Abstract
Pea, one of the founder crops from the Near East, has two wild species: Pisum sativum subsp. elatius, with a wide distribution centered in the Mediterranean, and P. fulvum, which is restricted to Syria, Lebanon, Israel, Palestine and Jordan. Using genome wide analysis of 11,343 polymorphic single nucleotide polymorphisms (SNPs) on a set of wild P. elatius (134) and P. fulvum (20) and 74 domesticated accessions (64 P. sativum landraces and 10 P. abyssinicum), we demonstrated that domesticated P. sativum and the Ethiopian pea (P. abyssinicum) were derived from different P. elatius genepools. Therefore, pea has at least two domestication events. The analysis does not support a hybrid origin of P. abyssinicum, which was likely introduced into Ethiopia and Yemen followed by eco-geographic adaptation. Both P. sativum and P. abyssinicum share traits that are typical of domestication, such as non-dormant seeds. Non-dormant seeds were also found in several wild P. elatius accessions which could be the result of crop to wild introgression or natural variation that may have been present during pea domestication. A sub-group of P. elatius overlaps with P. sativum landraces. This may be a consequence of bidirectional gene-flow or may suggest that this group of P. elatius is the closest extant wild relative of P. sativum.
1. Introduction
About 10,000 years ago, humans began the domestication of crops and animals, initiating one of the largest changes in human history. There are two broad views of the origin of plant cultivation and domestication, especially in the Near East. One proposes that crops’ origin was singular, rather fast (hundreds of years) and took place in the so-called small core area of south-eastern Turkey and adjoining parts of Syria [1], followed by further evolution of domesticated crops that improved their quality [2]. The alternative view is that domestication was a protracted process with multiple origins of crops that went on slowly and in parallel in multiple locations [3,4,5,6,7,8]. Recent studies propose that the use of wild cereals and pulses considerably predate their domestication [3,4,9]; similarly, the gathering of pea from ‘gardens’ by pre-agricultural peoples has been discussed [10].
The Near Eastern center of domestication has been extensively studied [11,12] and plant domestication in this region has provided a large number of crops that are important today: cereals: einkorn wheat (Triticum monococcum L.), emmer wheat (Triticum dicoccum (Schrank) Schuebl), barley (Hordeum vulgare L.), fibre and oil crop: flax (Linum usitatissimum L.) and legumes: lentil (Lens culinaris Medic), chickpea (Cicer arietinum L.), bitter vetch (Vicia ervilia L.) Willd., faba bean (Vicia faba L.) and pea (Pisum sativum L.) [1,9,12,13,14,15,16,17,18]. In early agriculture and until the widespread use of N-fertilizer from the Haber-Bosch process [19,20], grain legumes accompanied cereals [2,9,13]. Indeed, the co-domestication of cereals and legumes was a recurring feature in the independent origins of agriculture [21]. Despite the crucial position of legumes as an important source of protein in the human diet, comparatively little is known about their domestication.
The legume family is one of the most abundantly represented among crops. Although cereals have a higher total production, there are at least as many legume crops. FAOstat lists 12 cereal crops and 18 legumes; cereal production in 2016 was ca. 3000 Mt and legume production was ca. 500 Mt [22]. There are 13 genera (in six legume tribes) that constitute major legume crops [23,24]. Pea (Pisum sativum L.) belongs to the tribe Fabeae [25], which contains: Lathyrus (grass pea); Lens (lentils); Pisum (peas), Vicia (vetches) and monotypic Vavilovia. Although Schaefer et al. [25] showed that Pisum and Vavilovia are imbedded within Lathyrus, here we keep the Linnean designation Pisum sativum L. For Pisum, many species and subspecies names have been used [26,27], however most commonly, two species, Pisum fulvum Sibth. & Sm. and P. sativum L. are recognized. The latter is divided into two subspecies, the domesticated pea P. sativum subsp. sativum and the wild form, P. sativum subsp. elatius (M. Bieb.) Asch. & Graebn [28,29]. Geographically, P. sativum subsp. elatius is distributed widely across the Mediterranean basin from Spain to the Middle East and north to Central Europe, the Caucasus and the Caspian Sea, whereas P. fulvum is found around its eastern edge (Syria, Lebanon, Israel, Palestine and Jordan) [29,30]. P. fulvum forms a distinct clade in all molecular diversity analyses and is the only good candidate in the genus for a distinct species [30,31]. The taxonomic status of the Ethiopian pea has been discussed extensively and has varied from subspecies (P. sativum subsp. abyssinicum (A.Br.) Berger) to species (Pisum abyssinicum A.Br.) [23,28,29,32,33,34]. Molecular analysis has proposed that P. abyssinicum is not a subgroup of cultivated P. sativum, however that it has probably been domesticated independently from a separate Pisum genepool [31,33,34,35,36,37,38]. Taken together, the genus Pisum is best described as “a species complex with multiple sub-species which interbreed to different degrees” [31,33,34].
Both morphological and genetic studies have identified P. sativum subsp. elatius and P. fulvum as wild germplasm in that they have dehiscent pods and seed dormancy (thick testa), occur in the wild and are undesirable as a domesticated annual crop. In contrast, P. sativum subsp. sativum (including varieties arvense, transcaucasicum and asiaticum) are diagnosed by characters that are selected during domestication, namely: non-dehiscing pods, absence of seed dormancy and seeds with a smooth, thin testa [2,39]. The Ethiopian pea shares these domestication characteristics and they are the main reason why it was previously considered to be a sub-group of P. sativum.
Archaeological evidence dates the origin of cultivated pea 10,000 BC in the Near East [12,40] and Central Asia [10,41]. The archaeological evidence further suggests that the cultivation of pea spread from the Fertile Crescent westwards through the Danube valley, ancient Greece and Rome into Europe. Linguistic evidence additionally backs this suggestion [42]. During this same period, pea also moved eastward to Persia (now Iran and Afghanistan), India and China [27,43]. This might explain the novel diversity of Afghan type and Chinese landrace peas [29,44,45], either through genetic drift or through natural selection in diverse environments [46]. Similarly, human selection for early flowering as a drought avoidance phenotype may have acted on the Ethiopian pea (P. abyssinicum).
An independent domestication of the Ethiopian (P. abyssinicum) pea has been proposed by several authors [31,32,33,35,36,37] and a chromosomal translocation [47,48] that differentiates P. sativum and P. abyssinicum karyotypes is consistent with a distinct origin. Wild peas have not been described in Ethiopia, suggesting that it is unlikely to be a native plant. It was most likely introduced to Ethiopia along with barley [33,37,49]. P. abyssinicum has been reported in both Ethiopia and Yemen [33]. Human population genetic analyses have suggested that there are southern Arabia (Yemen) and Ethiopia blend African and Eurasian lineages [50]. The major episodes in the peopling of Arabia took place from north to south in the Late Glacial Maximum and, to a lesser extent, the immediate post-glacial/Neolithic ages [50]. Genetic connection between contemporary Ethiopians and Anatolian people, as well as archaeological evidence, dates the arrival of Near Eastern crop domesticates to the same time period as this late phase of the human history of Ethiopia (circa 3000 years ago). This suggests that the direct descendants of the farmers that earlier brought agriculture into Europe may have also played a role in the introduction of these crops to the Horn of Africa [51].
Pea genetic diversity that is conserved in genebanks has been extensively studied over the past two decades (reviewed in [52,53]). There are 25 large collections preserving pea diversity, together holding around 72,000 accessions. A further 27,000 accessions are distributed over 146 collections worldwide [23,29,52]. However, few (less than 1%) of these are wild pea relatives [52]. Molecular analysis of pea diversity preserved in germplasm collections has been carried out using various methods [30,31,33,35,54,55,56,57,58,59,60]. Recently, genotyping-by-sequencing was applied [61] to a set of 431 P. sativum (sensu lato) accessions, including 11 P. sativum subsp. elatius, 2 P. abyssinicum and 25 P. fulvum accessions, and a 13k singles nucleotide polymorphism (SNP) panel of mapped genes to 917 accessions, including 50 wild peas [55]. The largest number of accessions analyzed so far (3020 and 4200) were dominated by cultivated types and had relatively few (45) markers (retrotransposon-based insertion polymorphisms [29,62]). Genome-wide next generation sequencing techniques have been used recently to study the diversity of wild pea [30], such as a study that comprised of 143 P. elatius and 18 P. fulvum accessions. All of these studies indicated that Pisum is very diverse and that the diversity is structured, showing a range of degrees of relatedness that partially reflect taxonomic identifiers, eco-geography and, in the case of cultivated material, breeding gene pools.
In contrast with other well studied crops such as rice [63], barley [64,65], wheat [66,67], maize [68], chickpea [69], groundnut [70], common bean [71] and soybean [72], the domestication scenario in pea is not well known. In order to fill gaps in our understanding of pea domestication, we asked the following questions: (1) What is the relationship between wild P. elatius and cultivated P. sativum and P. abyssinicum? (2) Was P. abyssinicum derived from domesticated P. sativum or was it domesticated independently? (3) Might P. abyssinicum originate from hybridization between P. fulvum and P. elatius?
2. Materials and Methods
2.1. Plant Material
Pisum taxonomy has frequently been revised without adding much clarity as to the partitioning of genetic variation within the genus. Here we adopted a scheme that treats the genus as monospecific and accepts the Linnean term Pisum sativum, while acknowledging that Schaefer et al. [25] clearly demonstrated that Pisum is embedded within Lathyrus (and Lathyrus within Vicia). For simplicity, we omitted the intermediate level species name because it is common to all taxa, thus, Pisum sativum corresponds to Pisum sativum subsp. sativum, P. elatius to Pisum sativum subsp. elatius. For cultivated pea, we used Pisum sativum L. as described by Linnaeus [73]; Pisum abyssinicum A.Braun [74] was used for the Ethiopian form of cultivated pea. For wild pea, we used Pisum elatius M. Bieb. [75] in the broad sense and Pisum fulvum Sibth. & Sm. [76].
A panel consisting of 154 wild P. elatius (134) and P. fulvum (20) from Smýkal et al. [30,77] and 74 domesticated (64 P. sativum landraces and 10 P. abyssinicum) accessions were sampled to maximize the geographic diversity of the wild and landrace pea habitats (Table S1). The set of P. sativum landraces (64) originated from several geographical regions: Middle East (Turkey, Syria, Cyprus, Lebanon) 12, Caucasus region (Georgia, Russia, Armenia) 13, Europe (Greece, Balkan, Italy, Spain and Central Europe) 10, Central Asia (Afghanistan, Iran, Pakistan, Tajikistan, Nepal) 10, Ethiopia and Northern Africa (Morocco, Libya) 10, China and India (10). Sampling was based on our previous studies [29,31]. The material originated from several major genebank collections (Table S1) and was largely of single-seed descent. In terms of wild (P. elatius) accessions, 134 covered the geographic range of the species [30]. Of these 90, they were from the Middle Eastern center (Turkey, Syria, Israel, Jordan), 33 were from Europe (Western, Eastern Mediterranean, Hungary, Italy), 8 were from the Caucasus (Russia, Armenia, Georgia) and 3 were from Northern Africa (Algeria, Morocco). Twenty P. fulvum were sampled: Israel (6), Syria (12), Jordan (1) and Turkey (1). All ten P. abyssinicum accessions were from Ethiopia. The latitude and longitude for the collection points of the wild pea samples were obtained from germplasm passport data and were processed using ArcGIS for Desktop (version 10.4, http://desktop.arcgis.com). All of the wild material was purified by single-seed descent for several generations to eliminate accession heterogeneity. Leaf samples were taken from a single plant per accession and were freeze dried before DNA isolation.
2.2. DArTseq Analysis
Genomic DNA was isolated from approximately 100 mg of dry leaf material using the Invisorb Plant Genomic DNA Isolation kit (Stratec Molecular, Berlin, Germany). Standard diversity array technology sequencing (DArTseq) analysis [30,77] was undertaken at Diversity Arrays Technology Ltd. (Canberra, Australia) using proprietary methodology [78,79,80].
In brief, DNA samples were processed in digestion/ligation reactions [30,77], however a single PstI-compatible adaptor was replaced with two adaptors. The PstI-compatible adapter was designed to include Illumina flowcell attachment sequence, sequencing primer sequence and barcode region. The reverse adapter contained flowcell attachment region and MseI-compatible sequence. Only mixed fragments (PstI-MseI) were effectively amplified in 30 rounds of PCR using the following reaction conditions: 94 °C for 1 min, 30 cycles of: 94 °C for 20 s, 58 °C for 30 s, 72 °C for 45 s and a final extension of 72 °C for 7 min. Afterwards, PCR equimolar amounts of amplification products from each sample were bulked and sequenced on Illumina Hiseq2500 (San Diego, CA, USA) which was run for 77 cycles.
2.3. Bioinformatic Data Processing
DArTseq analysis of 228 pea samples, each with 75,862 DArTseq fragments, yielded 0.98, 2.19, 1.24 and 0.02 million scores for -, 0, 1 and 2, respectively. The “-” are missing data (22%), “0” and “1” are alternative sequences for a given fragment and “2” indicates that both the “0” and “1” sequences were detected. Taking into account the single seed descent of most of the material and based on our previous experience using the same DARTseq procedure on a recombinant inbred lines (RIL) mapping population (F6) and wild-collected material [75], we concluded that “2” (0.45% of total markers) most likely represents sequencing error or paralogous sequences. Accordingly, both “-” and “2” scores were treated as missing data.
These 75,862 DARTseq fragments were filtered to maximum 20% missing data (Table S2). We used 11,343 polymorphic high-quality SNPs (HQ SNPs), with one SNP per locus mapped to a pea genome assembly (BioProject: PRJNA432052, GenBank assembly: GCA_003013575.1), thereby avoiding an uneven representation of markers by genomic location.
2.3.1. Genetic Structure and Diversity Analysis
To describe diversity and differentiation, we used a series of complimentary analyses to examine the relationships among accessions in a step-wise fashion beginning with ordination analyses, which have few assumptions, and progressing to model based admixture analyses. We first used Principal Coordinate-Model Clustering (PCO-MC), a principal coordinates based density clustering procedure, to identify distinct genotypic clusters [81]. To perform the ordination, a simple band sharing coefficient was used to calculate the necessary genetic distance matrix from homozygous co-dominant DArTseq data [81]. We then constructed a reticulate network using the NeighborNet algorithm [82] that was implemented in SplitsTree 4.14.4 [83]. Phylogenetic networks are generalizations of phylogenetic trees that represent conflicting signals in data derived from non-tree like processes, such as hybridization followed by introgression. The network displays relative evolutionary distances between taxa as well as uncertainty in the groupings in the form of “splits” (or “webbing”) of internal branches. In addition, we applied a discriminant analysis of principle components (DAPC) that was implemented in the adegenet package in R [84] for visual exploration of the data. The number of clusters for the DAPC analysis was set to 10, according to the value of Bayesian Information Criterion (BIC). DAPC clustering was performed using five Principal Component Analysis (PCA) axes and three Discriminant Analysis (DA) axes. Finally, a coancestry matrix among accessions was inferred using the Markov Chain Monte Carlo (MCMC) clustering algorithm of RADpainter and fineRADstructure v0.3.2 [85] which has been developed for large SNP datasets (specifically, RADseq). The fineRADstructure analysis used 500,000 burn in steps and 500,000 sample iterations to infer population structure.
2.3.2. Seed Phenotyping
Seeds were harvested from 3–5 plants per pot and accession was grown in five litre pots with peat-sand (90:10) Florcom Profi substrate mix (BB Com Ltd., Letohrad, Czech Republic) in glasshouse conditions (January–May 2016 and 2017) and a natural photoperiod (Palacky University campus, Olomouc, Czech Republic). After harvest, mature seeds were cleaned from the pods, were dried at room temperature and were packed in paper bags. Germination of intact seeds was tested at 25/15 °C in the dark at 14/10 h (day/night) on water saturated filter papers (Whatman Grade 1, Sigma, Prague, Czech Republic) in 90 mm Petri dishes (P-Lab, Prague, Czech Republic) in temperature-controlled chambers (Laboratory Incubator ST4, BioTech, Prague, Czech Republic). Then, 25 seeds per accession were incubated in duplicate. Seeds were monitored at 24 h intervals for a total of 28 days. Seeds were considered germinated when the radicle protruded from the testa. The cumulative percentage of germinated seeds was calculated [86]. Samples were classified as “dormant” if by the end of the test period (28 days), fewer than 50% of the seeds had germinated and, conversely, were classified “non-dormant” if over 80% had germinated. Seed batches with 50–80% germination were classified as intermediate. Photographs of ten individual seeds per genotype were taken with an Olympus SZ61 stereomicroscope (Olympus Corp., Tokyo, Japan) that was equipped with an Olympus E-410 digital camera (Olympus Corp., Tokyo, Japan). The photographs were processed by QuickPHOTO MICRO 3.0, which was supplemented by the Deep Focus 3.3 module (PROMICRA, Prague, Czech Republic). Accordingly, the seed coat was classified either as smooth or rough and either as pigmented or not pigmented [87].
3. Results
3.1. Diversity Array Technology Analysis and Bioinformatic Processing
A total of 75,862 different sequences were obtained by DArTseq; of these, 66,910 were polymorphic. Further, 72,679 (95.9%) sequenced fragments could be mapped to a shotgun pea genome (GCA_003013575.1) and, of these, 22,013 (61.9%) could be mapped to the pea RNA atlas; these correspond to 8405 unique sequences on the pea Infinium BeadChip (Illumina, SanDiego, CA, USA), known as the Genopea 13.2K SNP Array. These Infinium BeadChip sequences corresponded to 3838 positions on the genetic map of Tayeh et al. [55] which were distributed across all the linkage groups. The distribution of these loci on the pea linkage groups is shown in Supplementary Table S2 and Figure S1. After filtering according to missing data and minor allele frequency cutoffs (see Materials and Methods), 11,343 SNPs remained for analysis (Figure 1).
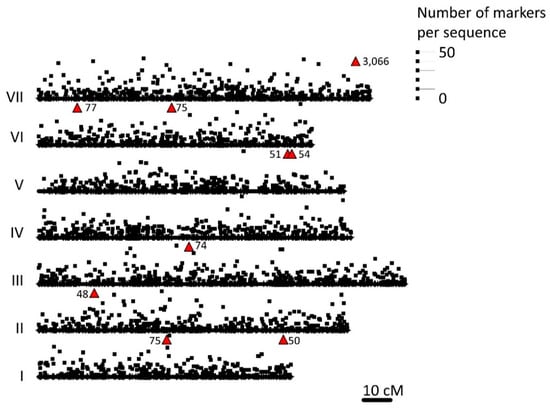
Figure 1.
The distribution of 1929 loci, corresponding to 3838 different sequences, mapped by Tayeh et al. [57] that match the diversity array technology (DArT) sequences scored in this study is presented on the x axis for each pea linkage group (I to VII). The y axis gives the number of DArTseq markers corresponding to each of these sequences corresponding to the scale on the right. Where the number of sequences would be off scale or obscured by those of another linkage group, the number is indicated adjacent to a red triangle.
3.2. Genetic Structure and Diversity Analysis
The PCO-MC analysis identified 6 distinct clusters within the SNP data (Figure 2). Both P. fulvum (cluster 1, red squares) and P. abyssinicum (cluster 2, yellow diamonds) formed distinct groups with high stability values, 96% and 71%, respectively. With this method, stability values > 15% generally mark true positive genetic clusters. Additionally, a fraction of the P. elatius accessions (in green) cluster into four distinct groups (3–6 in Figure 2). Group 3 comprised of 11 P. elatius accessions from eastern Turkey (T14-2, T14-9, T15-1, T15-5, T15-11, W6-26109, W6-26127, JI261, P013, PI344001, PI344002) and two from Israel (711, PI560059), group 4 comprised of 16 P. elatius accessions largely from Israel (7), Jordan (1) and Turkey (3), however three from Italy, Spain and Algeria, group 5 comprised of 22 P. elatius accessions largely of European origin and, finally, group 6 comprised of 20 P. elatius, however also two P. sativum (PI343972, TG2548) accessions. The remaining 63 P. elatius (green) and 62 P. sativum accessions (in blue) form a continuum with accessions that were interspersed with one another in the PCO bi-plot.
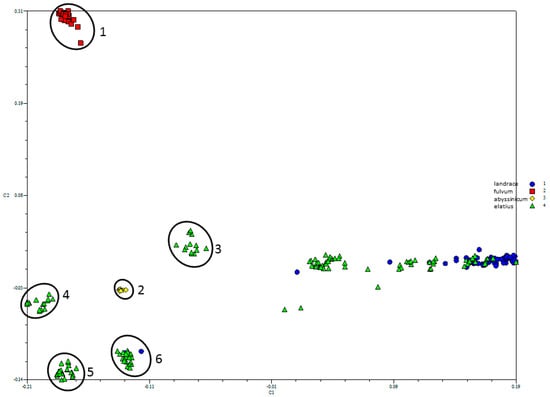
Figure 2.
Six distinct genetic clusters identified using Principal Coordinate-Model Clustering (PCO-MC), a principal coordinates-based clustering procedure. Clusters 1 and 2 contain exclusively P. fulvum (red squares) and P. abyssinicum (yellow circles), respectively. The remainder contains distinct segregates of Pisum elatius, with the exception of cluster 6 which also includes P. sativum landrace accessions TG2548 and PI343972. An intergrading cloud of P. elatius (green triangles) and P. sativum landrace (in blue) material remained unassigned.
NeighborNet was used to construct a phylogenetic network (Figure 3). The narrow reticulation leading to P. abyssinicum showed its high genetic uniformity and its broad base indicated its affinity with two P. elatius groups (colored dark blue and brown) as well as P. fulvum (in dark green). A group of P. fulvum was well separated with an internal bifurcation. There were seven separate groups of P. elatius, in agreement with DAPC analysis (Figure 4). Four of them correspond to PCO-MC groups 3 to 6 (Figure 2), while NeighborNet analysis further separated 3 additional groups of P. elatius, one overlapping with P. sativum group. Domesticated P. sativum landraces have a complex reticulate structure and are not separated from the central reticulation by a long branch.
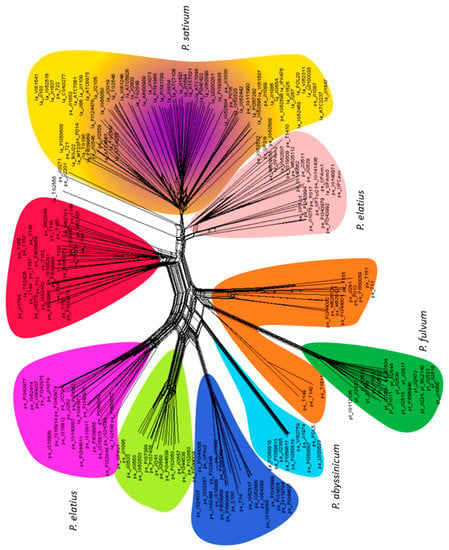
Figure 3.
NeighborNet network shows reticulate genetic relationship between pea accessions. Color coding corresponds to the discriminant analysis of principle components (DAPC) K10 clusters of Figure 4.
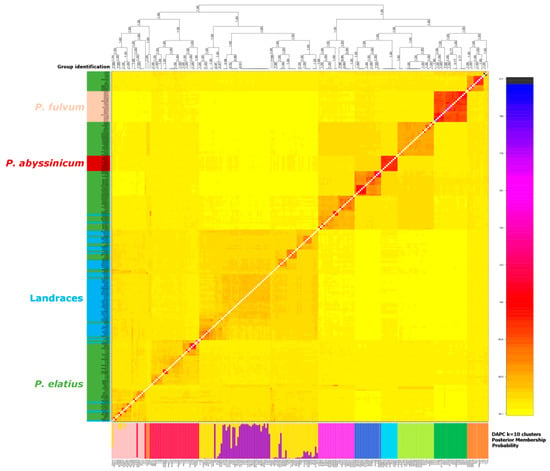
Figure 4.
Coancestry matrix for 228 individuals based on HQ SNPs (high-quality single nucleotide polymorphism) dataset calculated by fineRADstructure. The heat map depicts the high resolution genetic relationship structure of individuals selected across Pisum. The lower bar plot shows the results of the DAPC analysis of 10 identified groups.
DAPC identified 10 clusters of accessions that were designated Group 1 to Group 10 in Figure 4. Group 1 (red) contained 30 accessions, all except for three from Turkey. All Group 2 (dark green) accessions were P. fulvum and all P. fulvum accessions were assigned to this group. Group 3 (yellow) was a mixture of P. elatius and landraces of P. sativum. Group 4 (dark blue) comprised P. elatius accessions, seven from Israel, six from south-eastern Turkey and one each from Jordan, Algeria and Italy. Group 5 (brown) comprised 12 accessions from eastern Turkey and two (711, PI560059) from Israel. Group 6 (purple) comprised seven P. elatius accessions from Georgia, two from Armenia one from southern Turkey, and one from Ethiopia. Group 7 (light blue) comprised all 10 P. abyssinicum accessions. Group 8 (magenta) comprised 19 accessions, all except for one (PI560057, from Portugal by passport data assigned as P. sativum landrace) that was of Middle East origin. Group 9 (light green) comprised 22 accessions of various geographical origins: Western and Central Mediterranean (nine), Eastern and Southern Europe (eight) and Middle East (four). Group 10 (pink) contained 16 P. elatius and two P. sativum (TG2568, UP-Arm3) accessions. Groups 4, 8, 9 and 10 correspond to PCO-MC identified groups 3 to 6, which are clearly separated.
fineRADstructure described the shared ancestry between accessions (Figure 4). The analysis shows blocks of related accessions as orange to red squares, and these have a nested structure. P. abyssinicum and P. fulvum are seen as distinct and relatively homogenous groups. It showed two large groups, one including P. fulvum, P. abyssinicum and four sub-groups of P. elatius accessions, corresponding to DAPC groups 1–4 and to group 4 which was identified by PCO-MC. The second large group contained two subgroups of P. sativum landraces and two subgroups of P. elatius accessions. In this case, the internal structure was noticeably graded. FineRADstructure analysis identified a group of 11 P. elatius accessions from eastern Turkey (T14-2, T14-9, T15-1, T15-5, T15-11, W6-26109, W6-26127, JI261, P013, PI344001, PI344002) and two from Israel (711, PI560059) which are ancestral to P. elatius and P. fulvum. There was an overall good correspondence between the groups as inferred from DAPC, the distance tree and from fineRADstructure (Figure 3 and Figure 4).
3.3. Seed Phenotyping
Seed dormancy, caused by a water impermeable seed coat, is typical of wild legume species, including wild pea. This trait has been lost during domestication. We analyzed mature dry seeds for seed coat pigmentation and surface properties. Wild pea seeds are typically rough (as, for example, determined by the gene Gritty), while seeds of cultivated pea usually have a smooth and thin testa. All 64 P. sativum landraces had non-dormant seeds (Table S1), germinating with the fraction 0.8 ± 0.057 germinating within 48 h of being exposed to water, and all except four (PI343972, IG52507 from Turkey and UP_Arm2, UP_Arm3 from Armenia) had a smooth testa (gritty). Forty accessions had a pigmented seed coat, while 24 did not. P. abyssinicum seeds were smooth, sometimes glossy, and were either pigmented or not. Importantly, these were also non-dormant and imbibed readily. Seeds of P. fulvum were all dormant and pigmented (usually black or dark brown). The seed coats of P. elatius accessions were mostly pigmented and rough as well as dormant (103 out of 134). Further, 15 accessions had smooth, pigmented testa—four of which were dormant, while 11 were non-dormant. Three accessions of P. elatius (JI1030, IG111992, JI1853) had a smooth testa, were non-pigmented and non-dormant—a combination that could be taken as indicative of a cultivated type. Two accessions (JI1075 from Turkey and JI2201 from Russia) had a rough testa, were non-pigmented and non-dormant (Table S1).
4. Discussion
Here, we have undertaken a comprehensive study of wild Pisum together with landrace material. This work gives a more comprehensive view of pea genetic diversity in relation to domestication than previous studies [29,30,31,61] by combining high density, genome wide, marker assays with a targeted set of accessions.
4.1. Pisum Taxonomy
Phylogenetic studies have layered new complexities in the classification of Pisum. Analysis using chloroplast genes nested a monophyletic Vicieae tribe within Trifolieae, with the genus Trifolium as sister to the Vicieae [25,88]. The genus Vicia was shown to be paraphyletic, with the genera Pisum, Vavilovia, Lathyrus and Lens nested within Vicia. Schaefer et al. [25] proposed that one solution to resolve the paraphyly of Vicia would include transferring Pisum and Vavilovia to Lathyrus. In their analysis, Pisum and Vavilovia were each monophyletic and together monophyletic within Lathyrus, noting that Vavilovia had previously been included within Pisum. If this scheme were to be accepted, then Vicia section Ervum must also be elevated to the status of a genus and the precedent would be to rename Pisum L. as Lathyrus oleraceus Lam. [89]. This classification has already been used [90] in accordance with the ‘International Code of Nomenclature of algae, fungi and plants’ (International Code, 2012). Subsequently, Kosterin [91] has renamed Pisum sativum as Lathyrus schaeferi Kosterin nomen novum pro Pisum abyssinicum A. Braun and Lathyrus fulvus (Sibthrop et Smith) Kosterin combinatio nova pro Pisum fulvum Sibthrop et Smith.
For the purpose of our study, we have retained the commonly understood Pisum L., as indicated in the Materials and Methods section, and in the absence of agreement on the wholesale reclassification of these taxa. As a direct result of its broad phenotypic diversity, a large number of different Latin names at different ranks have been proposed for various forms of pea [29,37]. The classification of Pisum L. based on morphology and karyology clearly delineates two species, P. fulvum Sibth. & Sm. and P. sativum L. [92,93]. P. sativum has been further divided into three taxa recognized either as subspecies or species: P. elatius Bieb. [75], P. humile Boiss. & Noe and P. syriacum Boiss. & Noe. [27]. P. elatius M. Bieb. was first described at the rank of species in 1808 [75] and was later reduced in rank to a subspecies [94], although many authors ascribe this to Ascheron and Graebner [95]. Pisum humile was described by Boissier and Noë [96], however their name was illegitimate because it is a later homonym of P. humile Miller [97], a form of cultivated pea. Berger [98] downgraded that to a subspecies and gave it a new name: P. sativum subsp. syriacum A. Berger, however its status was raised again to species by Lehmann [99] as P. syriacum (A. Berger). C.O. Lehm., though this nomenclatural change remained unsupported. The work of Ben-Ze’ev and Zohary [47] has become the standard text for pea species relationships and was based on classical species definitions using hybridization barriers along with ecological aspects of distribution. Their work followed the taxonomy of Boissier [96], which recognized three wild pea species: P. fulvum, P. sativum subsp. elatius Bieb. and P. humile Boiss & Noë (=P. syriacum (A. Berger) C.O. Lehm.), and the domesticated pea P. sativum. These two wild groups of P. sativum subsp. elatius Bieb. and P. humile Boiss & Noë (=P. syriacum (A. Berger) C.O. Lehm.) were described as being morphologically, ecologically and genetically distinct [25,45]. Recently, Ladizinsky and Abbo [100] also recognized two groups: subsp. elatius and subsp. humile and further described two varieties of subsp. humile, a “southern” and a “northern” form, based on geographic distribution. They delimited the southern form as subsp. humile var. humile (Boiss et Noë) Ladizinsky, and the northern form as subsp. humile var. syriacum (A. Berger) Ladizinsky. Recent comprehensive genome wide analysis of 150 P. elatius and P. fulvum samples [30], however, did not support this geographical or morphological division, although P. elatius diversity was structured into five groups.
Based on the results of this study, we propose to use Pisum sativum L., as described by Linnaeus [73], for cultivated pea, Pisum abyssinicum A. Braun for the Ethiopian form of cultivated pea and for wild pea to retain Pisum fulvum Sibth. & Sm. and to use Pisum elatius M. Bieb. in the broader sense that includes the rest of the genus.
4.2. Pisum Genetic Diversity
The current study included the samples used by Smýkal et al. [30], however added P. abyssinicum and P. sativum cultivated material. It should be noted that the marker set was slightly different as this analysis included the combination of two DARTseq results which were newly computationally processed. This was likely one of the reasons why 47 out of 161 samples that were common in both studies were assigned to different genetic clusters (Table S1). The second reason could be that the addition (or removal) of any samples affected the analysis and the use of slightly different analytical methods.
In the PCO analysis (Figure 2), P. fulvum and P. abyssinicum were assigned to single distinct groups, while wild P. elatius formed four distinct groups plus residual, un-assigned samples that overlap with P. sativum landrace material, which is suggestive of a shared history between P. sativum and P. elatius. P. abyssinicum was clearly distinct from all P. sativum. There was good agreement in the grouping of P. elatius assignments between this and a prior study [30] in three of the P. elatius clusters (Table S1), while the remainder P. elatius were differently assigned.
We have previously [30] analyzed the spatial distribution of the geographical pattern to the genetic structure of wild P. elatius accessions by the centroid approach and found the clusters were mostly all overlapping without clear isolation by distance. In the Middle East, the center of origin of the Fabeae tribe origin [25], the genetic diversity of Pisum accessions, was greater than elsewhere [30]. In Europe, one sample from Portugal and one from the Balearic Islands were distinct from all other European P. elatius which belong to two groups, one with affinity to domesticated P. sativum and the other a distinct subgroup of P. elatius (Figure 4 and Figure 5). There was small (12 accessions) DAPC group 5 of P. elatius, which all except for one originated from eastern Turkey and had a significant proportion of P. fulvum alleles. This group corresponds to Q6 in our previous study [30]. This material, with the exception of JI261 [31], was not included in previous studies [55,56,61,62]. The complete plastid genome sequence of accession W6-26109 from this group 5 was recently shown to match P. fulvum [101]. It was suggested that this accession belongs to an ancestral group from which P. elatius and P. fulvum were derived. In this case, both the plastid [101] and nuclear [this study] genomes suggest the same conclusion and likely reflects a shared ancestry. The other accessions that group 5 (Figure 5, Table S1) were collected from was north-eastern Turkey outside the past and present [30] geographical distribution of P. fulvum. This is newly identified diversity in Pisum genus and indicates that there is potential for further collecting new diversity.
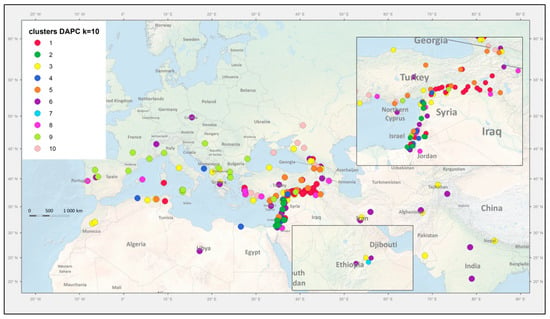
Figure 5.
Geographic distribution of collection sites. Markers are coloured according to the assignment to respective genetic groups identified in Figure 4. P. sativum landraces (in green) are placed centrally in their respective country of origin.
In our study, we used fewer cultivated accessions than Jing et al. [31], however the selected 64 landraces in this study came from a broad geographical area, yet they were grouped together. DAPC identified two groups of P. sativum accessions, one with 30 and the other with 27 accessions, however the groups were not geographically distinct. The remaining seven P. sativum accessions were incorporated into DAPC groups that mostly comprised P. elatius. These possibly correspond to mis-identification in the passport data (as mentioned in [62]), or P. elatius-P. sativum introgression (similar to introgression in chickpea, [102]), however this could also be the signature of additional domestication events. The accessions TG2548, TG2426, TG2558, from Georgia, have been described as morphologically distinct having a vetch-like appearance and were assigned to a subspecies: P. sativum subsp. transcaucasicum [26]. Except for TG2548, they have smooth seed coats and non-dormant seeds.
Our data showed no clear separation of subgroups within P. sativum in contrast with findings of Siol et al. [55] using an Illumina SNP assay. Siol et al. [55] observed three large genetically distinct clusters: (1) wild peas and landraces from the Middle East and Asia, (2) winter peas and (3) spring peas varieties; in addition to these three groups, these authors identified distinct groups from the Far East and China, as well as Central Asia (Afghanistan, Pakistan, Nepal). Similarly, using a genotyping by sequencing (GBS) based analysis [61] of 431 largely cultivated types, a separate group from Central Asia (defined as Afghanistan, China, India, Nepal and Pakistan) which were closer to P. elatius was identified.
4.3. Pea Domestication
The ever-intriguing question of crop domestication is whether it happened only once or multiple times. For pea, conclusions must be drawn from extant material as archeological samples cannot yet be examined at a comparable level of detail. In the case of domesticated plants, multiple origins might be masked by hybridization which may bring independently domesticated stocks together. In several crops, there is evidence of one (maize, [68], chickpea, [69], soybean, [72], groundnut, [70]) or two (common bean, [71]) domestication events. It was recently shown that multiple origins might be revealed as genome mosaics (barley, [64,65], rice, [63] and emmer wheat, [67]). Multiple domestications where gene flow occurred early in the process have also been reported [65,103]. In most of these studies, a strong genetic bottleneck was detected.
A description of domestication for several legume species is only beginning to emerge (chickpea [69], groundnut [70] or common bean [71] and soybean [72]). One of the best studied legume genera is common bean (Phaseolus), with P. vulgaris having two gene pools and two independent domestication events that contributed to the modern common bean crop (reviewed in [71]). On the other hand, while the two types of cultivated chickpea, desi and kabuli, display large genomic differences, they were derived from a single domestication event [104,105]. In soybean, a combination of archaeological and molecular data suggested that a prolonged period of low-intensity management or semi-cultivation of wild soybeans at multiple locations preceded domestication [72]. These pre-domesticates may have been assimilated within wild soybeans or were integrated into the domesticated soybean [72]. This might also be the case in pea, being first gathered from wild stands, then cultivated [106], although the morphological distinction between the wild and domesticated plants remains problematic. Reticulate evolution during the domestication of emmer wheat has been proposed [67].
Our data showing overlap between domesticated and wild pea suggests a similar scenario. In contrast to wild cereals which occur in large stands [64,65,66], wild peas occur in scattered patches within which the populations are likely closely related [77]. During the pre-domestication period, it is possible that pea seeds were collected over a large area and were brought to a common point, likely creating mixed stands. In spite of open-pollination being rare in pea [77], in mixed stands, considerable diversity may remain during the domestication process due to sporadic gene-flow between lineages.
It has been argued that seed dormancy would have been a substantial barrier to the efficient cultivation of legumes during their domestication process, so it has been proposed that non-dormant types existed in wild populations that were selected by humans [11]. Results from the experimental cultivation of wild peas suggest that the crucial trait in pea domestication was the loss of the seed dormancy [107]. It has been proposed that at early stages of domestication, selection acted on standing variation, with further refinement of specific traits narrowed during and post-domestication [2]. Until now, we have had very little information on domestication genes in legumes (reviewed in [108]). Weeden [109] suggested that at least 15 genes are critical for pea domestication and our recent study of pod dehiscence and seed dormancy identified several candidates for these traits [87]. Here, we identified a group of (wild) P. elatius from Armenia, Georgia and south-eastern Turkey that are the most closely related wild peas to the cultivated P. sativum genepool (Figure 3, Table S1). Either these are genetically the closest extant progenitors of domesticated pea, or they represent early escapes from cultivation with a reversion to the wild type. Wang et al. [110] showed that analyses including “feral” rice i.e., wild rice that carries a causative domestication allele [111] alters the way wild rice and domesticated rice accessions are clustered [110]. Interestingly, feral origin for P. sativum subsp. humile var. humile was suggested [112]. However, Ben-Ze´ev and Zohary [47] noted that subsp. humile differs by one reciprocal chromosomal translocation from domesticated pea, which is not consistent with the claim of Abbo et al. [112]. Ladizinsky and Abbo [100] later proposed that pea domestication involved more than one cytotype. Jing et al. [31] and Siol et al. [55] proposed that the high genetic diversity in cultivated pea indicates a relatively weak domestication bottleneck and that genetic diversity may have been maintained because of the diversity of uses and the wide range of environmental conditions in which it is grown. Alternatively, this might indicate either multiple domestication events and/or prolonged wild to crop gene flow as has been shown in barley [65], emmer wheat [67] and chickpea [102]. Even today, in many areas of Middle East dryland, cropping is carried out in small patches of open woodland, which very often is in close vicinity to wild crop progenitors (Smykal, Berger, personal observations), thus enabling gene flow in both directions. This could contribute to the observed admixture both in wild P. elatius and cultivated P. sativum landraces groups. The Central Asian peas, sometimes called Afghan types or P. sativum subsp. asiaticum (Govorov) as defined by Govorov [26] and Makasheva [27], was discussed by Jing et al. [31] who supported the proposal of Young and Matthews [113] that this group is not solely characterized by the nodulation phenotype. In our analysis, these were represented by eight accessions (CGN3277, IPK476, JI86, JI103, JI2019, PI124478, VIR1246, PI639969) all of which, except for IPK476 and VIR1246, formed a distinct branch in SplitsTree (Figure 3). This is in agreement with previous studies [31,55,61] showing distinction of this group. In contrast, Chinese origin landraces, which were stated to be distinct [45], were distributed among several different branches within the group that contained P. sativum (Figure 3 and Figure 4, Table S1).
4.4. Independent Domestication of the Ethiopian (Pisum abyssinicum) Pea
P. abyssinicum was first described [74] at a species rank (the type specimen was collected in 1840 by Schimper W.H and is stored at the herbarium of National Botanic Garden of Belgium (BR, BR0000006255831) and this was adopted by many authors [26,28,29,31,33,34], while others considered it a subspecies of the common pea P. sativum subsp. abyssinicum (A. Br.) Berger [27,98], or only a form [48,110,114]. Ethiopian pea (P. abyssinicum) called Dekoko (‘minute seeded’, in Amharic) is considered endemic to Ethiopia and southern Yemen. It was first described in Tigray and Amhara regions in northern Ethiopia. According to a report [115], it used to be more common and, currently, its cultivation is restricted to South Tigray and North Wollo provinces and Southern Yemen where it accompanies the main cereal crops, chickpea, linseed and grasspea. Common pea (P. sativum) is also grown in Ethiopia, however the Ethiopian pea (P. abyssinicum) is valued for its higher nutritional quality [115].
The intriguing question is: was Ethiopian pea domesticated independently? And if so, was it domesticated in place or brought from the Middle East? An independent domestication of the Ethiopian pea (P. abyssinicum) has been proposed by several authors [31,32,33,34,36,37] and is supported by it having a distinct karyotype [47,48,114]. The P. abyssinicum genome is reportedly slightly (ca. 8%) larger than that of P. sativum [116]. Serological studies of Pisum taxa [117] suggested that P. abyssinicum might have originated from hybridization between P. sativum subsp. elatius and P. fulvum. Retrotransposon based diversity analysis showed substantial marker sharing between both [33,34]. Similarly, an extended study [31] showed that P. abyssinicum shares several phenotypic traits and a significant proportion of molecular marker alleles with P. fulvum and tends to occupy an intermediate position between the latter and P. elatius in molecular diversity plots, which is consistent with the possibility that progenitor of P. abyssinicum shared some ancestry with what is now P. fulvum and also with P. elatius. Since P. fulvum and P. elatius are sympatric in Israel [100], it might be that a naturally occurring hybrid was recognized by humans and was moved to Ethiopia and southern Arabia. Although wild pea is unlikely to be native to Ethiopia, the possibility of wild pea species having occurred in North Africa is supported by an intriguing record of P. elatius seeds (JI254) from a market in Ethiopia [118]. P. abyssinicum might have its origin within its present range, which is itself a known center of origin for cultivated plants [15]. The distribution of wild Pisum species may have also changed since the time of domestication. The desertification of the Sahara is known to have occurred in two abrupt phases, the last of these possibly as recent as 3500BP [119]. However, it seems most likely that domesticated P. abyssinicum was introduced via human migration [51]. The earliest known remains of P. abyssinicum (from present day Eritrea) are dated back to approximately 400AD [32] and are considerably more recent than the archaeological finds of P. sativum. Our study confirms prior studies suggesting that P. abyssinicum is a distinct genetic lineage [31,32,33,34,120]. The very low genetic diversity present in P. abyssinicum is the result of a severe genetic bottleneck and its allelic composition suggests rather ancient divergence of P. abyssinicum from other Pisum lineages. It is likely that specific allele composition contributes to the narrow eco-geographical range of P. abyssinicum. Although both species are commonly grown in Ethiopia, a reproductive barrier prevented gene flow between them [47,91,121]. Recently, the issue of the Ethiopian pea was revisited [38] using 54 gene sequences on a set of 76 cultivated pea (P. sativum subsp. sativum), two wild pea (P. sativum subsp. elatius), 11 P. fulvum and one P.s. subsp. abyssinicum sample, which demonstrated a close relationship among the three P. sativum subspecies and rejected the hypothesis that P.s. subsp. abyssinicum was formed by hybridization between one of the P. sativum subspecies and P. fulvum. This study [38] supported its status as a distinct subspecies, P. sativum subsp. abyssinicum. Crosses between P. abyssinicum and cultivated P. sativum showed some segregation of pod dehiscence, a domestication trait, suggesting that this might be governed by different genes in the two groups [118]. These populations will provide valuable tools to test the domestication scenario further once the respective domestication genes are identified. Identification of the genes underlying key domestication traits in pea [87,108] as well as the availability of the pea genome sequence should shed light on the number of domestication events in pea.
5. Conclusions
The results indicate that P. abyssinicum and P. sativum were derived from different genepools, thus representing two independent domestication events. The data does not support the hypothesis of a hybrid origin of P. abyssinicum nor of it being derived from P. sativum. P. fulvum is genetically distinct, while P. elatius diversity is structured to seven identified groups. P. sativum diversity shows partial overlap with P. elatius and is not geographically structured. A group of P. elatius accessions was identified as the suggested group from which P. fulvum and P. abyssinicum arose.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/11/535/s1, Table S1: List and description of analyzed material, Table S2: DARTseq dataset.
Author Contributions
Formal analysis, O.T., J.B., I.H., A.R., R.R.D., P.K., P.R., C.R. and P.S.; Funding acquisition, P.S.; Investigation, I.H.; Methodology, O.T. and P.S.; Resources, C.J.C. and P.S.; Writing—original draft, O.T., C.J.C., P.R., C.R. and P.S.; Writing—review & editing, J.B., A.R., R.R.D., P.K., C.J.C., P.R. and P.S. All authors read and approved the final manuscript.
Funding
This research was funded by the Grant Agency of Czech Republic grant 16-21053S. O.T. is partially supported by institutional funding on the long-term conceptual development of the Agricultural Research, Ltd. organization.
Acknowledgments
We are thankful to Mike Ambrose and Bob Redden, who were genebanks curators for years, for stimulating discussion, sharing their knowledge and selection of pea germplasm. We are also very thankful to TH Noel Ellis for his great help with this study and manuscript writing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lev-Yadun, S.; Gopher, A.; Abbo, S. Archaeology. The cradle of agriculture. Science 2000, 288, 1602–1603. [Google Scholar] [CrossRef] [PubMed]
- Abbo, S.; Pinhasi van-Oss, R.; Gopher, A.; Saranga, Y.; Ofner, I.; Peleg, Z. Plant domestication versus crop evolution: A conceptual framework for cereals and grain legumes. Trends Plant Sci. 2014, 19, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Willcox, G.; Fornite, S.; Herveux, L. Early holocene cultivation before domestication in northern Syria. Veget. Hist. Archaeobot. 2008, 17, 313–325. [Google Scholar] [CrossRef]
- Tanno, K.-I.; Willcox, G. How fast was wild wheat domesticated? Science 2006, 311, 1886. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.; Kislev, M.E.; Hartmann, A. Anthropology: Autonomous cultivation before domestication. Science 2006, 312, 1608–1610. [Google Scholar] [CrossRef] [PubMed]
- Allaby, R.G.; Fuller, D.Q.; Brown, T.A. The genetic expectations of a protracted model for the origins of domesticated crops. Proc. Natl. Acad. Sci. USA 2008, 105, 13982. [Google Scholar] [CrossRef] [PubMed]
- Fuller, D.Q. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann. Bot. 2007, 100, 903–924. [Google Scholar] [CrossRef] [PubMed]
- Fuller, D.Q.; Asouti, E.; Purugganan, M.D. Cultivation as slow evolutionary entanglement: Comparative data on rate and sequence of domestication. Veget. Hist. Archaeobot. 2012, 21, 131–147. [Google Scholar] [CrossRef]
- Abbo, S.; Saranga, Y.; Peleg, Z.; Kerem, Z.; Lev-Yadun, S.; Gopher, A. Reconsidering domestication of legumes versus cereals in the ancient near east. Q. Rev. Biol. 2009, 84, 29–50. [Google Scholar] [CrossRef] [PubMed]
- Mithen, S. After the Ice: A Global Human History 20,000-5,000 BC; Weidenfield & Nicholson: London, UK, 2003; ISBN 13 978-0674019997. [Google Scholar]
- Ladizinsky, G. Plant Evolution under Domestication; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1998; ISBN 10 0412822105. [Google Scholar]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin, 4th ed.; Oxford University Press: Oxford, UK, 2012; ISBN 9780199549061. [Google Scholar]
- Zohary, D.; Hopf, M. Domestication of Pulses in the Old World: Legumes were companions of wheat and barley when agriculture began in the Near East. Science 1973, 182, 887–894. [Google Scholar] [CrossRef] [PubMed]
- De Candolle, A. Origin of Cultivated Plants; Kessinger Publishing: Whitefish, UK, 2006; ISBN 9781116495034. [Google Scholar]
- Vavilov, N.I. The Origin, Variation, Immunity and Breeding of Cultivated Plants; Translated from the Russian by K. Starchester; Chronica Botanica: Leyden, The Netherlands, 1951; Volume 13, pp. 1–364. [Google Scholar]
- Smartt, J. Grain Legumes: Evolution and Genetic Resources; Cambridge University Press: Cambridge, UK, 1990. [Google Scholar]
- Ambrose, M.J. From Near East centre of origin the prized pea migrates thorough world. Diversity 1995, 11, 118–119. [Google Scholar]
- Kislev, M.E.; Bar-Yosef, O. The legumes: The earliest domesticated plants in the Near East? Curr. Anthropol. 1988, 29, 175–179. [Google Scholar] [CrossRef]
- Holtgrieve, G.W.; Schindler, D.E.; Hobbs, W.O.; Leavitt, P.R.; Ward, E.J.; Bunting, L.; Chen, G.; Finney, B.P.; Gregory-Eaves, I.; Holmgren, S.; et al. A coherent signature of anthropogenic nitrogen deposition to remote watersheds of the Northern Hemisphere. Science 2011, 334, 1545–1548. [Google Scholar] [CrossRef] [PubMed]
- Erisman, J.W.; Sutton, M.A.; Galloway, J.; Klimont, Z.; Winiwarter, W. How a century of ammonia synthesis changed the world. Nat. Geosci. 2008, 1, 636–639. [Google Scholar] [CrossRef]
- Diamond, J. Evolution, consequences and future of plant and animal domestication. Nature 2002, 418, 700–707. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT 2016. Available online: http://www.fao.org/faostat/en/ (accessed on 26 September 2018).
- Smýkal, P.; Coyne, C.; Ambrose, M.; Maxted, N.; Schaefer, H.; Blair, M.; Berger, J.; Greene, S.L.; Nelson, M.; Besharat, N.; et al. Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant Sci. 2014, 33, 43–104. [Google Scholar] [CrossRef]
- Lewis, G.; Schrire, B.; Mackinder, B.; Lock, M. Legumes of the World; Royal Botanic Gardens: London, UK, 2005; ISBN 1900347806. [Google Scholar]
- Schaefer, H.; Hechenleitner, P.; Santos-Guerra, A.; Menezes de Sequeira, M.; Pennington, R.T.; Kenicer, G.; Carine, M.A. Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evol. Biol. 2012, 12, 250. [Google Scholar] [CrossRef] [PubMed]
- Govorov, L. Pisum. In Flora of Cultivated Plants IV, Grain Leguminosae; Vavilovk, N.I., Wulff, E.V., Eds.; State Agricultural Publishing Company: Moscow/Leningrad, Russia, 1937; pp. 231–336. [Google Scholar]
- Makasheva, R.K. Gorokh (pea). In Kulturnaya Flora SSR; Korovina, O.N., Ed.; Kolos Publishers: Leningrad, Russia, 1979; pp. 1–324. [Google Scholar]
- Maxted, N.; Ambrose, M. Peas (Pisum L.). In Plant Genetic Resources of Legumes in the Mediterranean; Maxted, N., Bennett, S.J., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2001; pp. 181–190. ISBN 0792367073. [Google Scholar]
- Smýkal, P.; Kenicer, G.; Flavell, A.J.; Corander, J.; Kosterin, O.; Redden, R.J.; Ford, R.; Coyne, C.J.; Maxted, N.; Ambrose, M.J.; et al. Phylogeny, phylogeography and genetic diversity of the Pisum genus. Plant Genet. Res. 2011, 9, 4–18. [Google Scholar] [CrossRef]
- Smýkal, P.; Hradilová, I.; Trněný, O.; Brus, J.; Rathore, A.; Bariotakis, M.; Das, R.R.; Bhattacharyya, D.; Richards, C.; Coyne, C.J.; et al. Genomic diversity and macroecology of the crop wild relatives of domesticated pea. Sci. Rep. 2017, 7, 17384. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Vershinin, A.; Grzebyta, J.; Shaw, P.; Smýkal, P.; Marshall, D.; Ambrose, M.J.; Ellis, T.H.N.; Flavell, A.J. The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol. Biol. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Butler, A. The ethiopian pea: Seeking the evidence for a separate domestication. In Food, Fuel and Fields: Progress in African Archaeobotany; African Archaeobotany: Frankfurt, Germany, 2003; pp. 37–47. ISBN 3927688207. [Google Scholar]
- Ellis, T.H.; Poyser, S.J.; Knox, M.R.; Vershinin, A.V.; Ambrose, M.J. Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol. Gen. Genet. 1998, 260, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Vershinin, A.V.; Allnutt, T.R.; Knox, M.R.; Ambrose, M.J.; Ellis, T.H.N. Transposable elements reveal the impact of introgression, rather than transposition, in Pisum diversity, evolution, and domestication. Mol. Biol. Evol. 2003, 20, 2067–2075. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Johnson, R.; Seres, A.; Kiss, G.; Ambrose, M.J.; Knox, M.R.; Ellis, T.H.N.; Flavell, A.J. Gene-based sequence diversity analysis of field pea (Pisum). Genetics 2007, 177, 2263–2275. [Google Scholar] [CrossRef] [PubMed]
- Polans, N.O.; Moreno, R.R. Microsatellite and ITS sequence variation in wild species and cultivars of pea. Pisum Genet. 2009, 41, 3–6. [Google Scholar]
- Ellis, T.H.N. Pisum. In Wild Crop Relatives, Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 237–248. ISBN 978-3-642-14387-8. [Google Scholar]
- Weeden, N.F. Domestication of pea (Pisum sativum L.): The case of the Abyssinian pea. Front. Plant Sci. 2018, 9, 515. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Vernoud, V.; Blair, M.W.; Soukup, A.; Thompson, R.D. The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 2014, 5, 351. [Google Scholar] [CrossRef] [PubMed]
- Baldev, B. Origin, distribution, taxonomy, and morphology. In Pulse Crops; Baldev, B., Ramanujam, S., Jain, H.K., Eds.; Oxford and IBH Publishing Co.: New Delhi, India, 1988; pp. 3–51. ISBN 9788120402782. [Google Scholar]
- Riehl, S.; Zeidi, M.; Conard, N.J. Emergence of agriculture in the foothills of the Zagros Mountains of Iran. Science 2013, 341, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Mikić, A.; Medović, A.; Jovanović, Ž.; Stanisavljević, N. Integrating archaeobotany, paleogenetics and historical linguistics may cast more light onto crop domestication: The case of pea (Pisum sativum). Genet. Resour. Crop Evol. 2014, 61, 887–892. [Google Scholar] [CrossRef]
- Chimwamurombe, P.M.; Khulbe, R.K. Domestication. In Biology and Breeding of Food Legumes; Pratap, A., Kumar, J., Eds.; CABI: Cambridge, UK, 2011; pp. 19–34. ISBN 978-1-84593-766-9. [Google Scholar]
- Zong, X.; Guan, J.P.; Wang, S.M.; Liu, Q.; Redden, R.; Ford, R. Genetic diversity and core collection of alien Pisum sativum L. germplasm. Acta Agron. Sin. 2008, 34, 1518–1528. [Google Scholar] [CrossRef]
- Zong, X.; Redden, R.J.; Liu, Q.; Wang, S.; Guan, J.; Liu, J.; Xu, Y.; Liu, X.; Gu, J.; Yan, L.; et al. Analysis of a diverse global Pisum sp. collection and comparison to a Chinese local P. sativum collection with microsatellite markers. Theor. Appl. Genet. 2009, 118, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Redden, R.J.; Zong, X.; Berger, J.D.; Bennett, S.J. Ecogeographic analysis of pea collection sites from China to determine potential sites with abiotic stresses. Genet. Resour. Crop Evol. 2013, 60, 1801–1815. [Google Scholar] [CrossRef]
- Ben-Ze’ev, N.; Zohary, D. Species relationships in the genus Pisum L. Isr. J. Bot. 1973, 22, 73–91. [Google Scholar]
- Errico, A.; Conicella, C.; Venora, G. Karyotype studies on Pisum fulvum and Pisum sativum, using a chromosome image analysis system. Genome 1991, 34, 105–108. [Google Scholar] [CrossRef]
- Pourkheirandish, M.; Hensel, G.; Kilian, B.; Senthil, N.; Chen, G.; Sameri, M.; Azhaguvel, P.; Sakuma, S.; Dhanagond, S.; Sharma, R.; et al. Evolution of the grain dispersal system in barley. Cell 2015, 162, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, V.; Triska, P.; Pereira, J.B.; Alshamali, F.; Rito, T.; Machado, A.; Fajkošová, Z.; Cavadas, B.; Černý, V.; Soares, P.; et al. Genetic stratigraphy of key demographic events in Arabia. PLoS ONE 2015, 10, e0118625. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Llorente, M.; Jones, E.R.; Eriksson, A.; Siska, V.; Arthur, K.W.; Arthur, J.W.; Curtis, M.C.; Stock, J.T.; Coltorti, M.; Pieruccini, P.; et al. Ancient Ethiopian genome reveals extensive Eurasian admixture throughout the African continent. Science 2015, 350, 820–822. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Coyne, C.; Redden, R.; Maxted, N. Peas. In Genetic and Genomic Resources of Grain Legume Improvement; Singh, M., Upadhyaya, H.D., Bisht, I.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; ISBN 9780123979353. [Google Scholar]
- Warkentin, T.; Smykal, P.; Coyne, C.; Weeden, N.; Domoney, C.; Bing, D.; Leonforte, T.; Xuxiao, Z.; Dixit, G.; Boros, L.; et al. Pea. In Grain Legumes. Handook of Plant Breeding; De Ron, A.M., Ed.; Springer: New York, NY, USA, 2015; Volume 10, pp. 37–85. ISBN 978-1-4939-2797-5. [Google Scholar]
- Tar’an, B.; Zhang, C.; Warkentin, T.; Tullu, A.; Vandenberg, A. Genetic diversity among varieties and wild species accessions of pea (Pisum sativum L.) based on molecular markers, and morphological and physiological characters. Genome 2005, 48, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Siol, M.; Jacquin, F.; Chabert-Martinello, M.; Smýkal, P.; Le Paslier, M.-C.; Aubert, G.; Burstin, J. Patterns of genetic structure and linkage disequilibrium in a large collection of pea germplasm. G3 (Bethesda) 2017, 7, 2461–2471. [Google Scholar] [CrossRef] [PubMed]
- Burstin, J.; Salloignon, P.; Chabert-Martinello, M.; Magnin-Robert, J.-B.; Siol, M.; Jacquin, F.; Chauveau, A.; Pont, C.; Aubert, G.; Delaitre, C.; et al. Genetic diversity and trait genomic prediction in a pea diversity panel. BMC Genom. 2015, 16, 105. [Google Scholar] [CrossRef] [PubMed]
- Tayeh, N.; Aluome, C.; Falque, M.; Jacquin, F.; Klein, A.; Chauveau, A.; Bérard, A.; Houtin, H.; Rond, C.; Kreplak, J.; et al. Development of two major resources for pea genomics: The GenoPea 13.2K SNP Array and a high-density, high-resolution consensus genetic map. Plant J. 2015, 84, 1257–1273. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, N.; Hao, J.; Hu, J.; Zhang, X.; Blair, M.W. Genetic diversity of Chinese and global pea (Pisum sativum L.) collections. Crop Sci. 2017, 57, 1–11. [Google Scholar] [CrossRef]
- Baranger, A.; Aubert, G.; Arnau, G.; Lainé, A.L.; Deniot, G.; Potier, J.; Weinachter, C.; Lejeune-Hénaut, I.; Lallemand, J.; Burstin, J. Genetic diversity within Pisum sativum using protein- and PCR-based markers. Theor. Appl. Genet. 2004, 108, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Tayeh, N.; Klein, A.; Le Paslier, M.-C.; Jacquin, F.; Houtin, H.; Rond, C.; Chabert-Martinello, M.; Magnin-Robert, J.-B.; Marget, P.; Aubert, G.; et al. Genomic prediction in pea: Effect of marker density and training population size and composition on prediction accuracy. Front. Plant Sci. 2015, 6, 941. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, W.L.; Gazave, E.; Cheng, P.; Myers, J.R.; Gore, M.A.; Coyne, C.J.; McGee, R.J.; Mazourek, M. A community resource for exploring and utilizing genetic diversity in the USDA pea single plant plus collection. Hort. Res. 2017, 4, 17017. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Ambrose, M.A.; Knox, M.R.; Smykal, P.; Hybl, M.; Ramos, Á.; Caminero, C.; Burstin, J.; Duc, G.; van Soest, L.J.M.; et al. Genetic diversity in European Pisum germplasm collections. Theor. Appl. Genet. 2012, 125, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Platts, A.E.; Fuller, D.Q.; Hsing, Y.-I.; Wing, R.A.; Purugganan, M.D. The rice paradox: Multiple origins but single domestication in Asian rice. Mol. Biol. Evol. 2017, 34, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Poets, A.M.; Fang, Z.; Clegg, M.T.; Morrell, P.L. Barley landraces are characterized by geographically heterogeneous genomic origins. Genome Biol. 2015, 16, 173. [Google Scholar] [CrossRef] [PubMed]
- Pankin, A.; Altmüller, J.; Becker, C.; von Korff, M. Targeted resequencing reveals genomic signatures of barley domestication. New Phytol. 2018, 218, 1247–1259. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.H.; Sun, D.; Nevo, E. Domestication evolution, genetics and genomics in wheat. Mol. Breed. 2011, 28, 281. [Google Scholar] [CrossRef]
- Civáň, P.; Ivaničová, Z.; Brown, T.A. Reticulated origin of domesticated emmer wheat supports a dynamic model for the emergence of agriculture in the Fertile Crescent. PLoS ONE 2013, 8, e81955. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Vigouroux, Y.; Goodman, M.M.; Sanchez, G.J.; Buckler, E.; Doebley, J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 2002, 99, 6080–6084. [Google Scholar] [CrossRef] [PubMed]
- von Wettberg, E.J.B.; Chang, P.L.; Başdemir, F.; Carrasquila-Garcia, N.; Korbu, L.B.; Moenga, S.M.; Bedada, G.; Greenlon, A.; Moriuchi, K.S.; Singh, V.; et al. Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nat. Commun. 2018, 9, 649. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Bitocchi, E.; Rau, D.; Bellucci, E.; Rodriguez, M.; Murgia, M.L.; Gioia, T.; Santo, D.; Nanni, L.; Attene, G.; Papa, R. Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front. Plant Sci. 2017, 8, 722. [Google Scholar] [CrossRef] [PubMed]
- Sedivy, E.J.; Wu, F.; Hanzawa, Y. Soybean domestication: The origin, genetic architecture and molecular bases. New Phytol. 2017, 214, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Linnaeus, C. Species Plantarum; Salvius: Stockholm, Sweden, 1753; Volume 2. [Google Scholar]
- Braun, A. Bemerkungen über die Flora von Abyssinien. Flora Oder Allgemeine Botanische Zeitung 1841, 1, 257–288. [Google Scholar]
- Bieberstein, M. Flora Taurico-Caucasica Exhibens Stirpes Phaenomagas in Chersoneso Taurica et regionibus Caucasicis Sponte Crescents; Typis Academicis: Charkouiae/Kharkov, Ukraine, 1808; Volume 2, p. 447. [Google Scholar]
- Sibthorp, J.M.D.; Smith, J.E. Flora Graeca Sibthorpiana; Richard Taylor: London, UK, 1813; Volume 2. [Google Scholar]
- Smýkal, P.; Trněný, O.; Brus, J.; Hanáček, P.; Rathore, A.; Roma, R.D.; Pechanec, V.; Duchoslav, M.; Bhattacharyya, D.; Bariotakis, M.; et al. Genetic structure of wild pea (Pisum sativum subsp elatius) populations in the northern part of the Fertile Crescent reflects moderate cross-pollination and strong effect of geographic but not environmental distance. PLoS ONE 2018, 13, e0194056. [Google Scholar] [CrossRef]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity arrays technology: A generic genome profiling technology on open platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Cruz, V.M.V.; Kilian, A.; Dierig, D.A. Development of DArT marker platforms and genetic diversity assessment of the U.S. collection of the new oilseed crop Lesquerella and related species. PLoS ONE 2013, 8, e64062. [Google Scholar] [CrossRef] [PubMed]
- Raman, H.; Raman, R.; Kilian, A.; Detering, F.; Carling, J.; Coombes, N.; Diffey, S.; Kadkol, G.; Edwards, D.; McCully, M.; et al. Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PLoS ONE 2014, 9, e101673. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.A.; Richards, C.M. Accurate inference of subtle population structure (and other genetic discontinuities) using principal coordinates. PLoS ONE 2009, 4, e4269. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.; Moulton, V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Malinsky, M.; Trucchi, E.; Lawson, D.J.; Falush, D. RADpainter and fineRADstructure: Population inference from RADseq data. Mol. Biol. Evol. 2018, 35, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Ranal, M.A.; Santana, D.G. How and why to measure the germination process? Braz. J. Bot. 2006, 29, 1–11. [Google Scholar] [CrossRef]
- Hradilová, I.; Trněný, O.; Válková, M.; Cechová, M.; Janská, A.; Prokešová, L.; Aamir, K.; Krezdorn, N.; Rotter, B.; Winter, P.; et al. A combined comparative transcriptomic, metabolomic, and anatomical analyses of two key domestication traits: Pod dehiscence and seed dormancy in pea (Pisum sp.). Front. Plant Sci. 2017, 8, 542. [Google Scholar] [CrossRef] [PubMed]
- Steele, K.P.; Wojciechowski, M.F. Phylogenetic analyses of tribes Trifolieae and Vicieae, based on sequences of the plastid gene matK (Papilionoideae: Leguminosae). In Advances in Legume Systematics, Part 10, Higher Level Systematics; Klitgaard, B.B., Bruneau, A., Eds.; Kew Royal Botanical Gardens: London, UK, 2003; pp. 355–370. [Google Scholar]
- de Lamarck, M.M. Flore Françoise, ou Description Succinte de Toutes des Plantes qui Croiffent Naturellement en France, disposées selon une nouvelle méthode d’analyse, et précédées par un exposé des principes élémentaires de la botanique, 3rd ed.; Desray: Paris, France, 1778. [Google Scholar]
- Coulot, P.; Rabaute, P. Monographie de Leguminosae de France. 4. Tribus des Fabeae, des Cicereae et des Genisteae. Bulletin de la Société Botanique du Centre-Ouest 2016, 46, 1–902. [Google Scholar]
- Kosterin, O. Abyssinian pea (Lathyrus schaeferi Kosterin nom. Nov. pro Pisum abyssinicum A. Br.) is a problematic taxon. Vavilov J. Genet. Breed. 2017, 21, 158–169. [Google Scholar] [CrossRef]
- Davis, P.H. Lathyrus. In Flora of Turkey; Davis, P.H., Ed.; Edinburgh University Press: Edinburgh, Scottland, UK, 1970; Volume 3, pp. 328–369. ISBN 10 0852245599. [Google Scholar]
- Kupicha, F.K. Vicieae (Adans.) DC. (1825) nom conserv prop. In Advances in Legume Systematics; Polhill, R.M., Raven, P.H., Eds.; Royal Botanical Gardens: Kew, London, UK, 1981; Volume 1, pp. 377–381. ISBN 9780855212247. [Google Scholar]
- Schmalhausen, I. Flora Srednei y Yuzhnoj Rossii, Kryma i Severnogo Kavkaza; Botanicheskii Institut (Akademiia nauk SSSR): Kiev, Russia, 1895; Volume 1, p. 468. [Google Scholar]
- Ascheron, P.; Graebner, P. Synopsis der Mitteleuropaischen Flora Bd 6, Abt 2, IV; W. Engelmann: Leipzig, Germany, 1910. [Google Scholar]
- Boissier, E. Diagnoses plantarum orientalum novarum. Lipsie 1856, 3, 125. [Google Scholar]
- Miller, P. The Gardener’s Dictionary; Containing the Methods of Cultivating and Improving the Kitchen, Fruit and Flower Garden, 8th ed.; Printed by J. and J. Rivington, 1768; Verlag von J. Cramer: Leutershausen, Germany; London, UK, 1969. [Google Scholar]
- Berger, A. Systematic botany of peas and their allies. Peas of New York. In The Vegetables of New York; Hedrick, J.B., Ed.; Lyon Company: Albany, NY, USA, 1928; pp. 1–132. [Google Scholar]
- Lehmann, C. Das morphologische system der saaterbsen (Pisum sativum L. sensu lato Gov. subsp. sativum). Der Züchter 1954, 24, 316–337. [Google Scholar]
- Ladizinsky, G.; Abbo, S. The Search for Wild Relatives of Cool Season Legumes; Springer: Heidelberg, Germany, 2015. [Google Scholar]
- Bogdanova, V.S.; Mglinets, A.V.; Shatskaya, N.V.; Kosterin, O.E.; Solovyev, V.I.; Vasiliev, G.V. Cryptic divergences in the genus Pisum L. (peas), as revealed by phylogenetic analysis of plastid genomes. Mol. Phyl. Evol. 2018, 129, 280–290. [Google Scholar] [CrossRef] [PubMed]
- van Oss, R.; Abbo, S.; Eshed, R.; Sherman, A.; Coyne, C.J.; Vandemark, G.J.; Zhang, H.B.; Peleg, Z. Genetic relationship in Cicer sp. expose evidence for geneflow between the cultigen and its wild progenitor. PLoS ONE 2015, 10, e0139789. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.S.; DuVal, A.E.; Jensen, H.R. Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New Phytol. 2012, 196, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Song, C.; Saxena, R.K.; Azam, S.; Yu, S.; Sharpe, A.G.; Cannon, S.; Baek, J.; Rosen, B.D.; Tar’an, B.; et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013, 31, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Parween, S.; Nawaz, K.; Roy, R.; Pole, A.K.; Venkata Suresh, B.; Misra, G.; Jain, M.; Yadav, G.; Parida, S.K.; Tyagi, A.K.; et al. An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.). Sci. Rep. 2015, 5, 12806. [Google Scholar] [CrossRef] [PubMed]
- Caracuta, V.; Vardi, J.; Paz, Y.; Boaretto, E. Farming legumes in the pre-pottery Neolithic: New discoveries from the site of Ahihud (Israel). PLoS ONE 2017, 12, e0177859. [Google Scholar] [CrossRef] [PubMed]
- Abbo, S.; Rachamim, E.; Zehavi, Y.; Zezak, I.; Lev-Yadun, S.; Gopher, A. Experimental growing of wild pea in Israel and its bearing on Near Eastern plant domestication. Ann. Bot. 2011, 107, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Nelson, M.; Berger, J.; Smýkal, P.; Nelson, M.N.; Berger, J.D.; von Wettberg, E.J.B. The impact of genetic changes during crop domestication. Agronomy 2018, 8, 119. [Google Scholar] [CrossRef]
- Weeden, N.F. Genetic changes accompanying the domestication of Pisum sativum: Is there a common genetic basis to the “domestication syndrome” for legumes? Ann. Bot. 2007, 100, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Vieira, F.G.; Crawford, J.E.; Chu, C.; Nielsen, R. Asian wild rice is a hybrid swarm with extensive gene flow and feralization from domesticated rice. Genome Res. 2017, 27, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Li, L.F.; Li, Y.L.; Jia, Y.; Caicedo, A.L.; Olsen, K.M. Signatures of adaptation in the weedy rice genome. Nat. Genet. 2017, 49, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Abbo, S.; Lev-Yadun, S.; Heun, M.; Gopher, A. On the “lost” crops of the neolithic Near East. J. Exp. Bot. 2013, 64, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Young, J.P.W.; Matthews, P. A distinct class of peas (Pisum sativum L.) from Afghanistan that show strain specificity for symbiotic Rhizobium. Heredity 1982, 48, 203. [Google Scholar] [CrossRef]
- Lamprecht, H. Die Enstehung der Arten und hohhren Kaategorien; Springer: Wien, Germany, 1966. [Google Scholar]
- Gebreslassie, B.; Abraha, B. Review: Distribution and productivity of dekoko (Pisum sativum var. abyssinicum A. Braun) in Ethiopia. Glob. J. Sci. Front. Res. C Biol. Sci. 2016, 16, 1–14. [Google Scholar]
- Baranyi, M.; Greilhuber, J. Flow cytometric analysis of genome size variation in cultivated and wild Pisum sativum (Fabaceae). Plant Syst. Evol. 1995, 194, 231–239. [Google Scholar] [CrossRef]
- Kloz, J. Serology of the Leguminosae. In Chemotaxonomy of the Leguminosae; Harborne, J.B., Boulter, D., Turner, B.L., Eds.; Academic Press: London, UK, 1971; pp. 309–365. ISBN 10-0123246520. [Google Scholar]
- Holden, D. Genetic studies of wide crosses between cultivated pea species, and the domestication of Pisum abyssinicum. Ph.D. Thesis, School of Biological Sciences, University of East Anglia, John Innes Centre & Unilever Research, Colworth, UK, 2009. [Google Scholar]
- Claussen, M.; Kubatzki, C.; Brovkin, V.; Ganopolski, A.; Hoelzmann, P.; Pachur, H.J. Simulation of an abrupt change in Saharan vegetation at the end of the Mid-Holocene. Geophys. Res. Lett. 1999, 24, 2037–2040. [Google Scholar] [CrossRef]
- Weeden, N.; Wolko, B. Allozyme analysis of Pisum sativum ssp. abyssinicum and the development of a genotype definition for this subspecies. Pisum Genet. 2001, 33, 21–25. [Google Scholar]
- Conicella, C.; Errico, A. Karyotype Variations in Pisum Sativum Ect. Abyssinicum. Caryologia 1990, 43, 87–97. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).