Hearing Loss in Alzheimer’s Disease Is Associated with Altered Serum Lipidomic Biomarker Profiles
Abstract
1. Introduction
2. Methods
2.1. Database
2.2. Lipid Analysis
2.3. Clinical Diagnosis and Hearing Loss Assessment
2.4. Statistical Methods
3. Results
3.1. Demographics
3.2. Lipidomic Biomarker Sets That Separate HL from NHL Subjects
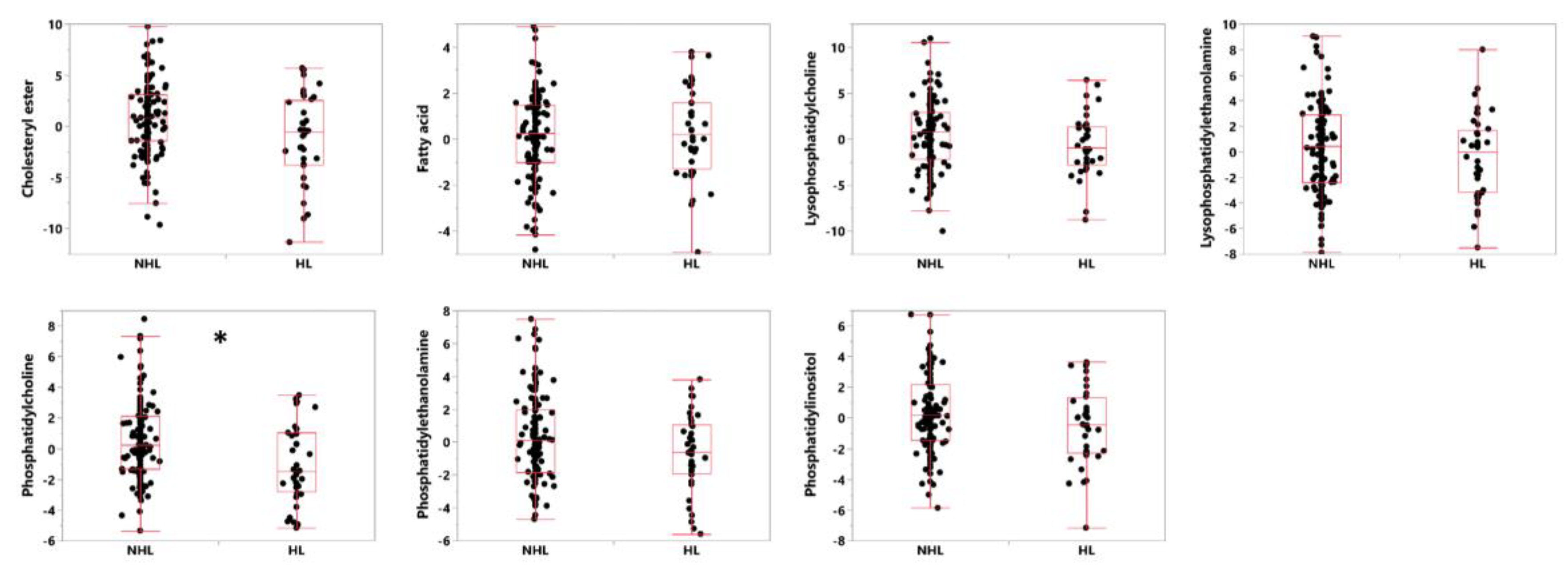
3.3. Lipid Classes and Individual Lipids That Separate HL from NHL Subjects
3.4. Analysis of Non-AD Subjects and Apolipoprotein E (APOE)
4. Discussion
4.1. Weaknesses in the Study
4.2. Phosphatidylcholine, Alzheimer’s Disease and Hearing Loss
4.3. Origins of Measured Lipids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nash, S.D.; Cruickshanks, K.J.; Klein, R.; Klein, B.E.; Nieto, F.J.; Huang, G.H.; Tweed, T.S. The prevalence of hearing impairment and associated risk factors: The Beaver Dam Offspring Study. Arch. Otolaryngol. Head Neck Surg. 2011, 137, 432–439. [Google Scholar] [CrossRef]
- Hebert, L.E.; Weuve, J.; Scherr, P.A.; Evans, D.A. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology 2013, 80, 1778–1783. [Google Scholar] [CrossRef]
- Ortman, J.M.; Velkoff, V.A.; Hogan, H. An Aging Nation: The Older Population in the United States; United States Census Bureau, Economics and Statistics Administration: Washington, DC, USA, 2014. [Google Scholar]
- Lin, F.R.; Ferrucci, L.; Metter, E.J.; An, Y.; Zonderman, A.B.; Resnick, S.M. Hearing loss and cognition in the Baltimore Longitudinal Study of Aging. Neuropsychology 2011, 25, 763–770. [Google Scholar] [CrossRef]
- Lin, F.R.; Metter, E.J.; O’Brien, R.J.; Resnick, S.M.; Zonderman, A.B.; Ferrucci, L. Hearing loss and incident dementia. Arch. Neurol. 2011, 68, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.R.; Thorpe, R.; Gordon-Salant, S.; Ferrucci, L. Hearing loss prevalence and risk factors among older adults in the United States. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2011, 66, 582–590. [Google Scholar] [CrossRef]
- Lin, F.R.; Yaffe, K.; Xia, J.; Xue, Q.-L.; Harris, T.B.; Purchase-Helzner, E.; Simonsick, E.M. Hearing loss and cognitive decline in older adults. JAMA Intern. Med. 2013, 173, 293–299. [Google Scholar] [CrossRef]
- Thomson, R.S.; Auduong, P.; Miller, A.T.; Gurgel, R.K. Hearing loss as a risk factor for dementia: A systematic review. Laryngoscope Investig. Otolaryngol. 2017, 2, 69–79. [Google Scholar] [CrossRef]
- Golub, J.S.; Luchsinger, J.A.; Manly, J.J.; Stern, Y.; Mayeux, R.; Schupf, N. Observed hearing loss and incident dementia in a multiethnic cohort. J. Am. Geriatr. Soc. 2017, 65, 1691–1697. [Google Scholar] [CrossRef]
- Panza, F.; Solfrizzi, V.; Logroscino, G. Age-related hearing impairment—A risk factor and frailty marker for dementia and AD. Nat. Rev. Neurol. 2015, 11, 166–175. [Google Scholar] [CrossRef]
- Ford, A.H.; Hankey, G.J.; Yeap, B.B.; Golledge, J.; Flicker, L.; Almeida, O.P. Hearing loss and the risk of dementia in later life. Maturitas 2018, 112, 1–11. [Google Scholar] [CrossRef]
- Nadhimi, Y.; Llano, D.A. Does hearing loss lead to dementia? A review of the literature. Hear. Res. 2020, 108038. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, W.; Li, L.; Perry, G.; Lee, H.-g.; Zhu, X. Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2014, 1842, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, C.; Yamasoba, T. Oxidative stresses and mitochondrial dysfunction in age-related hearing loss. Oxid. Med. Cell. Longev. 2014, 2014, 582849. [Google Scholar] [CrossRef] [PubMed]
- Someya, S.; Prolla, T.A. Mitochondrial oxidative damage and apoptosis in age-related hearing loss. Mech. Ageing Dev. 2010, 131, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Someya, S.; Tanokura, M.; Weindruch, R.; Prolla, T.A.; Yamasoba, T. Effects of caloric restriction on age-related hearing loss in rodents and rhesus monkeys. Curr. Aging Sci. 2010, 3, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Someya, S.; Xu, J.; Kondo, K.; Ding, D.; Salvi, R.J.; Yamasoba, T.; Tanokura, M. Age-related hearing loss in C57BL/6J mice is mediated by Bak-dependent mitochondrial apoptosis. Proc. Natl. Acad. Sci. USA 2009, 106, 19432–19437. [Google Scholar] [CrossRef] [PubMed]
- Someya, S.; Yu, W.; Hallows, W.C.; Xu, J.; Vann, J.M.; Leeuwenburgh, C.; Prolla, T.A. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 2010, 143, 802–812. [Google Scholar] [CrossRef]
- Menardo, J.; Tang, Y.; Ladrech, S.; Lenoir, M.; Casas, F.; Michel, C.; Maurice, T. Oxidative stress, inflammation, and autophagic stress as the key mechanisms of premature age-related hearing loss in SAMP8 mouse Cochlea. Antioxid. Redox Signal. 2012, 16, 263–274. [Google Scholar] [CrossRef]
- Campbell, K.C.; Rybak, L.P.; Khardori, R. Sensorineural hearing loss and dyslipidemia. Am. J. Audiol. 1996, 5, 11–14. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, C.; Li, J.-Q.; Tan, C.-C.; Cao, X.-P.; Tan, L.; Yu, J.-T.; Alzheimer’s Disease Neuroimaging Initiative. Age-related hearing loss accelerates cerebrospinal fluid tau levels and brain atrophy: A longitudinal study. Aging 2019, 11, 3156–3169. [Google Scholar] [CrossRef]
- Kao, Y.-C.; Ho, P.-C.; Tu, Y.-K.; Jou, I.; Tsai, K.-J. Lipids and Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 1505. [Google Scholar] [CrossRef] [PubMed]
- Barupal, D.K.; Baillie, R.; Fan, S.; Saykin, A.J.; Meikle, P.J.; Arnold, M.; Nho, K.; Fiehn, O.; Kaddurah-Daouk, R.; Alzheimer Disease Metabolomics Consortium. Sets of coregulated serum lipids are associated with Alzheimer’s disease pathophysiology. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2019, 11, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Gutierrez, E.; Muñoz-Arenas, G.; Treviño, S.; Espinosa, B.; Chavez, R.; Rojas, K.; Guevara, J. Alzheimer’s disease and metabolic syndrome: A link from oxidative stress and inflammation to neurodegeneration. Synapse 2017, 71, e21990. [Google Scholar] [CrossRef]
- Verdile, G.; Keane, K.N.; Cruzat, V.F.; Medic, S.; Sabale, M.; Rowles, J.; Newsholme, P. Inflammation and oxidative stress: The molecular connectivity between insulin resistance, obesity, and Alzheimer’s disease. Mediat. Inflamm. 2015, 2015, 105828. [Google Scholar] [CrossRef] [PubMed]
- Llano, D.; Li, J.; Waring, J.; Ellis, T.; Devanarayan, V.; Witte, D.; Lenz, R. Cerebrospinal fluid cytokine dynamics differ between Alzheimer’s Disease patients and elderly controls. Alzheimer Dis. Assoc. Disord. 2011, 26, 322–328. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fan, S.; Wancewicz, B.; Cajka, T.; Sa, M.; Showalter, M.R.; Kaddurah-Daouk, R. Generation and quality control of lipidomics data for the alzheimer’s disease neuroimaging initiative cohort. Sci. Data 2018, 5, 180263. [Google Scholar] [CrossRef]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R. On testing the significance of sets of genes. Ann. Appl. Stat. 2007, 1, 107–129. [Google Scholar] [CrossRef]
- Kamil, R.J.; Genther, D.J.; Lin, F.R. Factors associated with the accuracy of subjective assessments of hearing impairment. Ear Hear. 2015, 36, 164–167. [Google Scholar] [CrossRef]
- Whiley, L.; Sen, A.; Heaton, J.; Proitsi, P.; García-Gómez, D.; Leung, R.; Mecocci, P. Evidence of altered phosphatidylcholine metabolism in Alzheimer’s disease. Neurobiol. Aging 2014, 35, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mejia, R.O.; Mucke, L. Phospholipase A2 and arachidonic acid in Alzheimer’s disease. Biochim. Biophys. Acta 2010, 1801, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, E.J.; Bongard, V.; Beiser, A.S.; Lamon-Fava, S.; Robins, S.J.; Au, R.; Wolf, P.A. Plasma phosphatidylcholine docosahexaenoic acid content and risk of dementia and Alzheimer disease: The Framingham Heart Study. Arch. Neurol. 2006, 63, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Zamroziewicz, M.K.; Zwilling, C.E.; Barbey, A.K. Inferior prefrontal cortex mediates the relationship between phosphatidylcholine and executive functions in healthy, older adults. Front. Aging Neurosci. 2016, 8, 226. [Google Scholar] [CrossRef]
- Seidman, M.D.; Khan, M.J.; Tang, W.X.; Quirk, W.S. Influence of lecithin on mitochondrial DNA and age-related hearing loss. Otolaryngol. Head Neck Surg. 2002, 127, 138–144. [Google Scholar] [CrossRef]
- Kurutas, E.B. The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: Current state. Nutr. J. 2015, 15, 1–22. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, S. The role of mitochondrial oxidative stress in hearing loss. Neurol. Disord. Ther. 2017, 1, 1–5. [Google Scholar] [CrossRef]
- Seidman, M.D.; Khan, M.J.; Dolan, D.F.; Quirk, W.S. Age-related differences in cochlear microcirculation and auditory brain stem response. Arch. Otolaryngol. Head Neck Surg. 1996, 122, 1221–1226. [Google Scholar] [CrossRef]
- Bruce, K.D.; Zsombok, A.; Eckel, R.H. Lipid processing in the brain: A key regulator of systemic metabolism. Front. Endocrinol. 2017, 8, 60. [Google Scholar] [CrossRef]
- Rapoport, S.I.; Chang, M.C.; Spector, A.A. Delivery and turnover of plasma-derived essential PUFAs in mammalian brain. J. Lipid Res. 2001, 42, 678–685. [Google Scholar]
- Malgrange, B.; Varela-Nieto, I.; de Medina, P.; Paillasse, M.R. Targeting cholesterol homeostasis to fight hearing loss: A new perspective. Front. Aging Neurosci. 2015, 7, 3. [Google Scholar] [CrossRef] [PubMed]
| Lipid Classes | Lipid Count |
|---|---|
| Acylcarnitine | 9 |
| Ceramide | 19 |
| Cholesterol | 1 |
| Cholesteryl ester | 8 |
| Diacylglycerol | 13 |
| Fatty acid | 29 |
| Galactoylceramide | 1 |
| Glucosylceramide | 6 |
| Lactosylceramide | 1 |
| Lysophosphatidylcholine | 22 |
| Lysophosphatidylethanolamine | 4 |
| Phosphatidylcholine | 82 |
| Phosphatidylethanolamine | 25 |
| Phosphatidylinositol | 11 |
| Sphingomyelin | 34 |
| Triacylglycerol | 84 |
| NHL | HL | ||
|---|---|---|---|
| n (# of AD subjects) | 145 | 40 | |
| Gender * (n) | F | 77 | 13 |
| M | 68 | 27 | |
| Age in years (Mean +/− SD) | 74.8 (7.7) | 77.2 (5.8) | |
| BMI in kg/m2 (Mean +/− SD) | 25.32 (3.8) | 26 (4.1) | |
| Use of lipid-lowering drugs (n) | No | 69 | 18 |
| Yes | 76 | 22 | |
| ADAS13 (Mean +/− SD) | 28.6 (7.6) | 30.4 (8) | |
| Lipid Set | Median (NHL) | Median (HL) | ROC AUC | p-Value (unadj.) | q-Value (FDR-BH) |
|---|---|---|---|---|---|
| Set.23 | 0.54 | −2.1 | 0.66 | 0.0006 | 0.0175 |
| Set.4 | 0.16 | −1.06 | 0.64 | 0.0032 | 0.0447 |
| Set.6 | −0.05 | 0.35 | 0.62 | 0.0148 | 0.1332 |
| Set.25 | 1 | −1.3 | 0.62 | 0.019 | 0.1332 |
| Set.3 | 0.37 | −0.74 | 0.6 | 0.0297 | 0.1664 |
| Set.16 | −0.16 | 0.27 | 0.59 | 0.0529 | 0.2171 |
| Set.10 | 0.26 | −1.18 | 0.6 | 0.0543 | 0.2171 |
| Set.14 | 0.51 | −1.16 | 0.6 | 0.0705 | 0.2467 |
| Set.11 | 0.55 | −1.21 | 0.61 | 0.0802 | 0.2495 |
| Set.27 | 0.6 | −1.61 | 0.58 | 0.1025 | 0.2871 |
| Set.19 | 0.2 | −0.35 | 0.58 | 0.1238 | 0.3014 |
| Set.7 | 0.06 | −0.74 | 0.58 | 0.1292 | 0.3014 |
| Set.8 | 0.31 | −0.26 | 0.57 | 0.1694 | 0.3467 |
| Set.28 | 0.4 | −0.12 | 0.56 | 0.1788 | 0.3467 |
| Set.15 | 0.71 | −0.31 | 0.56 | 0.1857 | 0.3467 |
| Set.20 | 0.22 | −0.45 | 0.56 | 0.2013 | 0.3523 |
| Set.13 | −0.67 | −1.3 | 0.56 | 0.2554 | 0.4206 |
| Set.17 | 0.34 | 0.2 | 0.47 | 0.2836 | 0.4387 |
| Set.24 | −0.4 | −1.43 | 0.56 | 0.3135 | 0.4387 |
| Set.2 | −0.2 | 0.25 | 0.47 | 0.3153 | 0.4387 |
| Set.9 | 0.54 | −0.33 | 0.55 | 0.329 | 0.4387 |
| Set.1 | −0.3 | −1.07 | 0.55 | 0.3879 | 0.4937 |
| Set.26 | 0.02 | 0.58 | 0.48 | 0.4248 | 0.5171 |
| Set.22 | −0.18 | −0.35 | 0.53 | 0.4606 | 0.5359 |
| Set.18 | −0.1 | −0.68 | 0.54 | 0.4785 | 0.5359 |
| Set.5 | 0.54 | −0.55 | 0.53 | 0.6265 | 0.6747 |
| Set.21 | −0.21 | 0.32 | 0.5 | 0.8784 | 0.9109 |
| Set.12 | 0.09 | 0.07 | 0.51 | 0.9827 | 0.9827 |
| Lipid Class | Median (NHL) | Median (HL) | ROC AUC | p-Value (Unadjusted) |
|---|---|---|---|---|
| Phosphatidylcholine | 0.25 | −1.48 | 0.63 | 0.0057 * |
| Phosphatidylethanolamine | 0.13 | −0.6 | 0.59 | 0.0216 |
| Cholesteryl ester | 0.58 | −0.53 | 0.62 | 0.0239 |
| Phosphatidylinositol | 0.16 | −0.45 | 0.58 | 0.1142 |
| Lysophosphatidylcholine | 0.48 | −0.93 | 0.58 | 0.1255 |
| Lysophosphatidylethanolamine | 0.35 | −0.02 | 0.55 | 0.3185 |
| Fatty acid | 0.12 | 0.22 | 0.54 | 0.3344 |
| Lipid ID | Lipid Class | Lipid Set | Median (NHL) | Median (HL) | Fold Change (HL/NHL) | p-Value |
|---|---|---|---|---|---|---|
| UCD.Lipid.162 | Phosphatidylcholine | Set-23 | 63,671 | 52,397 | 0.82 | 0.0003 |
| UCD.Lipid.163 | Phosphatidylcholine | Set-23 | 29,367 | 24,419.5 | 0.83 | 0.0006 |
| UCD.Lipid.148 | Phosphatidylcholine | Set-4 | 5,056,746 | 4,380,450.5 | 0.87 | 0.0010 |
| UCD.Lipid.161 | Phosphatidylcholine | Set-23 | 46,893 | 39,148.5 | 0.83 | 0.0014 |
| UCD.Lipid.164 | Phosphatidylcholine | Set-23 | 20,525 | 16,734.5 | 0.82 | 0.0033 |
| UCD.Lipid.17 | Cholesteryl ester | Set-4 | 254,617 | 198,944.5 | 0.78 | 0.0055 |
| UCD.Lipid.150 | Phosphatidylcholine | Set-4 | 58,706 | 49,712 | 0.85 | 0.0069 |
| UCD.Lipid.406 | Phosphatidylcholine | Set-4 | 130,587 | 116,420 | 0.89 | 0.0079 |
| UCD.Lipid.128 | Lysophosphatidylcholine | Set-4 | 45,190 | 37,318.5 | 0.83 | 0.0130 |
| UCD.Lipid.451 | Phosphatidylethanolamine | Set-23 | 5726.33 | 4968 | 0.87 | 0.0149 |
| UCD.Lipid.143 | Phosphatidylcholine | Set-4 | 23,780 | 19,794 | 0.83 | 0.0150 |
| UCD.Lipid.409 | Phosphatidylcholine | Set-4 | 69,527 | 57,894 | 0.83 | 0.0163 |
| UCD.Lipid.149 | Phosphatidylcholine | Set-4 | 35,516.5 | 27,777 | 0.78 | 0.0166 |
| UCD.Lipid.462 | Phosphatidylinositol | Set-4 | 9408 | 8256 | 0.88 | 0.0183 |
| UCD.Lipid.447 | Phosphatidylethanolamine | Set-23 | 12,761 | 11,301.5 | 0.89 | 0.0197 |
| UCD.Lipid.450 | Phosphatidylethanolamine | Set-23 | 12,513.79 | 10,888.5 | 0.87 | 0.0217 |
| UCD.Lipid.410 | Phosphatidylcholine | Set-4 | 7910 | 7552 | 0.95 | 0.0310 |
| UCD.Lipid.145 | Phosphatidylcholine | Set-4 | 22,891 | 19,311 | 0.84 | 0.0329 |
| UCD.Lipid.126 | Lysophosphatidylcholine | Set-4 | 14,172.5 | 12,585 | 0.89 | 0.0428 |
| UCD.Lipid.399 | Phosphatidylcholine | Set-4 | 98,283 | 79,011 | 0.80 | 0.0661 |
| UCD.Lipid.16 | Cholesteryl ester | Set-4 | 179,126 | 125,757 | 0.70 | 0.0846 |
| UCD.Lipid.442 | Phosphatidylethanolamine | Set-4 | 1817.5 | 1492 | 0.82 | 0.1016 |
| UCD.Lipid.381 | Lysophosphatidylethanolamine | Set-4 | 5577.5 | 5206 | 0.93 | 0.2087 |
| UCD.Lipid.517 | Fatty acid | Set-4 | 111,966 | 100,323 | 0.90 | 0.5032 |
| UCD.Lipid.513 | Fatty acid | Set-4 | 22,307 | 21,665 | 0.97 | 0.7021 |
| MCI Subjects | |||||
| Lipid Set | Median (HN) | Median (HL) | ROC AUC | p-Value (unadj.) | q-Value (FDR-BH) |
| Set.13 | −0.91 | −0.42 | 0.54 | 0.0538 | 0.9348 |
| Set.22 | 0.03 | 0.82 | 0.53 | 0.0775 | 0.9348 |
| Set.14 | −0.26 | 0.17 | 0.53 | 0.1122 | 0.9348 |
| Set.25 | 0.19 | −0.08 | 0.49 | 0.1824 | 0.9468 |
| Set.16 | −0.22 | 0.26 | 0.56 | 0.23 | 0.9468 |
| Set.18 | −1.34 | −1.33 | 0.51 | 0.3203 | 0.9468 |
| Set.27 | 0.31 | −0.15 | 0.55 | 0.363 | 0.9468 |
| Set.8 | 0.04 | 0.32 | 0.51 | 0.4406 | 0.9468 |
| Set.17 | 0.03 | −0.1 | 0.5 | 0.4798 | 0.9468 |
| Set.24 | −1.51 | −0.39 | 0.51 | 0.5114 | 0.9468 |
| Subjects without Memory Loss or Complaint | |||||
| Lipid Set | Median (HN) | Median (HL) | ROC AUC | p-Value (unadj.) | q-Value (FDR-BH) |
| Set.22 | 0.4 | 1.7 | 0.59 | 0.0092 | 0.2293 |
| Set.17 | 0.48 | −0.84 | 0.6 | 0.0502 | 0.4158 |
| Set.14 | 0.37 | 0.54 | 0.54 | 0.0632 | 0.4158 |
| Set.13 | 0.14 | 1.03 | 0.57 | 0.0818 | 0.4158 |
| Set.3 | −0.1 | −0.07 | 0.53 | 0.0832 | 0.4158 |
| Set.7 | −0.33 | −0.18 | 0.54 | 0.1062 | 0.4163 |
| Set.8 | −0.37 | 0.74 | 0.57 | 0.1166 | 0.4163 |
| Set.11 | −0.49 | −0.24 | 0.54 | 0.1438 | 0.4495 |
| Set.20 | −0.14 | −0.17 | 0.55 | 0.2283 | 0.5529 |
| Set.23 | 1.25 | 1.24 | 0.48 | 0.2393 | 0.5529 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llano, D.A.; Issa, L.K.; Devanarayan, P.; Devanarayan, V.; Alzheimer’s Disease Neuroimaging Initiative (ADNI). Hearing Loss in Alzheimer’s Disease Is Associated with Altered Serum Lipidomic Biomarker Profiles. Cells 2020, 9, 2556. https://doi.org/10.3390/cells9122556
Llano DA, Issa LK, Devanarayan P, Devanarayan V, Alzheimer’s Disease Neuroimaging Initiative (ADNI). Hearing Loss in Alzheimer’s Disease Is Associated with Altered Serum Lipidomic Biomarker Profiles. Cells. 2020; 9(12):2556. https://doi.org/10.3390/cells9122556
Chicago/Turabian StyleLlano, Daniel A., Lina K. Issa, Priya Devanarayan, Viswanath Devanarayan, and Alzheimer’s Disease Neuroimaging Initiative (ADNI). 2020. "Hearing Loss in Alzheimer’s Disease Is Associated with Altered Serum Lipidomic Biomarker Profiles" Cells 9, no. 12: 2556. https://doi.org/10.3390/cells9122556
APA StyleLlano, D. A., Issa, L. K., Devanarayan, P., Devanarayan, V., & Alzheimer’s Disease Neuroimaging Initiative (ADNI). (2020). Hearing Loss in Alzheimer’s Disease Is Associated with Altered Serum Lipidomic Biomarker Profiles. Cells, 9(12), 2556. https://doi.org/10.3390/cells9122556





