Tissue-Specific Impact of Autophagy Genes on the Ubiquitin–Proteasome System in C. elegans
Abstract
1. Introduction
2. Material and Methods
2.1. C. elegans and Growth Conditions
2.2. C. elegans RNA Interference (RNAi)
2.3. Microscopy and Image Analysis
2.4. Quantitative Real-Time PCR
2.5. In-Gel Proteasome Activity Assay and Western Blotting
2.6. Immunohistochemical Analysis
2.7. Statistical Analysis
3. Results
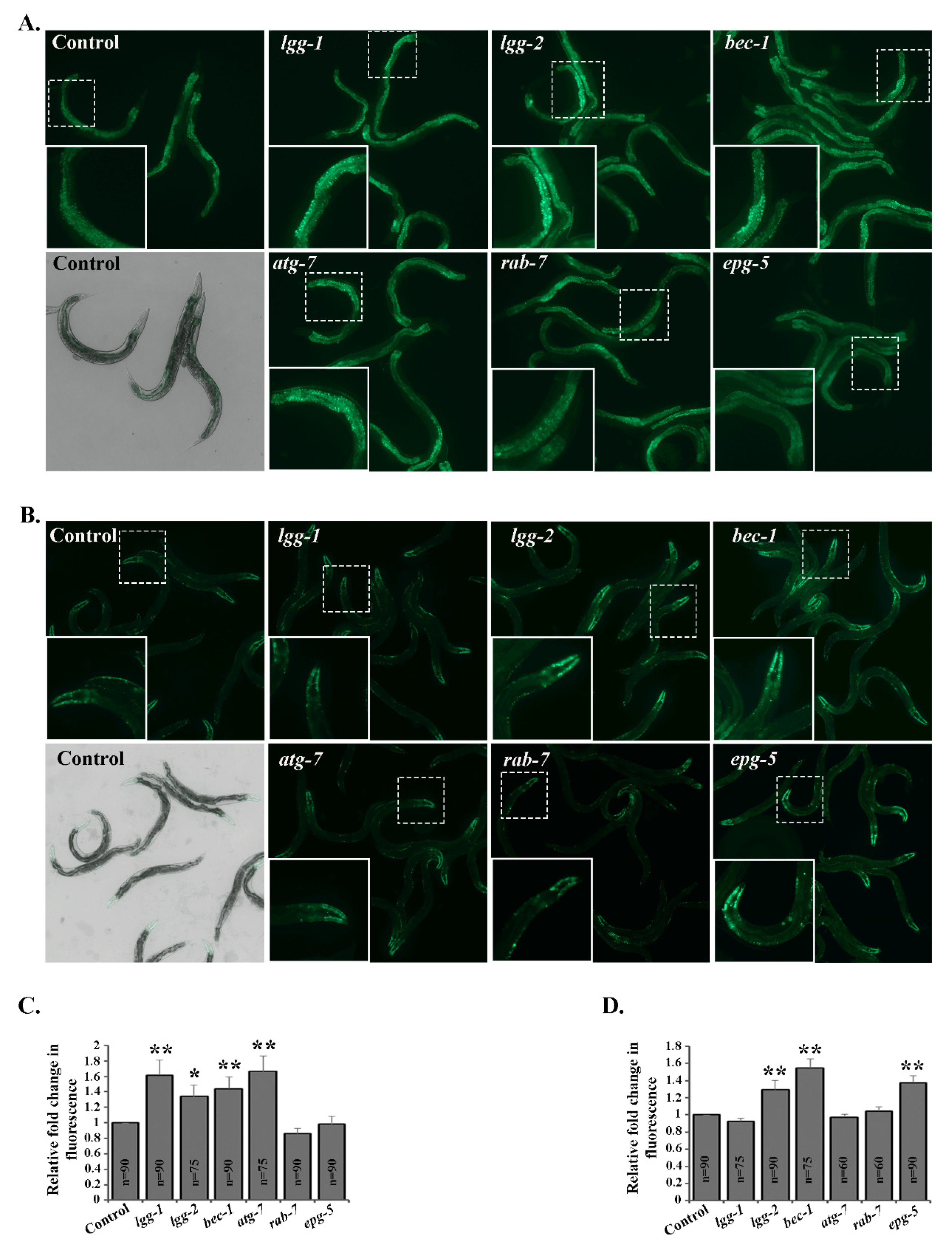
3.1. Downregulation of Autophagy Genes Affects the Accumulation of Polyubiquitinated Proteins in a Tissue-Specific Manner In Vivo
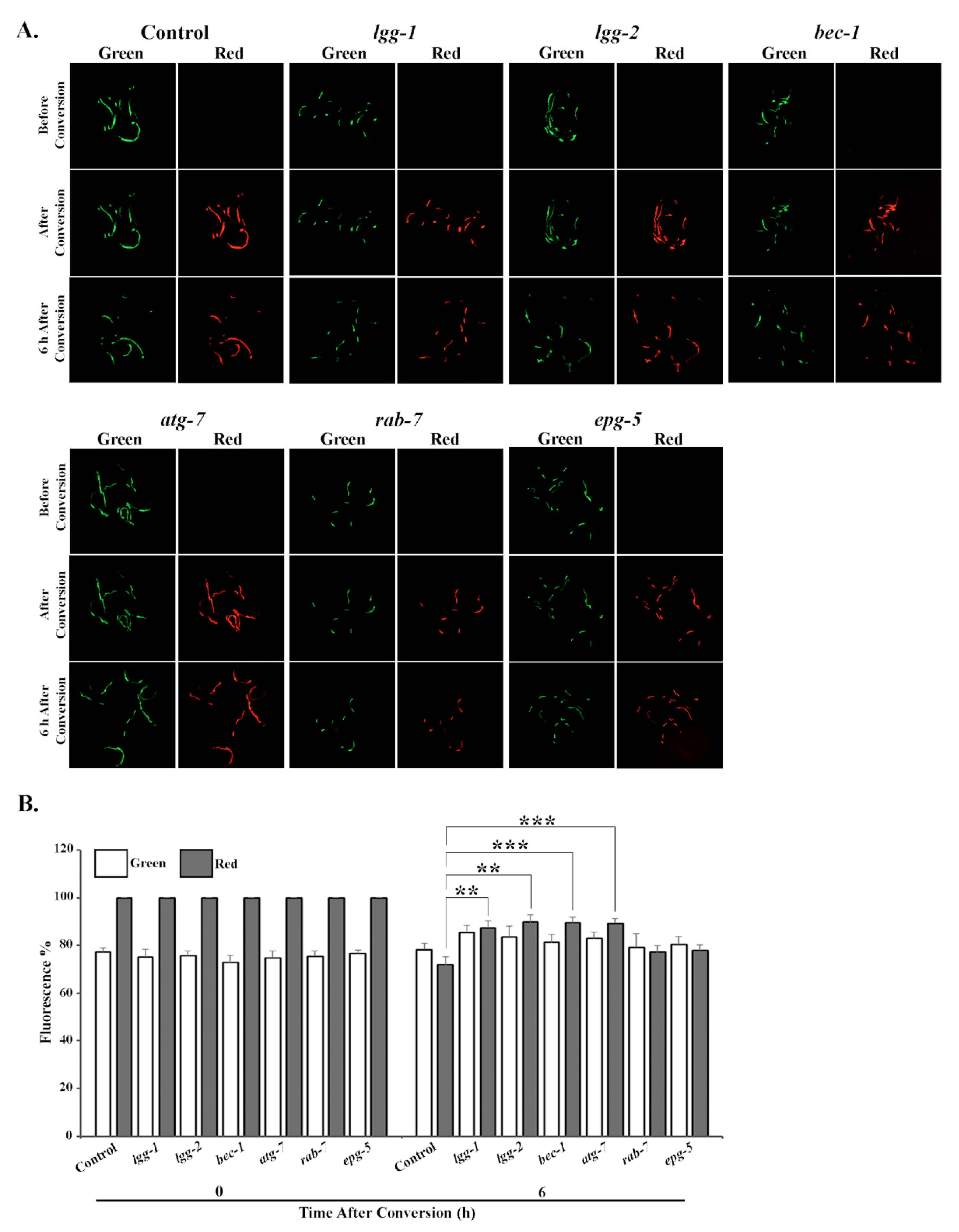
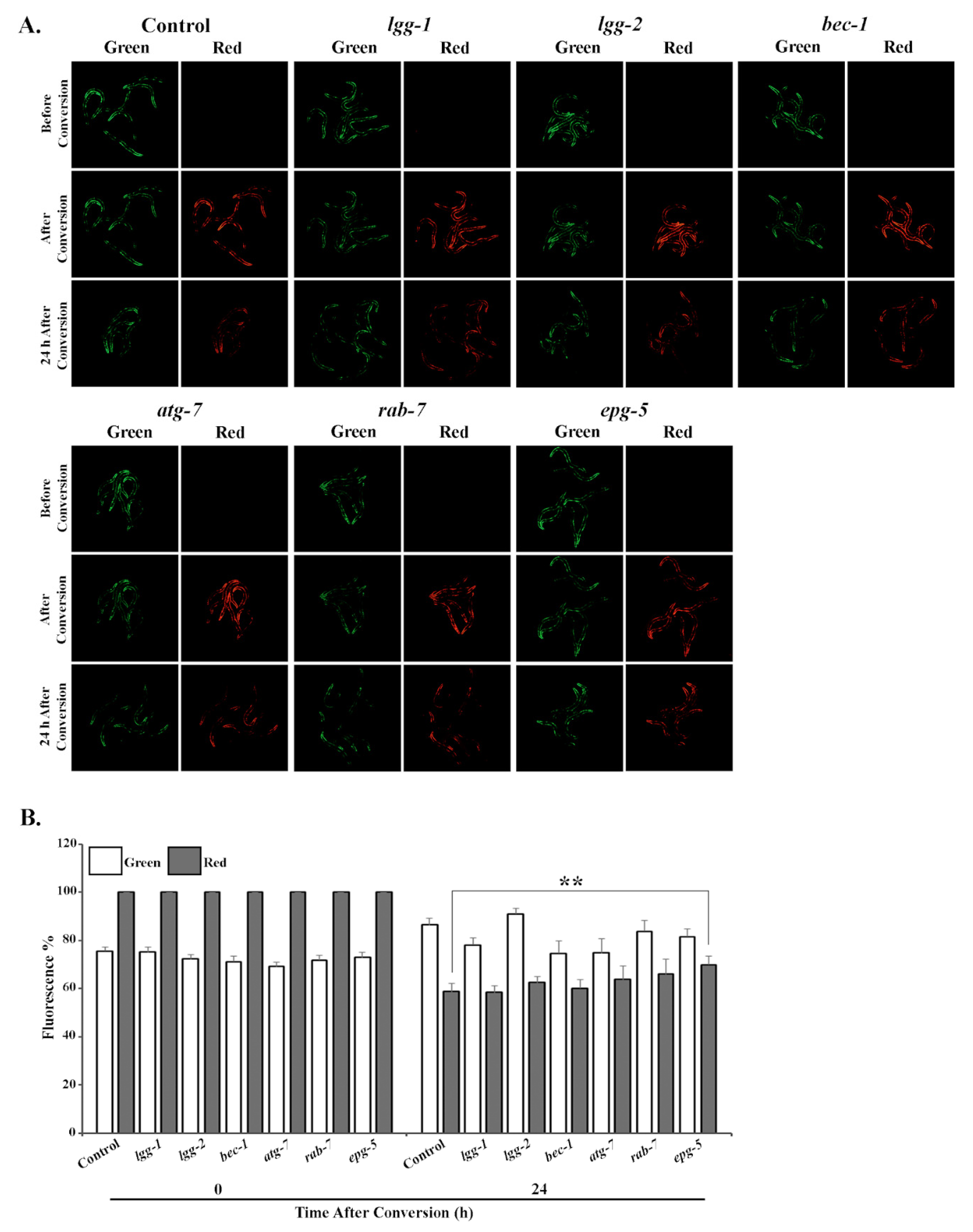
3.2. Tissue-Specific Differences in UPS Activity In Vivo Upon Knockdown of Autophagy Genes
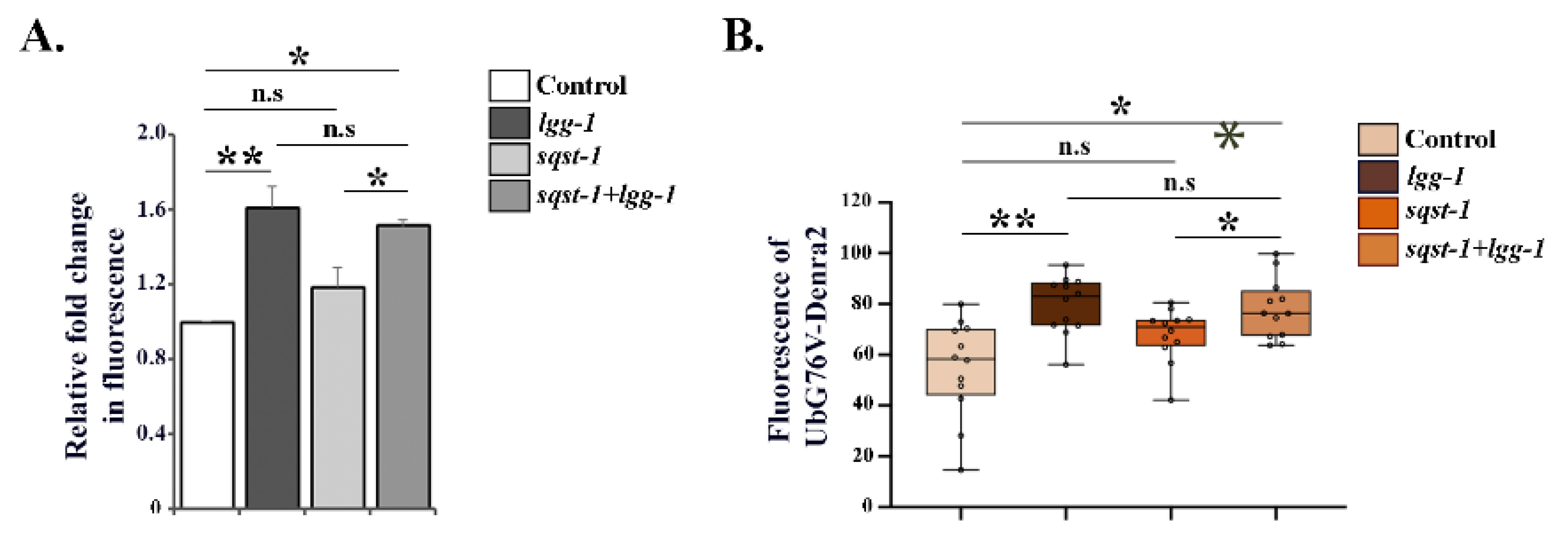
3.3. Downregulation of lgg-1 Affects UPS Function Independently of p62/SQST-1 Accumulation
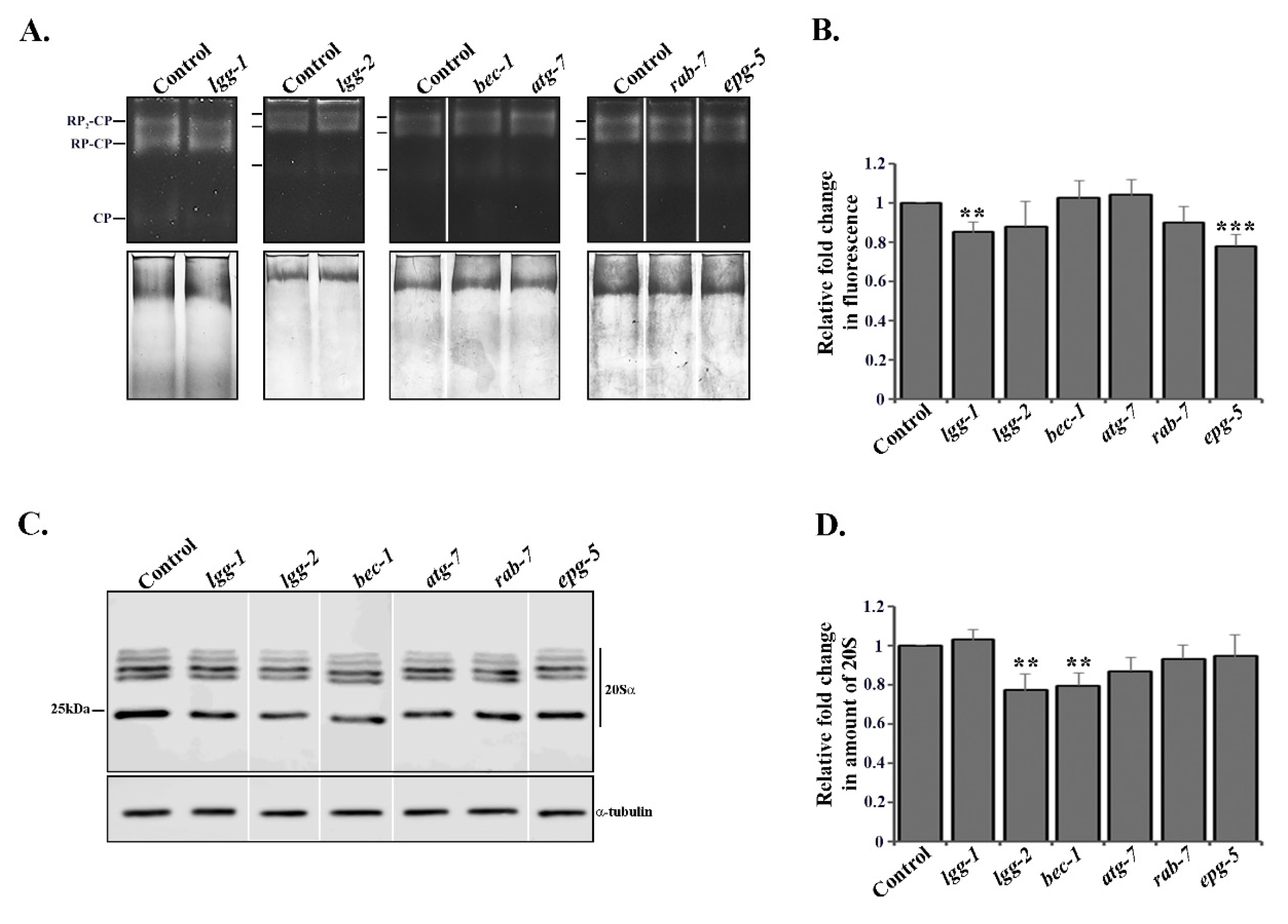
3.4. Depletion of Autophagy Genes Affects Proteasome Activity or Expression
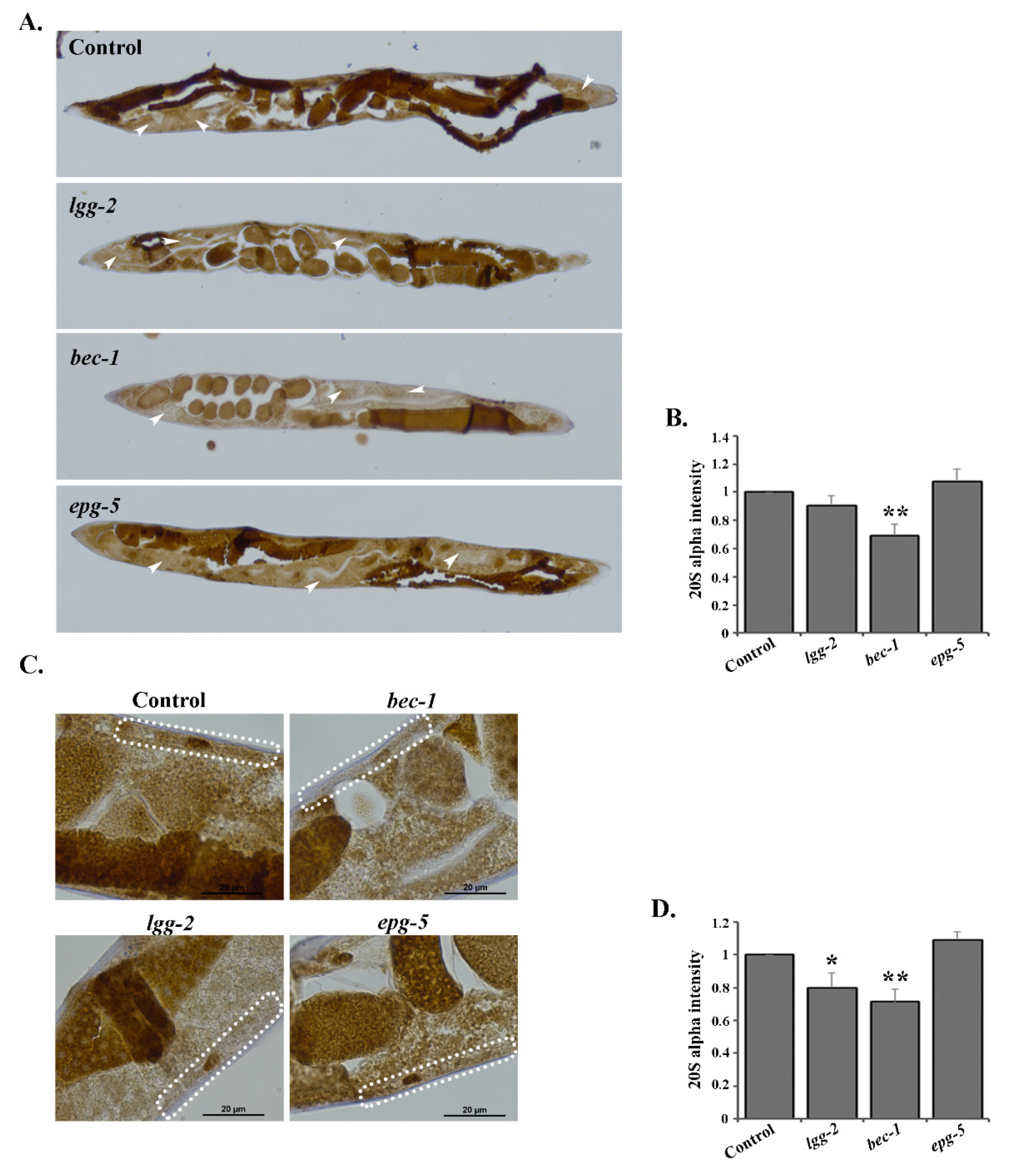
3.5. RNAi of lgg-2 or bec-1 Causes Distinct Responses in Proteasome Tissue Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 2014, 1843, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Coll-Martínez, B.; Crosas, B. How the 26S Proteasome Degrades Ubiquitinated Proteins in the Cell. Biomolecules 2019, 9, 395. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Klionsky, D.J. At a glance: A history of autophagy and cancer. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Nam, T.; Han, J.H.; Devkota, S.; Lee, H.W. Emerging Paradigm of Crosstalk between Autophagy and the Ubiquitin-Proteasome System. Mol. Cells 2017, 40, 897–905. [Google Scholar]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef]
- Nakatogawa, H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 2013, 55, 39–50. [Google Scholar]
- Meng, Y.C.; Lou, X.L.; Yang, L.Y.; Li, D.; Hou, Y.Q. Role of the autophagy-related marker LC3 expression in hepatocellular carcinoma: A meta-analysis. J. Cancer Res. Clin. Oncol. 2020, 146, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.G.; Munafo, D.B.; Beron, W.; Colombo, M.I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 2004, 117, 2687–2697. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Li, Z.; Hu, W.; Ren, H.; Tian, E.; Zhao, Y.; Lu, Q.; Huang, X.; Yang, P.; Li, X.; et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 2010, 141, 1042–1055. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, Y.G.; Wang, X.; Xu, L.; Miao, L.; Feng, D.; Chen, Q.; Kovacs, A.L.; Fan, D.; Zhang, H. Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J. Cell Biol. 2013, 200, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef]
- Fan, T.; Huang, Z.; Chen, L.; Wang, W.; Zhang, B.; Xu, Y.; Pan, S.; Mao, Z.; Hu, H.; Geng, Q. Associations between autophagy, the ubiquitin-proteasome system and endoplasmic reticulum stress in hypoxia-deoxygenation or ischemia-reperfusion. Eur. J. Pharmacol. 2016, 791, 157–167. [Google Scholar] [CrossRef]
- Shen, Y.F.; Tang, Y.; Zhang, X.J.; Huang, K.X.; Le, W.D. Adaptive changes in autophagy after UPS impairment in Parkinson’s disease. Acta Pharmacol. Sin. 2013, 34, 667–673. [Google Scholar] [CrossRef]
- Zheng, Q.; Su, H.; Tian, Z.; Wang, X. Proteasome malfunction activates macroautophagy in the heart. Am. J. Cardiovasc. Dis. 2011, 1, 214–226. [Google Scholar]
- Li, C.; Wang, X.; Li, X.; Qiu, K.; Jiao, F.; Liu, Y.; Kong, Q.; Liu, Y.; Wu, Y. Proteasome Inhibition Activates Autophagy-Lysosome Pathway Associated with TFEB Dephosphorylation and Nuclear Translocation. Front. Cell Dev. Biol. 2019, 7, 170. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Park, S.; Lee, J.H.; Mun, J.Y.; Choi, W.H.; Yun, Y.; Lee, J.; Kim, J.H.; Kang, M.J.; Lee, M.J. Dual Function of USP14 Deubiquitinase in Cellular Proteasomal Activity and Autophagic Flux. Cell Rep. 2018, 24, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Mansilla, A.; Menzies, F.M.; Rubinsztein, D.C. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell 2009, 33, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Yu, J.; Wong, S.H.; Cheng, A.S.; Chan, F.K.; Ng, S.S.; Cho, C.H.; Sung, J.J.; Wu, W.K. A novel crosstalk between two major protein degradation systems: Regulation of proteasomal activity by autophagy. Autophagy 2013, 9, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Zhang, J. Inhibition of lysosomal functions reduces proteasomal activity. Neurosci. Lett. 2009, 456, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Scarcelli, V.; Legouis, R. Approaches for Studying Autophagy in Caenorhabditis elegans. Cells 2017, 6, 27. [Google Scholar] [CrossRef]
- Palmisano, N.J.; Meléndez, A. Autophagy in C. elegans development. Dev. Biol. 2019, 447, 103–125. [Google Scholar] [CrossRef]
- Papaevgeniou, N.; Chondrogianni, N. The ubiquitin proteasome system in Caenorhabditis elegans and its regulation. Redox Biol. 2014, 2, 333–347. [Google Scholar] [CrossRef]
- Chang, J.T.; Kumsta, C.; Hellman, A.B.; Adams, L.M.; Hansen, M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. eLife 2017, 6. [Google Scholar] [CrossRef]
- Chapin, H.C.; Okada, M.; Merz, A.J.; Miller, D.L. Tissue-specific autophagy responses to aging and stress in C. elegans. Aging (Albany NY) 2015, 7, 419–434. [Google Scholar] [CrossRef]
- Hamer, G.; Matilainen, O.; Holmberg, C.I. A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat. Methods 2010, 7, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Matilainen, O.; Arpalahti, L.; Rantanen, V.; Hautaniemi, S.; Holmberg, C.I. Insulin/IGF-1 signaling regulates proteasome activity through the deubiquitinating enzyme UBH-4. Cell Rep. 2013, 3, 1980–1995. [Google Scholar] [CrossRef] [PubMed]
- Mikkonen, E.; Haglund, C.; Holmberg, C.I. Immunohistochemical analysis reveals variations in proteasome tissue expression in C. elegans. PLoS ONE 2017, 12, e0183403. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [PubMed]
- Pispa, J.; Matilainen, O.; Holmberg, C.I. Tissue-specific effects of temperature on proteasome function. Cell Stress Chaperones 2020, 25, 563–572. [Google Scholar] [CrossRef]
- Timmons, L.; Court, D.L.; Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001, 263, 103–112. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Elsasser, S.; Schmidt, M.; Finley, D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005, 398, 353–363. [Google Scholar]
- Matilainen, O.; Jha, S.; Holmberg, C.I. Fluorescent Tools for In Vivo Studies on the Ubiquitin-Proteasome System. Methods Mol. Biol. 2016, 1449, 215–222. [Google Scholar]
- Li, X.; Matilainen, O.; Jin, C.; Glover-Cutter, K.M.; Holmberg, C.I.; Blackwell, T.K. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011, 7, e1002119. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.X.; Ni, H.M.; Gao, W.; Yoshimori, T.; Stolz, D.B.; Ron, D.; Yin, X.M. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am. J. Pathol. 2007, 171, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Rideout, H.J.; Lang-Rollin, I.; Stefanis, L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int. J. Biochem. Cell Biol. 2004, 36, 2551–2562. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Feleciano, D.R.; Juenemann, K.; Iburg, M.; Brás, I.C.; Holmberg, C.I.; Kirstein, J. Crosstalk Between Chaperone-Mediated Protein Disaggregation and Proteolytic Pathways in Aging and Disease. Front. Aging Neurosci. 2019, 11, 9. [Google Scholar] [CrossRef]
- Seibenhener, M.L.; Babu, J.R.; Geetha, T.; Wong, H.C.; Krishna, N.R.; Wooten, M.W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 2004, 24, 8055–8068. [Google Scholar] [CrossRef]
- Kumsta, C.; Chang, J.T.; Lee, R.; Tan, E.P.; Yang, Y.; Loureiro, R.; Choy, E.H.; Lim, S.H.Y.; Saez, I.; Springhorn, A.; et al. The autophagy receptor p62/SQST-1 promotes proteostasis and longevity in C. elegans by inducing autophagy. Nat. Commun. 2019, 10, 5648. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jha, S.; Holmberg, C.I. Tissue-Specific Impact of Autophagy Genes on the Ubiquitin–Proteasome System in C. elegans. Cells 2020, 9, 1858. https://doi.org/10.3390/cells9081858
Jha S, Holmberg CI. Tissue-Specific Impact of Autophagy Genes on the Ubiquitin–Proteasome System in C. elegans. Cells. 2020; 9(8):1858. https://doi.org/10.3390/cells9081858
Chicago/Turabian StyleJha, Sweta, and Carina I. Holmberg. 2020. "Tissue-Specific Impact of Autophagy Genes on the Ubiquitin–Proteasome System in C. elegans" Cells 9, no. 8: 1858. https://doi.org/10.3390/cells9081858
APA StyleJha, S., & Holmberg, C. I. (2020). Tissue-Specific Impact of Autophagy Genes on the Ubiquitin–Proteasome System in C. elegans. Cells, 9(8), 1858. https://doi.org/10.3390/cells9081858





