuracy | Training Loss | Val Loss | PGD Attack and FGSM Attack | OOD |
|---|---|---|---|---|---|---|
| None | 0.96 | 0.95 | 0.017 | 0.001 | No effect | 0.0015 |
| 20 | 0.95 | 0.96 | 0.12 | 0.025 | No effect | 0.05 |
| 50 | 0.82 | 0.84 | 0.25 | 0.21 | Less effect | 0.3 |
| 70 | 0.77 | 0.715 | 0.5 | 0.1 | There is an effect | 0.40 |
| Method | Pretext | Best Val Ac (50%) | PGD Attack and FGSM Attack | OOD |
|---|---|---|---|---|
| Gaussian Noise | supervised | 99.9 | Highly robust | 0.04 |
| Rotation | Supervised | 80 | Poor robustness beyond 20% data reduction | 0.34 |
| Generative | Unsupervised | 84 | Moderate robustness | 0.30 |
| SimCLR | Unsupervised | 97.7 | Moderate robustness | 0.29 |
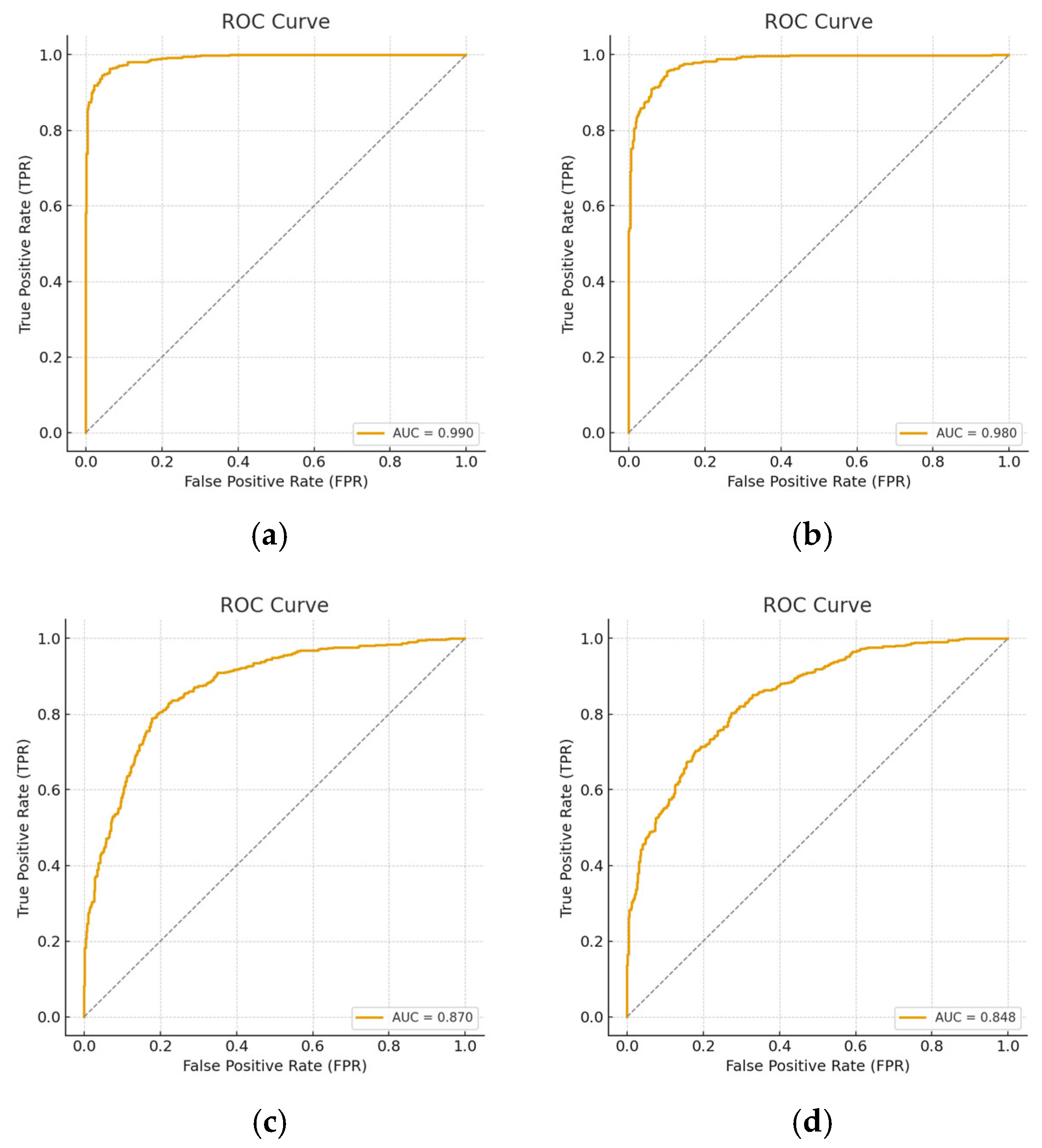
| Model | Reduction | Sensitivity | Specificity | Precision | F-Score | AUC |
|---|---|---|---|---|---|---|
| GAUSE | Full | 0.98 | 0.99 | 0.99 | 0.99 | 1 |
| 50 | 0.98 | 0.98 | 0.99 | 0.98 | 1 | |
| 20 | 0.99 | 0.98 | 0.98 | 0.98 | 1 | |
| 70 | 0.97 | 0.96 | 0.97 | 0.98 | 1 | |
| ROTATION | Full | 0.98 | 0.96 | 0.95 | 0.97 | 0.99 |
| 50 | 0.97 | 0.75 | 0.79 | 0.81 | 0.87 | |
| 20 | 0.97 | 0.96 | 0.95 | 0.97 | 0.98 | |
| 70 | 0.95 | 0.94 | 0.96 | 0.90 | 0.85 | |
| GEN-MODEL | Full | 0.97 | 0.97 | 0.96 | 0.96 | 0.99 |
| 50 | 0.95 | 0.95 | 0.97 | 0.96 | 0.99 | |
| 20 | 0.96 | 0.94 | 0.93 | 0.95 | 0.99 | |
| 70 | 0.99 | 0.90 | 0.97 | 0.90 | 0.96 |
| Model | Reduction | Sensitivity | Specificity | Precision | F-Score | AUC |
|---|---|---|---|---|---|---|
| GAUSE | Full | 0.94 | 0.94 | 0.93 | 0.94 | 0.95 |
| 50 | 0.95 | 0.93 | 0.93 | 0.94 | 0.94 | |
| 20 | 0.94 | 0.94 | 0.93 | 0.93 | 0.95 | |
| 70 | 0.93 | 0.92 | 0.91 | 0.94 | 0.94 | |
| ROTATION | Full | 0.93 | 0.93 | 0.90 | 0.93 | 0.93 |
| 50 | 0.92 | 0.86 | 0.85 | 0.88 | 0.90 | |
| 20 | 0.94 | 0.91 | 0.92 | 0.91 | 0.93 | |
| 70 | 0.93 | 0.94 | 0.90 | 0.92 | 0.88 | |
| GEN-MODEL | Full | 0.94 | 0.90 | 0.91 | 0.92 | 0.95 |
| 50 | 0.93 | 0.92 | 0.91 | 0.93 | 0.94 | |
| 20 | 0.94 | 0.91 | 0.89 | 0.90 | 0.94 | |
| 70 | 0.95 | 0.88 | 0.92 | 0.88 | 0.92 |
| Source | SS | df | MS |
|---|---|---|---|
| Between-Evaluation Metric | 0.0108 | 2 | 0.058 |
| Within-Evaluation Metric | 0.0025 | 18 | 0.0002 |
| Total | 0.0133 | 20 |
| HSD0.05 = 0.0208 HSD0.01 = 0.0273 | Q0.05 = 3.8576 Q0.01 = 4.7895 | ||
|---|---|---|---|
| B1:B2 | M1 = 0.98 M2 = 0.97 | 0.05 | Q = 9.07 (p = 0.0000) |
| B1:B3 | M1 = 0.98 M3 = 0.99 | 0.01 | Q = 0.87 (p = 0.7415) |
| B1:B4 | M1 = 0.98 M4 = 0.95 | 0.01 | Q = 2.96 (p = 0.0760) |
| B2:B3 | M2 = 0.97 M3 = 0.99 | 0.05 | Q = 10.63 (p = 0.0000) |
| B2:B4 | M2 = 0.97 M4 = 0.95 | 0.07 | Q = 15.18 (p = 0.0000) |
| B3:B4 | M3 = 0.99 M4 = 0.95 | 0.01 | Q = 2.01 (p = 0.3108) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, U.; Kaya, M.; Alhajj, R. Improving Coronary Artery Disease Diagnosis in Cardiac MRI with Self-Supervised Learning. Diagnostics 2025, 15, 2618. https://doi.org/10.3390/diagnostics15202618
Khalid U, Kaya M, Alhajj R. Improving Coronary Artery Disease Diagnosis in Cardiac MRI with Self-Supervised Learning. Diagnostics. 2025; 15(20):2618. https://doi.org/10.3390/diagnostics15202618
Chicago/Turabian StyleKhalid, Usman, Mehmet Kaya, and Reda Alhajj. 2025. "Improving Coronary Artery Disease Diagnosis in Cardiac MRI with Self-Supervised Learning" Diagnostics 15, no. 20: 2618. https://doi.org/10.3390/diagnostics15202618
APA StyleKhalid, U., Kaya, M., & Alhajj, R. (2025). Improving Coronary Artery Disease Diagnosis in Cardiac MRI with Self-Supervised Learning. Diagnostics, 15(20), 2618. https://doi.org/10.3390/diagnostics15202618







