Heterochiasmy and Sexual Dimorphism: The Case of the Barn Swallow (Hirundo rustica, Hirundinidae, Aves)
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Chromosome Spreading and Staining
2.3. Chromosome Measurements and Generation of Recombination Maps
3. Results
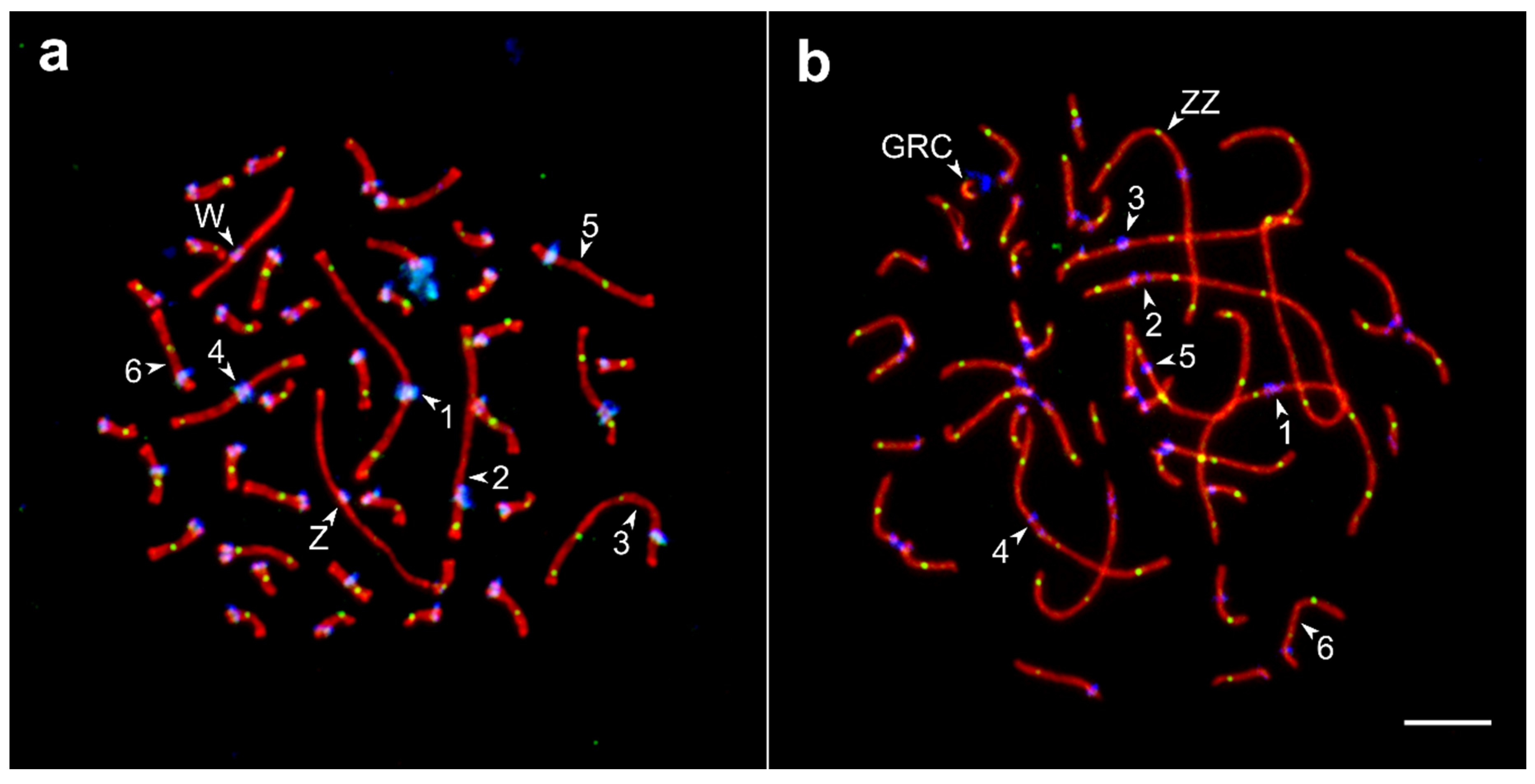
3.1. Pachytene Karyotype of the Barn Swallow
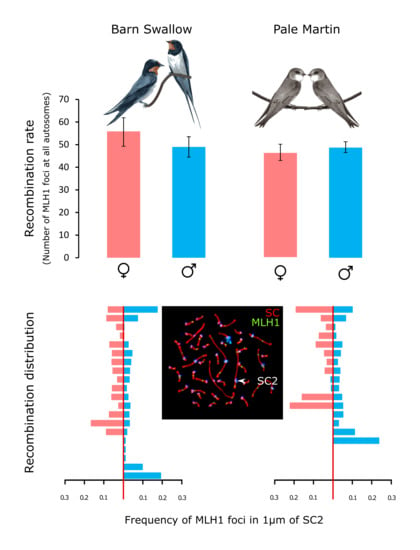
3.2. Recombination Rate
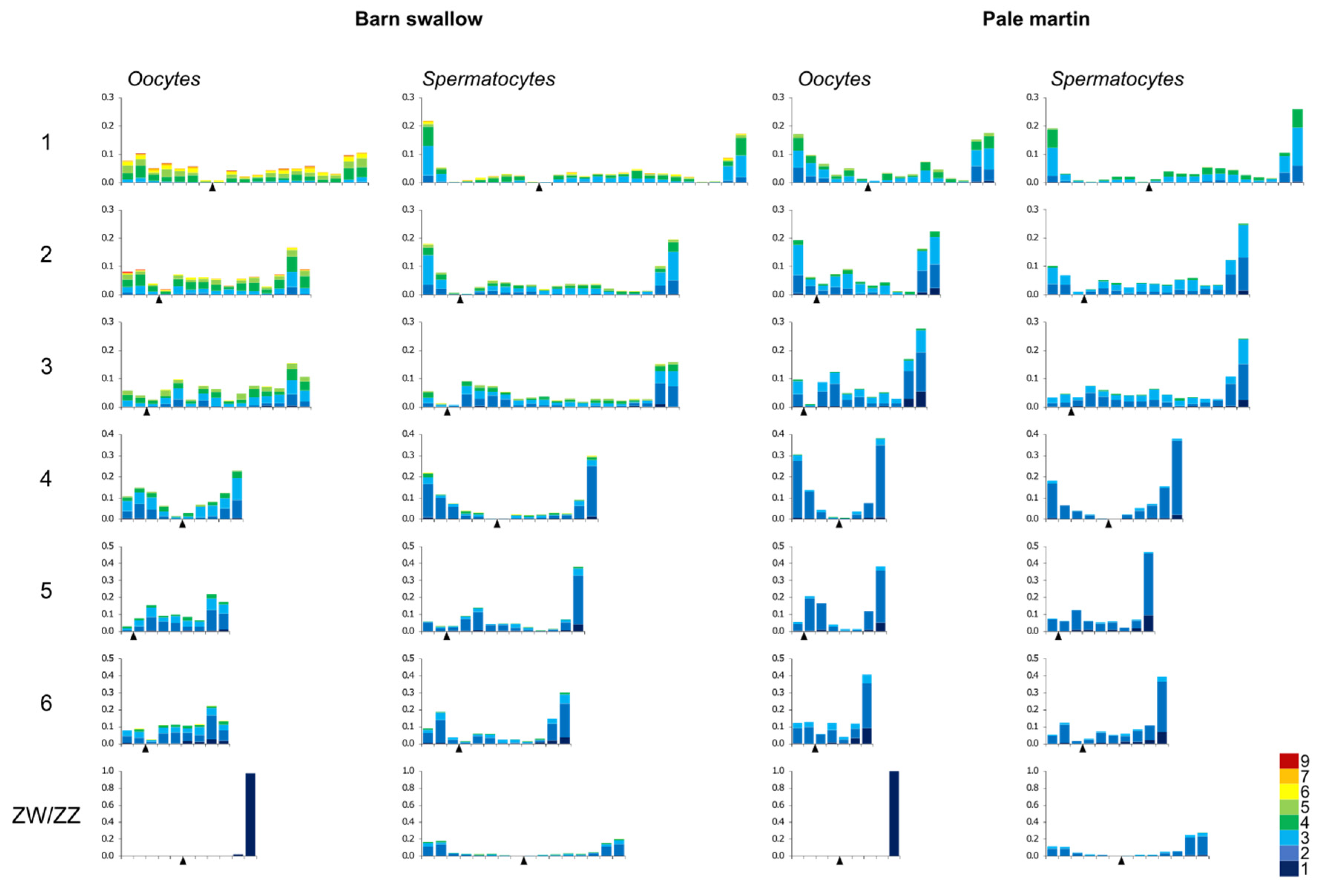
3.3. Recombination Distribution along the Macrochromosomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ritz, K.R.; Noor, M.A.F.; Singh, N.D. Variation in recombination rate: Adaptive or not? Trends Genet. 2017, 33, 364–374. [Google Scholar] [CrossRef]
- Stapley, J.; Feulner, P.G.D.; Johnston, S.E.; Santure, A.W.; Smadja, C.M. Variation in recombination frequency and distribution across eukaryotes: Patterns and processes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160455. [Google Scholar] [CrossRef]
- Lenormand, T.; Engelstädter, J.; Johnston, S.E.; Wijnker, E.; Haag, C.R. Evolutionary mysteries in meiosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Burt, A.; Bell, G.; Harvey, P.H. Sex-differences in recombination. J. Evol. Biol. 1991, 4, 259–277. [Google Scholar] [CrossRef]
- Lenormand, T.; Dutheil, J. Recombination difference between sexes: A role for haploid selection. PLoS Biol. 2005, 3, e63. [Google Scholar] [CrossRef]
- Brandvain, Y.; Coop, G. Scrambling eggs: Meiotic drive and the evolution of female recombination rates. Genetics 2012, 190, 709–723. [Google Scholar] [CrossRef]
- Gruhn, J.R.; Rubio, C.; Broman, K.W.; Hunt, P.A.; Hassold, T. Cytological studies of human meiosis: Sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS ONE 2013, 8, e85075. [Google Scholar] [CrossRef]
- Sardell, J.M.; Kirkpatrick, M. Sex differences in the recombination landscape. Am. Nat. 2020, 195, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Petkov, P.M.; Broman, K.W.; Szatkiewicz, J.P.; Paigen, K. Crossover interference underlies sex differences in recombination rates. Trends Genet. 2007, 23, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Haldane, J.B.S. Sex-ratio and unisexual sterility in hybrid animals. J. Genet. 1922, 12, 101. [Google Scholar] [CrossRef]
- Lenormand, T. The evolution of sex dimorphism in recombination. Genetics 2003, 163, 811. [Google Scholar] [PubMed]
- Mank, J.E. The evolution of heterochiasmy: The role of sexual selection and sperm competition in determining sex-specific recombination rates in eutherian mammals. Genet. Res. 2009, 91, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Haig, D. Games in tetrads: Segregation, recombination, and meiotic drive. Am. Nat. 2010, 176, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Trivers, R. Sex differences in rates of recombination and sexual selection. In The Evolution of Sex: An Examination of Current Ideas; Michod, R.E., Levin, B.R., Eds.; Sinauer Press: Sunderland, MA, USA, 1988; pp. 270–286. [Google Scholar]
- Hassold, T.; Sherman, S.; Hunt, P. Counting cross-overs: Characterizing meiotic recombination in mammals. Hum. Mol. Genet. 2000, 9, 2409–2419. [Google Scholar] [CrossRef] [PubMed]
- Torgasheva, A.A.; Borodin, P.M. Immunocytological analysis of meiotic recombination in the gray goose (Anser anser). Cytogenet. Genome Res. 2017, 151, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Møller, A.P. Sexual Selection and the Barn Swallow. Oxford Series in Ecology and Evolution; Oxford University Press: Oxford, UK, 1994; ISBN 9780198540281. [Google Scholar]
- Romano, A.; Costanzo, A.; Rubolini, D.; Saino, N.; Møller, A.P. Geographical and seasonal variation in the intensity of sexual selection in the barn swallow Hirundo rustica: A meta-analysis. Biol. Rev. 2017, 92, 1582–1600. [Google Scholar] [CrossRef]
- Pavlova, A.; Zink, R.M.; Drovetski, S.V.; Rohwer, S. Pleistocene evolution of closely related sand martins Riparia riparia and R. diluta. Mol. Phylogenet. Evol. 2008, 48, 61–73. [Google Scholar] [CrossRef]
- Sheldon, F.H.; Whittingham, L.A.; Moyle, R.G.; Slikas, B.; Winkler, D.W. Phylogeny of swallows (Aves: Hirundinidae) estimated from nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2005, 35, 254–270. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. Bold: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. Available online: http://www.barcodinglife.org (accessed on 4 September 2020). [CrossRef]
- Peters, A.H.; Plug, A.W.; van Vugt, M.J.; de Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosom. Res. 1997, 5, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.K.; Reeves, A.; Webb, L.M.; Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 1999, 151, 1569–1579. [Google Scholar] [PubMed]
- Reeves, A. MicroMeasure: A new computer program for the collection and analysis of cytogenetic data. Genome 2001, 44, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Malinovskaya, L.P.; Zadesenets, K.S.; Karamysheva, T.V.; Akberdina, E.A.; Kizilova, E.A.; Romanenko, M.V.; Shnaider, E.P.; Scherbakova, M.M.; Korobitsyn, I.G.; Rubtsov, N.B.; et al. Germline-restricted chromosome (GRC) in the sand martin and the pale martin (Hirundinidae, Aves): Synapsis, recombination and copy number variation. Sci. Rep. 2020, 10, 1058. [Google Scholar] [CrossRef]
- Torgasheva, A.A.; Malinovskaya, L.P.; Zadesenets, K.S.; Karamysheva, T.V.; Kizilova, E.A.; Akberdina, E.A.; Pristyazhnyuk, I.E.; Shnaider, E.P.; Volodkina, V.A.; Saifitdinova, A.F.; et al. Germline-restricted chromosome (GRC) is widespread among songbirds. Proc. Natl. Acad. Sci. USA 2019, 116, 11650–11845. [Google Scholar] [CrossRef]
- Owens, I.P.F.; Hartley, I.R. Sexual dimorphism in birds: Why are there so many different forms of dimorphism? Proc. R. Soc. Lond. Ser. B Biol. Sci. 1998, 265, 397–407. [Google Scholar] [CrossRef]
- Romano, A.; Saino, N.; Møller, A.P. Viability and expression of sexual ornaments in the barn swallow Hirundo rustica: A meta-analysis. J. Evol. Biol. 2017, 30, 1929–1935. [Google Scholar] [CrossRef]
- Parés-Casanova, P.M. An analysis of sexual size dimorphism in goose. Br. Poult. Sci. 2014, 55, 143–147. [Google Scholar] [CrossRef]
- Aslam, M.L.; Bastiaansen, J.W.M.; Crooijmans, R.P.M.A.; Vereijken, A.; Megens, H.J.; Groenen, M.A.M. A SNP based linkage map of the turkey genome reveals multiple intrachromosomal rearrangements between the Turkey and Chicken genomes. BMC Genom. 2010, 11, 647. [Google Scholar] [CrossRef]
- Hammond, J.C.; Marsden, S.J. Sexing Turkeys from Hatching to Maturity. Poult. Sci. 1937, 16, 287–288. [Google Scholar] [CrossRef]
- Pigozzi, M.I. Distribution of MLH1 foci on the synaptonemal complexes of chicken oocytes. Cytogenet. Cell Genet. 2001, 95, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Remes, V.; Szekely, T. Domestic chickens defy Rensch’s rule: Sexual size dimorphism in chicken breeds. J. Evol. Biol. 2010, 23, 2754–2759. [Google Scholar] [CrossRef]
- Groenen, M.A.M.; Wahlberg, P.; Foglio, M.; Cheng, H.H.; Megens, H.J.; Crooijmans, R.P.M.A.R.P.; Besnier, F.; Lathrop, M.; Muir, W.M.; Wong, G.K.S.; et al. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009, 19, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Calderon, P.L.; Pigozzi, M.I.; Calderón, P.L.; Pigozzi, M.I. MLH1-focus mapping in birds shows equal recombination between sexes and diversity of crossover patterns. Chromosom. Res. 2006, 14, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Huss, D.; Poynter, G.; Lansford, R. Japanese quail (Coturnix japonica) as a laboratory animal model. Lab Anim. 2008, 37, 513–519. [Google Scholar] [CrossRef]
- Pigozzi, M.I.; Solari, A.J. Equal frequencies of recombination nodules in both sexes of the pigeon suggest a basic difference with eutherian mammals. Genome 1999, 42, 315–321. [Google Scholar] [CrossRef]
- Horng, Y.-M.; Wu, C.-P.; Wang, Y.-C.; Huang, M.-C. A novel molecular genetic marker for gender determination of pigeons. Theriogenology 2006, 65, 1759–1768. [Google Scholar] [CrossRef]
- Jaari, S.; Li, M.-H.; Merilä, J. A first-generation microsatellite-based genetic linkage map of the Siberian jay (Perisoreus infaustus): Insights into avian genome evolution. BMC Genom. 2009, 10, 1. [Google Scholar] [CrossRef]
- Ekman, J.; Bylin, A.; TegelstrÎm, H. Increased lifetime reproductive success for Siberian jay (Perisoreus infaustus) males with delayed dispersal. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1999, 266, 911–915. [Google Scholar] [CrossRef]
- Møller, A.P. Sexual selection in the barn swallow (Hirundo rustica). IV. Patterns of fluctuating asymmetry and selection against asymmetry. Evolution 1994, 48, 658. [Google Scholar] [CrossRef]
- Åkesson, M.; Hansson, B.; Hasselquist, D.; Bensch, S. Linkage mapping of AFLP markers in a wild population of great reed warblers: Importance of heterozygosity and number of genotyped individuals. Mol. Ecol. 2007, 16, 2189–2202. [Google Scholar] [CrossRef] [PubMed]
- Forstmeier, W. Repertoire size, sexual selection, and offspring viability in the great reed warbler: Changing patterns in space and time. Behav. Ecol. 2004, 15, 555–563. [Google Scholar] [CrossRef]
- Hansson, B.; Ljungqvist, M.; Dawson, D.; Mueller, J.C.; Olano-Marin, J.; Ellegren, H.; Nilsson, J. Avian genome evolution: Insights from a linkage map of the blue tit (Cyanistes caeruleus). Heredity 2010, 104, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Andersson, S.; Örnborg, J.; Andersson, M. Ultraviolet sexual dimorphism and assortative mating in blue tits. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1998, 265, 445–450. [Google Scholar] [CrossRef]
- Backström, N.; Karaiskou, N.; Leder, E.H.; Gustafsson, L.; Primmer, C.R.; Qvarnström, A.; Ellegren, H. A gene-based genetic linkage map of the collared flycatcher (Ficedula albicollis) reveals extensive synteny and gene-order conservation during 100 million years of avian evolution. Genetics 2008, 179, 1479–1495. [Google Scholar] [CrossRef] [PubMed]
- Siitari, H. Individual color variation and male quality in pied flycatchers (Ficedula hypoleuca): A role of ultraviolet reflectance. Behav. Ecol. 2002, 13, 737–741. [Google Scholar] [CrossRef]
- Mak, S.-S.; Wrabel, A.; Nagai, H.; Ladher, R.K.; Sheng, G. Zebra finch as a developmental model. Genesis 2015, 53, 669–677. [Google Scholar] [CrossRef]
| Species | Sex | N Specimens | N Nuclei | MLH1 Foci Number | SC Length (µm) |
|---|---|---|---|---|---|
| Barn swallow | female | 3 | 182 | 55.6 ± 6.3 *§ | 184.8 ± 32.3 *§ |
| Barn swallow | male | 5 | 275 | 49.0 ± 4.5 § | 215.5 ± 33.8 § |
| Pale martin | female | 3 | 145 | 46.6 ± 3.6 *§ | 169.3 ± 22.2 *§ |
| Pale martin | male | 6 | 293 | 48.9 ± 2.4 | 210.3 ± 28.9 |
| MLH1 Foci Number | SC Length (µm) | |||||||
|---|---|---|---|---|---|---|---|---|
| SC | Barn Swallow | Pale Martin | Barn Swallow | Pale Martin | ||||
| Female | Male | Female | Male | Female | Male | Female | Male | |
| 1 | 4.4 ± 1.2 *§ | 3.4 ± 1.0 § | 2.9 ± 0.9 | 2.9 ± 0.8 | 18.6 ± 3.6 *§ | 25.3 ± 4.6 § | 15.1 ± 2.4 * | 19.8 ± 5.1 |
| 2 | 3.6 ± 1.1 *§ | 2.9 ± 0.9 § | 2.5 ± 0.8 | 2.4 ± 0.7 | 14.8 ± 2.8 *§ | 20.1 ± 3.9 § | 12.2 ± 2.1 * | 15.9 ± 4.0 |
| 3 | 3.1 ± 1.1 *§ | 2.6 ± 0.9 § | 2.1 ± 0.7 | 2.3 ± 0.7 | 14.3 ± 2.9 *§ | 19.8 ± 4.2 § | 11.5 ± 1.8 * | 15.9 ± 4.5 |
| 4 | 2.6 ± 0.8 *§ | 2.2 ± 0.7 § | 2.0 ± 0.4 | 2.0 ± 0.4 | 10.0 ± 1.7 *§ | 13.9 ± 3.3 § | 8.5 ± 1.1 * | 10.9 ± 2.1 |
| 5 | 2.3 ± 0.7 *§ | 2.0 ± 0.6 | 1.9 ± 0.4 | 1.8 ± 0.5 | 9.5 ± 1.6 *§ | 12.6 ± 2.9 § | 7.8 ± 1.1 * | 9.5 ± 1.7 |
| 6 | 2.1 ± 0.8 | 2.0 ± 0.6 § | 1.9 ± 0.6 | 1.8 ± 0.5 | 8.9 ± 1.6 *§ | 11.6 ± 2.5 § | 7.5 ± 1.0 * | 9.5 ± 1.7 |
| ZW/ZZ | 1.0 ± 0.0 * | 2.3 ± 0.7 | 1.0 ± 0.0* | 2.1 ± 0.5 | 13.1 ± 1.8 *§ | 15.7 ± 3.0 § | 10.2 ± 2.8 * | 13.6 ± 3.3 |
| Species | Genetic Map Length (cM) | References | Heterochiasmy Index | Sexual Dimorphism | |
|---|---|---|---|---|---|
| Female | Male | ||||
| Domestic goose (Anser anser) a | 3655 | 3030 | [16] | 0.19 | low (weight) [30] |
| Turkey (Meleagris gallopavo) b | 2077 | 2431 | [31] | −0.16 | high (weight, plumage) [32] |
| Domestic chicken (Gallus domesticus) a | 3310 | 3285 | [33] | 0.01 | high (weight, plumage) [34] |
| Domestic chicken (Gallus domesticus) b | 3098 | 3145 | [35] | −0.02 | high (weight, plumage) [34] |
| Japanese quail (Coturnix japonica) a | 2815 | 2815 | [36] | 0.00 | high (weight, plumage) [37] |
| Pigeon (Columba livia) c | 3135 | 3235 | [38] | −0.03 | low (size) [39] |
| Siberian jay (Perisoreus infaustus) b | 998 | 774 | [40] | 0.25 | none [41] |
| Pale martin (Riparia diluta) a | 2380 | 2450 | this paper | −0.03 | none [19] |
| Barn swallow (Hirundo rustica) a | 2815 | 2430 | this paper | 0.15 | moderate (tail length) [42] |
| Great reed warbler (Acrocephalus arundinaceus) b | 858 | 552 | [43] | 0.43 | none [44] |
| Blue tit (Parus caeruleus) b | 1046 | 887 b | [45] | 0.16 | low (plumage) [46] |
| Collared flycatcher (Ficedula albicollis) b | 1627 | 1982 | [47] | −0.20 | low (plumage) [48] |
| Zebra finch (Taeniopygia guttata) a | 2335 | 2310 | [36] | 0.01 | high (plumage) [49] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malinovskaya, L.P.; Tishakova, K.; Shnaider, E.P.; Borodin, P.M.; Torgasheva, A.A. Heterochiasmy and Sexual Dimorphism: The Case of the Barn Swallow (Hirundo rustica, Hirundinidae, Aves). Genes 2020, 11, 1119. https://doi.org/10.3390/genes11101119
Malinovskaya LP, Tishakova K, Shnaider EP, Borodin PM, Torgasheva AA. Heterochiasmy and Sexual Dimorphism: The Case of the Barn Swallow (Hirundo rustica, Hirundinidae, Aves). Genes. 2020; 11(10):1119. https://doi.org/10.3390/genes11101119
Chicago/Turabian StyleMalinovskaya, Lyubov P., Katerina Tishakova, Elena P. Shnaider, Pavel M. Borodin, and Anna A. Torgasheva. 2020. "Heterochiasmy and Sexual Dimorphism: The Case of the Barn Swallow (Hirundo rustica, Hirundinidae, Aves)" Genes 11, no. 10: 1119. https://doi.org/10.3390/genes11101119
APA StyleMalinovskaya, L. P., Tishakova, K., Shnaider, E. P., Borodin, P. M., & Torgasheva, A. A. (2020). Heterochiasmy and Sexual Dimorphism: The Case of the Barn Swallow (Hirundo rustica, Hirundinidae, Aves). Genes, 11(10), 1119. https://doi.org/10.3390/genes11101119