Membrane Transporters for Amino Acids as Players of Cancer Metabolic Rewiring
Abstract
1. Introduction
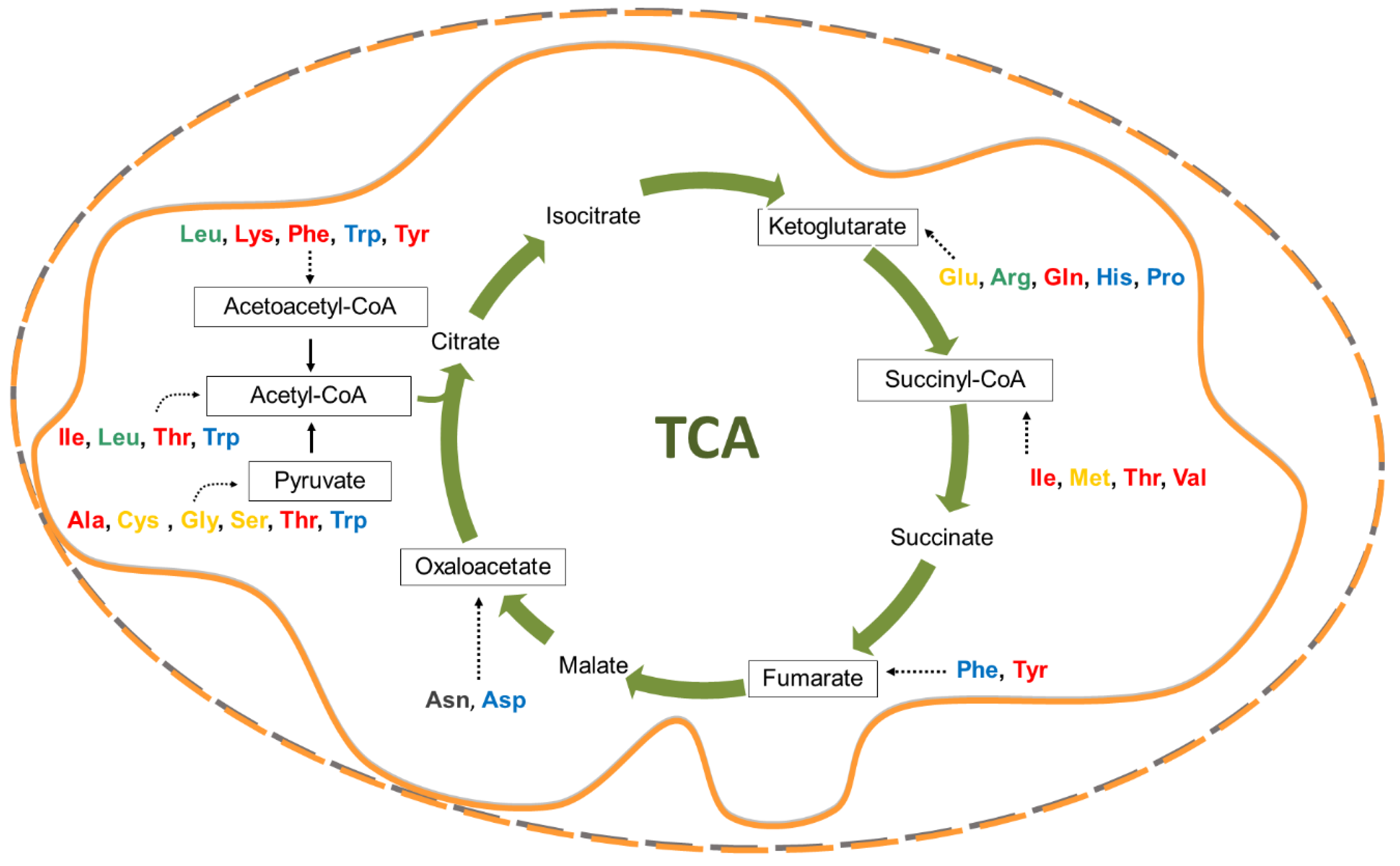
2. Crucial Amino Acids for Cancer Cells
2.1. Glutamine and Glutamate
2.2. Asparagine and Aspartate
2.3. Leucine and Other Essential Amino Acids
2.4. Cysteine, Methionine, Serine, Glycine and the One-Carbon Atom Metabolism
2.5. Proline as the Bridge Between TCA and Urea Cycle
2.6. Aspects of Amino Acids in Cancer Nutrition
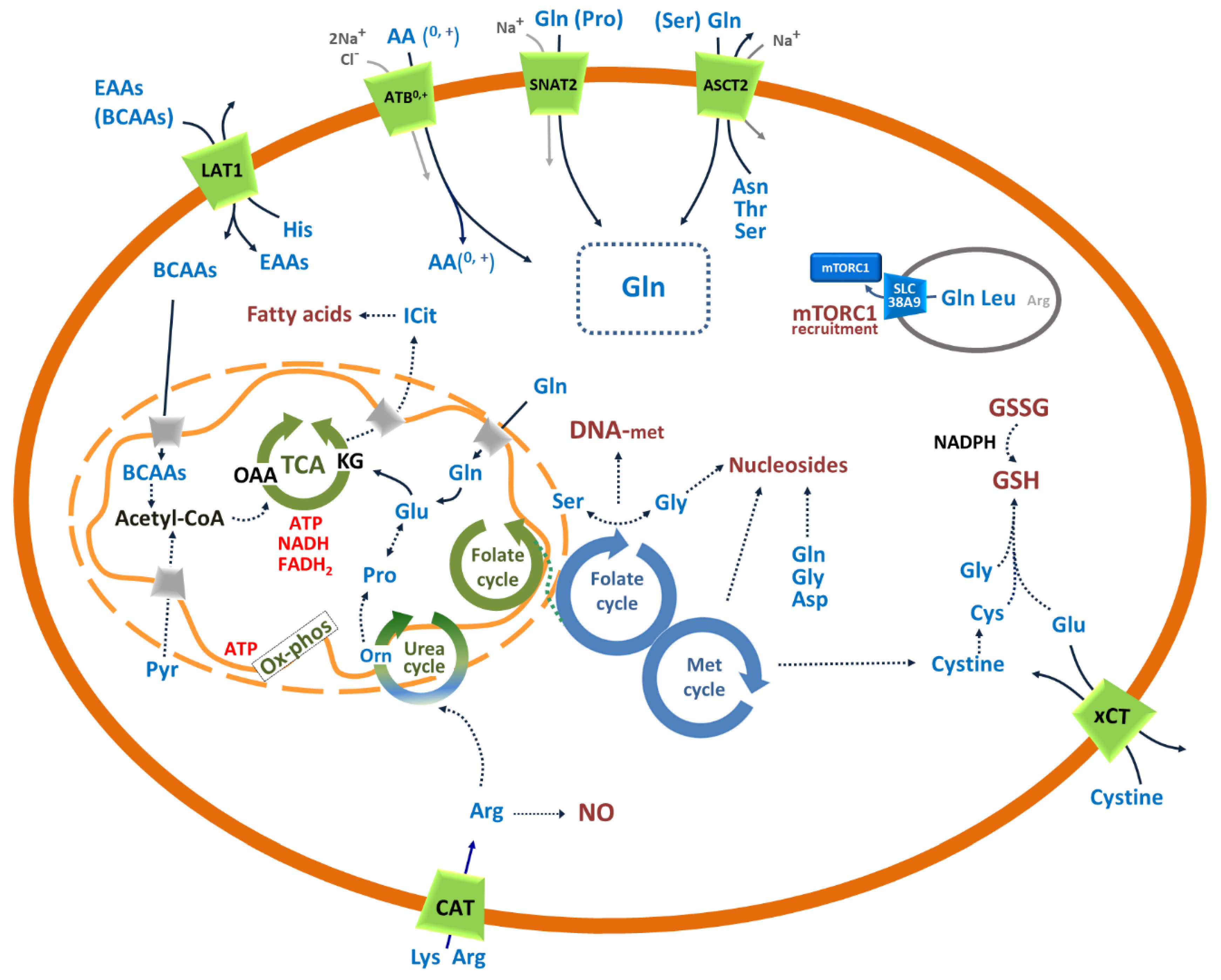
3. Amino Acid Transporters in Cancer
3.1. SLC1A5: Role in the Traffic of Glutamine and Other Neutral Amino Acids in Cancer
3.2. SLC6A14: Concentrative Transporter for Amino Acids in Cancer
3.3. SLC7A1, SLC7A2 and SLC7A3: Role in the Traffic of Cationic Amino Acids in Cancer
3.4. SLC7A5: Role in the Traffic of Essential Amino Acids in Cancer
3.5. xCT: Role in Managing the Redox Stress of Cancer Cells
3.6. SLC38A2: Concentrative Transporter of Glutamine and Other Neutral Amino Acids in Cancer
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ganapathy, V.; Thangaraju, M.; Prasad, P.D. Nutrient transporters in cancer: Relevance to Warburg hypothesis and beyond. Pharmacol. Ther. 2009, 121, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Alberghina, L.; Gaglio, D.; Moresco, R.M.; Gilardi, M.C.; Messa, C.; Vanoni, M. A systems biology road map for the discovery of drugs targeting cancer cell metabolism. Curr. Pharm. Des. 2014, 20, 2648–2666. [Google Scholar] [CrossRef]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar] [PubMed]
- Chen, J.Q.; Russo, J. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochim. Biophys. Acta 2012, 1826, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Vettore, L.; Westbrook, R.L.; Tennant, D.A. New aspects of amino acid metabolism in cancer. Br. J. Cancer 2020, 122, 150–156. [Google Scholar] [CrossRef]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef]
- Tsun, Z.Y.; Possemato, R. Amino acid management in cancer. Semin. Cell Dev. Biol. 2015, 43, 22–32. [Google Scholar] [CrossRef]
- Zhang, L.; Sui, C.; Yang, W.; Luo, Q. Amino acid transporters: Emerging roles in drug delivery for tumor-targeting therapy. Asian J. Pharm. Sci. 2020, 15, 192–206. [Google Scholar] [CrossRef]
- Bhutia, Y.D.; Ganapathy, V. Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim. Biophys. Acta 2016, 1863, 2531–2539. [Google Scholar] [CrossRef]
- Kandasamy, P.; Gyimesi, G.; Kanai, Y.; Hediger, M.A. Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 2018, 43, 752–789. [Google Scholar] [CrossRef]
- Broer, S.; Palacin, M. The role of amino acid transporters in inherited and acquired diseases. Biochem. J. 2011, 436, 193–211. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, S.M.; Gao, X.; Dai, Z.; Locasale, J.W. Methionine metabolism in health and cancer: A nexus of diet and precision medicine. Nat. Rev. Cancer 2019, 19, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Procopio, J.; Lima, M.M.; Pithon-Curi, T.C.; Curi, R. Glutamine and glutamate—their central role in cell metabolism and function. Cell Biochem. Funct. 2003, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bode, B.P. Recent molecular advances in mammalian glutamine transport. J. Nutr. 2001, 131, 2475–2485. [Google Scholar] [CrossRef]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. C-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef]
- Fung, M.K.L.; Chan, G.C. Drug-induced amino acid deprivation as strategy for cancer therapy. J. Hematol. Oncol. 2017, 10, 144. [Google Scholar] [CrossRef]
- Indiveri, C.; Abruzzo, G.; Stipani, I.; Palmieri, F. Identification and purification of the reconstitutively active glutamine carrier from rat kidney mitochondria. Biochem. J. 1998, 333, 285–290. [Google Scholar] [CrossRef]
- Yoo, H.C.; Park, S.J.; Nam, M.; Kang, J.; Kim, K.; Yeo, J.H.; Kim, J.K.; Heo, Y.; Lee, H.S.; Lee, M.Y.; et al. A Variant of SLC1A5 Is a Mitochondrial Glutamine Transporter for Metabolic Reprogramming in Cancer Cells. Cell Metab 2020, 31, 267–283.e212. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Galluccio, M.; Console, L.; Indiveri, C. Glutamine Transport and Mitochondrial Metabolism in Cancer Cell Growth. Front. Oncol. 2017, 7, 306. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011, 481, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Rohrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bai, C.; Ruan, Y.; Liu, M.; Chu, Q.; Qiu, L.; Yang, C.; Li, B. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat. Commun. 2019, 10, 201. [Google Scholar] [CrossRef]
- Krall, A.S.; Xu, S.; Graeber, T.G.; Braas, D.; Christofk, H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 2016, 7, 11457. [Google Scholar] [CrossRef]
- Lopes, A.M.; Oliveira-Nascimento, L.; Ribeiro, A.; Tairum, C.A., Jr.; Breyer, C.A.; Oliveira, M.A.; Monteiro, G.; Souza-Motta, C.M.; Magalhaes, P.O.; Avendano, J.G.; et al. Therapeutic l-asparaginase: Upstream, downstream and beyond. Crit. Rev. Biotechnol. 2017, 37, 82–99. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, J.; Venneti, S.; Cross, J.R.; Takagi, T.; Bhinder, B.; Djaballah, H.; Kanai, M.; Cheng, E.H.; Judkins, A.R.; et al. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol. Cell 2014, 56, 205–218. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef]
- Albano, F.; Chiurazzi, F.; Mimmi, S.; Vecchio, E.; Pastore, A.; Cimmino, C.; Frieri, C.; Iaccino, E.; Pisano, A.; Golino, G.; et al. The expression of inhibitor of bruton’s tyrosine kinase gene is progressively up regulated in the clinical course of chronic lymphocytic leukaemia conferring resistance to apoptosis. Cell Death Dis. 2018, 9, 13. [Google Scholar] [CrossRef]
- Rebsamen, M.; Pochini, L.; Stasyk, T.; de Araujo, M.E.; Galluccio, M.; Kandasamy, R.K.; Snijder, B.; Fauster, A.; Rudashevskaya, E.L.; Bruckner, M.; et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015, 519, 477–481. [Google Scholar] [CrossRef]
- Scalise, M.; Galluccio, M.; Pochini, L.; Cosco, J.; Trotta, M.; Rebsamen, M.; Superti-Furga, G.; Indiveri, C. Insights into the transport side of the human SLC38A9 transceptor. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Sabatini, D.M. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab. 2017, 26, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Broer, S.; Broer, A. Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem. J. 2017, 474, 1935–1963. [Google Scholar] [CrossRef] [PubMed]
- Yoneshiro, T.; Wang, Q.; Tajima, K.; Matsushita, M.; Maki, H.; Igarashi, K.; Dai, Z.; White, P.J.; McGarrah, R.W.; Ilkayeva, O.R.; et al. BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature 2019, 572, 614–619. [Google Scholar] [CrossRef]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef]
- Scalise, M.; Galluccio, M.; Console, L.; Pochini, L.; Indiveri, C. The Human SLC7A5 (LAT1): The Intriguing Histidine/Large Neutral Amino Acid Transporter and Its Relevance to Human Health. Front. Chem. 2018, 6, 243. [Google Scholar] [CrossRef]
- Panagiotidis, C.A.; Artandi, S.; Calame, K.; Silverstein, S.J. Polyamines alter sequence-specific DNA-protein interactions. Nucleic Acids Res. 1995, 23, 1800–1809. [Google Scholar] [CrossRef]
- Cinelli, M.A.; Do, H.T.; Miley, G.P.; Silverman, R.B. Inducible nitric oxide synthase: Regulation, structure, and inhibition. Med. Res. Rev. 2020, 40, 158–189. [Google Scholar] [CrossRef]
- Chanthaphavong, R.S.; Loughran, P.A.; Lee, T.Y.; Scott, M.J.; Billiar, T.R. A role for cGMP in inducible nitric-oxide synthase (iNOS)-induced tumor necrosis factor (TNF) alpha-converting enzyme (TACE/ADAM17) activation, translocation, and TNF receptor 1 (TNFR1) shedding in hepatocytes. J. Biol. Chem. 2012, 287, 35887–35898. [Google Scholar] [CrossRef]
- Lejeune, P.; Lagadec, P.; Onier, N.; Pinard, D.; Ohshima, H.; Jeannin, J.F. Nitric oxide involvement in tumor-induced immunosuppression. J. Immunol. 1994, 152, 5077–5083. [Google Scholar]
- Greene, L.I.; Bruno, T.C.; Christenson, J.L.; D’Alessandro, A.; Culp-Hill, R.; Torkko, K.; Borges, V.F.; Slansky, J.E.; Richer, J.K. A Role for Tryptophan-2,3-dioxygenase in CD8 T-cell Suppression and Evidence of Tryptophan Catabolism in Breast Cancer Patient Plasma. Mol. Cancer Res. 2019, 17, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, M.L.; Warren, M.B.; Toro, J.R.; Matrosova, V.; Glenn, G.; Turner, M.L.; Duray, P.; Merino, M.; Choyke, P.; Pavlovich, C.P.; et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell 2002, 2, 157–164. [Google Scholar] [CrossRef]
- Farber, S.; Diamond, L.K. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 1948, 238, 787–793. [Google Scholar] [CrossRef]
- Newman, A.C.; Maddocks, O.D.K. One-carbon metabolism in cancer. Br. J. Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, O.D.; Berkers, C.R.; Mason, S.M.; Zheng, L.; Blyth, K.; Gottlieb, E.; Vousden, K.H. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013, 493, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Labuschagne, C.F.; van den Broek, N.J.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Mastroberardino, L.; Spindler, B.; Pfeiffer, R.; Skelly, P.J.; Loffing, J.; Shoemaker, C.B.; Verrey, F. Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature 1998, 395, 288–291. [Google Scholar] [CrossRef]
- Pramod, A.B.; Foster, J.; Carvelli, L.; Henry, L.K. SLC6 transporters: Structure, function, regulation, disease association and therapeutics. Mol. Asp. Med. 2013, 34, 197–219. [Google Scholar] [CrossRef] [PubMed]
- Stover, P.; Schirch, V. Serine hydroxymethyltransferase catalyzes the hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate. J. Biol. Chem. 1990, 265, 14227–14233. [Google Scholar] [PubMed]
- Tsun, Z.Y.; Bar-Peled, L.; Chantranupong, L.; Zoncu, R.; Wang, T.; Kim, C.; Spooner, E.; Sabatini, D.M. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 2013, 52, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Hine, C.; Harputlugil, E.; Zhang, Y.; Ruckenstuhl, C.; Lee, B.C.; Brace, L.; Longchamp, A.; Trevino-Villarreal, J.H.; Mejia, P.; Ozaki, C.K.; et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 2015, 160, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Phang, J.M.; Liu, W.; Hancock, C.N.; Fischer, J.W. Proline metabolism and cancer: Emerging links to glutamine and collagen. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 71–77. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, C.; Patriarca, E.J.; Phang, J.M.; Minchiotti, G. Proline Metabolism in Tumor Growth and Metastatic Progression. Front. Oncol. 2020, 10, 776. [Google Scholar] [CrossRef]
- Scadden, D.T. Nice neighborhood: Emerging concepts of the stem cell niche. Cell 2014, 157, 41–50. [Google Scholar] [CrossRef]
- Weijin, F.; Zhibin, X.; Shengfeng, Z.; Xiaoli, Y.; Qijian, D.; Jiayi, L.; Qiumei, L.; Yilong, C.; Hua, M.; Deyun, L.; et al. The clinical significance of PYCR1 expression in renal cell carcinoma. Medicine 2019, 98, e16384. [Google Scholar] [CrossRef]
- Kanarek, N.; Petrova, B.; Sabatini, D.M. Dietary modifications for enhanced cancer therapy. Nature 2020, 579, 507–517. [Google Scholar] [CrossRef]
- Christensen, H.N. Role of amino acid transport and countertransport in nutrition and metabolism. Physiol. Rev. 1990, 70, 43–77. [Google Scholar] [CrossRef]
- Pochini, L.; Scalise, M.; Galluccio, M.; Indiveri, C. Membrane transporters for the special amino acid glutamine: Structure/function relationships and relevance to human health. Front. Chem. 2014, 2, 61. [Google Scholar] [CrossRef] [PubMed]
- Chiotellis, A.; Muller, A.; Weyermann, K.; Leutwiler, D.S.; Schibli, R.; Ametamey, S.M.; Kramer, S.D.; Mu, L. Synthesis and preliminary biological evaluation of O-2((2-[(18)F]fluoroethyl)methylamino)ethyltyrosine ([(18)F]FEMAET) as a potential cationic amino acid PET tracer for tumor imaging. Amino Acids 2014, 46, 1947–1959. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hardie, R.A.; Hoy, A.J.; van Geldermalsen, M.; Gao, D.; Fazli, L.; Sadowski, M.C.; Balaban, S.; Schreuder, M.; Nagarajah, R.; et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol. 2015, 236, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Sakamoto, S.; Matsushima, J.; Kimura, T.; Ueda, T.; Mizokami, A.; Kanai, Y.; Ichikawa, T. Up-Regulation of LAT1 during Antiandrogen Therapy Contributes to Progression in Prostate Cancer Cells. J. Urol. 2016, 195, 1588–1597. [Google Scholar] [CrossRef]
- Yanagisawa, N.; Satoh, T.; Hana, K.; Ichinoe, M.; Nakada, N.; Endou, H.; Okayasu, I.; Murakumo, Y. L-amino acid transporter 1 may be a prognostic marker for local progression of prostatic cancer under expectant management. Cancer Biomark. 2015, 15, 365–374. [Google Scholar] [CrossRef]
- Wang, Q.; Bailey, C.G.; Ng, C.; Tiffen, J.; Thoeng, A.; Minhas, V.; Lehman, M.L.; Hendy, S.C.; Buchanan, G.; Nelson, C.C.; et al. Androgen receptor and nutrient signaling pathways coordinate the demand for increased amino acid transport during prostate cancer progression. Cancer Res. 2011, 71, 7525–7536. [Google Scholar] [CrossRef]
- Lin, J.; Yang, T.; Peng, Z.; Xiao, H.; Jiang, N.; Zhang, L.; Ca, D.; Wu, P.; Pan, Q. SLC1A5 Silencing Inhibits Esophageal Cancer Growth via Cell Cycle Arrest and Apoptosis. Cell Physiol. Biochem. 2018, 48, 397. [Google Scholar] [CrossRef]
- Gupta, N.; Miyauchi, S.; Martindale, R.G.; Herdman, A.V.; Podolsky, R.; Miyake, K.; Mager, S.; Prasad, P.D.; Ganapathy, M.E.; Ganapathy, V. Upregulation of the amino acid transporter ATB0,+ (SLC6A14) in colorectal cancer and metastasis in humans. Biochim. Biophys. Acta 2005, 1741, 215–223. [Google Scholar] [CrossRef]
- Dai, R.; Peng, F.; Xiao, X.; Gong, X.; Jiang, Y.; Zhang, M.; Tian, Y.; Xu, Y.; Ma, J.; Li, M.; et al. Hepatitis B virus X protein-induced upregulation of CAT-1 stimulates proliferation and inhibits apoptosis in hepatocellular carcinoma cells. Oncotarget 2017, 8, 60962–60974. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, W.; Wang, J.; Yang, C.; Mao, H.; Fu, X.; Wu, Y.; Cai, J.; Han, J.; Xu, Z.; et al. Overexpression of arginine transporter CAT-1 is associated with accumulation of L-arginine and cell growth in human colorectal cancer tissue. PLoS ONE 2013, 8, e73866. [Google Scholar] [CrossRef]
- Iino, I.; Kikuchi, H.; Miyazaki, S.; Hiramatsu, Y.; Ohta, M.; Kamiya, K.; Kusama, Y.; Baba, S.; Setou, M.; Konno, H. Effect of miR-122 and its target gene cationic amino acid transporter 1 on colorectal liver metastasis. Cancer Sci. 2013, 104, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, W.; Liu, F.; Wang, Q.; Song, M.; Yu, Q.; Tang, K.; Teng, T.; Wu, D.; Wang, X.; et al. IMCA Induces Ferroptosis Mediated by SLC7A11 through the AMPK/mTOR Pathway in Colorectal Cancer. Oxid. Med. Cell Longev. 2020, 2020, 14. [Google Scholar] [CrossRef]
- Sugano, K.; Maeda, K.; Ohtani, H.; Nagahara, H.; Shibutani, M.; Hirakawa, K. Expression of xCT as a predictor of disease recurrence in patients with colorectal cancer. Anticancer Res. 2015, 35, 677–682. [Google Scholar] [PubMed]
- Coburn, L.A.; Singh, K.; Asim, M.; Barry, D.P.; Allaman, M.M.; Al-Greene, N.T.; Hardbower, D.M.; Polosukhina, D.; Williams, C.S.; Delgado, A.G.; et al. Loss of solute carrier family 7 member 2 exacerbates inflammation-associated colon tumorigenesis. Oncogene 2019, 38, 1067–1079. [Google Scholar] [CrossRef]
- Kou, L.; Yao, Q.; Sivaprakasam, S.; Luo, Q.; Sun, Y.; Fu, Q.; He, Z.; Sun, J.; Ganapathy, V. Dual targeting of l-carnitine-conjugated nanoparticles to OCTN2 and ATB(0,+) to deliver chemotherapeutic agents for colon cancer therapy. Drug Deliv. 2017, 24, 1338–1349. [Google Scholar] [CrossRef]
- Namikawa, M.; Kakizaki, S.; Kaira, K.; Tojima, H.; Yamazaki, Y.; Horiguchi, N.; Sato, K.; Oriuchi, N.; Tominaga, H.; Sunose, Y.; et al. Expression of amino acid transporters (LAT1, ASCT2 and xCT) as clinical significance in hepatocellular carcinoma. Hepatol. Res. 2015, 45, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, H.; Okabe, H.; Beppu, T.; Chikamoto, A.; Hayashi, H.; Imai, K.; Mima, K.; Nakagawa, S.; Ishimoto, T.; Miyake, K.; et al. Cystine/glutamic acid transporter is a novel marker for predicting poor survival in patients with hepatocellular carcinoma. Oncol. Rep. 2013, 29, 685–689. [Google Scholar] [CrossRef]
- Hassanein, M.; Qian, J.; Hoeksema, M.D.; Wang, J.; Jacobovitz, M.; Ji, X.; Harris, F.T.; Harris, B.K.; Boyd, K.L.; Chen, H.; et al. Targeting SLC1a5-mediated glutamine dependence in non-small cell lung cancer. Int. J. Cancer 2015, 137, 1587–1597. [Google Scholar] [CrossRef]
- Kaira, K.; Kawashima, O.; Endoh, H.; Imaizumi, K.; Goto, Y.; Kamiyoshihara, M.; Sugano, M.; Yamamoto, R.; Osaki, T.; Tanaka, S.; et al. Expression of amino acid transporter (LAT1 and 4F2hc) in pulmonary pleomorphic carcinoma. Hum. Pathol. 2019, 84, 142–149. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, X.; Zhao, Z.; Shan, X. LncRNA SLC7A11-AS1 Contributes to Lung Cancer Progression Through Facilitating TRAIP Expression by Inhibiting miR-4775. OncoTargets Ther. 2020, 13, 6295–6302. [Google Scholar] [CrossRef]
- Ji, X.; Qian, J.; Rahman, S.M.J.; Siska, P.J.; Zou, Y.; Harris, B.K.; Hoeksema, M.D.; Trenary, I.A.; Heidi, C.; Eisenberg, R.; et al. xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene 2018, 37, 5007–5019. [Google Scholar] [CrossRef] [PubMed]
- Babu, E.; Bhutia, Y.D.; Ramachandran, S.; Gnanaprakasam, J.P.; Prasad, P.D.; Thangaraju, M.; Ganapathy, V. Deletion of the amino acid transporter Slc6a14 suppresses tumour growth in spontaneous mouse models of breast cancer. Biochem. J. 2015, 469, 17–23. [Google Scholar] [CrossRef] [PubMed]
- McCracken, A.N.; Edinger, A.L. Targeting cancer metabolism at the plasma membrane by limiting amino acid access through SLC6A14. Biochem. J. 2015, 470, e17–e19. [Google Scholar] [CrossRef] [PubMed]
- van Geldermalsen, M.; Wang, Q.; Nagarajah, R.; Marshall, A.D.; Thoeng, A.; Gao, D.; Ritchie, W.; Feng, Y.; Bailey, C.G.; Deng, N.; et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 2016, 35, 3201–3208. [Google Scholar] [CrossRef] [PubMed]
- El Ansari, R.; Craze, M.L.; Miligy, I.; Diez-Rodriguez, M.; Nolan, C.C.; Ellis, I.O.; Rakha, E.A.; Green, A.R. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res. 2018, 20, 21. [Google Scholar] [CrossRef]
- Sikder, M.O.F.; Yang, S.; Ganapathy, V.; Bhutia, Y.D. The Na(+)/Cl(-)—Coupled, Broad-Specific, Amino Acid Transporter SLC6A14 (ATB(0,+)): Emerging Roles in Multiple Diseases and Therapeutic Potential for Treatment and Diagnosis. AAPS J. 2017, 20, 12. [Google Scholar] [CrossRef]
- Abdelmagid, S.A.; Rickard, J.A.; McDonald, W.J.; Thomas, L.N.; Too, C.K. CAT-1-mediated arginine uptake and regulation of nitric oxide synthases for the survival of human breast cancer cell lines. J. Cell Biochem. 2011, 112, 1084–1092. [Google Scholar] [CrossRef]
- Habib, E.; Linher-Melville, K.; Lin, H.X.; Singh, G. Expression of xCT and activity of system xc(-) are regulated by NRF2 in human breast cancer cells in response to oxidative stress. Redox Biol. 2015, 5, 33–42. [Google Scholar] [CrossRef]
- Morotti, M.; Bridges, E.; Valli, A.; Choudhry, H.; Sheldon, H.; Wigfield, S.; Gray, N.; Zois, C.E.; Grimm, F.; Jones, D.; et al. Hypoxia-induced switch in SNAT2/SLC38A2 regulation generates endocrine resistance in breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 12452–12461. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Khelifa, S.; Ratnikov, B.; Scott, D.A.; Feng, Y.; Parisi, F.; Ruller, C.; Lau, E.; Kim, H.; Brill, L.M.; et al. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell 2015, 27, 354–369. [Google Scholar] [CrossRef]
- Chen, M.C.; Hsu, L.L.; Wang, S.F.; Hsu, C.Y.; Lee, H.C.; Tseng, L.M. ROS Mediate xCT-Dependent Cell Death in Human Breast Cancer Cells under Glucose Deprivation. Cells 2020, 9, 1598. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Di, X.; Zhang, S.; Kan, Q.; Liu, H.; Lu, T.; Wang, Y.; Fu, Q.; Sun, J.; He, Z. Large amino acid transporter 1 mediated glutamate modified docetaxel-loaded liposomes for glioma targeting. Colloids Surf. B Biointerfaces 2016, 141, 260–267. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Lu, X.; Zhao, W.; Sun, T.; Zhang, Y.; Lu, Y.; Jiang, C. Amino Acid Metabolism Abnormity and Microenvironment Variation Mediated Targeting and Controlled Glioma Chemotherapy. Small 2016, 12, 5633–5645. [Google Scholar] [CrossRef] [PubMed]
- Marshall, A.D.; van Geldermalsen, M.; Otte, N.J.; Lum, T.; Vellozzi, M.; Thoeng, A.; Pang, A.; Nagarajah, R.; Zhang, B.; Wang, Q.; et al. ASCT2 regulates glutamine uptake and cell growth in endometrial carcinoma. Oncogenesis 2017, 6, e367. [Google Scholar] [CrossRef]
- Marshall, A.D.; van Geldermalsen, M.; Otte, N.J.; Anderson, L.A.; Lum, T.; Vellozzi, M.A.; Zhang, B.K.; Thoeng, A.; Wang, Q.; Rasko, J.E.; et al. LAT1 is a putative therapeutic target in endometrioid endometrial carcinoma. Int. J. Cancer 2016, 139, 2529–2539. [Google Scholar] [CrossRef]
- Bjersand, K.; Seidal, T.; Sundstrom-Poromaa, I.; Akerud, H.; Skirnisdottir, I. The clinical and prognostic correlation of HRNPM and SLC1A5 in pathogenesis and prognosis in epithelial ovarian cancer. PLoS ONE 2017, 12, e0179363. [Google Scholar] [CrossRef]
- Sun, T.; Bi, F.; Liu, Z.; Yang, Q. SLC7A2 serves as a potential biomarker and therapeutic target for ovarian cancer. Aging 2020, 12, 13281–13296. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, L.; An, H.; Chang, Y.; Zhang, W.; Zhu, Y.; Xu, L.; Xu, J. High expression of Solute Carrier Family 1, member 5 (SLC1A5) is associated with poor prognosis in clear-cell renal cell carcinoma. Sci. Rep. 2015, 5, 16954. [Google Scholar] [CrossRef]
- Liu, W.; Chen, H.; Wong, N.; Haynes, W.; Baker, C.M.; Wang, X. Pseudohypoxia induced by miR-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Lett. 2017, 394, 65–75. [Google Scholar] [CrossRef]
- Coothankandaswamy, V.; Cao, S.; Xu, Y.; Prasad, P.D.; Singh, P.K.; Reynolds, C.P.; Yang, S.; Ogura, J.; Ganapathy, V.; Bhutia, Y.D. Amino acid transporter SLC6A14 is a novel and effective drug target for pancreatic cancer. Br. J. Pharmacol. 2016, 173, 3292–3306. [Google Scholar] [CrossRef]
- Penheiter, A.R.; Erdogan, S.; Murphy, S.J.; Hart, S.N.; Felipe Lima, J.; Rakhshan Rohakhtar, F.; O’Brien, D.R.; Bamlet, W.R.; Wuertz, R.E.; Smyrk, T.C.; et al. Transcriptomic and Immunohistochemical Profiling of SLC6A14 in Pancreatic Ductal Adenocarcinoma. Biomed. Res. Int. 2015, 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.J.; Amendola, C.R.; Hollinshead, K.E.R.; Yu, Q.; Yamamoto, K.; Encarnacion-Rosado, J.; Rose, R.E.; LaRue, M.M.; Sohn, A.S.W.; Biancur, D.E.; et al. Selective Alanine Transporter Utilization Creates a Targetable Metabolic Niche in Pancreatic Cancer. Cancer Discov. 2020, 10, 1018–1037. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.H.; De Mello, R.A.; Yan, Q.L.; Wang, J.W.; Chen, Y.; Ye, Q.H.; Wang, Z.J.; Tang, H.J.; Huang, T. MiR-139-5p/SLC7A11 inhibits the proliferation, invasion and metastasis of pancreatic carcinoma via PI3K/Akt signaling pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165747. [Google Scholar] [CrossRef]
- Lo, M.; Ling, V.; Wang, Y.Z.; Gout, P.W. The xc- cystine/glutamate antiporter: A mediator of pancreatic cancer growth with a role in drug resistance. Br. J. Cancer 2008, 99, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, M.; Tao, Z.; Gao, S.; Li, Y.; Cao, Y.; Lu, C.; Zou, X. Effects of targeting SLC1A5 on inhibiting gastric cancer growth and tumor development in vitro and in vivo. Oncotarget 2017, 8, 76458–76467. [Google Scholar] [CrossRef] [PubMed]
- Kasai, N.; Sasakawa, A.; Hosomi, K.; Poh, T.W.; Chua, B.L.; Yong, W.P.; So, J.; Chan, S.L.; Soong, R.; Kono, K.; et al. Anti-tumor efficacy evaluation of a novel monoclonal antibody targeting neutral amino acid transporter ASCT2 using patient-derived xenograft mouse models of gastric cancer. Am. J. Transl. Res. 2017, 9, 3399–3410. [Google Scholar]
- Hayashi, K.; Anzai, N. Novel therapeutic approaches targeting L-type amino acid transporters for cancer treatment. World J. Gastrointest. Oncol. 2017, 9, 21–29. [Google Scholar] [CrossRef]
- Wang, J.; Fei, X.; Wu, W.; Chen, X.; Su, L.; Zhu, Z.; Zhou, Y. SLC7A5 Functions as a Downstream Target Modulated by CRKL in Metastasis Process of Gastric Cancer SGC-7901 Cells. PLoS ONE 2016, 11, e0166147. [Google Scholar] [CrossRef]
- Muto, Y.; Furihata, T.; Kaneko, M.; Higuchi, K.; Okunushi, K.; Morio, H.; Reien, Y.; Uesato, M.; Matsubara, H.; Anzai, N. Different Response Profiles of Gastrointestinal Cancer Cells to an L-Type Amino Acid Transporter Inhibitor, JPH203. Anticancer Res. 2019, 39, 159–165. [Google Scholar] [CrossRef]
- Li, N.; Ouyang, Y.; Chen, S.; Peng, C.; He, C.; Hong, J.; Yang, X.; Zhu, Y.; Lu, N.H. Integrative Analysis of Differential lncRNA/mRNA Expression Profiling in Helicobacter pylori Infection-Associated Gastric Carcinogenesis. Front. Microbiol. 2020, 11, 880. [Google Scholar] [CrossRef]
- Papin-Michault, C.; Bonnetaud, C.; Dufour, M.; Almairac, F.; Coutts, M.; Patouraux, S.; Virolle, T.; Darcourt, J.; Burel-Vandenbos, F. Study of LAT1 Expression in Brain Metastases: Towards a Better Understanding of the Results of Positron Emission Tomography Using Amino Acid Tracers. PLoS ONE 2016, 11, e0157139. [Google Scholar] [CrossRef]
- Gupta, N.; Prasad, P.D.; Ghamande, S.; Moore-Martin, P.; Herdman, A.V.; Martindale, R.G.; Podolsky, R.; Mager, S.; Ganapathy, M.E.; Ganapathy, V. Up-regulation of the amino acid transporter ATB(0,+) (SLC6A14) in carcinoma of the cervix. Gynecol. Oncol. 2006, 100, 8–13. [Google Scholar] [CrossRef]
- Broer, A.; Rahimi, F.; Broer, S. Deletion of Amino Acid Transporter ASCT2 (SLC1A5) Reveals an Essential Role for Transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to Sustain Glutaminolysis in Cancer Cells. J. Biol. Chem. 2016, 291, 13194–13205. [Google Scholar] [CrossRef] [PubMed]
- Cetindis, M.; Biegner, T.; Munz, A.; Teriete, P.; Reinert, S.; Grimm, M. Glutaminolysis and carcinogenesis of oral squamous cell carcinoma. Eur. Arch. Otorhinolaryngol. 2016, 273, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Honjo, H.; Kaira, K.; Miyazaki, T.; Yokobori, T.; Kanai, Y.; Nagamori, S.; Oyama, T.; Asao, T.; Kuwano, H. Clinicopathological significance of LAT1 and ASCT2 in patients with surgically resected esophageal squamous cell carcinoma. J. Surg. Oncol. 2016, 113, 381–389. [Google Scholar] [CrossRef]
- Nikkuni, O.; Kaira, K.; Toyoda, M.; Shino, M.; Sakakura, K.; Takahashi, K.; Tominaga, H.; Oriuchi, N.; Suzuki, M.; Iijima, M.; et al. Expression of Amino Acid Transporters (LAT1 and ASCT2) in Patients with Stage III/IV Laryngeal Squamous Cell Carcinoma. Pathol. Oncol. Res. 2015, 21, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.R.; Roh, J.L.; Lee, S.M.; Park, Y.; Cho, K.J.; Choi, S.H.; Nam, S.Y.; Kim, S.Y. Overexpression of cysteine-glutamate transporter and CD44 for prediction of recurrence and survival in patients with oral cavity squamous cell carcinoma. Head Neck 2018, 40, 2340–2346. [Google Scholar] [CrossRef]
- Hayashi, K.; Jutabha, P.; Maeda, S.; Supak, Y.; Ouchi, M.; Endou, H.; Fujita, T.; Chida, M.; Anzai, N. LAT1 acts as a crucial transporter of amino acids in human thymic carcinoma cells. J. Pharmacol. Sci. 2016, 132, 201–204. [Google Scholar] [CrossRef]
- Shimizu, A.; Kaira, K.; Kato, M.; Yasuda, M.; Takahashi, A.; Tominaga, H.; Oriuchi, N.; Nagamori, S.; Kanai, Y.; Oyama, T.; et al. Prognostic significance of L-type amino acid transporter 1 (LAT1) expression in cutaneous melanoma. Melanoma Res. 2015, 25, 399–405. [Google Scholar] [CrossRef]
- Galvan, I.; Inacio, A.; Danino, M.; Corbi-Llopis, R.; Monserrat, M.T.; Bernabeu-Wittel, J. High SLC7A11 expression in normal skin of melanoma patients. Cancer Epidemiol. 2019, 62. [Google Scholar] [CrossRef]
- Rosilio, C.; Nebout, M.; Imbert, V.; Griessinger, E.; Neffati, Z.; Benadiba, J.; Hagenbeek, T.; Spits, H.; Reverso, J.; Ambrosetti, D.; et al. L-type amino-acid transporter 1 (LAT1): A therapeutic target supporting growth and survival of T-cell lymphoblastic lymphoma/T-cell acute lymphoblastic leukemia. Leukemia 2015, 29, 1253–1266. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.; Pieh, D.; Echchannaoui, H.; Rupp, J.; Rajalingam, K.; Theobald, M.; Closs, E.I.; Munder, M. Cationic Amino Acid Transporter-1-Mediated Arginine Uptake Is Essential for Chronic Lymphocytic Leukemia Cell Proliferation and Viability. Front. Oncol. 2019, 9, 1268. [Google Scholar] [CrossRef] [PubMed]
- Mussai, F.; Egan, S.; Higginbotham-Jones, J.; Perry, T.; Beggs, A.; Odintsova, E.; Loke, J.; Pratt, G.; U, P.K.; Lo, A.; et al. Arginine dependence of acute myeloid leukemia blast proliferation: A novel therapeutic target. Blood 2015, 125, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Cimen Bozkus, C.; Elzey, B.D.; Crist, S.A.; Ellies, L.G.; Ratliff, T.L. Expression of Cationic Amino Acid Transporter 2 Is Required for Myeloid-Derived Suppressor Cell-Mediated Control of T Cell Immunity. J. Immunol. 2015, 195, 5237–5250. [Google Scholar] [CrossRef] [PubMed]
- Shiozaki, A.; Iitaka, D.; Ichikawa, D.; Nakashima, S.; Fujiwara, H.; Okamoto, K.; Kubota, T.; Komatsu, S.; Kosuga, T.; Takeshita, H.; et al. xCT, component of cysteine/glutamate transporter, as an independent prognostic factor in human esophageal squamous cell carcinoma. J. Gastroenterol. 2014, 49, 853–863. [Google Scholar] [CrossRef]
- Sorensen, M.F.; Heimisdottir, S.B.; Sorensen, M.D.; Mellegaard, C.S.; Wohlleben, H.; Kristensen, B.W.; Beier, C.P. High expression of cystine-glutamate antiporter xCT (SLC7A11) is an independent biomarker for epileptic seizures at diagnosis in glioma. J. Neurooncol. 2018, 138, 49–53. [Google Scholar] [CrossRef]
- Keen, J.R.; Swanger, S.A.; Traynelis, S.F.; Olson, J.J. The role of glutamate transport and SLC7A11 expression in tumor-associate seizures and survival in patients with malignant gliomas. Ann. Transl. Med. 2016, 4, S18. [Google Scholar] [CrossRef]
- Zhou, X.W.; Wang, X.; Yang, Y.; Luo, J.W.; Dong, H.; Liu, Y.H.; Mao, Q. Biomarkers related with seizure risk in glioma patients: A systematic review. Clin. Neurol. Neurosurg. 2016, 151, 113–119. [Google Scholar] [CrossRef]
- Robert, S.M.; Buckingham, S.C.; Campbell, S.L.; Robel, S.; Holt, K.T.; Ogunrinu-Babarinde, T.; Warren, P.P.; White, D.M.; Reid, M.A.; Eschbacher, J.M.; et al. SLC7A11 expression is associated with seizures and predicts poor survival in patients with malignant glioma. Sci. Transl. Med. 2015, 7, 289ra286. [Google Scholar] [CrossRef]
- Shen, L.; Qian, C.; Cao, H.; Wang, Z.; Luo, T.; Liang, C. Upregulation of the solute carrier family 7 genes is indicative of poor prognosis in papillary thyroid carcinoma. World J. Surg. Oncol. 2018, 16, 235. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, H.; Lian, M.; Yue, C.; Dong, G.; Jin, Y.; Li, R.; Wan, H.; Wang, R.; Wang, Y.; et al. SLC7A11, a component of cysteine/glutamate transporter, is a novel biomarker for the diagnosis and prognosis in laryngeal squamous cell carcinoma. Oncol. Rep. 2017, 38, 3019–3029. [Google Scholar] [CrossRef] [PubMed]
- Polewski, M.D.; Reveron-Thornton, R.F.; Cherryholmes, G.A.; Marinov, G.K.; Aboody, K.S. SLC7A11 Overexpression in Glioblastoma Is Associated with Increased Cancer Stem Cell-Like Properties. Stem Cells Dev. 2017, 26, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Polewski, M.D.; Reveron-Thornton, R.F.; Cherryholmes, G.A.; Marinov, G.K.; Cassady, K.; Aboody, K.S. Increased Expression of System xc- in Glioblastoma Confers an Altered Metabolic State and Temozolomide Resistance. Mol. Cancer Res. 2016, 14, 1229–1242. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, M.; Kaira, K.; Ohshima, Y.; Ishioka, N.S.; Shino, M.; Sakakura, K.; Takayasu, Y.; Takahashi, K.; Tominaga, H.; Oriuchi, N.; et al. Prognostic significance of amino-acid transporter expression (LAT1, ASCT2, and xCT) in surgically resected tongue cancer. Br. J. Cancer 2014, 110, 2506–2513. [Google Scholar] [CrossRef] [PubMed]
- Januchowski, R.; Zawierucha, P.; Andrzejewska, M.; Rucinski, M.; Zabel, M. Microarray-based detection and expression analysis of ABC and SLC transporters in drug-resistant ovarian cancer cell lines. Biomed. Pharmacother. 2013, 67, 240–245. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Console, L.; Losso, M.A.; Indiveri, C. The Human SLC1A5 (ASCT2) Amino Acid Transporter: From Function to Structure and Role in Cell Biology. Front. Cell Dev. Biol. 2018, 6, 96. [Google Scholar] [CrossRef]
- Kekuda, R.; Prasad, P.D.; Fei, Y.J.; Torres-Zamorano, V.; Sinha, S.; Yang-Feng, T.L.; Leibach, F.H.; Ganapathy, V. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J. Biol. Chem. 1996, 271, 18657–18661. [Google Scholar] [CrossRef]
- Broer, A.; Brookes, N.; Ganapathy, V.; Dimmer, K.S.; Wagner, C.A.; Lang, F.; Broer, S. The astroglial ASCT2 amino acid transporter as a mediator of glutamine efflux. J. Neurochem. 1999, 73, 2184–2194. [Google Scholar]
- Oppedisano, F.; Pochini, L.; Galluccio, M.; Cavarelli, M.; Indiveri, C. Reconstitution into liposomes of the glutamine/amino acid transporter from renal cell plasma membrane: Functional characterization, kinetics and activation by nucleotides. Biochim. Biophys. Acta 2004, 1667, 122–131. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Panni, S.; Pingitore, P.; Hedfalk, K.; Indiveri, C. Transport mechanism and regulatory properties of the human amino acid transporter ASCT2 (SLC1A5). Amino Acids 2014, 46, 2463–2475. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Pingitore, P.; Hedfalk, K.; Indiveri, C. Cysteine is not a substrate but a specific modulator of human ASCT2 (SLC1A5) transporter. FEBS Lett. 2015, 589, 3617–3623. [Google Scholar] [CrossRef] [PubMed]
- Deitmer, J.W.; Rose, C.R. pH regulation and proton signalling by glial cells. Prog. Neurobiol. 1996, 48, 73–103. [Google Scholar] [CrossRef]
- Oppedisano, F.; Pochini, L.; Galluccio, M.; Indiveri, C. The glutamine/amino acid transporter (ASCT2) reconstituted in liposomes: Electrical nature of the glutamine/glutamate antiport. Ital. J. Biochem. 2007, 56, 275–278. [Google Scholar] [PubMed]
- Scalise, M.; Mazza, T.; Pappacoda, G.; Pochini, L.; Cosco, J.; Rovella, F.; Indiveri, C. The Human SLC1A5 Neutral Amino Acid Transporter Catalyzes a pH-Dependent Glutamate/Glutamine Antiport, as Well. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Freidman, N.; Chen, I.; Wu, Q.; Briot, C.; Holst, J.; Font, J.; Vandenberg, R.; Ryan, R. Amino Acid Transporters and Exchangers from the SLC1A Family: Structure, Mechanism and Roles in Physiology and Cancer. Neurochem. Res. 2020, 45, 1268–1286. [Google Scholar] [CrossRef]
- Garaeva, A.A.; Oostergetel, G.T.; Gati, C.; Guskov, A.; Paulino, C.; Slotboom, D.J. Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nat. Struct. Mol. Biol. 2018, 25, 515–521. [Google Scholar] [CrossRef]
- Garaeva, A.A.; Slotboom, D.J. Elevator-type mechanisms of membrane transport. Biochem. Soc. Trans. 2020, 48, 1227–1241. [Google Scholar] [CrossRef]
- Console, L.; Scalise, M.; Tarmakova, Z.; Coe, I.R.; Indiveri, C. N-linked glycosylation of human SLC1A5 (ASCT2) transporter is critical for trafficking to membrane. Biochim. Biophys. Acta 2015, 1853, 1636–1645. [Google Scholar] [CrossRef]
- Bungard, C.I.; McGivan, J.D. Identification of the promoter elements involved in the stimulation of ASCT2 expression by glutamine availability in HepG2 cells and the probable involvement of FXR/RXR dimers. Arch. Biochem. Biophys. 2005, 443, 53–59. [Google Scholar] [CrossRef]
- Amaral, J.S.; Pinho, M.J.; Soares-da-Silva, P. Genomic regulation of intestinal amino acid transporters by aldosterone. Mol. Cell Biochem. 2008, 313, 1–10. [Google Scholar] [CrossRef]
- Reynolds, M.R.; Lane, A.N.; Robertson, B.; Kemp, S.; Liu, Y.; Hill, B.G.; Dean, D.C.; Clem, B.F. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene 2014, 33, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Ge, M.; Hu, J.; Li, X.; Che, L.; Sun, K.; Cheng, L.; Huang, Y.; Pilo, M.G.; Cigliano, A.; et al. A functional mammalian target of rapamycin complex 1 signaling is indispensable for c-Myc-driven hepatocarcinogenesis. Hepatology 2017, 66, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Broer, A.; Gauthier-Coles, G.; Rahimi, F.; van Geldermalsen, M.; Dorsch, D.; Wegener, A.; Holst, J.; Broer, S. Ablation of the ASCT2 (SLC1A5) gene encoding a neutral amino acid transporter reveals transporter plasticity and redundancy in cancer cells. J. Biol. Chem. 2019, 294, 4012–4026. [Google Scholar] [CrossRef] [PubMed]
- Bolzoni, M.; Chiu, M.; Accardi, F.; Vescovini, R.; Airoldi, I.; Storti, P.; Todoerti, K.; Agnelli, L.; Missale, G.; Andreoli, R.; et al. Dependence on glutamine uptake and glutamine addiction characterize myeloma cells: A new attractive target. Blood 2016, 128, 667–679. [Google Scholar] [CrossRef]
- Nakanishi, T.; Tamai, I. Solute carrier transporters as targets for drug delivery and pharmacological intervention for chemotherapy. J. Pharm. Sci. 2011, 100, 3731–3750. [Google Scholar] [CrossRef] [PubMed]
- Sloan, J.L.; Mager, S. Cloning and functional expression of a human Na(+) and Cl(−)—dependent neutral and cationic amino acid transporter B(0+). J. Biol. Chem. 1999, 274, 23740–23745. [Google Scholar] [CrossRef]
- Nakanishi, T.; Kekuda, R.; Fei, Y.J.; Hatanaka, T.; Sugawara, M.; Martindale, R.G.; Leibach, F.H.; Prasad, P.D.; Ganapathy, V. Cloning and functional characterization of a new subtype of the amino acid transport system N. Am. J. Physiol. Cell Physiol. 2001, 281, C1757–C1768. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Palazzolo, L.; Paravicini, C.; Laurenzi, T.; Adobati, S.; Saporiti, S.; Guerrini, U.; Gianazza, E.; Indiveri, C.; Anderson, C.M.H.; Thwaites, D.T.; et al. SLC6A14, a Pivotal Actor on Cancer Stage: When Function Meets Structure. SLAS Discov. 2019, 24, 928–938. [Google Scholar] [CrossRef]
- Samluk, L.; Czeredys, M.; Nalecz, K.A. Regulation of amino acid/carnitine transporter B 0,+ (ATB 0,+) in astrocytes by protein kinase C: Independent effects on raft and non-raft transporter subpopulations. J. Neurochem. 2010, 115, 1386–1397. [Google Scholar] [CrossRef]
- Vale, N.; Ferreira, A.; Matos, J.; Fresco, P.; Gouveia, M.J. Amino Acids in the Development of Prodrugs. Molecules 2018, 23, 2318. [Google Scholar] [CrossRef]
- Closs, E.I.; Boissel, J.P.; Habermeier, A.; Rotmann, A. Structure and function of cationic amino acid transporters (CATs). J. Membr. Biol. 2006, 213, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Fotiadis, D.; Kanai, Y.; Palacin, M. The SLC3 and SLC7 families of amino acid transporters. Mol. Asp. Med. 2013, 34, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Closs, E.I.; Graf, P.; Habermeier, A.; Cunningham, J.M.; Forstermann, U. Human cationic amino acid transporters hCAT-1, hCAT-2A, and hCAT-2B: Three related carriers with distinct transport properties. Biochemistry 1997, 36, 6462–6468. [Google Scholar] [CrossRef] [PubMed]
- Kavanaugh, M.P. Voltage dependence of facilitated arginine flux mediated by the system y+ basic amino acid transporter. Biochemistry 1993, 32, 5781–5785. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.; Sawamura, T.; Masaki, T.; MacLeod, C.L. Increased Cat3-mediated cationic amino acid transport functionally compensates in Cat1 knockout cell lines. J. Biol. Chem. 1998, 273, 14663–14666. [Google Scholar] [CrossRef]
- Bhattacharyya, S.N.; Habermacher, R.; Martine, U.; Closs, E.I.; Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006, 125, 1111–1124. [Google Scholar] [CrossRef]
- Lowman, X.H.; Hanse, E.A.; Yang, Y.; Ishak Gabra, M.B.; Tran, T.Q.; Li, H.; Kong, M. p53 Promotes Cancer Cell Adaptation to Glutamine Deprivation by Upregulating Slc7a3 to Increase Arginine Uptake. Cell Rep. 2019, 26, 3051–3060.e3054. [Google Scholar] [CrossRef]
- Duelli, R.; Enerson, B.E.; Gerhart, D.Z.; Drewes, L.R. Expression of large amino acid transporter LAT1 in rat brain endothelium. J. Cereb. Blood Flow Metab. 2000, 20, 1557–1562. [Google Scholar] [CrossRef]
- Ohgaki, R.; Ohmori, T.; Hara, S.; Nakagomi, S.; Kanai-Azuma, M.; Kaneda-Nakashima, K.; Okuda, S.; Nagamori, S.; Kanai, Y. Essential Roles of L-Type Amino Acid Transporter 1 in Syncytiotrophoblast Development by Presenting Fusogenic 4F2hc. Mol. Cell Biol. 2017, 37. [Google Scholar] [CrossRef]
- Milkereit, R.; Persaud, A.; Vanoaica, L.; Guetg, A.; Verrey, F.; Rotin, D. LAPTM4b recruits the LAT1-4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat. Commun. 2015, 6, 7250. [Google Scholar] [CrossRef]
- Napolitano, L.; Scalise, M.; Galluccio, M.; Pochini, L.; Albanese, L.M.; Indiveri, C. LAT1 is the transport competent unit of the LAT1/CD98 heterodimeric amino acid transporter. Int. J. Biochem. Cell Biol. 2015, 67, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Tarlungeanu, D.C.; Deliu, E.; Dotter, C.P.; Kara, M.; Janiesch, P.C.; Scalise, M.; Galluccio, M.; Tesulov, M.; Morelli, E.; Sonmez, F.M.; et al. Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder. Cell 2016, 167, 1481–1494.e1418. [Google Scholar] [CrossRef] [PubMed]
- Cascio, L.; Chen, C.F.; Pauly, R.; Srikanth, S.; Jones, K.; Skinner, C.D.; Stevenson, R.E.; Schwartz, C.E.; Boccuto, L. Abnormalities in the genes that encode Large Amino Acid Transporters increase the risk of Autism Spectrum Disorder. Mol. Genet. Genomic Med. 2020, 8, e1036. [Google Scholar] [CrossRef] [PubMed]
- Pantham, P.; Rosario, F.J.; Weintraub, S.T.; Nathanielsz, P.W.; Powell, T.L.; Li, C.; Jansson, T. Down-Regulation of Placental Transport of Amino Acids Precedes the Development of Intrauterine Growth Restriction in Maternal Nutrient Restricted Baboons. Biol. Reprod. 2016, 95, 98. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, E.; Sato, M.; Yang, H.; Miyagawa, F.; Harasaki, M.; Tomita, K.; Matsuoka, S.; Noma, A.; Iwai, K.; Minato, N. 4F2 (CD98) heavy chain is associated covalently with an amino acid transporter and controls intracellular trafficking and membrane topology of 4F2 heterodimer. J. Biol. Chem. 1999, 274, 3009–3016. [Google Scholar] [CrossRef]
- Cantor, J.M.; Ginsberg, M.H. CD98 at the crossroads of adaptive immunity and cancer. J. Cell Sci. 2012, 125, 1373–1382. [Google Scholar] [CrossRef]
- Napolitano, L.; Galluccio, M.; Scalise, M.; Parravicini, C.; Palazzolo, L.; Eberini, I.; Indiveri, C. Novel insights into the transport mechanism of the human amino acid transporter LAT1 (SLC7A5). Probing critical residues for substrate translocation. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 727–736. [Google Scholar] [CrossRef]
- Yan, R.; Zhao, X.; Lei, J.; Zhou, Q. Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 2019, 568, 127–130. [Google Scholar] [CrossRef]
- Lee, Y.; Wiriyasermkul, P.; Jin, C.; Quan, L.; Ohgaki, R.; Okuda, S.; Kusakizako, T.; Nishizawa, T.; Oda, K.; Ishitani, R.; et al. Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc. Nat. Struct. Mol. Biol. 2019, 26, 510–517. [Google Scholar] [CrossRef]
- Dickens, D.; Chiduza, G.N.; Wright, G.S.; Pirmohamed, M.; Antonyuk, S.V.; Hasnain, S.S. Modulation of LAT1 (SLC7A5) transporter activity and stability by membrane cholesterol. Sci. Rep. 2017, 7, 43580. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Sawa, R.; Wake, I.; Morimoto, A.; Okimura, Y. Glucose-mediated inactivation of AMP-activated protein kinase reduces the levels of L-type amino acid transporter 1 mRNA in C2C12 cells. Nutr. Res. 2017, 47, 13–20. [Google Scholar] [CrossRef]
- Matsuyama, R.; Tomi, M.; Akanuma, S.; Tabuchi, A.; Kubo, Y.; Tachikawa, M.; Hosoya, K. Up-regulation of L-type amino acid transporter 1 (LAT1) in cultured rat retinal capillary endothelial cells in response to glucose deprivation. Drug Metab. Pharmacokinet. 2012, 27, 317–324. [Google Scholar] [CrossRef]
- Kobayashi, N.; Okazaki, S.; Sampetrean, O.; Irie, J.; Itoh, H.; Saya, H. Author Correction: CD44 variant inhibits insulin secretion in pancreatic beta cells by attenuating LAT1-mediated amino acid uptake. Sci. Rep. 2020, 10, 6084. [Google Scholar] [CrossRef]
- Shin, G.; Kang, T.W.; Yang, S.; Baek, S.J.; Jeong, Y.S.; Kim, S.Y. GENT: Gene expression database of normal and tumor tissues. Cancer Inform. 2011, 10, 149–157. [Google Scholar] [CrossRef]
- Hayashi, K.; Jutabha, P.; Endou, H.; Anzai, N. c-Myc is crucial for the expression of LAT1 in MIA Paca-2 human pancreatic cancer cells. Oncol. Rep. 2012, 28, 862–866. [Google Scholar] [CrossRef]
- Janpipatkul, K.; Suksen, K.; Borwornpinyo, S.; Jearawiriyapaisarn, N.; Hongeng, S.; Piyachaturawat, P.; Chairoungdua, A. Downregulation of LAT1 expression suppresses cholangiocarcinoma cell invasion and migration. Cell Signal. 2014, 26, 1668–1679. [Google Scholar] [CrossRef]
- Shi, L.; Luo, W.; Huang, W.; Huang, S.; Huang, G. Downregulation of L-type amino acid transporter 1 expression inhibits the growth, migration and invasion of gastric cancer cells. Oncol. Lett. 2013, 6, 106–112. [Google Scholar] [CrossRef]
- Sato, H.; Tamba, M.; Ishii, T.; Bannai, S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 1999, 274, 11455–11458. [Google Scholar] [CrossRef]
- Combs, J.A.; DeNicola, G.M. The Non-Essential Amino Acid Cysteine Becomes Essential for Tumor Proliferation and Survival. Cancers 2019, 11, 678. [Google Scholar] [CrossRef]
- Bannai, S. Exchange of cystine and glutamate across plasma membrane of human fibroblasts. J. Biol. Chem. 1986, 261, 2256–2263. [Google Scholar]
- Lewerenz, J.; Hewett, S.J.; Huang, Y.; Lambros, M.; Gout, P.W.; Kalivas, P.W.; Massie, A.; Smolders, I.; Methner, A.; Pergande, M.; et al. The cystine/glutamate antiporter system x(c)(-) in health and disease: From molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 2013, 18, 522–555. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascon, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Ye, P.; Mimura, J.; Okada, T.; Sato, H.; Liu, T.; Maruyama, A.; Ohyama, C.; Itoh, K. Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition. Mol. Cell Biol. 2014, 34, 3421–3434. [Google Scholar] [CrossRef] [PubMed]
- Shih, A.Y.; Erb, H.; Sun, X.; Toda, S.; Kalivas, P.W.; Murphy, T.H. Cystine/glutamate exchange modulates glutathione supply for neuroprotection from oxidative stress and cell proliferation. J. Neurosci. 2006, 26, 10514–10523. [Google Scholar] [CrossRef]
- Banjac, A.; Perisic, T.; Sato, H.; Seiler, A.; Bannai, S.; Weiss, N.; Kolle, P.; Tschoep, K.; Issels, R.D.; Daniel, P.T.; et al. The cystine/cysteine cycle: A redox cycle regulating susceptibility versus resistance to cell death. Oncogene 2008, 27, 1618–1628. [Google Scholar] [CrossRef]
- Lien, E.C.; Ghisolfi, L.; Geck, R.C.; Asara, J.M.; Toker, A. Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef]
- Koppula, P.; Zhang, Y.; Zhuang, L.; Gan, B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. 2018, 38. [Google Scholar] [CrossRef]
- Timmerman, L.A.; Holton, T.; Yuneva, M.; Louie, R.J.; Padro, M.; Daemen, A.; Hu, M.; Chan, D.A.; Ethier, S.P.; van’t Veer, L.J.; et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 2013, 24, 450–465. [Google Scholar] [CrossRef]
- Muir, A.; Danai, L.V.; Gui, D.Y.; Waingarten, C.Y.; Lewis, C.A.; Vander Heiden, M.G. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. Elife 2017, 6. [Google Scholar] [CrossRef]
- Curthoys, N.P.; Watford, M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995, 15, 133–159. [Google Scholar] [CrossRef]
- Sasaki, H.; Sato, H.; Kuriyama-Matsumura, K.; Sato, K.; Maebara, K.; Wang, H.; Tamba, M.; Itoh, K.; Yamamoto, M.; Bannai, S. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 2002, 277, 44765–44771. [Google Scholar] [CrossRef]
- Sato, H.; Nomura, S.; Maebara, K.; Sato, K.; Tamba, M.; Bannai, S. Transcriptional control of cystine/glutamate transporter gene by amino acid deprivation. Biochem. Biophys. Res. Commun. 2004, 325, 109–116. [Google Scholar] [CrossRef]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef]
- Martin, L.; Gardner, L.B. Stress-induced inhibition of nonsense-mediated RNA decay regulates intracellular cystine transport and intracellular glutathione through regulation of the cystine/glutamate exchanger SLC7A11. Oncogene 2015, 34, 4211–4218. [Google Scholar] [CrossRef]
- Liu, X.X.; Li, X.J.; Zhang, B.; Liang, Y.J.; Zhou, C.X.; Cao, D.X.; He, M.; Chen, G.Q.; He, J.R.; Zhao, Q. MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett. 2011, 585, 1363–1367. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, X.; Song, B.; Qiu, X.; Zhao, J. MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med. 2017, 6, 1686–1697. [Google Scholar] [CrossRef]
- Drayton, R.M.; Dudziec, E.; Peter, S.; Bertz, S.; Hartmann, A.; Bryant, H.E.; Catto, J.W. Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin. Cancer Res. 2014, 20, 1990–2000. [Google Scholar] [CrossRef]
- Gu, Y.; Albuquerque, C.P.; Braas, D.; Zhang, W.; Villa, G.R.; Bi, J.; Ikegami, S.; Masui, K.; Gini, B.; Yang, H.; et al. mTORC2 Regulates Amino Acid Metabolism in Cancer by Phosphorylation of the Cystine-Glutamate Antiporter xCT. Mol. Cell 2017, 67, 128–138. [Google Scholar] [CrossRef]
- Schioth, H.B.; Roshanbin, S.; Hagglund, M.G.; Fredriksson, R. Evolutionary origin of amino acid transporter families SLC32, SLC36 and SLC38 and physiological, pathological and therapeutic aspects. Mol. Asp. Med. 2013, 34, 571–585. [Google Scholar] [CrossRef]
- Broer, S. The SLC38 family of sodium-amino acid co-transporters. Pflugers Arch. 2014, 466, 155–172. [Google Scholar] [CrossRef]
- Mackenzie, B.; Erickson, J.D. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Arch. 2004, 447, 784–795. [Google Scholar] [CrossRef]
- Yao, D.; Mackenzie, B.; Ming, H.; Varoqui, H.; Zhu, H.; Hediger, M.A.; Erickson, J.D. A novel system A isoform mediating Na+/neutral amino acid cotransport. J. Biol. Chem. 2000, 275, 22790–22797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gameiro, A.; Grewer, C. Highly conserved asparagine 82 controls the interaction of Na+ with the sodium-coupled neutral amino acid transporter SNAT2. J. Biol. Chem. 2008, 283, 12284–12292. [Google Scholar] [CrossRef]
- Shaffer, P.L.; Goehring, A.; Shankaranarayanan, A.; Gouaux, E. Structure and mechanism of a Na+—Independent amino acid transporter. Science 2009, 325, 1010–1014. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J.; Cai, R.; Yuan, Y.; Guo, Z.; Grewer, C.; Zhang, Z. Identification of a Disulfide Bridge in Sodium-Coupled Neutral Amino Acid Transporter 2(SNAT2) by Chemical Modification. PLoS ONE 2016, 11, e0158319. [Google Scholar] [CrossRef]
- Solbu, T.T.; Bjorkmo, M.; Berghuis, P.; Harkany, T.; Chaudhry, F.A. SAT1, A Glutamine Transporter, is Preferentially Expressed in GABAergic Neurons. Front. Neuroanat. 2010, 4, 1. [Google Scholar] [CrossRef]
- Jenstad, M.; Chaudhry, F.A. The Amino Acid Transporters of the Glutamate/GABA-Glutamine Cycle and Their Impact on Insulin and Glucagon Secretion. Front. Endocrinol. 2013, 4, 199. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Palii, S.S.; Chen, H.; Kilberg, M.S. Transcriptional control of the human sodium-coupled neutral amino acid transporter system A gene by amino acid availability is mediated by an intronic element. J. Biol. Chem. 2004, 279, 3463–3471. [Google Scholar] [CrossRef] [PubMed]
- Franchi-Gazzola, R.; Gaccioli, F.; Bevilacqua, E.; Visigalli, R.; Dall’Asta, V.; Sala, R.; Varoqui, H.; Erickson, J.D.; Gazzola, G.C.; Bussolati, O. The synthesis of SNAT2 transporters is required for the hypertonic stimulation of system A transport activity. Biochim. Biophys. Acta 2004, 1667, 157–166. [Google Scholar] [CrossRef][Green Version]
- Schulte, M.L.; Fu, A.; Zhao, P.; Li, J.; Geng, L.; Smith, S.T.; Kondo, J.; Coffey, R.J.; Johnson, M.O.; Rathmell, J.C.; et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat. Med. 2018, 24, 194–202. [Google Scholar] [CrossRef]
- Oda, K.; Hosoda, N.; Endo, H.; Saito, K.; Tsujihara, K.; Yamamura, M.; Sakata, T.; Anzai, N.; Wempe, M.F.; Kanai, Y.; et al. L-type amino acid transporter 1 inhibitors inhibit tumor cell growth. Cancer Sci. 2010, 101, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, N.; Tang, T.; Feng, F.; Sun, H.; Qu, W. Target the human Alanine/Serine/Cysteine Transporter 2(ASCT2): Achievement and Future for Novel Cancer Therapy. Pharmacol. Res. 2020, 158. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Xu, J.; Sun, J.; Huang, H.; Zhang, Z.; Ma, W.; Wan, Z.; Liu, Y.; Pardeshi, A.; Li, S. Co-delivery of 2-Deoxyglucose and a glutamine metabolism inhibitor V9302 via a prodrug micellar formulation for synergistic targeting of metabolism in cancer. Acta Biomater. 2020, 105, 239–252. [Google Scholar] [CrossRef]
- Okano, N.; Naruge, D.; Kawai, K.; Kobayashi, T.; Nagashima, F.; Endou, H.; Furuse, J. First-in-human phase I study of JPH203, an L-type amino acid transporter 1 inhibitor, in patients with advanced solid tumors. Invest. New Drugs 2020. [Google Scholar] [CrossRef]
- Scalise, M.; Console, L.; Galluccio, M.; Pochini, L.; Indiveri, C. Chemical Targeting of Membrane Transporters: Insights into Structure/Function Relationships. ACS Omega 2020, 5, 2069–2080. [Google Scholar] [CrossRef]
| SLC1A5 (ASCT2) | SLC6A14 (ATB0,+) | SLC7A1-2-3 (CAT 1-2-3) | SLC7A5 (LAT1) | SLC7A11 (xCT) | SLC38A2 (SNAT2) | References |
|---|---|---|---|---|---|---|
| PCA | PCA | PCA | PCA | [19,61,62,63,64,65,66,67] | ||
| CRC | CRC | CRC (C1-2) | CRC | CRC | [19,61,67,68,69,70,71,72,73,74,75] | |
| HCC | HCC (C1) | HCC | HCC | [61,69,76,77] | ||
| LC | LC | LC | [19,61,67,78,79,80,81] | |||
| BC | BC | BC (C1) | BC | BC | BC | [19,26,61,62,67,69,79,82,83,84,85,86,87,88,89,90,91] |
| N & G | N & G | [61,92,93] | ||||
| EC | EC | [61,94,95] | ||||
| OC | OC | OC | [19,61,96,97] | |||
| RCC | RCC | [19,61,98,99] | ||||
| P & BC | P & BC | PC | PC | [61,100,101,102,103,104] | ||
| GC | GC (C1) | GC | [61,105,106,107,108,109,110] | |||
| BT | [111] | |||||
| PM (C1) | [61] | |||||
| CC | CC | CC | CC | [19,86,112,113] | ||
| OSCC | OSCC | OSCC | [114,115,116,117] | |||
| TC (C1) | [118] | |||||
| M (C1) | M | [119,120] | ||||
| L (C1-2) | L | [121,122,123,124] | ||||
| ESCA | ESCA | ESCA | [115,125] | |||
| G | [88,126,127,128,129] | |||||
| THCA | [130] | |||||
| LSCC | [131] | |||||
| GBM | [132,133] | |||||
| THCA (C3) | [130] | |||||
| OC (C3) | TSCC | [134,135] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scalise, M.; Console, L.; Rovella, F.; Galluccio, M.; Pochini, L.; Indiveri, C. Membrane Transporters for Amino Acids as Players of Cancer Metabolic Rewiring. Cells 2020, 9, 2028. https://doi.org/10.3390/cells9092028
Scalise M, Console L, Rovella F, Galluccio M, Pochini L, Indiveri C. Membrane Transporters for Amino Acids as Players of Cancer Metabolic Rewiring. Cells. 2020; 9(9):2028. https://doi.org/10.3390/cells9092028
Chicago/Turabian StyleScalise, Mariafrancesca, Lara Console, Filomena Rovella, Michele Galluccio, Lorena Pochini, and Cesare Indiveri. 2020. "Membrane Transporters for Amino Acids as Players of Cancer Metabolic Rewiring" Cells 9, no. 9: 2028. https://doi.org/10.3390/cells9092028
APA StyleScalise, M., Console, L., Rovella, F., Galluccio, M., Pochini, L., & Indiveri, C. (2020). Membrane Transporters for Amino Acids as Players of Cancer Metabolic Rewiring. Cells, 9(9), 2028. https://doi.org/10.3390/cells9092028










