Abstract
GOX (3QVR), glucose oxidase, is an oxidoreductase enzyme, which has found many applications in biotechnology and modern diagnostics with typical assays including biosensors useful in the determination of free glucose in body fluids. PEI (polyethylenimines) are polymer molecules made up of amine groups and two aliphatic carbons, which are cyclically repeated. PEI are transfection reagents which, using positively charged units, bind well to anionic DNA residues. During the studies on GOX, PEI were used both in their linear and branched structures. Rhombellanes, RBL, are structures decorated with rhombs/squares. The aim of the paper is to study the interactions of two kinds of linear ligands: PEIs (Polyethylenimines) and CHRs (ethers of Hexahydroxy-cyclohexane) with the glucose oxidase enzyme, GOX (3QVR). To understand the structure-activity relationship between the GOX enzyme and the linear ligands PEI and CHR, two steps of docking simulation were performed; mapping the whole area of the 3QVR enzyme and docking on the first and second surface of the enzyme, separately. The studied ligands interacted with amino acids of GOX inside the protein and on its surface, with stronger and shorter bonds inside of the protein. However, long chain ligands can only interact with amino acids on the external protein surface. After the study, two domains of the enzyme were clearly evidenced; the external surface domain more easily creates interactions with ligands, particularly with CHR ligands.
1. Introduction
GOX, glucose oxidase, is an oxidoreductase that catalyzes the oxidation of β-d-glucose to d-glucono-β-lactone, which is non-enzymatically hydrolyzed to gluconic acid [1]. In these reactions, FADH2 is formed as the reduction product of the FAD ring of GOX. FADH2 is then re-oxidized by using molecular oxygen to obtain hydrogen peroxide (H2O2). GOX has found many applications in biotechnology and modern diagnostics. Typical assays utilize it as a biosensor useful in determination of free glucose in body fluids.
There are several types of PEI [2]: The branched PEI (BPEI), linear PEI (LPEI), and dendrymer PEI (DPEI). Branched PEI have all types of amino groups, while linear PEI contain primary and secondary amino groups.
BPEI is liquid at room temperature, while LPEI is solid because its melting point is about 73–75 °C. It is well soluble in water (hot with low pH), methanol, ethanol, and chloroform.
PEI binds to anionic residues of DNA by its positively charged units [3]. Despite high toxicity, PEI has many applications [4] due to its polycationic character. PEI has been used in studies on GOX, both in its linear and branched structures [5,6,7,8].
Rhombellanes, RBL, are structures built of rings in the form of rhombs or squares (Figure 1, left); they have recently been proposed by Diudea [9,10].
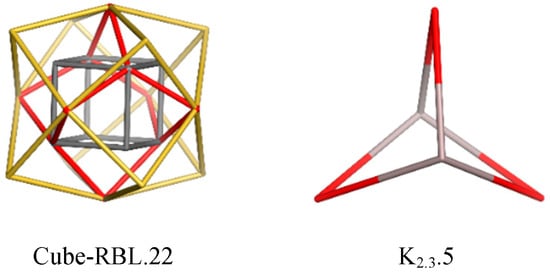
Figure 1.
Rhombellane basic structures [9,10].
Rombellanes are organic molecules, C5H6, structurally related to [1,1,1] propellane, which contains only triangles [11]. Its reduced form, C5H8, is named bicyclo [1.1.1]pentane and is made of rhomb rings or square rings (Figure 1, right). [1,1,1] Propellane undergoes spontaneous polymerization to [n] staffanes (Figure 2, left) [12,13], which are rigid, linear structures. The number suffixing a molecule name stands for the number of its (heavy) atoms.
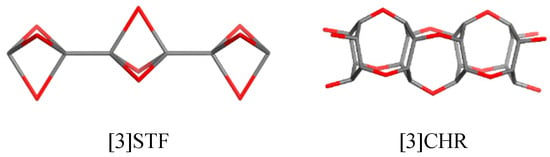
Figure 2.
Linear oligomers related to rhombellanes [9,10].
Rhombellanes have five common features [14,15,16], but foremost they are structures made of rhomb rings or square rings, where vertex classes consist of all non-connected vertices. The cube-rhombellane is presented in Figure 1. Rhombellanes are, in general, designed by the “rhombellation” operation [9,10].
By analogy to staffanes [n]STF [5], (Figure 2, left), a linear rod-like polymer (a poly-ether of 1,2,3,4,5,6-hexahydroxy-cyclohexane, [n] CHR, (Figure 2, right) was proposed by Diudea [17]. This idea came from cube-rhombellane (Figure 1, right) [9,10], with vertex/atom connectivity 6 and 3, respectively. For its realization as a molecule, it was proposed [10] to use hexahydroxy-cyclohexane for connectivity 6 (connectivity 3 being more accessible).
2. Materials and Methods
In the first step of the docking simulation, the whole area of the 3QVR enzyme was mapped step by step (Figure 3). The protein was downloaded from Brookhaven Protein Database PDB [18,19].
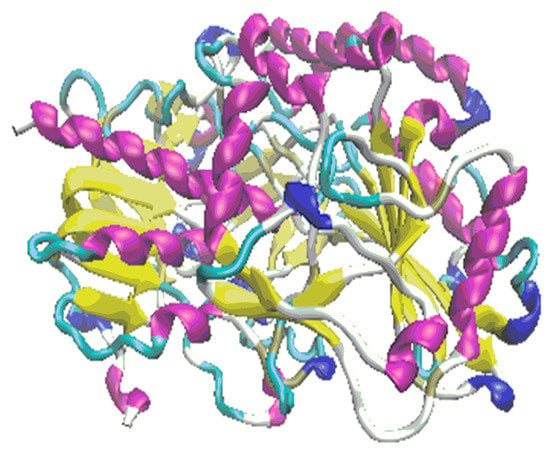
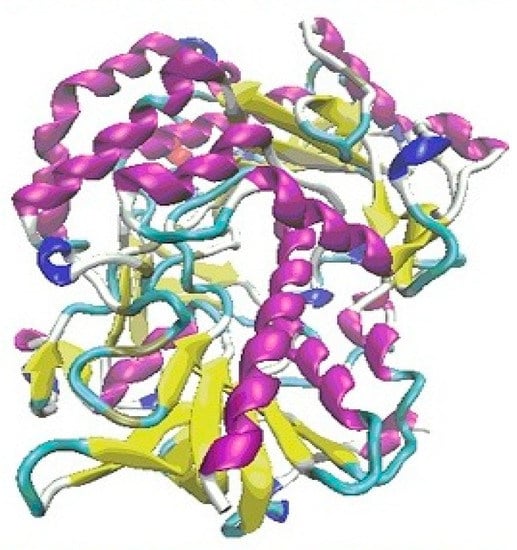
Figure 3.
The protein 3QVR/GOX, with the inside pocket (top) and external binding domain (bottom) docking surface.
To understand the structure–activity relationship of the ligand–protein complex (Supplementary Tables S1 and S2), molecular docking was carried out.
Two kinds of linear structures were used as ligands: PEI.C2+nN2+m; n = 2, 4, 6…; m = 1, 2, 3… (Figure 4) and [n]CHR.24 + 15n; n = 0, 1, 2…, respectively (Figure 4).
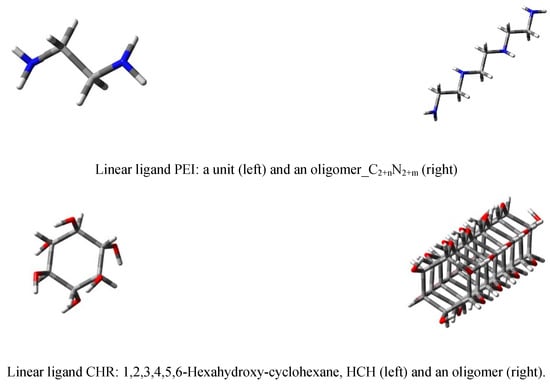
Figure 4.
Linear ligands PEI (top row) and CHR (bottom row).
All structures used during the molecular docking stage, including the ligands and proteins, were created with the use of the AutoDockTools package and non-polar hydrogen atoms were removed from each molecule. The dimensions of the grid box were fitted to the size of the protein; while during modeling, all possible interactions of the ligand with the whole protein surface were evaluated. The docking procedure was realized with the use of AutoDockVina [20,21], using the Lamarckian genetic algorithm [22]. The calculations were realized with exhaustiveness parameter equal 20, with the chosen value being a compromise between good reproducibility of calculations and computation time cost. Finally, the validity of the used algorithm was verified [23,24].
3. Results
3.1. The First Step of the Study
By performing the docking procedure, it is clear that ligands can interact with amino acids of GOX both inside of protein (Figure 5, top) and on its surface (Figure 5, bottom). Selected values of the free energy of binding to the proteins active site are given in Supplementary Tables S1 and S2 for ligands PEI and CHR, respectively.
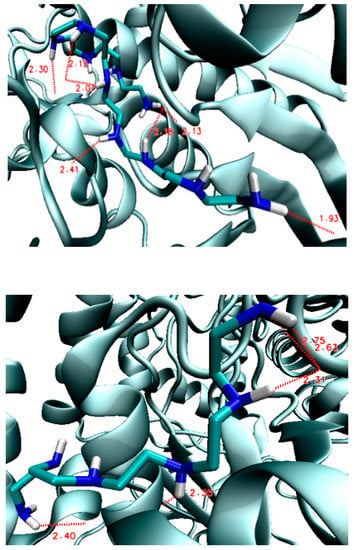
Figure 5.
Hydrogen bond lengths (in Å) in the PEI–GOX complexes: Inside the protein (3QVR–PEI.C18N10.01.0Lin2, top) and on its surface (3QVR–PEI.C10N6.01.0Lin3, bottom).
However, the bonds created inside are stronger and shorter, compared with the bonds formed on the outside surface of enzyme, which is what could be expected (see Supplementary Tables S1 and S2; Supplementary Figures S1 and S2. The structural analysis of the formed complexes clearly shows that the ligand affinity values on the external surface are lower compared to the values of affinity inside of enzyme (see also Supplementary Figures S1–S3; Supplementary Tables S1 and S2.
With the increasing chain length of PEI and CHR ligands, the values of ligand–protein affinity also increase (Supplementary Figure S4; Supplementary Tables S1 and S2. This is caused by the fact that increasing the structural elongation of the ligand increases the number of atoms participating in the ligand–enzyme hydrogen bond formation (Figure 6).
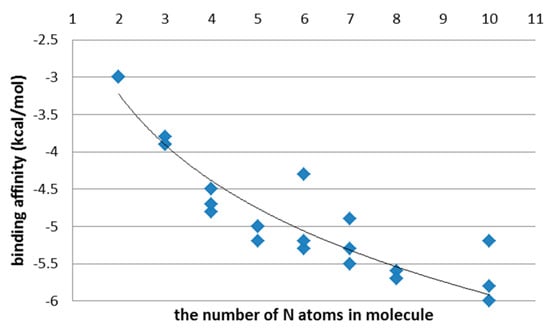
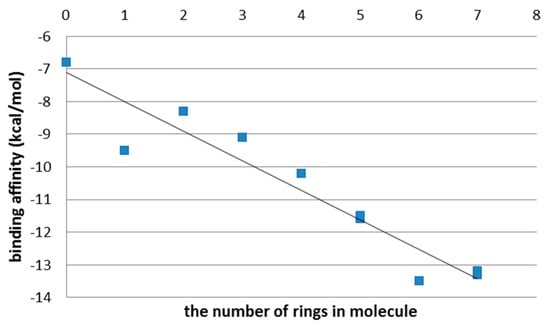
Figure 6.
Binding affinity (kcal/mol) in PEI (top) and CHR (bottom) ligands, after the first step of the docking procedure.
Hydrogen bonds of ligand and protein are detailed in Supplementary Figures S1 and S2 (amino acids are hidden for the sake of image clarity).
The ligand affinity depends on the number of hydrogen bonds and the strength of the interactions between ligand and protein (Figure 7; Supplementary Table S3). It also depends on whether these interactions occur on the external surface of the enzyme or inside it.
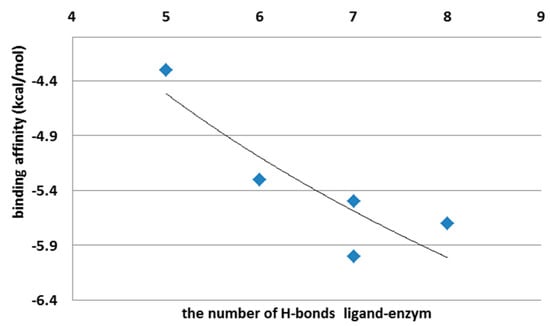
Figure 7.
Binding affinity (kcal/mol) vs. the number of H-bonds created between ligand and enzyme.
Jeffrey [25] categorizes H bonds by donor–acceptor distances. Based on such categorization, H-bonds created with the protein by PEI and CHR ligands are strong and moderate (Supplementary Table S3). The number and strength of the hydrogen bonds between the ligand and protein will, finally, determine the values of binding affinity, and consequently, the ligand–protein overall distance. Thus, a high value of ligand–protein binding affinity is directly related to the formation of many strong hydrogen bonds.
In the case of PEI ligands, all the tested molecules, except for PEI.C10N6.01.0Lin3, form H-bonds inside of the protein. In the case of CHR molecules, only those with a short chain (namely [0]CHR.24 and [1]CHR.39) find room to interact with amino acids inside of the enzyme. Ligands with a longer chain ([n]CHR.24 + 15n; n = 3 to 7) can only embed on the external binding domain of the enzyme surface (Figure 3).
That is why, in the second step of this study, the docking was made on the surface of the enzyme, where the studied ligands can create H-bonds with the protein in an easy and quick way.
3.2. The Second Step of the Study
Again, with the growing chain length of PEI and CHR molecules, the values of ligand–protein affinity also increase (Figure 8), by increasing the number of hydrogen bonds created between the enzyme and ligand.
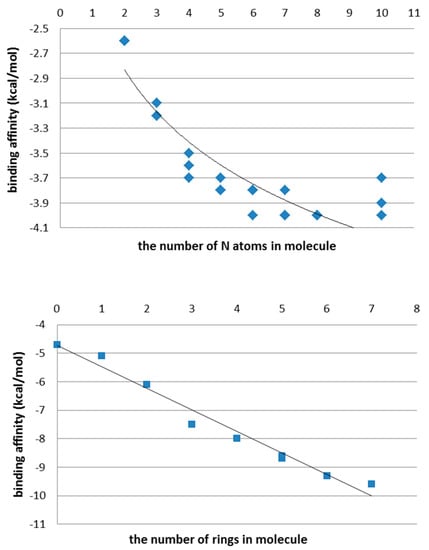
Figure 8.
Binding affinity (kcal/mol) in PEI (top) and CHR molecules (bottom) after docking of ligands on the external surface of protein.
One can see that the values of ligand–enzyme affinity on the external domain (Supplementary Tables S4 and S5) are much smaller than in the inside domain (Supplementary Tables S6 and S7), which suggests the creation of longer and weaker hydrogen bonds.
4. Discussion
When comparing the energy of hydrogen bonds created between ligands and GOX enzyme, it can be found that differences were around 1% in favor of the external domain of docking (Figure 4 and Figure 9, top) for PEI ligands; while in the case of CHR ligands, the differences ranged from 40% to 70%, also in favor of the external domain (Figure 9, bottom).
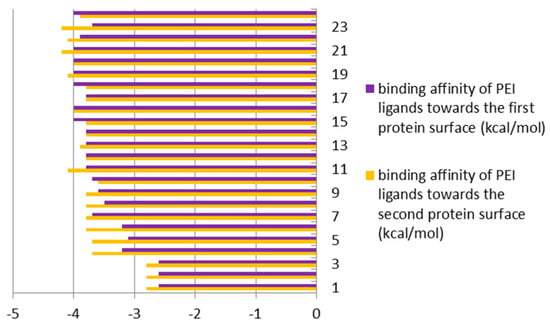
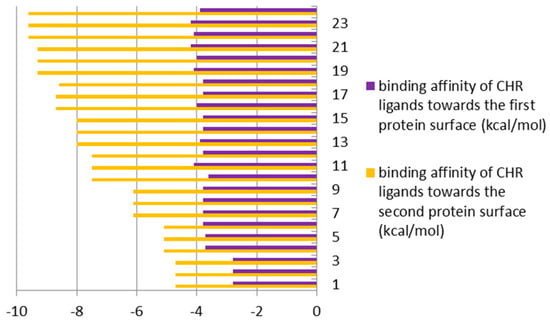
Figure 9.
Binding affinities (in kcal/mol) of enzyme (GOX protein)–ligands (PEI (top) and CHR (bottom)), bound inside (first protein surface) and outside/externally (second protein surface), respectively.
All of the above means that the external surface domain of enzyme (Figure 3, bottom) interacts more easily with ligands, particularly with CHR molecules. Here, the larger differences are in the case of longer chain molecules (Figure 9, bottom).
Detailed structural analysis of the ligand-protein complexes, formed after the docking procedure, confirmed the observation regarding the existing differences in the strength of hydrogen bonds, on the inside and outside domains of GOX enzyme, in docking of the ligands PEI and CHR, respectively (Figure 9). The number and strength of hydrogen bonds created between PEI and amino acids were comparable on the two docking domains of the protein. Contrarily, in the case of CHR ligands, the external surface domain provided more room to form a large number of interactions with the protein, in comparison to the internal pocket of enzyme (Figure 3).
5. Conclusions
To understand the structure–activity relationship between the GOX (3QVR) enzyme and linear ligands PEI and CHR, in the first step of this docking study, the whole area of the enzyme was mapped. It was found that ligands can interact with amino acids of GOX in two domains, namely, the inside of the protein and on its external surface. The bonds created in the inside domain are stronger and shorter, compared with the bonds formed on the external surface of enzyme. Also, in the latter, the bonding energy values were lower compared with the values recorded for the complexes formed with the inside docking domain of enzyme. Despite this, with the increasing chain length of PEI and CHR ligands, the values of ligand–protein affinity also increased, which is what is connected to the increasing ability to create more ligand–protein hydrogen bonds. The binding affinity depends on the number and strength of interactions between ligand and protein, and also depends on whether these interactions occur on the external surface of the enzyme or inside it. The longer ligand chains can only find room for docking with the amino acids lying on the external surface of the protein.
Comparison between the two docking sites of GOX revealed differences in binding affinity in favor of the external domain of docking; around 1% in the case of PEI ligands and from 40% to 70% in the case of CHR molecules.
The number and strength of hydrogen bonds created between PEI and GOX amino acids are comparable on the two sites of protein docking; while in case of CHR ligands, the docking is favored on the external domain of the protein surface. This means that the CHR ligands act selectively on the external binding site of the GOX enzyme.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-8994/11/7/901/s1, Figure S1: The length of hydrogen bonds in Å formed between ligands PEI and GOX enzyme. Table S1: The final lamarckian genetic algorithm docked state—Binding energy of PEI ligand to the active sites of type of 3QVR during the nine explored conformations. The number 1,2,3 in the ligand name means the use of another random variable in the docking procedure.
Funding
This research was supported by PL-Grid Infrastructure http://www.plgrid.pl/en.
Acknowledgments
This work was supported by GEMNS project granted in the European Union’s Seventh Framework Programme under the frame of the ERA-NET EuroNanoMed II (European Innovative Research and Technological Development Projects in Nanomedicine).
Conflicts of Interest
The author declares no conflict of interest.
References
- Leskovac, V.; Trivić, S.; Wohlfahrt, G.; Kandrac, J.; Pericin, D. Glucose oxidase from Aspergillus niger: The mechanism of action with molecular oxygen, quinines and one electron acceptors. Int. J. Biochem. Cell Biol. 2005, 37, 731–750. [Google Scholar] [CrossRef] [PubMed]
- Yemul, O.; Imae, T. Synthesis and characterization of poly(ethyleneimine) dendrimers. Colloid Polym. Sci. 2008, 286, 747–752. [Google Scholar] [CrossRef]
- Rudolph, C.; Lausier, J.; Naundorf, S.; Müller, R.H.; Rosenecker, J. In vivo gene delivery to the lung using polyethylenimine and fractured polyamidoamine dendrimers. J. Gene Med. 2000, 2, 269–278. [Google Scholar] [CrossRef]
- Akinc, A.; Thomas, M.; Klibanov, A.M.; Langer, R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2004, 7, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Szefler, B.; Diudea, M.V.; Grudziński, I.P. Nature of polyethylene imine-glucose oxidase interactions. Studia UBB_Chemia 2016, 61, 249–260. [Google Scholar]
- Lungu, C.N.; Diudea, M.V.; Putz, M.V.; Grudziński, I.P. Linear and branched PEIs (Polyethylenimines) and their properties space. Int. J. Mol. Sci. 2016, 17, 555. [Google Scholar] [CrossRef] [PubMed]
- Szefler, B.; Diudea, M.V.; Putz, M.V.; Grudziński, I.P. Molecular Dynamic Studies of the Complex Polyethylenimine and Glucose Oxidase. Int. J. Mol. Sci. 2016, 17, 1796. [Google Scholar] [CrossRef]
- Lungu, C.N.; Diudea, M.V.; Putz, M.V.; Grudzinski, I.P. FAD molecular adaptability among surrounding amino acids and its catalytic role in glucose oxidase and related flavoproteins. Res. J. Life Sci. Bioinf. Pharm. Chem. Sci. 2017, 3. [Google Scholar] [CrossRef]
- Diudea, M.V. Rhombellanes—A new class of structures. Int. J. Chem. Model. 2017, 9, 91–96. [Google Scholar]
- Diudea, M.V. Cube-Rhombellane: From graph to molecule. Int. J. Chem. Model. 2017, 9, 97–103. [Google Scholar]
- Wiberg, K.B.; Walker, F.H. [1.1.1] Propellane. J. Am. Chem. Soc. 1982, 104, 5239–5240. [Google Scholar] [CrossRef]
- Kazynsky, P.; Michl, J. [n] Staffanes: A molecular-size tinkertoy construction set for nanotechnology. Preparation of end-functionalized telomers and a polymer of [1.1.1] propellane. J. Am. Chem. Soc. 1988, 110, 5225–5226. [Google Scholar] [CrossRef]
- Dilmaç, A.; Spuling, E.; Meijere, A.; Bräse, S. Propellanes—From a chemical curiosity to “explosive” materials and natural products. Angew. Chem. Int. Ed. 2017, 56, 5684–5718. [Google Scholar] [CrossRef] [PubMed]
- Diudea, M.V. Hypercube related polytopes. Iran. J. Math. Chem. 2018, 9, 1–8. [Google Scholar]
- Diudea, M.V. Rhombellanic crystals and quasicrystals. Iran. J. Math. Chem. 2018, 9, 167–178. [Google Scholar]
- Szefler, B.; Czeleń, P.; Diudea, M.V. Docking of indolizine derivatives on cube rhombellane functionalized homeomorphs. Studia Universitatis Babes-Bolyai Chemia 2018, 63, 7–18. [Google Scholar] [CrossRef]
- Diudea, M.V.; Medeleanu, M.; Khalaj, Z.; Ashrafi, A.R. Spongy Diamond. Iran. J. Math. Chem. 2019, 10, 1–9. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDockVina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comp. Chem. 2010, 31, 455–461. [Google Scholar]
- Bertrand, J.A.; Thieffine, S.; Vulpetti, A.; Cristiani, C.; Valsasina, B.; Knapp, S.; Kalisz, H.M.; Flocco, M. Structural characterization of the GSK-3beta active site using selective and non-selective ATP-mimetic inhibitors. J. Mol. Biol. 2003, 333, 393–407. [Google Scholar] [CrossRef]
- Shoichet, B.K.; Kuntz, I.D.; Bodian, D.L. Molecular docking using shape descriptors. J. Comput. Chem. 2004, 13, 380–397. [Google Scholar] [CrossRef]
- Dhananjayan, K.; Kalathil, K.; Sumathy, A.; Sivanandy, P. A computational study on binding affinity of bio-flavonoids on the crystal structure of 3-hydroxy-3-methyl-glutaryl-CoA reductase—An insilico molecular docking approach. Der Pharma Chemica 2014, 6, 378–387. [Google Scholar]
- Abagyan, R.; Totrov, M. High-throughput docking for lead generation. Curr. Opin. Chem. Biol. 2001, 5, 375–382. [Google Scholar] [CrossRef]
- Shen, M.; Zhou, S.; Li, Y.; Pan, P.; Zhang, L.; Hou, T. Discovery and optimization of triazine derivatives as ROCK1 inhibitors: Molecular docking, molecular dynamics simulations and free energy calculations. Mol. Biosyst. 2013, 9, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Yu, H.; Li, Y.; Li, P.; Pan, P.; Zhou, S.; Zhang, L.; Li, S.; Lee, S.M.Y.; Hou, T. Discovery of Rho-kinase inhibitors by docking-based virtual screening. Mol. Biosyst. 2013, 9, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, G.A. An Introduction to Hydrogen Bonding; Oxford University Press: New York, NY, USA, 1997; Volume 12, p. 228. [Google Scholar]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).