Deciphering the Enigma of the Histone H2A.Z-1/H2A.Z-2 Isoforms: Novel Insights and Remaining Questions
Abstract
1. Introduction
1.1. Preamble
1.2. Histone H2A.Z in Brief
1.3. Rediscovery of the Two H2A.Z Isoforms
1.4. What We Currently Know
2. Materials and Methods
2.1. Structure Prediction
2.2. Phylogenetic Inference
2.3. Nuclear Magnetic Resonance Spectroscopy
2.4. Tissues, Cell Lines, Preparation of Nuclei, and Chromatin Fractionation
2.5. Cell Cycle Dependence
2.6. Electrophoretic Analysis
2.7. Western Blotting
2.8. RNA Extraction and cDNA Preparation of Liver and Brain
2.9. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.10. Graphic Representation and Statistical Analysis
3. Results
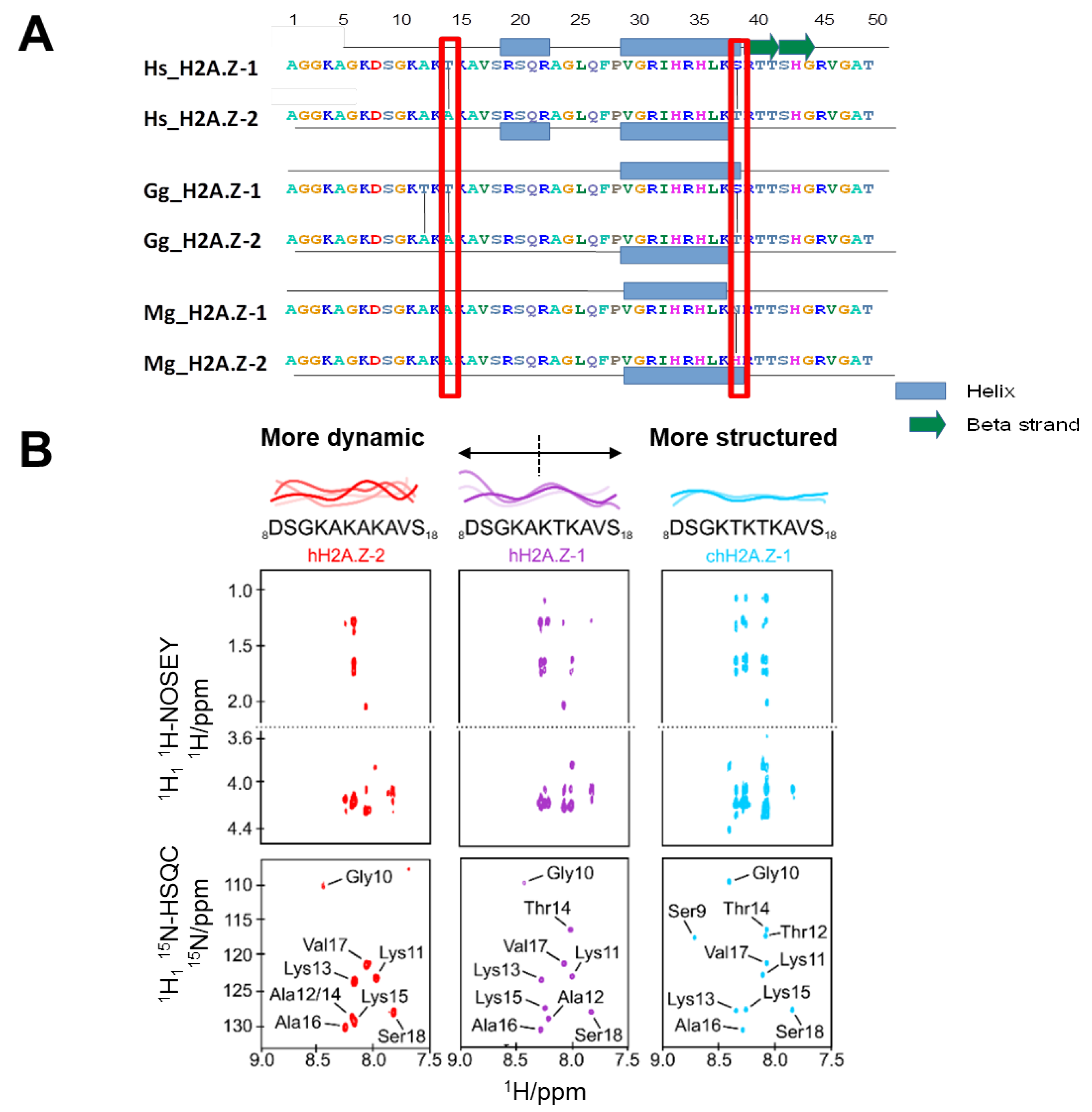
3.1. NMR Studies Reveal the Degree of N-terminal H2A.Z-1 and H2A.Z-2 Structure
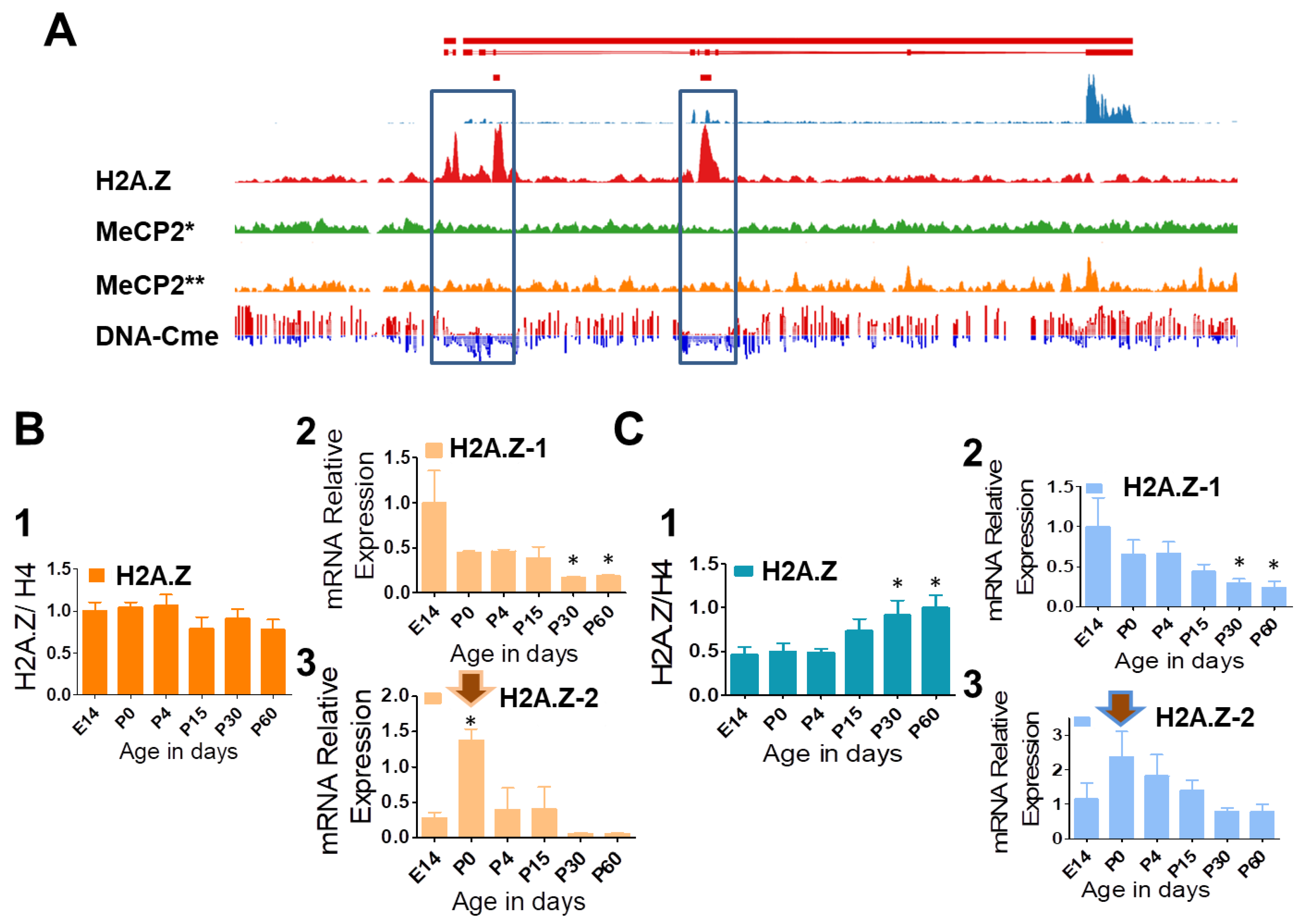
3.2. DNA Methylation, H2A.Z Exclusion, and Developmental Expression Changes of H2A.Z-1 and H2A.Z-2 in Mouse Liver and Brain
3.3. Changes in H2A.Z-1/H2A.Z-2 Composition in Different Cell Types and during the Cell Cycle
3.4. A Specific Role for One of the H2A.Z Isoforms in Spermatogenesis
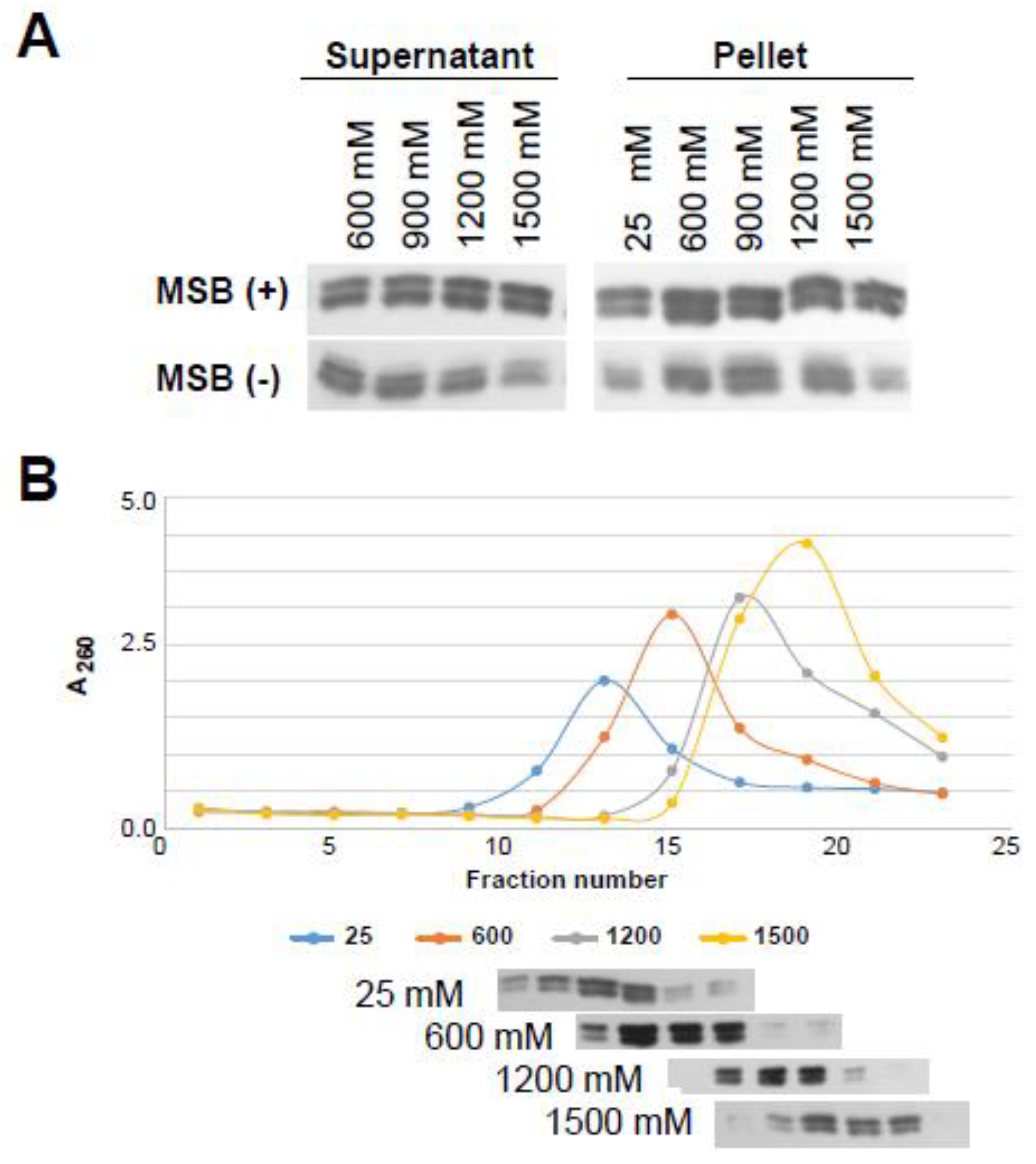
3.5. Histone Isoforms H2A.Z-1 and H2A.Z-2 Confer Identical Ionic Strength-Dependent Chromatin Affinity and Nucleosome Stability In Situ
4. Discussion
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Van Holde, K.E. Chromatin; Springer: New York, NY, USA, 1988. [Google Scholar]
- Arents, G.; Moudrianakis, E.N. The histone fold: A ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 1995, 92, 11170–11174. [Google Scholar] [CrossRef] [PubMed]
- Dominski, Z.; Marzluff, W.F. Formation of the 3’ end of histone mRNA. Gene 1999, 239, 1–14. [Google Scholar] [CrossRef]
- Cheema, M.S.; Ausio, J. The Structural Determinants behind the Epigenetic Role of Histone Variants. Genes (Basel) 2015, 6, 685–713. [Google Scholar] [CrossRef] [PubMed]
- West, M.H.; Bonner, W.M. Histone 2A, a heteromorphous family of eight protein species. Biochemistry 1980, 19, 3238–3245. [Google Scholar] [CrossRef]
- Hatch, C.L.; Bonner, W.M. Sequence of cDNAs for mammalian H2A.Z, an evolutionarily diverged but highly conserved basal histone H2A isoprotein species. Nucleic Acids Res. 1988, 16, 1113–1124. [Google Scholar] [CrossRef]
- Harvey, R.P.; Whiting, J.A.; Coles, L.S.; Krieg, P.A.; Wells, J.R. H2A.F: An extremely variant histone H2A sequence expressed in the chicken embryo. Proc. Natl. Acad. Sci. USA 1983, 80, 2819–2823. [Google Scholar] [CrossRef]
- Guillemette, B.; Gaudreau, L. Reuniting the contrasting functions of H2A.Z. Biochem. Cell Biol. 2006, 84, 528–535. [Google Scholar] [CrossRef]
- Venkatasubrahmanyam, S.; Hwang, W.W.; Meneghini, M.D.; Tong, A.H.; Madhani, H.D. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc. Natl. Acad. Sci. USA 2007, 104, 16609–16614. [Google Scholar] [CrossRef]
- Giaimo, B.D.; Ferrante, F.; Herchenrother, A.; Hake, S.B.; Borggrefe, T. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 2019, 12, 37. [Google Scholar] [CrossRef]
- Hu, G.; Cui, K.; Northrup, D.; Liu, C.; Wang, C.; Tang, Q.; Ge, K.; Levens, D.; Crane-Robinson, C.; Zhao, K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2013, 12, 180–192. [Google Scholar] [CrossRef]
- Draker, R.; Cheung, P. Transcriptional and epigenetic functions of histone variant H2A.Z. Biochem. Cell Biol. 2009, 87, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Draker, R.; Ng, M.K.; Sarcinella, E.; Ignatchenko, V.; Kislinger, T.; Cheung, P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012, 8, e1003047. [Google Scholar] [CrossRef] [PubMed]
- Draker, R.; Sarcinella, E.; Cheung, P. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 2011, 39, 3529–3542. [Google Scholar] [CrossRef] [PubMed]
- van Daal, A.; Elgin, S.C. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell 1992, 3, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Clarkson, M.J.; Wells, J.R.; Gibson, F.; Saint, R.; Tremethick, D.J. Regions of variant histone His2AvD required for Drosophila development. Nature 1999, 399, 694–697. [Google Scholar] [CrossRef]
- van Daal, A.; White, E.M.; Gorovsky, M.A.; Elgin, S.C. Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res. 1988, 16, 7487–7497. [Google Scholar] [CrossRef] [PubMed]
- Dryhurst, D.; Ishibashi, T.; Rose, K.L.; Eirin-Lopez, J.M.; McDonald, D.; Silva-Moreno, B.; Veldhoen, N.; Helbing, C.C.; Hendzel, M.J.; Shabanowitz, J.; et al. Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 2009, 7, 86. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Environmental responses mediated by histone variants. Trends Cell Biol. 2014, 24, 642–650. [Google Scholar] [CrossRef]
- Georgoulis, A.; Vorgias, C.E.; Chrousos, G.P.; Rogakou, E.P. Genome Instability and gammaH2AX. Int. J. Mol. Sci. 2017, 18, 1979. [Google Scholar] [CrossRef]
- Morrison, A.J.; Shen, X. Chromatin remodelling beyond transcription: The INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 2009, 10, 373–384. [Google Scholar] [CrossRef]
- Xu, M.; Chen, S.; Zhu, B. Investigating the cell cycle-associated dynamics of histone modifications using quantitative mass spectrometry. Methods Enzym. 2012, 512, 29–55. [Google Scholar] [CrossRef]
- Nishibuchi, I.; Suzuki, H.; Kinomura, A.; Sun, J.; Liu, N.A.; Horikoshi, Y.; Shima, H.; Kusakabe, M.; Harata, M.; Fukagawa, T.; et al. Reorganization of damaged chromatin by the exchange of histone variant H2A.Z-2. Int J. Radiat Oncol. Biol. Phys. 2014, 89, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Horigome, C.; Oma, Y.; Konishi, T.; Schmid, R.; Marcomini, I.; Hauer, M.H.; Dion, V.; Harata, M.; Gasser, S.M. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol. Cell 2014, 55, 626–639. [Google Scholar] [CrossRef] [PubMed]
- March-Diaz, R.; Garcia-Dominguez, M.; Lozano-Juste, J.; Leon, J.; Florencio, F.J.; Reyes, J.C. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant. J. 2008, 53, 475–487. [Google Scholar] [CrossRef]
- Kumar, S.V.; Wigge, P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Sura, W.; Kabza, M.; Karlowski, W.M.; Bieluszewski, T.; Kus-Slowinska, M.; Paweloszek, L.; Sadowski, J.; Ziolkowski, P.A. Dual Role of the Histone Variant H2A.Z in Transcriptional Regulation of Stress-Response Genes. Plant. Cell 2017, 29, 791–807. [Google Scholar] [CrossRef]
- Jarillo, J.A.; Pineiro, M. H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. Plant. J. 2015, 83, 96–109. [Google Scholar] [CrossRef]
- Kawashima, T.; Lorkovic, Z.J.; Nishihama, R.; Ishizaki, K.; Axelsson, E.; Yelagandula, R.; Kohchi, T.; Berger, F. Diversification of histone H2A variants during plant evolution. Trends Plant. Sci. 2015, 20, 419–425. [Google Scholar] [CrossRef]
- Gomez-Zambrano, A.; Merini, W.; Calonje, M. The repressive role of Arabidopsis H2A.Z in transcriptional regulation depends on AtBMI1 activity. Nat. Commun. 2019, 10, 2828. [Google Scholar] [CrossRef]
- Zilberman, D.; Coleman-Derr, D.; Ballinger, T.; Henikoff, S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 2008, 456, 125–129. [Google Scholar] [CrossRef]
- Deal, R.B.; Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant. Cell 2005, 17, 2633–2646. [Google Scholar] [CrossRef] [PubMed]
- March-Diaz, R.; Reyes, J.C. The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant. 2009, 2, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Kugou, K.; Ding, D.Q.; Fujita, Y.; Hiraoka, Y.; Murakami, H.; Ohta, K.; Yamada, T. The conserved histone variant H2A.Z illuminates meiotic recombination initiation. Curr. Genet. 2018, 64, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Rangasamy, D.; Greaves, I.; Tremethick, D.J. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct Mol. Biol. 2004, 11, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Zhang, L.; Lv, M.; Wen, Z.; Zhang, W.; Chen, X.; Zhang, P.; Li, T.; Chang, L.; Jin, C.; et al. H2A.Z facilitates licensing and activation of early replication origins. Nature 2020, 577, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Suto, R.K.; Clarkson, M.J.; Tremethick, D.J.; Luger, K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000, 7, 1121–1124. [Google Scholar]
- Lee, K.M.; Hayes, J.J. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proc. Natl. Acad. Sci. USA 1997, 94, 8959–8964. [Google Scholar] [CrossRef]
- Eirin-Lopez, J.M.; Gonzalez-Romero, R.; Dryhurst, D.; Ishibashi, T.; Ausio, J. The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol. Biol. 2009, 9, 31. [Google Scholar] [CrossRef]
- Coon, J.J.; Ueberheide, B.; Syka, J.E.; Dryhurst, D.D.; Ausio, J.; Shabanowitz, J.; Hunt, D.F. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 2005, 102, 9463–9468. [Google Scholar] [CrossRef]
- Rivera-Casas, C.; Gonzalez-Romero, R.; Vizoso-Vazquez, A.; Cheema, M.S.; Cerdan, M.E.; Mendez, J.; Ausio, J.; Eirin-Lopez, J.M. Characterization of mussel H2A.Z.2: A new H2A.Z variant preferentially expressed in germinal tissues from Mytilus. Biochem. Cell Biol. 2016, 94, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Park, C.; Lee, J.; Oh, M.; Noh, B.; Lee, I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 2007, 134, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, A. On the possibility of constructive neutral evolution. J. Mol. Evol. 1999, 49, 169–181. [Google Scholar] [CrossRef]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar]
- Semon, M.; Wolfe, K.H. Preferential subfunctionalization of slow-evolving genes after allopolyploidization in Xenopus laevis. Proc. Natl. Acad. Sci. USA 2008, 105, 8333–8338. [Google Scholar] [CrossRef] [PubMed]
- Horikoshi, N.; Sato, K.; Shimada, K.; Arimura, Y.; Osakabe, A.; Tachiwana, H.; Hayashi-Takanaka, Y.; Iwasaki, W.; Kagawa, W.; Harata, M.; et al. Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2. Acta Cryst. D Biol Cryst. 2013, 69, 2431–2439. [Google Scholar] [CrossRef]
- Matsuda, R.; Hori, T.; Kitamura, H.; Takeuchi, K.; Fukagawa, T.; Harata, M. Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 2010, 38, 4263–4273. [Google Scholar] [CrossRef]
- Latrick, C.M.; Marek, M.; Ouararhni, K.; Papin, C.; Stoll, I.; Ignatyeva, M.; Obri, A.; Ennifar, E.; Dimitrov, S.; Romier, C.; et al. Molecular basis and specificity of H2A.Z-H2B recognition and deposition by the histone chaperone YL1. Nat. Struct. Mol. Biol. 2016, 23, 309–316. [Google Scholar] [CrossRef]
- Obri, A.; Ouararhni, K.; Papin, C.; Diebold, M.L.; Padmanabhan, K.; Marek, M.; Stoll, I.; Roy, L.; Reilly, P.T.; Mak, T.W.; et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 2014, 505, 648–653. [Google Scholar] [CrossRef]
- Vardabasso, C.; Hasson, D.; Ratnakumar, K.; Chung, C.Y.; Duarte, L.F.; Bernstein, E. Histone variants: Emerging players in cancer biology. Cell Mol. Life Sci. 2013. [Google Scholar] [CrossRef]
- Dryhurst, D.; McMullen, B.; Fazli, L.; Rennie, P.S.; Ausio, J. Histone H2A.Z prepares the prostate specific antigen (PSA) gene for androgen receptor-mediated transcription and is upregulated in a model of prostate cancer progression. Cancer Lett. 2012, 315, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.D.; Kim, P.J.; Eun, J.W.; Shen, Q.; Kim, H.S.; Shin, W.C.; Ahn, Y.M.; Park, W.S.; Lee, J.Y.; Nam, S.W. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget 2016, 7, 11412–11423. [Google Scholar] [CrossRef] [PubMed]
- Min, C.; Eddy, S.F.; Sherr, D.H.; Sonenshein, G.E. NF-kappaB and epithelial to mesenchymal transition of cancer. J. Cell Biochem. 2008, 104, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Vardabasso, C.; Gaspar-Maia, A.; Hasson, D.; Punzeler, S.; Valle-Garcia, D.; Straub, T.; Keilhauer, E.C.; Strub, T.; Dong, J.; Panda, T.; et al. Histone Variant H2A.Z.2 Mediates Proliferation and Drug Sensitivity of Malignant Melanoma. Mol. Cell 2015, 59, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Vardabasso, C.; Hake, S.B.; Bernstein, E. Histone variant H2A.Z.2: A novel driver of melanoma progression. Mol. Cell Oncol. 2016, 3, e1073417. [Google Scholar] [CrossRef][Green Version]
- Cadet, J.; Douki, T. Formation of UV-induced DNA damage contributing to skin cancer development. Photochem. Photobiol. Sci. 2018, 17, 1816–1841. [Google Scholar] [CrossRef]
- Kauffmann, A.; Rosselli, F.; Lazar, V.; Winnepenninckx, V.; Mansuet-Lupo, A.; Dessen, P.; van den Oord, J.J.; Spatz, A.; Sarasin, A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene 2008, 27, 565–573. [Google Scholar] [CrossRef]
- Fukuto, A.; Ikura, M.; Ikura, T.; Sun, J.; Horikoshi, Y.; Shima, H.; Igarashi, K.; Kusakabe, M.; Harata, M.; Horikoshi, N.; et al. SUMO modification system facilitates the exchange of histone variant H2A.Z-2 at DNA damage sites. Nucleus 2018, 9, 87–94. [Google Scholar] [CrossRef]
- Dunn, C.J.; Sarkar, P.; Bailey, E.R.; Farris, S.; Zhao, M.; Ward, J.M.; Dudek, S.M.; Saha, R.N. Histone Hypervariants H2A.Z.1 and H2A.Z.2 Play Independent and Context-Specific Roles in Neuronal Activity-Induced Transcription of Arc/Arg3.1 and Other Immediate Early Genes. eNeuro 2017, 4. [Google Scholar] [CrossRef]
- Greenberg, R.S.; Long, H.K.; Swigut, T.; Wysocka, J. Single Amino Acid Change Underlies Distinct Roles of H2A.Z Subtypes in Human Syndrome. Cell 2019, 178, 1421–1436.e24. [Google Scholar] [CrossRef]
- Gretzinger, T.L.; Tyagi, M.; Fontaine, C.J.; Cheema, M.S.; Gonzalez-Perez, M.; Freeman, M.E.; Christie, B.R.; Ausio, J. Fetal alcohol spectrum disorder (FASD) affects the hippocampal levels of histone variant H2A.Z-2. Biochem. Cell Biol. 2019, 97, 431–436. [Google Scholar] [CrossRef]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. Nmr 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Martínez de Paz, A.; Khajavi, L.; Martin, H.; Claveria-Gimeno, R.; Tom Dieck, S.; Cheema, M.S.; Sanchez-Mut, J.V.; Moksa, M.M.; Carles, A.; Brodie, N.I.; et al. MeCP2-E1 isoform is a dynamically expressed, weakly DNA-bound protein with different protein and DNA interactions compared to MeCP2-E2. Epigenetics Chromatin 2019, 12, 63. [Google Scholar] [CrossRef]
- Ausió, J.; Dong, F.; van Holde, K.E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 1989, 206, 451–463. [Google Scholar] [CrossRef]
- Wang, X.; Moore, S.C.; Laszckzak, M.; Ausió, J. Acetylation increases the alpha-helical content of the histone tails of the nucleosome. J. Biol. Chem. 2000, 275, 35013–35020. [Google Scholar] [CrossRef]
- Ausió, J.; Seger, D.; Eisenberg, H. Nucleosome core particle stability and conformational change. Effect of temperature, particle and NaCl concentrations, and crosslinking of histone H3 sulfhydryl groups. J. Mol. Biol. 1984, 176, 77–104. [Google Scholar] [CrossRef]
- Ishibashi, T.; Dryhurst, D.; Rose, K.L.; Shabanowitz, J.; Hunt, D.F.; Ausio, J. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry 2009, 48, 5007–5017. [Google Scholar] [CrossRef] [PubMed]
- Thambirajah, A.A.; Ng, M.K.; Frehlick, L.J.; Li, A.; Serpa, J.J.; Petrotchenko, E.V.; Silva-Moreno, B.; Missiaen, K.K.; Borchers, C.H.; Adam Hall, J.; et al. MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Res. 2012, 40, 2884–2897. [Google Scholar] [CrossRef]
- Laemmli, U.K.; Amos, L.A.; Klug, A. Correlation between structural transformation and cleavage of the major head protein of T4 bacteriophage. Cell 1976, 7, 191–203. [Google Scholar] [CrossRef]
- Kasinsky, H.E.; Frehlick, L.J.; Su, H.W.; Ausio, J. Protamines in the internally fertilizing neobatrachian frog Eleutherodactylus coqui. Mol. Reprod. Dev. 2005, 70, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Cheema, M.S.; Dryhurst, D.; Eskiw, C.H.; Ausio, J. Metformin alters H2A.Z dynamics and regulates androgen dependent prostate cancer progression. Oncotarget 2018, 9, 37054–37068. [Google Scholar] [CrossRef]
- Martinez de Paz, A.; Vicente Sanchez-Mut, J.; Samitier-Marti, M.; Petazzi, P.; Saez, M.; Szczesna, K.; Huertas, D.; Esteller, M.; Ausio, J. Circadian cycle-dependent MeCP2 and brain chromatin changes. PLoS ONE 2015, 10, e0123693. [Google Scholar] [CrossRef]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef]
- Coleman-Derr, D.; Zilberman, D. DNA Methylation, H2A.Z, and the Regulation of Constitutive Expression. Cold Spring Harb Symp Quant. Biol. 2012, 77, 144–154. [Google Scholar] [CrossRef]
- Skene, P.J.; Illingworth, R.S.; Webb, S.; Kerr, A.R.; James, K.D.; Turner, D.J.; Andrews, R.; Bird, A.P. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 2010, 37, 457–468. [Google Scholar] [CrossRef]
- Kinde, B.; Wu, D.Y.; Greenberg, M.E.; Gabel, H.W. DNA methylation in the gene body influences MeCP2-mediated gene repression. Proc. Natl. Acad. Sci. USA 2016, 113, 15114–15119. [Google Scholar] [CrossRef]
- Gabel, H.W.; Kinde, B.; Stroud, H.; Gilbert, C.S.; Harmin, D.A.; Kastan, N.R.; Hemberg, M.; Ebert, D.H.; Greenberg, M.E. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 2015, 522, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Olson, C.O.; Zachariah, R.M.; Ezeonwuka, C.D.; Liyanage, V.R.; Rastegar, M. Brain Region-Specific Expression of MeCP2 Isoforms Correlates with DNA Methylation within Mecp2 Regulatory Elements. PLoS ONE 2014, 9, e90645. [Google Scholar] [CrossRef]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef] [PubMed]
- Pina, B.; Suau, P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 1987, 123, 51–58. [Google Scholar] [CrossRef]
- Stefanelli, G.; Azam, A.B.; Walters, B.J.; Brimble, M.A.; Gettens, C.P.; Bouchard-Cannon, P.; Cheng, H.M.; Davidoff, A.M.; Narkaj, K.; Day, J.J.; et al. Learning and Age-Related Changes in Genome-wide H2A.Z Binding in the Mouse Hippocampus. Cell Rep. 2018, 22, 1124–1131. [Google Scholar] [CrossRef]
- Kusakabe, M.; Oku, H.; Matsuda, R.; Hori, T.; Muto, A.; Igarashi, K.; Fukagawa, T.; Harata, M. Genetic complementation analysis showed distinct contributions of the N-terminal tail of H2A.Z to epigenetic regulations. Genes Cells 2016, 21, 122–135. [Google Scholar] [CrossRef]
- Lee, K.M.; Sif, S.; Kingston, R.E.; Hayes, J.J. hSWI/SNF disrupts interactions between the H2A N-terminal tail and nucleosomal DNA. Biochemistry 1999, 38, 8423–8429. [Google Scholar] [CrossRef]
- de Mateo, S.; Castillo, J.; Estanyol, J.M.; Ballesca, J.L.; Oliva, R. Proteomic characterization of the human sperm nucleus. Proteomics 2011, 11, 2714–2726. [Google Scholar] [CrossRef]
- Sharma, U.; Stefanova, D.; Holmes, S.G. Histone variant H2A.Z functions in sister chromatid cohesion in Saccharomyces cerevisiae. Mol. Cell Biol. 2013, 33, 3473–3481. [Google Scholar] [CrossRef]
- Yamada, S.; Kugou, K.; Ding, D.Q.; Fujita, Y.; Hiraoka, Y.; Murakami, H.; Ohta, K.; Yamada, T. The histone variant H2A.Z promotes initiation of meiotic recombination in fission yeast. Nucleic Acids Res. 2018, 46, 609–620. [Google Scholar] [CrossRef]
- Shang, E.; Salazar, G.; Crowley, T.E.; Wang, X.; Lopez, R.A.; Wang, X.; Wolgemuth, D.J. Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene Expr. Patterns 2004, 4, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Denis, G.V.; McComb, M.E.; Faller, D.V.; Sinha, A.; Romesser, P.B.; Costello, C.E. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J. Proteome Res. 2006, 5, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Thambirajah, A.A.; Dryhurst, D.; Ishibashi, T.; Li, A.; Maffey, A.H.; Ausio, J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 2006, 281, 20036–20044. [Google Scholar] [CrossRef] [PubMed]
- Colino-Sanguino, Y.; Cornett, E.M.; Moulder, D.; Smith, G.C.; Hrit, J.; Cordeiro-Spinetti, E.; Vaughan, R.M.; Krajewski, K.; Rothbart, S.B.; Clark, S.J.; et al. A Read/Write Mechanism Connects p300 Bromodomain Function to H2A.Z Acetylation. iScience 2019, 21, 773–788. [Google Scholar] [CrossRef]
- Chen, P.; Wang, Y.; Li, G. Dynamics of histone variant H3.3 and its coregulation with H2A.Z at enhancers and promoters. Nucleus 2014, 5, 21–27. [Google Scholar] [CrossRef]
- Jin, C.; Zang, C.; Wei, G.; Cui, K.; Peng, W.; Zhao, K.; Felsenfeld, G. H3.3/H2A.Z double variant-containing nucleosomes mark ’nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009, 41, 941–945. [Google Scholar] [CrossRef]
- Lamaa, A.; Humbert, J.; Aguirrebengoa, M.; Cheng, X.; Nicolas, E.; Cote, J.; Trouche, D. Integrated analysis of H2A.Z isoforms function reveals a complex interplay in gene regulation. eLife 2020, 9. [Google Scholar] [CrossRef]
- Bagchi, D.N.; Battenhouse, A.M.; Park, D.; Iyer, V.R. The histone variant H2A.Z in yeast is almost exclusively incorporated into the +1 nucleosome in the direction of transcription. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Belotti, E.; Lacoste, N.; Simonet, T.; Papin, C.; Padmanabhan, K.; Scionti, I.; Gangloff, Y.G.; Ramos, L.; Dalkara, D.; Hamiche, A.; et al. H2A.Z is dispensable for both basal and activated transcription in post-mitotic mouse muscles. Nucleic Acids Res. 2020. [Google Scholar] [CrossRef]
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. Embo Rep. 2014, 15, 244–253. [Google Scholar] [CrossRef]
- Yamada, Y.; Haga, H.; Yamada, Y. Concise review: Dedifferentiation meets cancer development: Proof of concept for epigenetic cancer. Stem Cells Transl Med. 2014, 3, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Santenard, A.; Torres-Padilla, M.E. Epigenetic reprogramming in mammalian reproduction: Contribution from histone variants. Epigenetics 2009, 4, 80–84. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Forward Primer Sequence | Reverse Primer Sequence | Annealing Temperature (°C) | References |
|---|---|---|---|---|
| H2A.Z-1 | CACCGCAGAGGTACTTGAGTT | TCCTTTCTTCCCGATCAGCG | 60 | |
| H2A.Z-2 | CAAGGCTAAGGCGGTGTCTC | CTGCTAACTCCAACACCTCAGC | 60 | Matsuda et al., 2010 [48] |
| GAPDH | AACGACCCCTTCATTGAC | TCCACGACATACTGAGCAC | 60 | |
| TBP | CCCCACAACTCTTCCATTCT | GCAGGAGTGATAGGGGTCAT | 60 | Martinez de Paz et al., 2015 [77] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheema, M.S.; Good, K.V.; Kim, B.; Soufari, H.; O’Sullivan, C.; Freeman, M.E.; Stefanelli, G.; Casas, C.R.; Zengeler, K.E.; Kennedy, A.J.; et al. Deciphering the Enigma of the Histone H2A.Z-1/H2A.Z-2 Isoforms: Novel Insights and Remaining Questions. Cells 2020, 9, 1167. https://doi.org/10.3390/cells9051167
Cheema MS, Good KV, Kim B, Soufari H, O’Sullivan C, Freeman ME, Stefanelli G, Casas CR, Zengeler KE, Kennedy AJ, et al. Deciphering the Enigma of the Histone H2A.Z-1/H2A.Z-2 Isoforms: Novel Insights and Remaining Questions. Cells. 2020; 9(5):1167. https://doi.org/10.3390/cells9051167
Chicago/Turabian StyleCheema, Manjinder S., Katrina V. Good, Bohyun Kim, Heddy Soufari, Connor O’Sullivan, Melissa E. Freeman, Gilda Stefanelli, Ciro Rivera Casas, Kristine E. Zengeler, Andrew J. Kennedy, and et al. 2020. "Deciphering the Enigma of the Histone H2A.Z-1/H2A.Z-2 Isoforms: Novel Insights and Remaining Questions" Cells 9, no. 5: 1167. https://doi.org/10.3390/cells9051167
APA StyleCheema, M. S., Good, K. V., Kim, B., Soufari, H., O’Sullivan, C., Freeman, M. E., Stefanelli, G., Casas, C. R., Zengeler, K. E., Kennedy, A. J., Eirin Lopez, J. M., Howard, P. L., Zovkic, I. B., Shabanowitz, J., Dryhurst, D. D., Hunt, D. F., Mackereth, C. D., & Ausió, J. (2020). Deciphering the Enigma of the Histone H2A.Z-1/H2A.Z-2 Isoforms: Novel Insights and Remaining Questions. Cells, 9(5), 1167. https://doi.org/10.3390/cells9051167







