Comparison of the Transcriptomic and Epigenetic Profiles of Gonadal Primordial Germ Cells of White Leghorn and Green-Legged Partridgelike Chicken Embryos
Abstract
1. Introduction
2. Materials and Methods
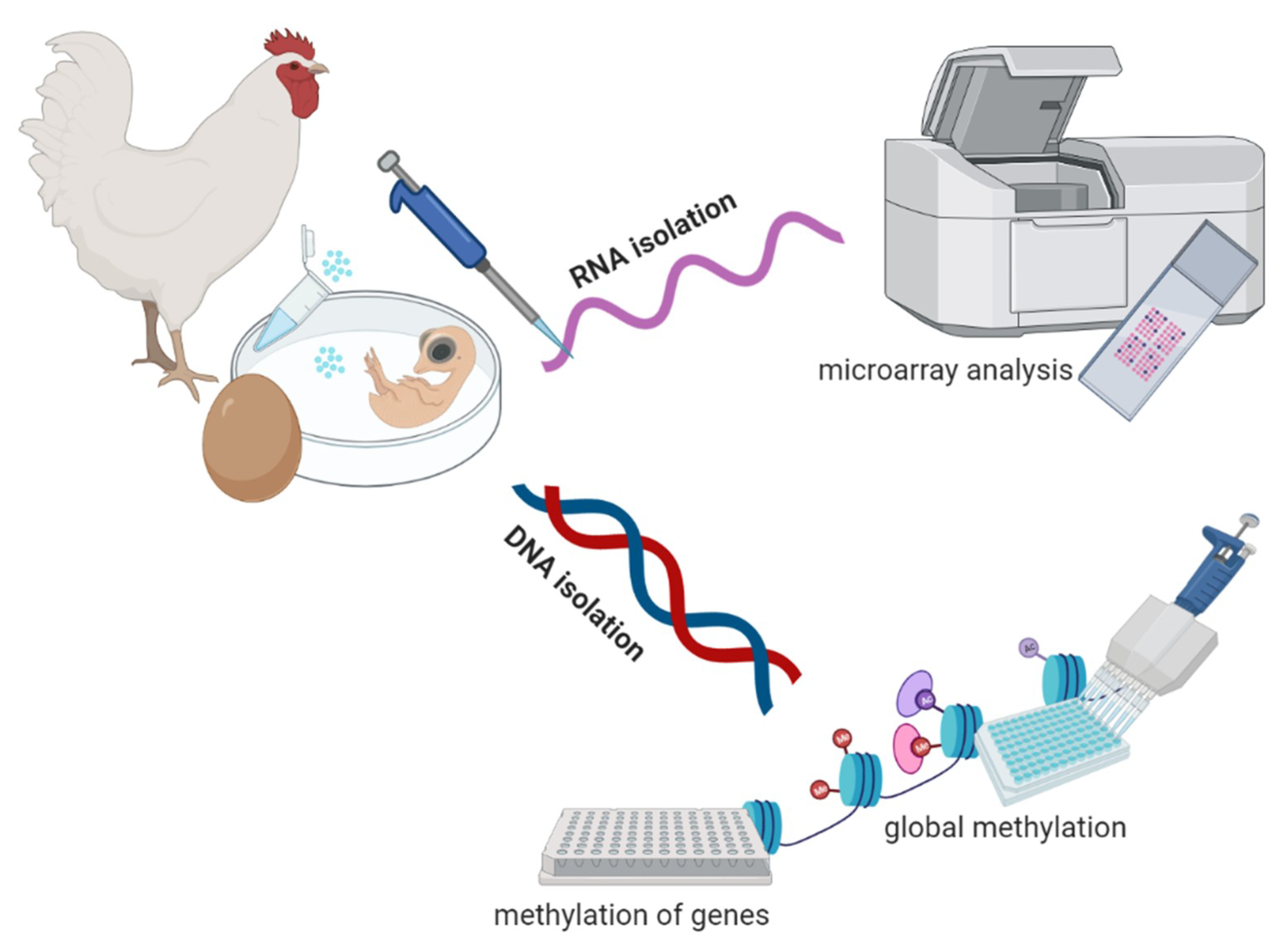
2.1. Gonadal PGC Isolation
2.2. RNA and DNA Isolation
2.3. Sex Determination of Embryos
2.4. Transcriptome Analysis
2.5. Global Methylation Analysis
2.6. Gene-Specific Methylation Analysis
3. Results
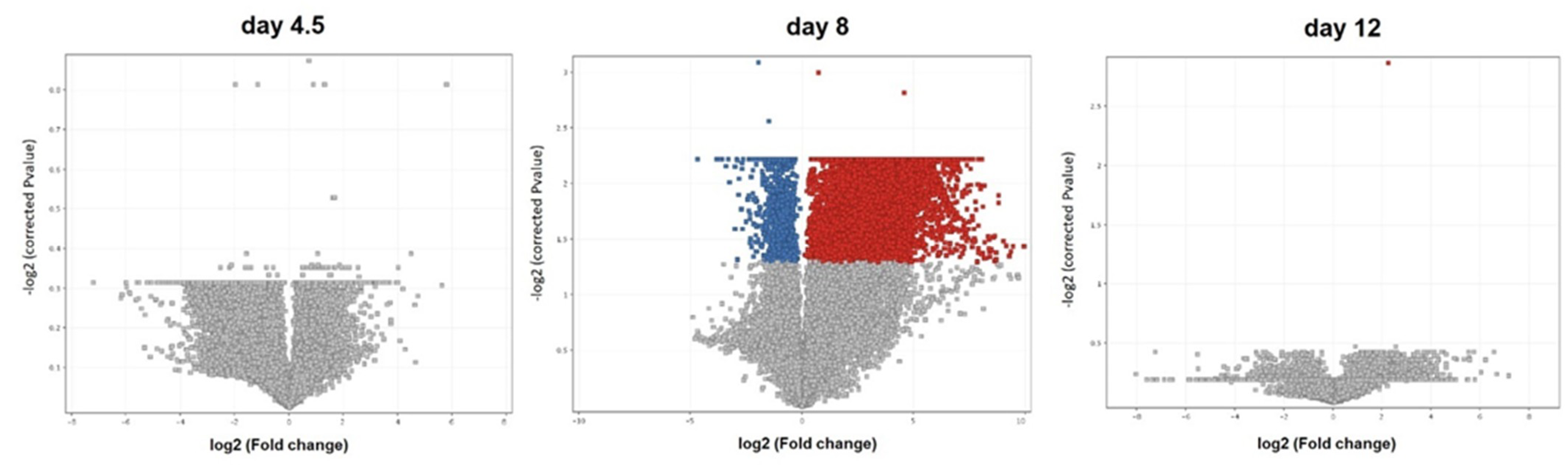
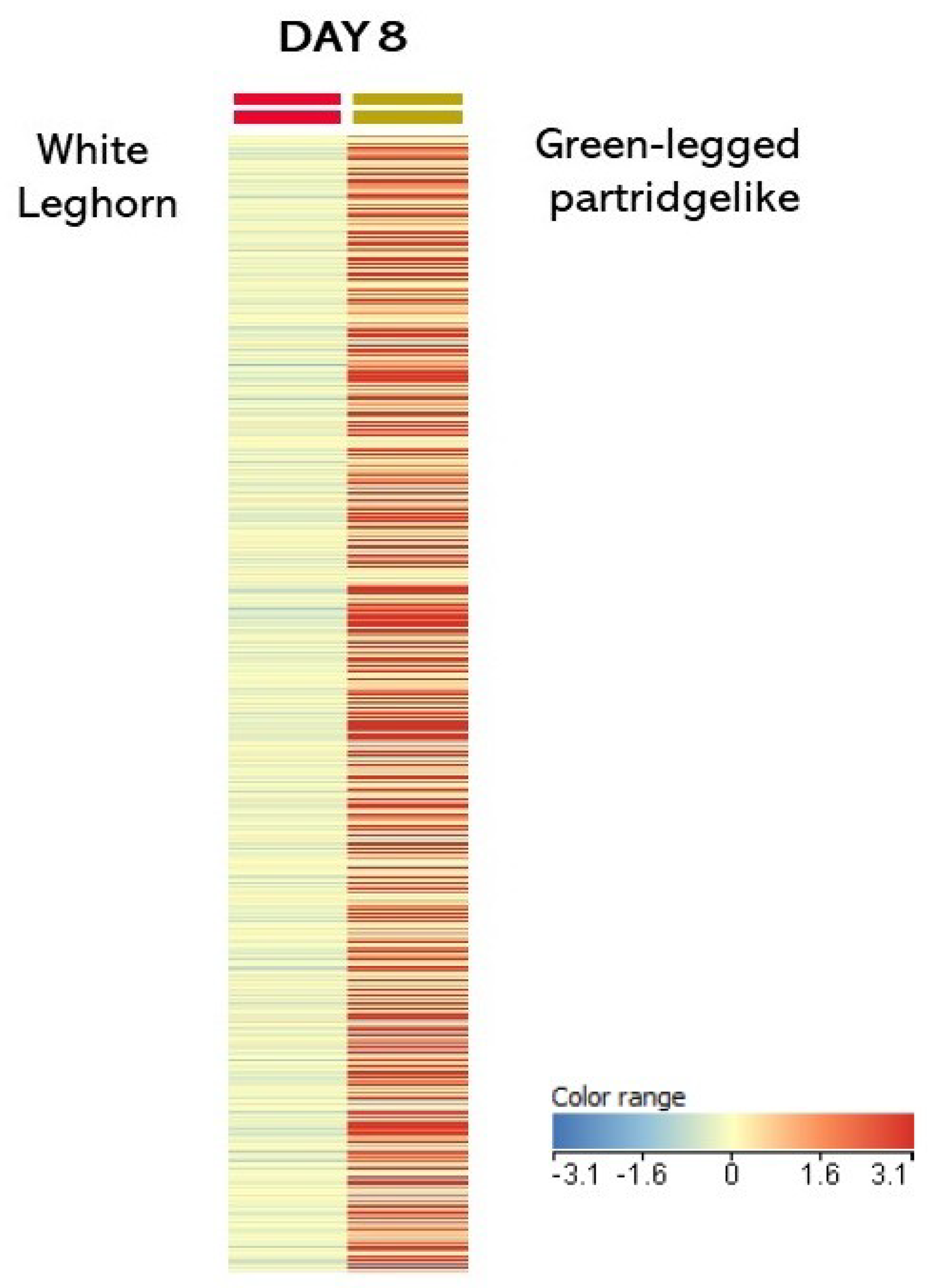
3.1. Transcriptome Analysis
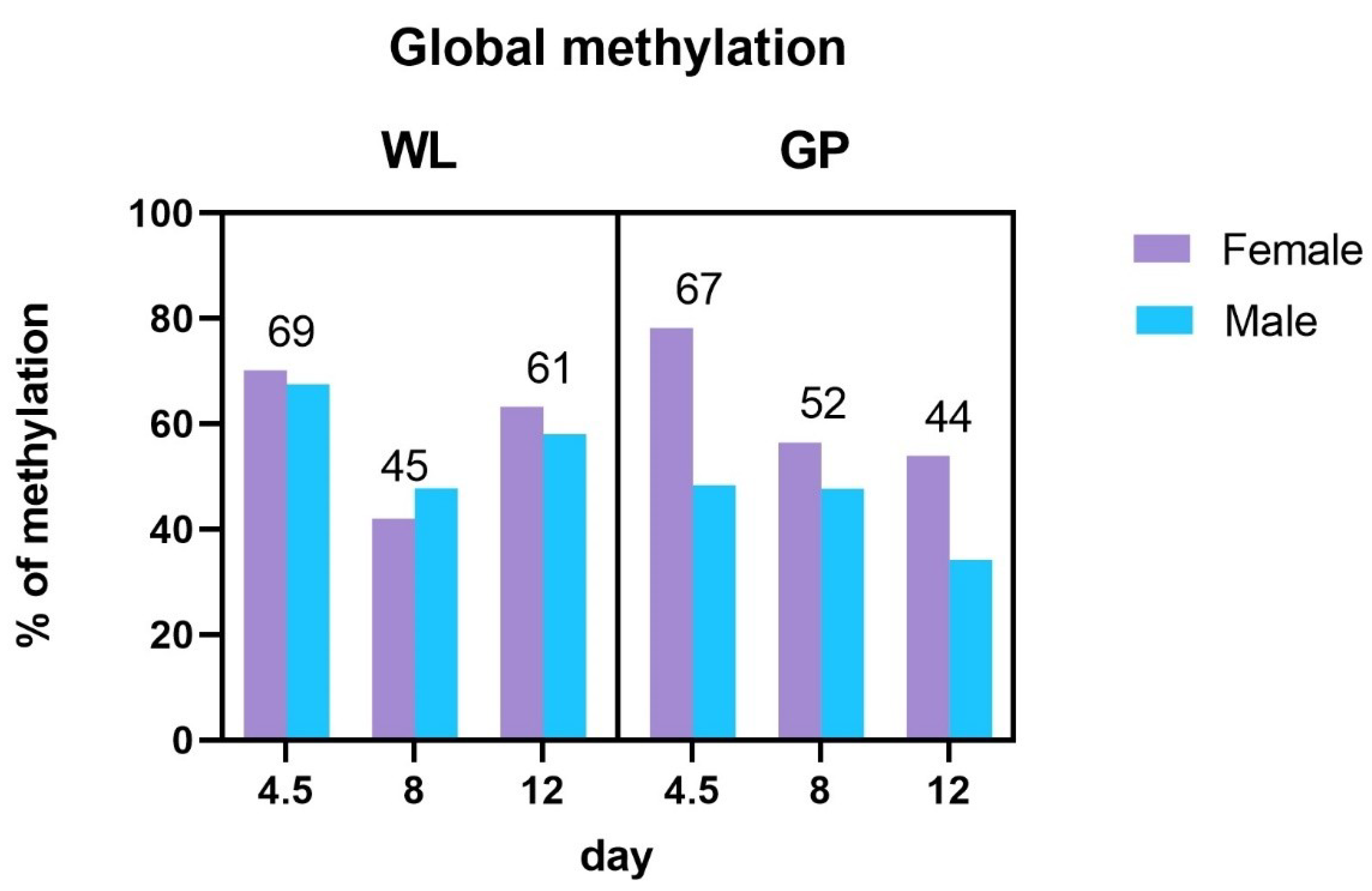
3.2. Global Methylation Analysis
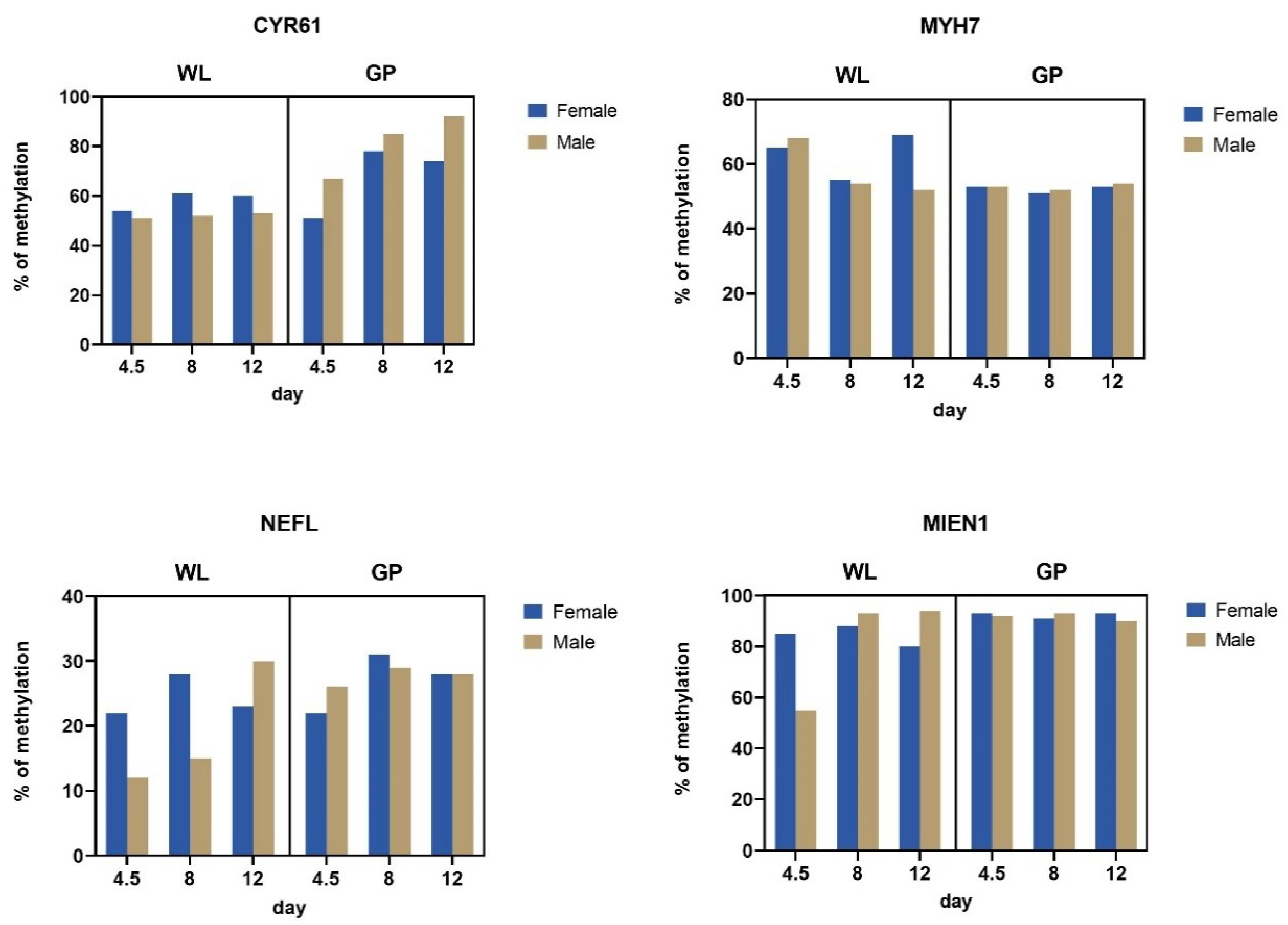
3.3. Methylation of Silenced Genes
4. Discussion
4.1. Transcriptome Analysis
4.2. Global Methylation Analysis
4.3. Gene-Specific Methylation Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buzała, M.; Janicki, B.; Czarnecki, R. Consequences of different growth rates in broiler breeder and layer hens on embryogenesis, metabolism and metabolic rate: A review. Poult. Sci. 2014, 94, 728–733. [Google Scholar] [CrossRef]
- Bednarczyk, M.; Rosinski, A. Comparison of egg hatchability and in vitro survival of goose embryos of various origins. Poult. Sci. 1999, 78, 579–585. [Google Scholar] [CrossRef]
- Sawicka, D.; Samek, K.; Chojnacka-Puchta, L.; Witkowski, A.; Knaga, S.; Debowska, M.; Bednarczyk, M. Changes in quail blastodermal cell status as a result of selection. Folia Biol. 2015, 63, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, R.; Pakdel, M.; Moradi Shabrbabak, M.; Moradi Shabrbabak, H. Divergent Selection Effect on Reproductive Trait in Japanese Quails. Res. J. Poult. Sci. 2013, 6, 18–22. [Google Scholar]
- Rashidi, H.; Sottile, V. The chick embryo: Hatching a model for contemporary biomedical research. BioEssays 2009, 31, 459–465. [Google Scholar] [CrossRef]
- Weeke-Klimp, A.; Bax, N.A.M.; Bellu, A.R.; Winter, E.M.; Vrolijk, J.; Plantinga, J.; Maas, S.; Brinker, M.; Mahtab, E.A.F.; Gittenberger-de Groot, A.C.; et al. Epicardium-derived cells enhance proliferation, cellular maturation and alignment of cardiomyocytes. J. Mol. Cell. Cardiol. 2010, 49, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Cogburn, L.A.; Wang, X.; Carre, W.; Rejto, L.; Porter, T.E.; Aggrey, S.E.; Simon, J. Systems-wide chicken DNA microarrays, gene expression profiling, and discovery of functional genes. Poult. Sci. 2003, 82, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D. Chicken chorioallantoic membrane angiogenesis model. Methods Mol. Biol. 2012, 843, 47–57. [Google Scholar]
- Dunislawska, A.; Slawinska, A.; Siwek, M. Hepatic DNA Methylation in Response to Early Stimulation of Microbiota with Lactobacillus Synbiotics in Broiler Chickens. Genes 2020, 11, 579. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, M.; Eyal-Giladi, H. Primordial Germ Cells of the Young Chick Blastoderm Originate from the Central Zone of the Area Pellucida Irrespective of the Embryo-Forming Process. Development 1987, 101, 209–219. [Google Scholar] [CrossRef]
- Chojnacka-Puchta, L.; Kasperczyk, K.; Płucienniczak, G.; Sawicka, D.; Bednarczyk, M. Primordial germ cells (PGCs) as a tool for creating transgenic chickens. Pol. J. Vet. Sci. 2012, 15, 181–188. [Google Scholar] [CrossRef]
- Mochizuki, K.; Matsui, Y. Epigenetic profiles in primordial germ cells: Global modulation and fine tuning of the epigenome for acquisition of totipotency. Dev. Growth Differ. 2010, 52, 517–525. [Google Scholar] [CrossRef]
- Dunislawska, A.; Szczerba, A.; Siwek, M.; Bednarczyk, M. Dynamics of the transcriptome during chicken embryo development based on primordial germ cells. BMC Res. Notes 2020, 13, 441. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Seo, H.W.; Lee, B.R.; Yoo, M.; Womack, J.E.; Han, J.Y. Gene expression and DNA methylation status of chicken primordial germ cells. Mol. Biotechnol. 2013, 54, 177–186. [Google Scholar] [CrossRef]
- Nakajima, Y.; Minematsu, T.; Naito, M.; Tajima, A. A New Method for Isolating Viable Gonadal Germ Cells from 7-day-old Chick Embryos. J. Poult. Sci. 2011, 48, 106–111. [Google Scholar] [CrossRef]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef] [PubMed]
- Clinton, M.; Haines, L.; Belloir, B.; McBride, D. Sexing chick embryos: A rapid and simple protocol. Br. Poult. Sci. 2001, 42, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Jawor, M.; Knaga, S.; Kozłowska, I.; Barna, J.; Váradi, É.; Kasperek, K.; Drobnyák, Á.; Bodzsár, N.; Várkonyi, E.P.; Jeżewska-Witkowska, G.; et al. Population Structure of Four Indigenous Chicken Breeds Undergoing In Situ Conservation. Anim. Sci. Pap. 2020, 38, 167–179. [Google Scholar]
- Szczerba, A.; Kuwana, T.; Bednarczyk, M. Concentration and total number of circulating primordial germ cells in Green-legged Partridgelike chicken embryos. Poult. Sci. 2021, 100, 319–324. [Google Scholar] [CrossRef]
- Siwek, M.; Wragg, D.; Sławińska, A.; Malek, M.; Hanotte, O.; Mwacharo, J.M. Insights into the genetic history of Green-legged Partridgelike fowl: MtDNA and genome-wide SNP analysis. Anim. Genet. 2013, 44, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Bednarczyk, M.; Lakota, P.; Slomski, R.; Plawski, A.; Lipinski, D.; Siemieniako, B.; Lisowski, M.; Czekalski, P.; Grajewski, B.; Dluzniewska, P. Reconstitution of a chicken breed by inter se mating of germline chimeric birds. Poult. Sci. 2002, 81, 1347–1353. [Google Scholar] [CrossRef]
- Kuwana, T.; Kawashima, T.; Naito, M.; Yamashita, H.; Matsuzaki, M.; Takano, T. Conservation of a Threatened Indigenous Fowl (Kureko Dori) Using the Germline Chimeras Transplanted from Primordial Germ Cells. J. Poult. Sci. 2006, 43, 60–66. [Google Scholar] [CrossRef][Green Version]
- Kostaman, T.; Yusuf, T.L.; Fahrudin, M.; Setiadi, M.A. Isolation and number of circulated primordial germ cells (circulated-PGCs) on stages of embryonic development of Gaok chicken. Jurnal Ilmu Ternak dan Veteriner 2013, 18, 54–62. [Google Scholar] [CrossRef]
- Sopiyana, S.; Setiadi, M.A.; Fahrudin, M.; Supriatna, I. Isolation and number of gonadal primordial germ cells (gonadal PGCs) on the stages of early embryonic development of KUB chicken. Media Peternak. 2017, 40, 1–6. [Google Scholar] [CrossRef][Green Version]
- Zhao, D.-F.; Yamashita, H.; Matsuzaki, M.; Takano, T.; Abe, S.-I.; Naito, M.; Kuwana, T. Genetic Factors Affect the Number of Circulating Primordial Germ Cells in Early Chick Embryos. J. Poult. Sci. 2003, 40, 101–113. [Google Scholar] [CrossRef][Green Version]
- Chojnacka-Puchta, L.; Sawicka, D.; Lakota, P.; Plucienniczak, G.; Bednarczyk, M.; Plucienniczak, A. Obtaining chicken primordial germ cells used for gene transfer: In vitro and in vivo results. J. Appl. Genet. 2015, 56, 493–504. [Google Scholar] [CrossRef]
- Kim, Y.M.; Han, J.Y. The early development of germ cells in chicken. Int. J. Dev. Biol. 2018, 62, 145–152. [Google Scholar] [CrossRef]
- Li, S.; Zhu, Y.; Zhi, L.; Han, X.; Shen, J.; Liu, Y.; Yao, J.; Yang, X. DNA Methylation Variation Trends during the Embryonic Development of Chicken. PLoS ONE 2016, 11, e0159230. [Google Scholar] [CrossRef]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef]
- Gryzinska, M.; Blaszczak, E.; Strachecka, A.; Jezewska-Witkowska, G. Analysis of age-related global DNA methylation in chicken. Biochem. Genet. 2013, 51, 554–563. [Google Scholar] [CrossRef]
- Romanov, G.A.; Vanyushin, B.F. Methylation of reiterated sequences in mammalian DNAs Effects of the tissue type, age, malignancy and hormonal induction. BBA Sect. Nucleic Acids Protein Synth. 1981, 653, 204–218. [Google Scholar] [CrossRef]
- Dunislawska, A.; Slawinska, A.; Gryzinska, M.; Siwek, M. Interaction between early in ovo stimulation of the gut microbiota and chicken host—Splenic changes in gene expression and methylation. J. Anim. Sci. Biotechnol. 2021, 12, 73. [Google Scholar] [CrossRef]
- Piprek, R.P.; Kolasa, M.; Podkowa, D.; Kloc, M.; Kubiak, J.Z. Transcriptional profiling validates involvement of extracellular matrix and proteinases genes in mouse gonad development. Mech. Dev. 2018, 149, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Dunislawska, A.; Slawinska, A.; Bednarczyk, M.; Siwek, M. Transcriptome modulation by in ovo delivered Lactobacillus synbiotics in a range of chicken tissues. Gene 2019, 698, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Martinsen, B.J. Reference guide to the stages of chick heart embryology. Dev. Dyn. 2005, 233, 1217–1237. [Google Scholar] [CrossRef] [PubMed]
- Karunanithi, S.; Venkatachalapathy, L.; Sudhalakshmi, Y. Comparative In Silico Analysis of Hypertrophic Cardiomyopathy Heart in Human and Normal Chicken Heart. J. Adv. Lab. Res. Biol. 2010, 1, 60–63. [Google Scholar]
- Xu, Z.; Che, T.; Li, F.; Tian, K.; Zhu, Q.; Mishra, S.K.; Dai, Y.; Li, M.; Li, D. The temporal expression patterns of brain transcriptome during chicken development and ageing. BMC Genom. 2018, 19, 917. [Google Scholar] [CrossRef] [PubMed]

| Gene (NCBI No.) | Primer Sequences | GC% | Amplicon Size | Reference | |
|---|---|---|---|---|---|
| CYR61 (429089) | M | F: TTTGGTTTTAGTGTTTAAAGACGT R: TTATATTTACCTTCAAAAAAACGTA | 58.33 44.00 | 150 | [9] |
| U | F: TTTTGGTTTTAGTGTTTAAAGATGT R: TATTTATATTTACCTTCAAAAAAACATA | 56.00 42.86 | 154 | ||
| MYH7 (395350) | M | F: AGGGTTTTGTTTCGTGTTTTATTC R: CTCCCCCATCTCTATAATAACGAT | 70.83 62.50 | 100 | This study |
| U | F: GGGTTTTGTTTTGTGTTTTATTTGT R: CCTCCCCCATCTCTATAATAACAAT | 76.00 64.00 | 100 | ||
| NEFL (419528) | M | F: TTTTTTGTATTCGGTGGATAGTTTC R: TAAAATCCTACAACTAAACCCGCT | 68.00 70.83 | 104 | This study |
| U | F: TTTTTGTATTTGGTGGATAGTTTTG R: TTAAAATCCTACAACTAAACCCACT | 68.00 68.00 | 104 | ||
| MIEN1 (100858225) | M | F: GGGGTAGTTGAGAGTTATACGT R:TACAAAATAATACAAAAAAAACGAC | 68.18 68.00 | 125 | This study |
| U | F: TGTGGGGTAGTTGAGAGTTATATGT R: TACAAAATAATACAAAAAAAACAAC | 64.00 68.00 | 128 | ||
| Breed | Day | CYR61 | MYH7 | NEFL | MIEN1 |
|---|---|---|---|---|---|
| WL | 8 vs. 4.5 | –3.26 | –2.37 | nd | nd |
| 12 vs. 4.5 | –2.34 | –3.91 | –4.19 | –2.52 | |
| GP | 8 vs. 4.5 | –1.78 | 1.30 | 0.37 | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunislawska, A.; Siwek, M.; Stadnicka, K.; Bednarczyk, M. Comparison of the Transcriptomic and Epigenetic Profiles of Gonadal Primordial Germ Cells of White Leghorn and Green-Legged Partridgelike Chicken Embryos. Genes 2021, 12, 1090. https://doi.org/10.3390/genes12071090
Dunislawska A, Siwek M, Stadnicka K, Bednarczyk M. Comparison of the Transcriptomic and Epigenetic Profiles of Gonadal Primordial Germ Cells of White Leghorn and Green-Legged Partridgelike Chicken Embryos. Genes. 2021; 12(7):1090. https://doi.org/10.3390/genes12071090
Chicago/Turabian StyleDunislawska, Aleksandra, Maria Siwek, Katarzyna Stadnicka, and Marek Bednarczyk. 2021. "Comparison of the Transcriptomic and Epigenetic Profiles of Gonadal Primordial Germ Cells of White Leghorn and Green-Legged Partridgelike Chicken Embryos" Genes 12, no. 7: 1090. https://doi.org/10.3390/genes12071090
APA StyleDunislawska, A., Siwek, M., Stadnicka, K., & Bednarczyk, M. (2021). Comparison of the Transcriptomic and Epigenetic Profiles of Gonadal Primordial Germ Cells of White Leghorn and Green-Legged Partridgelike Chicken Embryos. Genes, 12(7), 1090. https://doi.org/10.3390/genes12071090






