Functional Characterisation of the Autophagy ATG12~5/16 Complex in Dictyostelium discoideum
Abstract
1. Introduction
2. Materials and Methods
2.1. Dictyostelium Strains, Growth and Development
2.2. Primary Antibodies
2.3. SDS-PAGE and Western Blotting
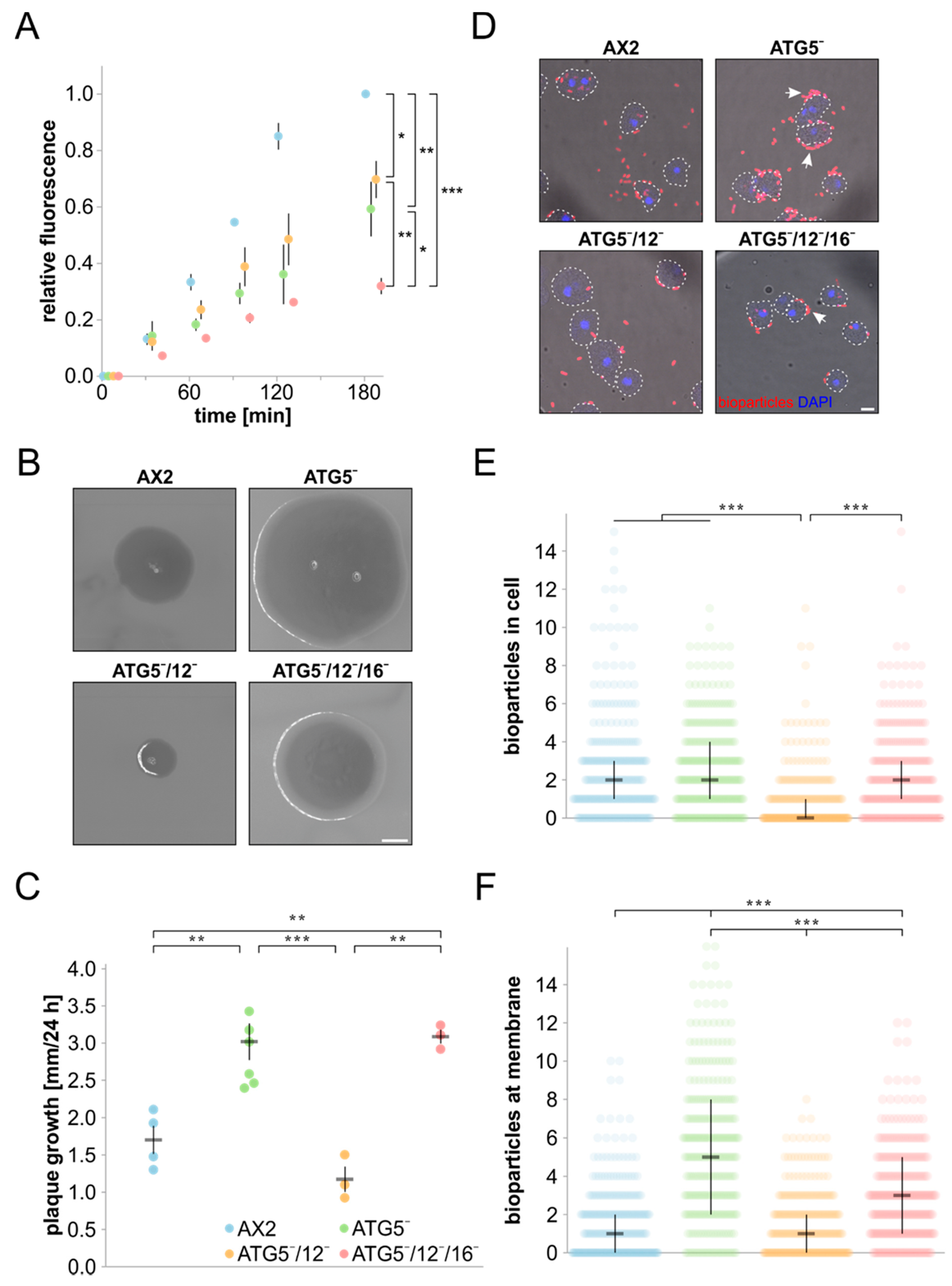
2.4. Macropinocytosis and Phagocytosis Assays
2.5. Random Cell Motility Analysis
2.6. Proteasomal Activity Analysis
2.7. Fluorescence Microscopy
2.8. Bioinformatics and RNAseq
2.9. Statistics and Reproducibility
3. Results
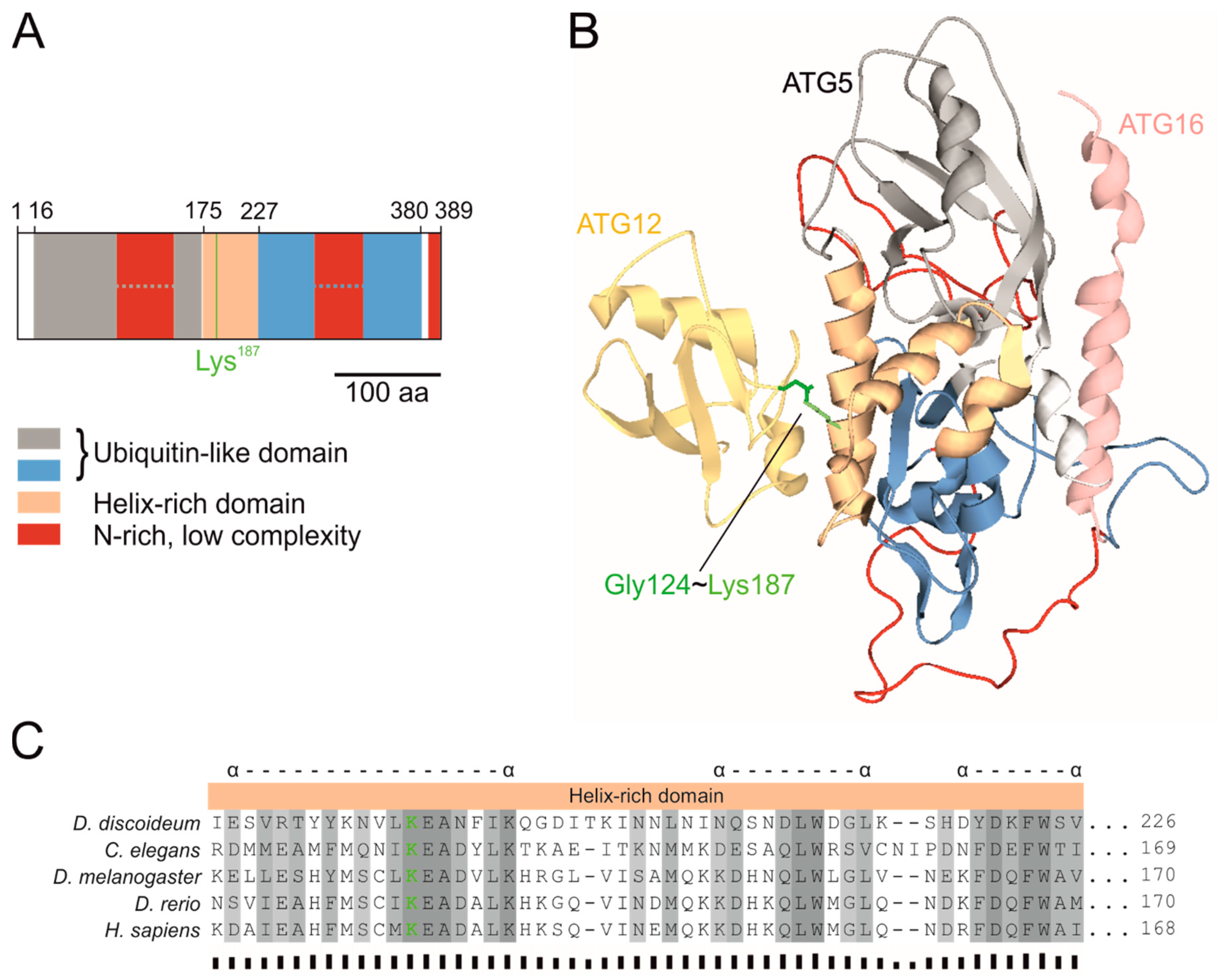
3.1. D. discoideum ATG5 Is highly Conserved
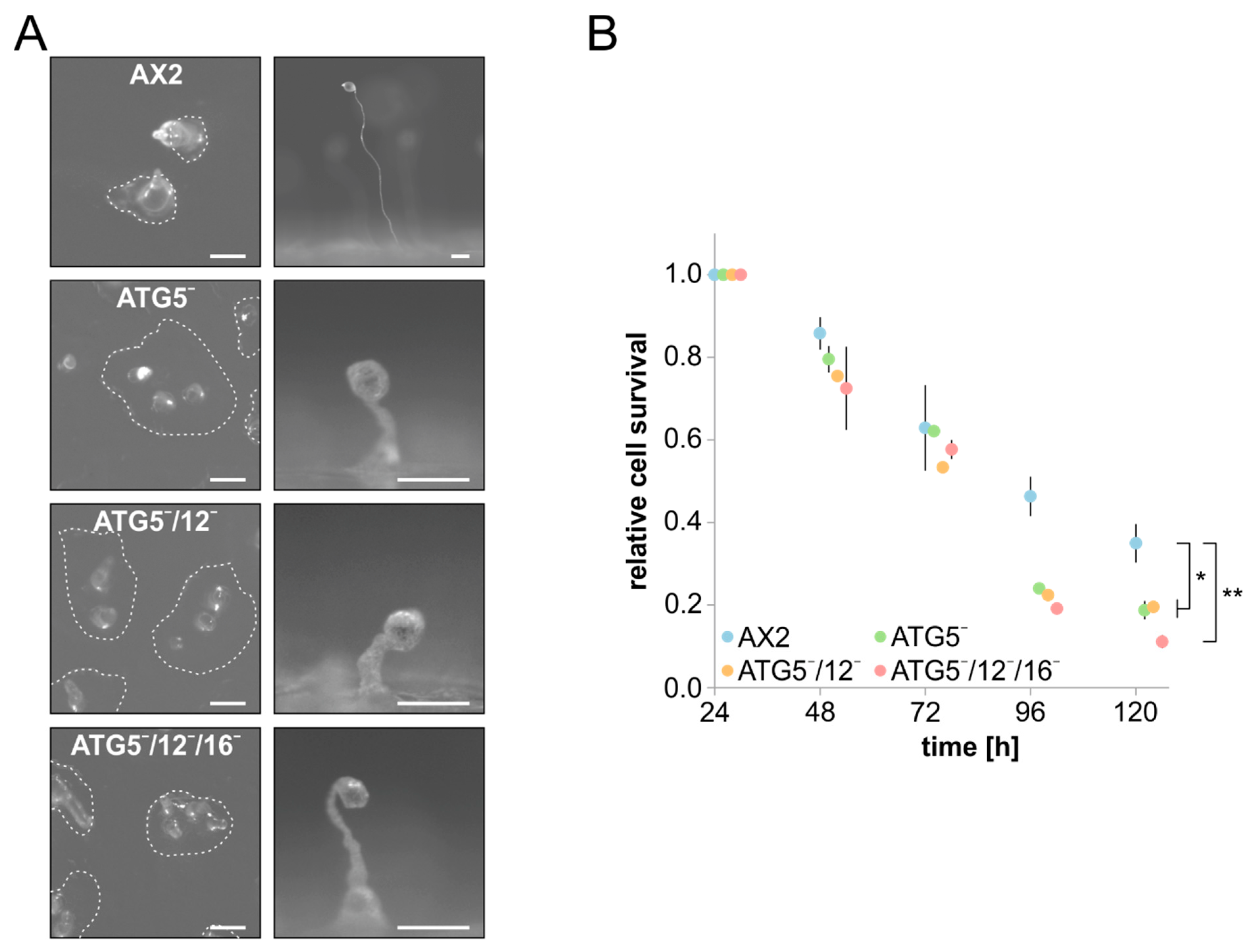
3.2. Cellular Processes Dependent on Canonical Autophagy Are Similarly Impaired in the Different atg5 Knock-Out Strains
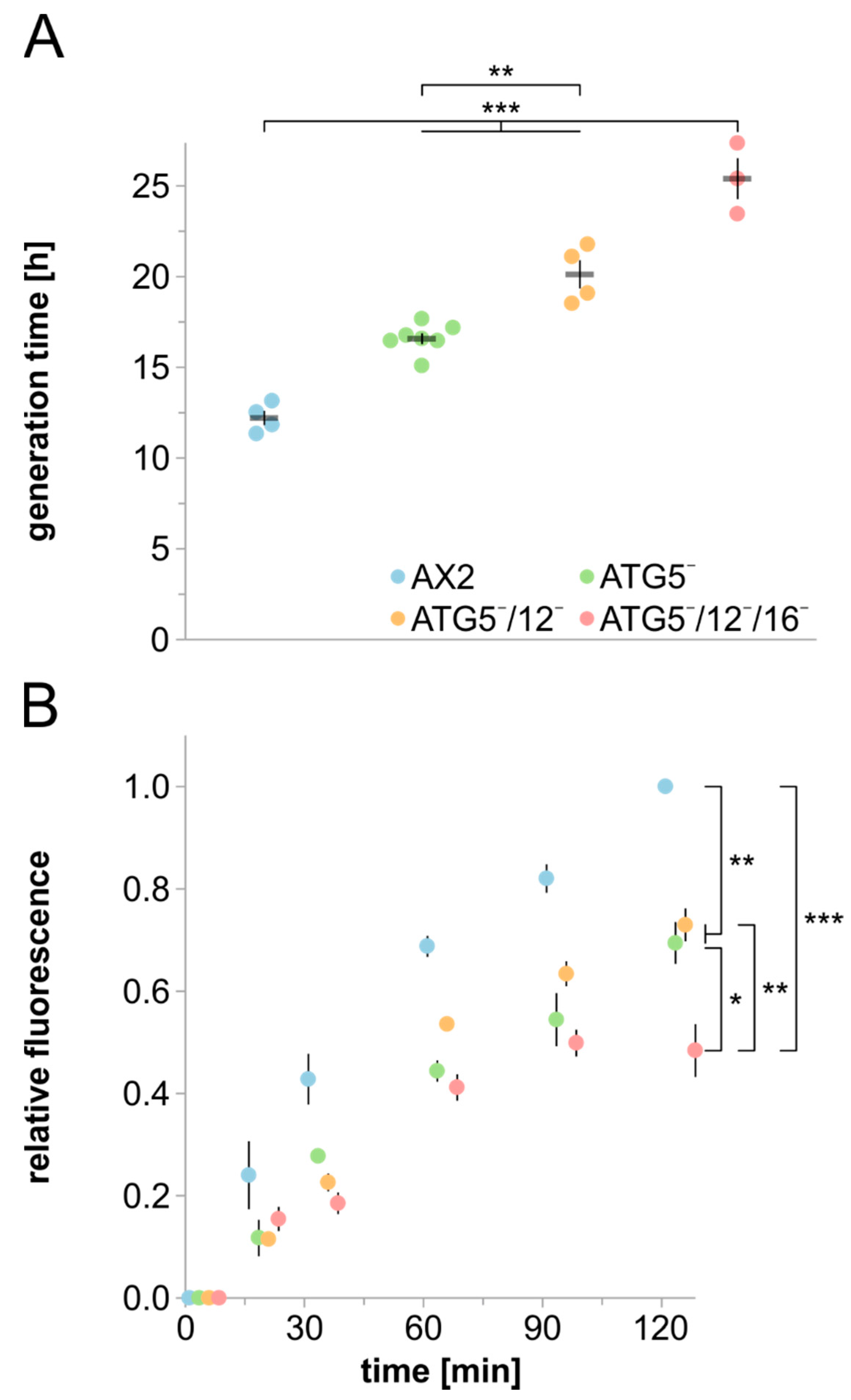
3.3. Cell Growth, Macropinocytosis and Phagocytosis Are Severely Affected in Strains Lacking ATG5
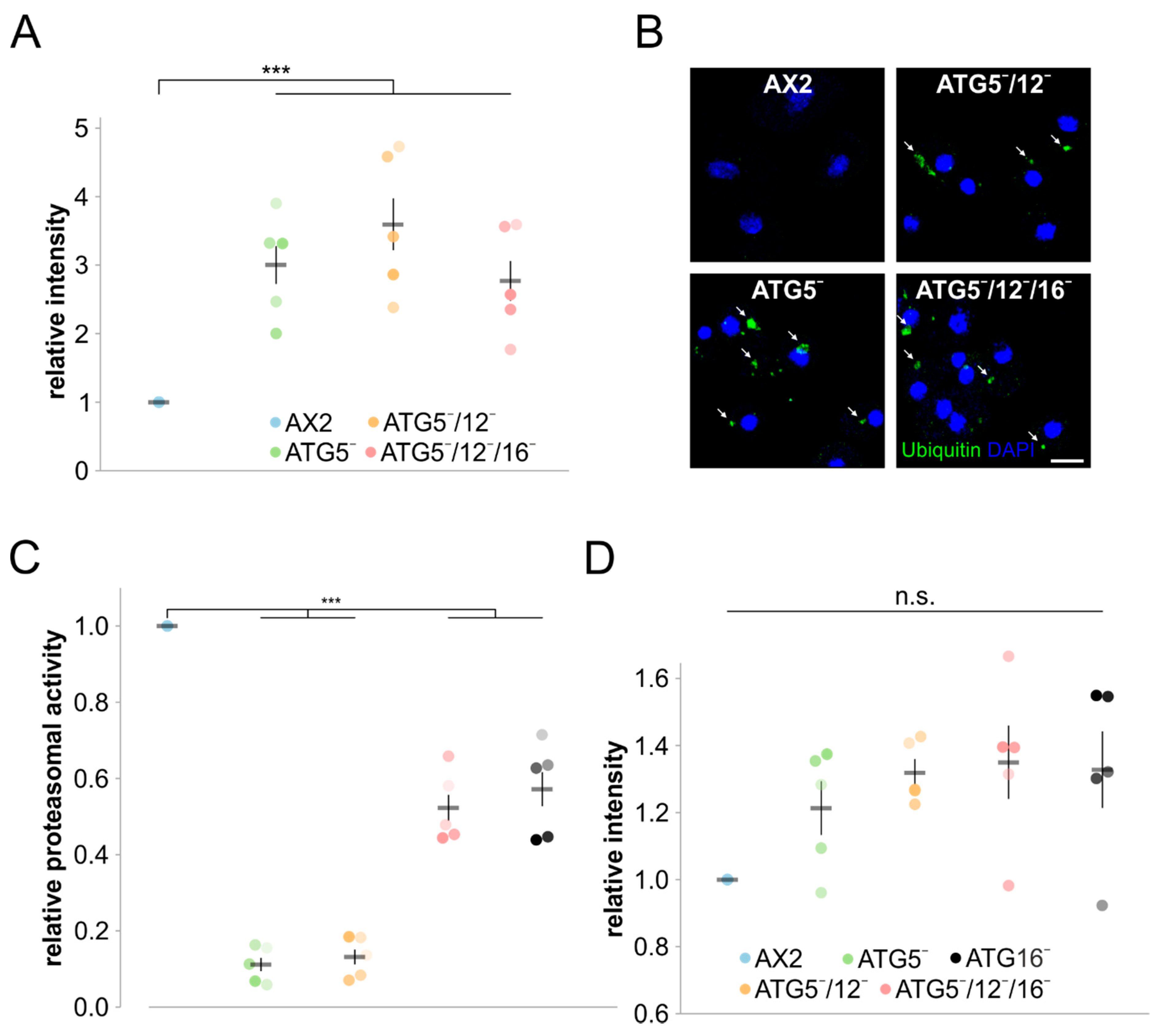
3.4. Protein Homeostasis Is Severely Disturbed in Mutant Strains
4. Discussion
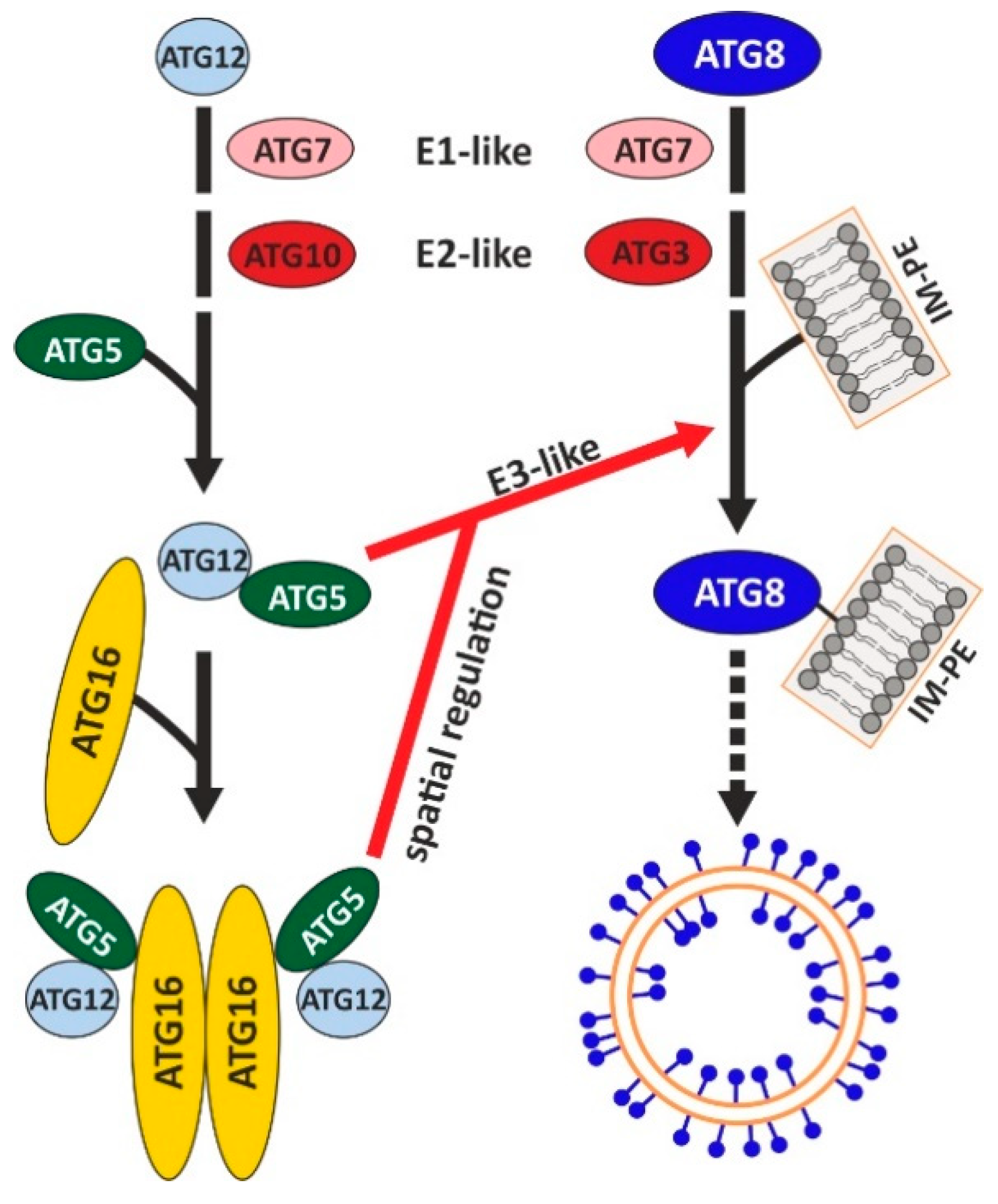
4.1. ATG5 Is Required for Conjugation of ATG8 to the Autophagosomal Membrane and for ATG12 Stability
4.2. The Dictyostelium ATG5¯, ATG5¯/12¯ and ATG5¯/12¯/16¯ Mutants Have Complex and Distinct Phenotypes
4.3. Protein Homeostasis Is Severely Impaired in ATG5 Deficient Strains
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stanley, R.E.; Ragusa, M.J.; Hurley, J.H. The beginning of the end: How scaffolds nucleate autophagosome biogenesis. Trends Cell Biol. 2014, 24, 73–81. [Google Scholar] [CrossRef]
- Mizushima, N.; Klionsky, D.J. Protein turnover via autophagy: Implications for metabolism. Annu. Rev. Nutr. 2007, 27, 19–40. [Google Scholar] [CrossRef]
- Mesquita, A.; Cardenal-Munoz, E.; Dominguez, E.; Munoz-Braceras, S.; Nunez-Corcuera, B.; Phillips, B.A.; Tabara, L.C.; Xiong, Q.; Coria, R.; Eichinger, L.; et al. Autophagy in dictyostelium: Mechanisms, regulation and disease in a simple biomedical model. Autophagy 2017, 13, 24–40. [Google Scholar] [CrossRef]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef]
- Schneider, J.L.; Cuervo, A.M. Autophagy and human disease: Emerging themes. Curr. Opin. Genet. Dev. 2014, 26, 16–23. [Google Scholar] [CrossRef]
- Kiriyama, Y.; Nochi, H. The function of autophagy in neurodegenerative diseases. Int. J. Mol. Sci. 2015, 16, 26797–26812. [Google Scholar] [CrossRef]
- Noda, N.N.; Inagaki, F. Mechanisms of autophagy. Annu. Rev. Biophys. 2015, 44, 101–122. [Google Scholar] [CrossRef]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef]
- Fischer, S.; Eichinger, L. Dictyostelium discoideum and autophagy—A perfect pair. Int. J. Dev. Biol. 2019, 63, 485–495. [Google Scholar] [CrossRef]
- Tanida, I.; Mizushima, N.; Kiyooka, M.; Ohsumi, M.; Ueno, T.; Ohsumi, Y.; Kominami, E. Apg7p/cvt2p: A novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell 1999, 10, 1367–1379. [Google Scholar] [CrossRef]
- Shintani, T.; Mizushima, N.; Ogawa, Y.; Matsuura, A.; Noda, T.; Ohsumi, Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999, 18, 5234–5241. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Noda, T.; Yoshimori, T.; Tanaka, Y.; Ishii, T.; George, M.D.; Klionsky, D.J.; Ohsumi, M.; Ohsumi, Y. A protein conjugation system essential for autophagy. Nature 1998, 395, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Noda, T.; Ohsumi, Y. Apg16p is required for the function of the apg12p–apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999, 18, 3888–3896. [Google Scholar] [CrossRef]
- Kabeya, Y.; Kamada, Y.; Baba, M.; Takikawa, H.; Sasaki, M.; Ohsumi, Y. Atg17 functions in cooperation with atg1 and atg13 in yeast autophagy. Mol. Biol. Cell 2005, 16, 2544–2553. [Google Scholar] [CrossRef]
- Kirisako, T.; Baba, M.; Ishihara, N.; Miyazawa, K.; Ohsumi, M.; Yoshimori, T.; Noda, T.; Ohsumi, Y. Formation process of autophagosome is traced with apg8/aut7p in yeast. J. Cell Biol. 1999, 147, 435–446. [Google Scholar] [CrossRef]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The reversible modification regulates the membrane-binding state of apg8/aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000, 151, 263–276. [Google Scholar] [CrossRef]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The atg12-atg5 conjugate has a novel e3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef]
- Romanov, J.; Walczak, M.; Ibiricu, I.; Schuchner, S.; Ogris, E.; Kraft, C.; Martens, S. Mechanism and functions of membrane binding by the atg5-atg12/atg16 complex during autophagosome formation. EMBO J. 2012, 31, 4304–4317. [Google Scholar] [CrossRef]
- Harada, K.; Kotani, T.; Kirisako, H.; Sakoh-Nakatogawa, M.; Oikawa, Y.; Kimura, Y.; Hirano, H.; Yamamoto, H.; Ohsumi, Y.; Nakatogawa, H. Two distinct mechanisms target the autophagy-related e3 complex to the pre-autophagosomal structure. eLife 2019, 8, e43088. [Google Scholar] [CrossRef]
- Nishimura, T.; Kaizuka, T.; Cadwell, K.; Sahani, M.H.; Saitoh, T.; Akira, S.; Virgin, H.W.; Mizushima, N. Fip200 regulates targeting of atg16l1 to the isolation membrane. EMBO Rep. 2013, 14, 284–291. [Google Scholar] [CrossRef]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. Wipi2 links lc3 conjugation with pi3p, autophagosome formation, and pathogen clearance by recruiting atg12-5-16l1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Juris, L.; Montino, M.; Rube, P.; Schlotterhose, P.; Thumm, M.; Krick, R. Pi3p binding by atg21 organises atg8 lipidation. EMBO J. 2015, 34, 955–973. [Google Scholar] [CrossRef]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The atg16l complex specifies the site of lc3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef] [PubMed]
- Kuma, A.; Hatano, M.; Matsui, M.; Yamamoto, A.; Nakaya, H.; Yoshimori, T.; Ohsumi, Y.; Tokuhisa, T.; Mizushima, N. The role of autophagy during the early neonatal starvation period. Nature 2004, 432, 1032–1036. [Google Scholar] [CrossRef]
- Muller-Taubenberger, A.; Kortholt, A.; Eichinger, L. Simple system--substantial share: The use of dictyostelium in cell biology and molecular medicine. Eur. J. Cell Biol. 2013, 92, 45–53. [Google Scholar] [CrossRef]
- Bozzaro, S.; Eichinger, L. The professional phagocyte dictyostelium discoideum as a model host for bacterial pathogens. Curr. Drug Targets 2011, 12, 942–954. [Google Scholar] [CrossRef]
- Williams, R.S.; Boeckeler, K.; Graf, R.; Muller-Taubenberger, A.; Li, Z.; Isberg, R.R.; Wessels, D.; Soll, D.R.; Alexander, H.; Alexander, S. Towards a molecular understanding of human diseases using dictyostelium discoideum. Trends Mol. Med. 2006, 12, 415–424. [Google Scholar] [CrossRef]
- Annesley, S.J.; Fisher, P.R. Dictyostelium discoideum—A model for many reasons. Mol. Cell. Biochem. 2009, 329, 73–91. [Google Scholar] [CrossRef]
- Dominguez-Martin, E.; Cardenal-Munoz, E.; King, J.S.; Soldati, T.; Coria, R.; Escalante, R. Methods to monitor and quantify autophagy in the social amoeba dictyostelium discoideum. Cells 2017, 6, 18. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef]
- Faix, J.; Linkner, J.; Nordholz, B.; Platt, J.L.; Liao, X.H.; Kimmel, A.R. The application of the cre-loxp system for generating multiple knock-out and knock-in targeted loci. Methods Mol. Biol. 2013, 983, 249–267. [Google Scholar]
- Fey, P.; Dodson, R.J.; Basu, S.; Chisholm, R.L. One stop shop for everything dictyostelium: Dictybase and the dicty stock center in 2012. Methods Mol. Biol. 2013, 983, 59–92. [Google Scholar]
- Calvo-Garrido, J.; Escalante, R. Autophagy dysfunction and ubiquitin-positive protein aggregates in dictyostelium cells lacking vmp1. Autophagy 2010, 6, 100–109. [Google Scholar] [CrossRef]
- Calvo-Garrido, J.; King, J.S.; Munoz-Braceras, S.; Escalante, R. Vmp1 regulates ptdins3p signaling during autophagosome formation in dictyostelium discoideum. Traffic 2014, 15, 1235–1246. [Google Scholar] [CrossRef]
- Munoz-Braceras, S.; Calvo, R.; Escalante, R. Tipc and the chorea-acanthocytosis protein vps13a regulate autophagy in dictyostelium and human hela cells. Autophagy 2015, 11, 918–927. [Google Scholar] [CrossRef]
- Messling, S.; Matthias, J.; Xiong, Q.; Fischer, S.; Eichinger, L. The two Dictyostelium discoideum autophagy 8 proteins have distinct autophagic functions. Eur. J. Cell Biol. 2017, 96, 312–324. [Google Scholar] [CrossRef]
- Otto, G.P.; Wu, M.Y.; Kazgan, N.; Anderson, O.R.; Kessin, R.H. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J. Biol. Chem. 2004, 279, 15621–15629. [Google Scholar] [CrossRef]
- Tung, S.M.; Unal, C.; Ley, A.; Pena, C.; Tunggal, B.; Noegel, A.A.; Krut, O.; Steinert, M.; Eichinger, L. Loss of dictyostelium atg9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of legionella pneumophila. Cell. Microbiol. 2010, 12, 765–780. [Google Scholar] [CrossRef]
- Fischer, S.; Rijal, R.; Frommolt, P.; Wagle, P.; Konertz, R.; Faix, J.; Messling, S.; Eichinger, L. Functional characterization of ubiquitin-like core autophagy protein atg12 in dictyostelium discoideum. Cells 2019, 8, 72. [Google Scholar] [CrossRef]
- Xiong, Q.; Unal, C.; Matthias, J.; Steinert, M.; Eichinger, L. The phenotypes of atg9, atg16 and atg9/16 knock-out mutants imply autophagy-dependent and -independent functions. Open Biol. 2015, 5, 150008. [Google Scholar] [CrossRef]
- Yamada, Y.; Schaap, P. The proppin bcas3 and its interactor kinkya localize to the early phagophore and regulate autophagy. Autophagy 2020, 1–16. [Google Scholar] [CrossRef]
- Watts, D.J.; Ashworth, J.M. Growth of myxamoebae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J. 1970, 119, 171–174. [Google Scholar] [CrossRef]
- Brink, M.; Gerisch, G.; Isenberg, G.; Noegel, A.A.; Segall, J.E.; Wallraff, E.; Schleicher, M. A dictyostelium mutant lacking an f-actin cross-linking protein, the 120-kd gelation factor. J. Cell Biol. 1990, 111, 1477–1489. [Google Scholar] [CrossRef]
- Sussman, M. Biochemical and genetic methods in the study of cellular slime mold development. Methods Cell Physiol. 1966, 2, 397–410. [Google Scholar]
- Williams, K.L.; Newell, P.C. A genetic study of aggregation in the cellular slime mould dictyostelium discoideum using complementation analysis. Genetics 1976, 82, 287–307. [Google Scholar]
- Raper, K.B. Dictyostelium discoideum, a new species of slime mold from decaying forest leaves. J. Agric. Res. 1935, 50, 135–147. [Google Scholar]
- Simpson, P.A.; Spudich, J.A.; Parham, P. Monoclonal antibodies prepared against dictyostelium actin: Characterization and interactions with actin. J. Cell Biol. 1984, 99, 287–295. [Google Scholar] [CrossRef]
- Schauer, T.M.; Nesper, M.; Kehl, M.; Lottspeich, F.; Muller-Taubenberger, A.; Gerisch, G.; Baumeister, W. Proteasomes from dictyostelium discoideum: Characterization of structure and function. J. Struct. Biol. 1993, 111, 135–147. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Matthias, J.; Messling, S.; Eichinger, L. The two dictyostelium autophagy eight proteins, atg8a and atg8b, associate with the autophagosome in succession. Eur. J. Cell Biol. 2016, 95, 15–25. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef]
- Strucksberg, K.H.; Tangavelou, K.; Schroder, R.; Clemen, C.S. Proteasomal activity in skeletal muscle: A matter of assay design, muscle type, and age. Anal. Biochem. 2010, 399, 225–229. [Google Scholar] [CrossRef]
- Arhzaouy, K.; Strucksberg, K.H.; Tung, S.M.; Tangavelou, K.; Stumpf, M.; Faix, J.; Schroder, R.; Clemen, C.S.; Eichinger, L. Heteromeric p97/p97r155c complexes induce dominant negative changes in wild-type and autophagy 9-deficient dictyostelium strains. PLoS ONE 2012, 7, e46879. [Google Scholar] [CrossRef]
- Chojnacki, S.; Cowley, A.; Lee, J.; Foix, A.; Lopez, R. Programmatic access to bioinformatics tools from embl-ebi update: 2017. Nucleic Acids Res. 2017, 45, W550–W553. [Google Scholar] [CrossRef]
- Capra, J.A.; Singh, M. Predicting functionally important residues from sequence conservation. Bioinformatics 2007, 23, 1875–1882. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the smart protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. Interpro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. Swiss-model: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. Swiss-model and the swiss-pdbviewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Otomo, C.; Metlagel, Z.; Takaesu, G.; Otomo, T. Structure of the human atg12~atg5 conjugate required for lc3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013, 20, 59–66. [Google Scholar] [CrossRef]
- Farbrother, P.; Wagner, C.; Na, J.; Tunggal, B.; Morio, T.; Urushihara, H.; Tanaka, Y.; Schleicher, M.; Steinert, M.; Eichinger, L. Dictyostelium transcriptional host cell response upon infection with legionella. Cell. Microbiol. 2006, 8, 438–456. [Google Scholar] [CrossRef] [PubMed]
- Wagle, P.; Nikolic, M.; Frommolt, P. Quickngs elevates next-generation sequencing data analysis to a new level of automation. BMC Genom. 2015, 16, 487. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, L.; Pachebat, J.A.; Glockner, G.; Rajandream, M.A.; Sucgang, R.; Berriman, M.; Song, J.; Olsen, R.; Szafranski, K.; Xu, Q.; et al. The genome of the social amoeba Dictyostelium discoideum. Nature 2005, 435, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- RCoreTeam. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Gastwirth, J.L.; Gel, Y.R.; Hui, W.L.W.; Lyubchich, V.; Miao, W.; Noguchi, K. Lawstat: Tools for Biostatistics, Public Policy, and Law. R package version 3.3. 2017. [Google Scholar]
- Dinno, A. Dunn.Test: Dunn’s Test of Multiple Comparisons using Rank Sums. R package version. 2017. [Google Scholar]
- Fox, J.; Weisberg, S. (Eds.) An R Companion to Applied Regression, 3rd ed.; Sage Publications: Thousand Oaks, CA, USA, 2018; p. 577. [Google Scholar]
- Wang, M.; Zhao, Y.; Zhang, B. Efficient test and visualization of multi-set intersections. Sci. Rep. 2015, 5, 16923. [Google Scholar] [CrossRef]
- Lew, M. Good statistical practice in pharmacology. Problem 2. Br. J. Pharmacol. 2007, 152, 299–303. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons using rank sums. Technometrics 1964, 6, 241–252. [Google Scholar] [CrossRef]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef]
- Matsushita, M.; Suzuki, N.N.; Obara, K.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. Structure of atg5.Atg16, a complex essential for autophagy. J. Biol. Chem. 2007, 282, 6763–6772. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Noda, N.N.; Yamamoto, H.; Shima, T.; Kumeta, H.; Kobashigawa, Y.; Akada, R.; Ohsumi, Y.; Inagaki, F. Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate. Structure 2012, 20, 1244–1254. [Google Scholar] [CrossRef]
- Soll, D.R.; Yarger, J.; Mirick, M. Stationary phase and the cell cycle of Dictyostelium discoideum in liquid nutrient medium. J. Cell Sci. 1976, 20, 513–523. [Google Scholar]
- Lystad, A.H.; Carlsson, S.R.; Simonsen, A. Toward the function of mammalian atg12-atg5-atg16l1 complex in autophagy and related processes. Autophagy 2019, 15, 1485–1486. [Google Scholar] [CrossRef]
- Mizushima, N. The atg conjugation systems in autophagy. Curr. Opin. Cell Biol. 2019, 63, 1–10. [Google Scholar] [CrossRef]
- Mizushima, N.; Yamamoto, A.; Hatano, M.; Kobayashi, Y.; Kabeya, Y.; Suzuki, K.; Tokuhisa, T.; Ohsumi, Y.; Yoshimori, T. Dissection of autophagosome formation using apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001, 152, 657–668. [Google Scholar] [CrossRef]
- Kishi-Itakura, C.; Koyama-Honda, I.; Itakura, E.; Mizushima, N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 2014, 127, 4089–4102. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Padman, B.S.; Usher, J.; Oorschot, V.; Ramm, G.; Lazarou, M. Atg8 family lc3/gabarap proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during pink1/parkin mitophagy and starvation. J. Cell Biol. 2016, 215, 857–874. [Google Scholar] [CrossRef]
- Sou, Y.S.; Waguri, S.; Iwata, J.; Ueno, T.; Fujimura, T.; Hara, T.; Sawada, N.; Yamada, A.; Mizushima, N.; Uchiyama, Y.; et al. The atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell 2008, 19, 4762–4775. [Google Scholar] [CrossRef]
- Tsuboyama, K.; Koyama-Honda, I.; Sakamaki, Y.; Koike, M.; Morishita, H.; Mizushima, N. The atg conjugation systems are important for degradation of the inner autophagosomal membrane. Science 2016, 354, 1036–1041. [Google Scholar] [CrossRef]
- Uemura, T.; Yamamoto, M.; Kametaka, A.; Sou, Y.S.; Yabashi, A.; Yamada, A.; Annoh, H.; Kametaka, S.; Komatsu, M.; Waguri, S. A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol. Cell. Biol. 2014, 34, 1695–1706. [Google Scholar] [CrossRef]
- Renna, M.; Rubinsztein, D.C. Macroautophagy without lc3 conjugation? Cell Res. 2017, 27, 5–6. [Google Scholar] [CrossRef][Green Version]
- Haller, M.; Hock, A.K.; Giampazolias, E.; Oberst, A.; Green, D.R.; Debnath, J.; Ryan, K.M.; Vousden, K.H.; Tait, S.W. Ubiquitination and proteasomal degradation of atg12 regulates its proapoptotic activity. Autophagy 2014, 10, 2269–2278. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Green, D.R. Autophagy-independent functions of the autophagy machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Warne, J.P.; Salas, E.; Xu, A.W.; Debnath, J. Loss of atg12, but not atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy 2015, 11, 145–154. [Google Scholar]
- Mauthe, M.; Langereis, M.; Jung, J.; Zhou, X.; Jones, A.; Omta, W.; Tooze, S.A.; Stork, B.; Paludan, S.R.; Ahola, T.; et al. An sirna screen for atg protein depletion reveals the extent of the unconventional functions of the autophagy proteome in virus replication. J. Cell Biol. 2016, 214, 619–635. [Google Scholar] [CrossRef]
- Nam, T.; Han, J.H.; Devkota, S.; Lee, H.W. Emerging paradigm of crosstalk between autophagy and the ubiquitin-proteasome system. Mol. Cells 2017, 40, 897–905. [Google Scholar]
- Jounai, N.; Takeshita, F.; Kobiyama, K.; Sawano, A.; Miyawaki, A.; Xin, K.Q.; Ishii, K.J.; Kawai, T.; Akira, S.; Suzuki, K.; et al. The atg5 atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14050–14055. [Google Scholar] [CrossRef]
- Fletcher, K.; Ulferts, R.; Jacquin, E.; Veith, T.; Gammoh, N.; Arasteh, J.M.; Mayer, U.; Carding, S.R.; Wileman, T.; Beale, R.; et al. The wd40 domain of atg16l1 is required for its non-canonical role in lipidation of lc3 at single membranes. EMBO J. 2018, 37, e97840. [Google Scholar] [CrossRef]
- Florey, O.; Kim, S.E.; Sandoval, C.P.; Haynes, C.M.; Overholtzer, M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 2011, 13, 1335–1343. [Google Scholar] [CrossRef]
- Lai, S.C.; Devenish, R.J. Lc3-associated phagocytosis (lap): Connections with host autophagy. Cells 2012, 1, 396–408. [Google Scholar] [CrossRef]
- Rai, S.; Arasteh, M.; Jefferson, M.; Pearson, T.; Wang, Y.; Zhang, W.; Bicsak, B.; Divekar, D.; Powell, P.P.; Naumann, R.; et al. The atg5-binding and coiled coil domains of atg16l1 maintain autophagy and tissue homeostasis in mice independently of the wd domain required for lc3-associated phagocytosis. Autophagy 2019, 15, 599–612. [Google Scholar] [CrossRef]
- Heckmann, B.L.; Green, D.R. Lc3-associated phagocytosis at a glance. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Lima, W.C.; Balestrino, D.; Forestier, C.; Cosson, P. Two distinct sensing pathways allow recognition of klebsiella pneumoniae by dictyostelium amoebae. Cell. Microbiol. 2014, 16, 311–323. [Google Scholar] [CrossRef]
- Nasser, W.; Santhanam, B.; Miranda, E.R.; Parikh, A.; Juneja, K.; Rot, G.; Dinh, C.; Chen, R.; Zupan, B.; Shaulsky, G.; et al. Bacterial discrimination by dictyostelid amoebae reveals the complexity of ancient interspecies interactions. Curr. Biol. 2013, 23, 862–872. [Google Scholar] [CrossRef]
- Gao, Z.; Gammoh, N.; Wong, P.-M.; Erdjument-Bromage, H.; Tempst, P.; Jiang, X. Processing of autophagic protein lc3 by the 20s proteasome. Autophagy 2010, 6, 126–137. [Google Scholar] [CrossRef]
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef]
- Xiong, Q.; Fischer, S.; Karow, M.; Muller, R.; Messling, S.; Eichinger, L. Atg16 mediates the autophagic degradation of the 19s proteasomal subunits psmd1 and psmd2. Eur. J. Cell Biol. 2018, 97, 523–532. [Google Scholar] [CrossRef]
- Ding, W.X.; Ni, H.M.; Gao, W.; Yoshimori, T.; Stolz, D.B.; Ron, D.; Yin, X.M. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am. J. Pathol. 2007, 171, 513–524. [Google Scholar] [CrossRef]
- Ji, C.H.; Kwon, Y.T. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells 2017, 40, 441–449. [Google Scholar]
- Korolchuk, V.I.; Menzies, F.M.; Rubinsztein, D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010, 584, 1393–1398. [Google Scholar] [CrossRef]
- Pandey, U.B.; Nie, Z.; Batlevi, Y.; McCray, B.A.; Ritson, G.P.; Nedelsky, N.B.; Schwartz, S.L.; DiProspero, N.A.; Knight, M.A.; Schuldiner, O.; et al. Hdac6 rescues neurodegeneration and provides an essential link between autophagy and the ups. Nature 2007, 447, 859–863. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Menzies, F.M.; Rubinsztein, D.C. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 2009, 5, 862–863. [Google Scholar] [CrossRef]
- Qiao, L.; Zhang, J. Inhibition of lysosomal functions reduces proteasomal activity. Neurosci. Lett. 2009, 456, 15–19. [Google Scholar] [CrossRef]
- Wang, X.; Terpstra, E.J. Ubiquitin receptors and protein quality control. J. Mol. Cell. Cardiol. 2013, 55, 73–84. [Google Scholar] [CrossRef]
- Fujita, N.; Saitoh, T.; Kageyama, S.; Akira, S.; Noda, T.; Yoshimori, T. Differential involvement of atg16l1 in crohn disease and canonical autophagy: Analysis of the organization of the atg16l1 complex in fibroblasts. J. Biol. Chem. 2009, 284, 32602–32609. [Google Scholar] [CrossRef]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26s proteasome is mediated by the dual atg8/ubiquitin receptor rpn10 in arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Marshall, R.S.; McLoughlin, F.; Vierstra, R.D. Autophagic turnover of inactive 26s proteasomes in yeast is directed by the ubiquitin receptor cue5 and the hsp42 chaperone. Cell Rep. 2016, 16, 1717–1732. [Google Scholar] [CrossRef]
- Waite, K.A.; De-La Mota-Peynado, A.; Vontz, G.; Roelofs, J. Starvation induces proteasome autophagy with different pathways for core and regulatory particles. J. Biol. Chem. 2016, 291, 3239–3253. [Google Scholar] [CrossRef]

| Strains | Summary | References |
|---|---|---|
| AX2 | Axenically growing derivate of wild isolate NC-4 | [46] |
| ATG5¯ | ATG5 null mutant | This work |
| ATG5¯/12¯ | ATG5/12 double null mutant | This work |
| ATG5¯/12¯/16¯ | ATG5/12/16 triple null mutant | This work |
| ATG12¯ | ATG12 null mutant | [39] |
| ATG16¯ | ATG16 null mutant | [40] |
| ATG12¯/16¯ | ATG12/16 double null mutant | [39] |
| Type | Cellular Process | Strains | ||
|---|---|---|---|---|
| ATG5¯ | ATG5¯/12¯ | ATG5¯/12¯/16¯ | ||
| (i) | Development | − | − | − |
| Cell viability | − | − | − | |
| Conjugation of ATG8 to PE | − | − | − | |
| (ii) | Generation time | − | −− | −−− |
| Macropinocytosis | − | − | −− | |
| Phagocytosis of yeast | − | − | −− | |
| (iii) | Growth on K. aerogenes | ++ | − | + |
| Phagocytosis/membrane association of bacteria | ++ | − | + | |
| (iv) | Proteasomal activity | −− | −− | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karow, M.; Fischer, S.; Meßling, S.; Konertz, R.; Riehl, J.; Xiong, Q.; Rijal, R.; Wagle, P.; S. Clemen, C.; Eichinger, L. Functional Characterisation of the Autophagy ATG12~5/16 Complex in Dictyostelium discoideum. Cells 2020, 9, 1179. https://doi.org/10.3390/cells9051179
Karow M, Fischer S, Meßling S, Konertz R, Riehl J, Xiong Q, Rijal R, Wagle P, S. Clemen C, Eichinger L. Functional Characterisation of the Autophagy ATG12~5/16 Complex in Dictyostelium discoideum. Cells. 2020; 9(5):1179. https://doi.org/10.3390/cells9051179
Chicago/Turabian StyleKarow, Malte, Sarah Fischer, Susanne Meßling, Roman Konertz, Jana Riehl, Qiuhong Xiong, Ramesh Rijal, Prerana Wagle, Christoph S. Clemen, and Ludwig Eichinger. 2020. "Functional Characterisation of the Autophagy ATG12~5/16 Complex in Dictyostelium discoideum" Cells 9, no. 5: 1179. https://doi.org/10.3390/cells9051179
APA StyleKarow, M., Fischer, S., Meßling, S., Konertz, R., Riehl, J., Xiong, Q., Rijal, R., Wagle, P., S. Clemen, C., & Eichinger, L. (2020). Functional Characterisation of the Autophagy ATG12~5/16 Complex in Dictyostelium discoideum. Cells, 9(5), 1179. https://doi.org/10.3390/cells9051179







