A Singular and Widespread Group of Mobile Genetic Elements: RNA Circles with Autocatalytic Ribozymes
Abstract
1. Introduction
2. The Discovery of the First CircRNAs
3. Genome-Encoded CircRNAs
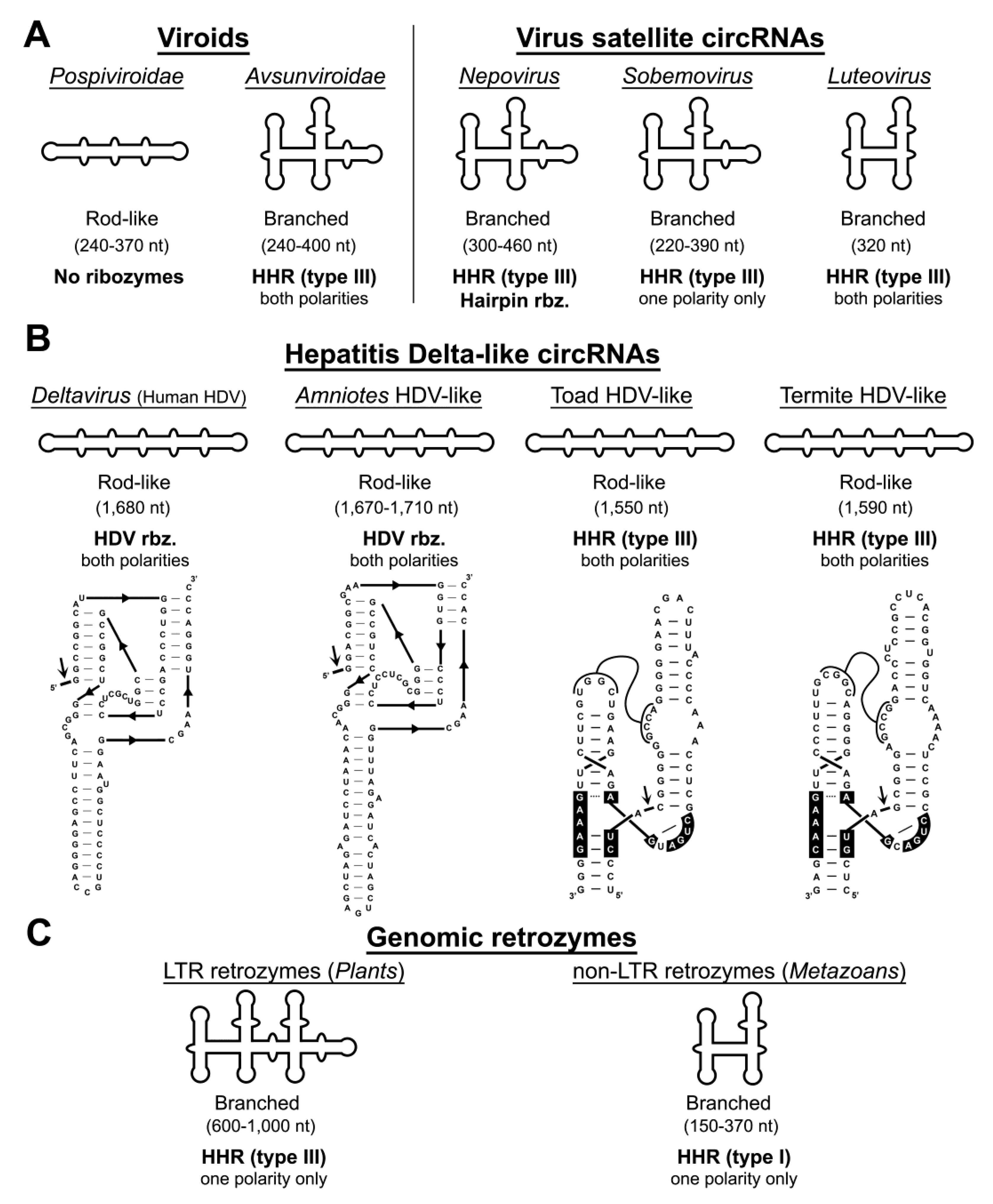
4. Autocatalytic RNA Cleavage in Diverse RNA Circles
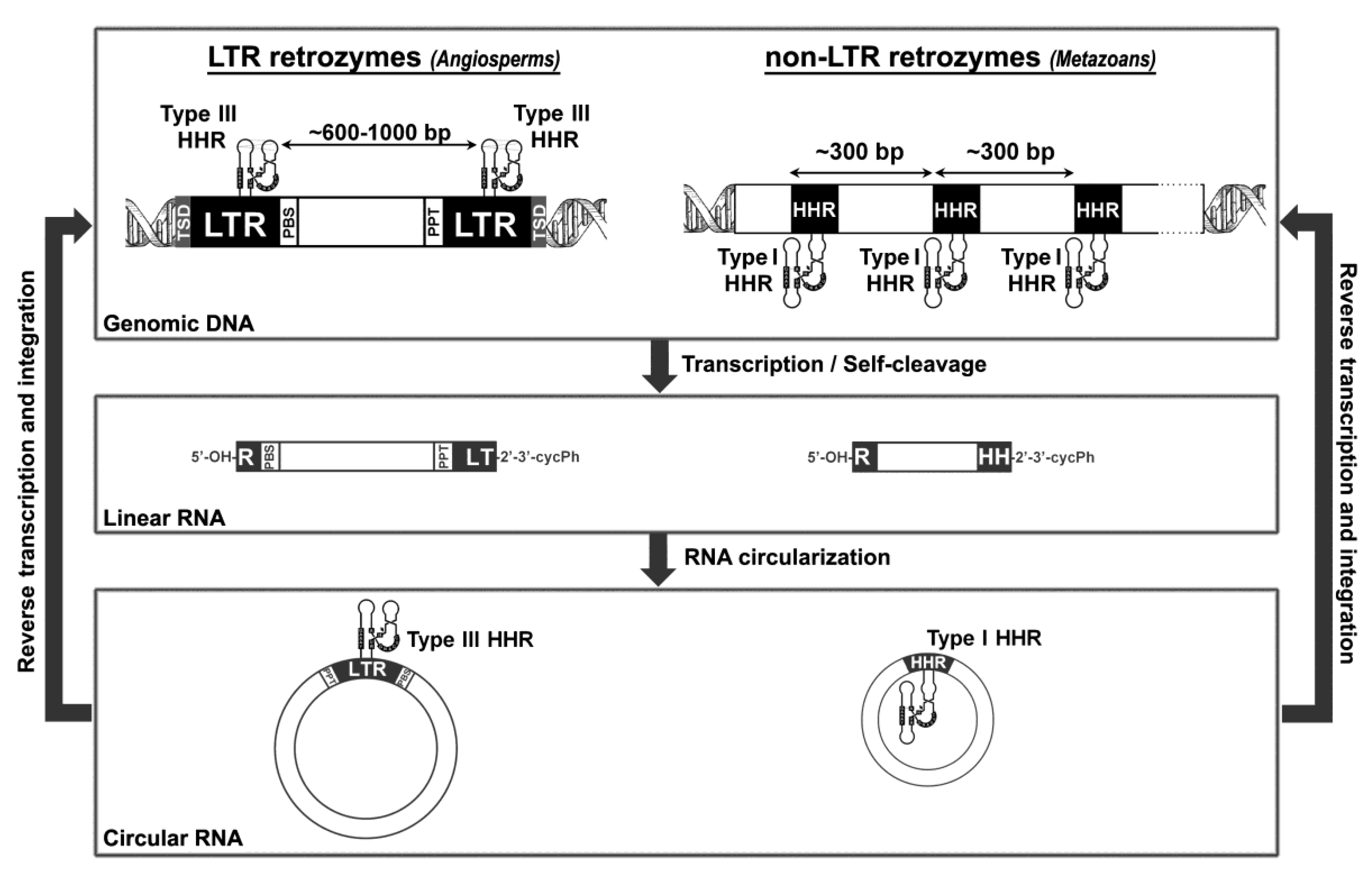
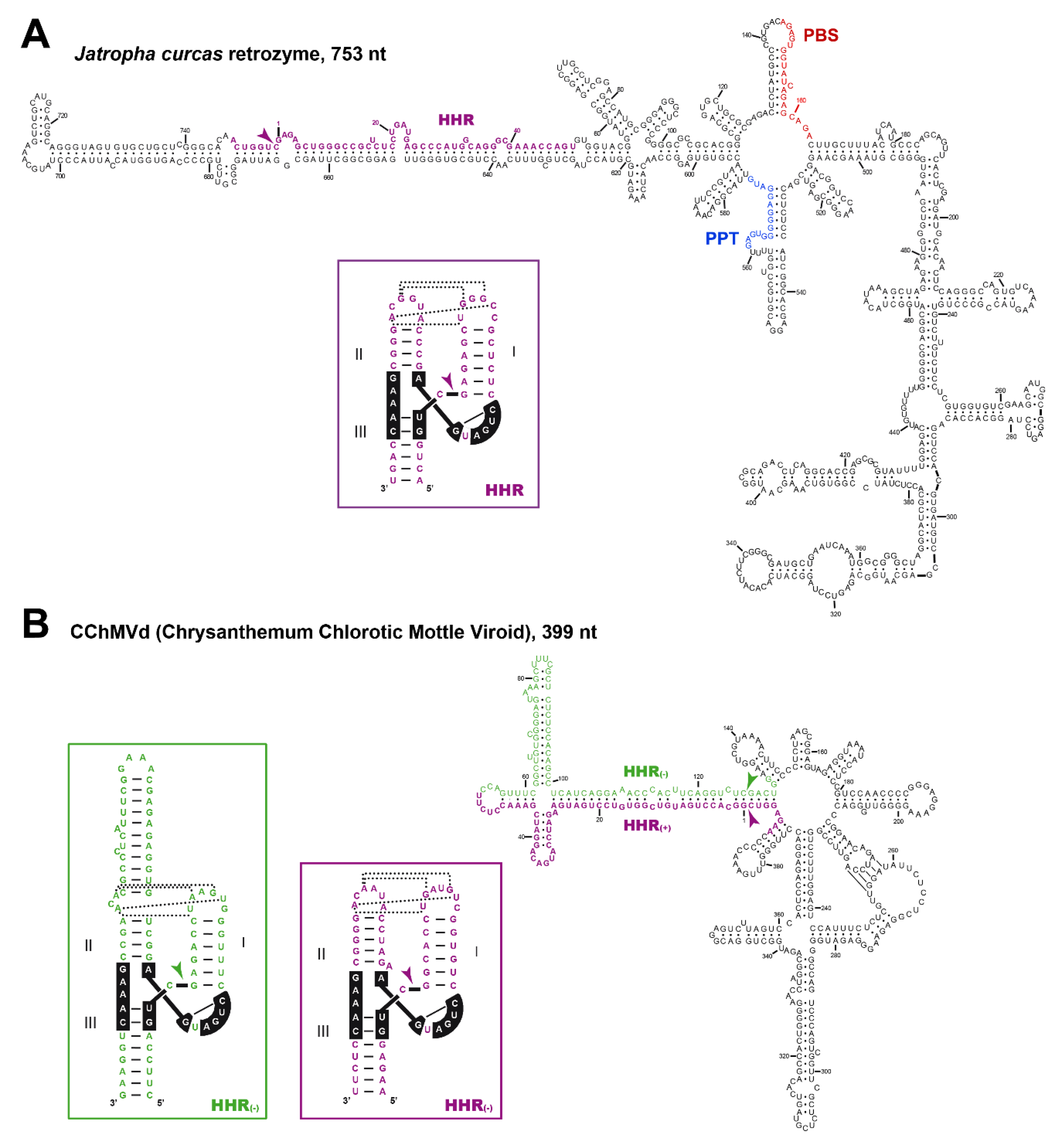
5. Hammerhead Ribozymes in the Genomes of Flowering Plants: The Family of LTR Retrozymes
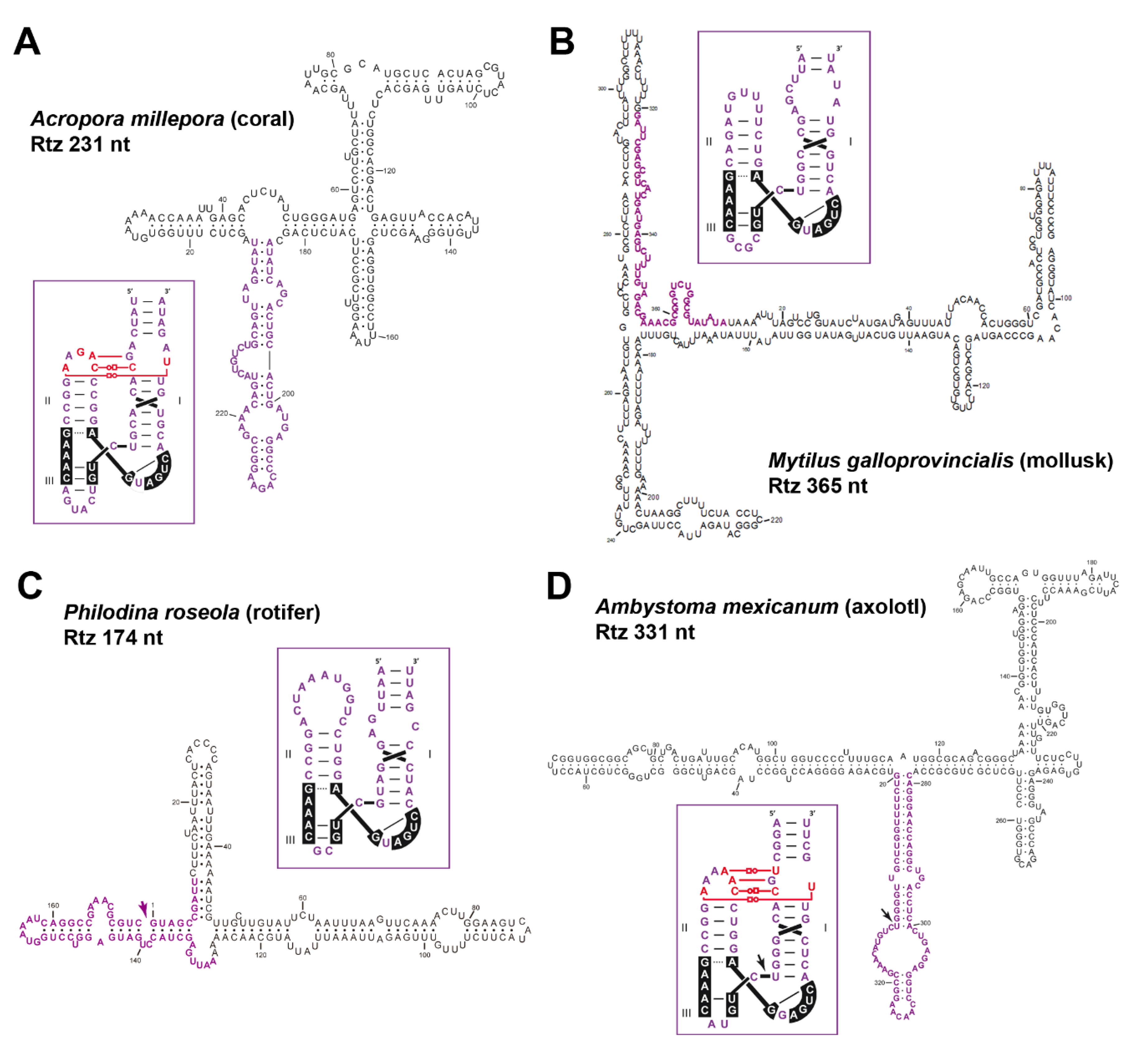
6. Diverse Type I HHRs Occur in Autonomous and in Non-autonomous Metazoan Retrotransposons
7. A general Model for Retrozyme Spreading in Plant and Animal Genomes
8. Concluding Remarks
Funding
Conflicts of Interest
References
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Randles, J.W.; Davies, C.; Hatta, T.; Gould, A.R.; Francki, R.I. Studies on encapsidated viroid-like RNA I. Characterization of velvet tobacco mottle virus. Virology 1981, 108, 111–122. [Google Scholar] [CrossRef]
- Daròs, J.A.; Flores, R. Identification of a retroviroid-like element from plants. Proc. Natl. Acad. Sci. USA 1995, 92, 6856–6860. [Google Scholar] [CrossRef]
- Hegedus, K.; Dallmann, G.; Balázs, E. The DNA form of a retroviroid-like element is involved in recombination events with itself and with the plant genome. Virology 2004, 325, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Minoia, S.; Carbonell, A.; Gisel, A.; Delgado, S.; Lopez-Carrasco, A.; Navarro, B.; Di Serio, F. Viroids, the simplest RNA replicons: How they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015, 209, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, R.M.; Polanco, J.A.; Lupták, A. Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 2015, 40, 648–661. [Google Scholar] [CrossRef]
- Makino, S.; Chang, M.F.; Shieh, C.K.; Kamahora, T.; Vannier, D.M.; Govindarajan, S.; Lai, M.M. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature 1987, 329, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Rizzetto, M. Hepatitis D virus: Introduction and epidemiology. Cold Spring Harb. Perspect. Med. 2015, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- AbouHaidar, M.G.; Venkataraman, S.; Golshani, A.; Liu, B.; Ahmad, T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc. Natl. Acad. Sci. USA 2014. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Netter, H.J.; Littlejohn, M.; Yuen, L.; Shi, M.; Eden, J.S.; Klaassen, M.; Holmes, E.C.; Hurt, A.C. A divergent hepatitis D-like agent in birds. Viruses 2018, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- Hetzel, U.; Szirovicza, L.; Smura, T.; Prähauser, B.; Vapalahti, O.; Kipar, A.; Hepojoki, J. Identification of a novel deltavirus in boa constrictors. MBio 2019, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, S.; Pirzer, F.; Goldmann, N.; Schmid, J.; Corman, V.M.; Gottula, L.T.; Schroeder, S.; Rasche, A.; Muth, D.; Drexler, J.F.; et al. Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus. Proc. Natl. Acad. Sci. USA 2020, 117, 17977–17983. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.-S.; Pettersson, J.H.-O.; Le Lay, C.; Shi, M.; Lo, N.; Wille, M.; Eden, J.-S.; Holmes, E.C. Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 2019. [Google Scholar] [CrossRef]
- Perez-Vargas, J.; Amirache, F.; Boson, B.; Mialon, C.; Freitas, N.; Sureau, C.; Fusil, F.; Cosset, F.L. Enveloped viruses distinct from HBV induce dissemination of hepatitis D virus in vivo. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Diener, T.O. Circular RNAs: Relics of precellular evolution? Proc. Natl. Acad. Sci. USA 1989, 86, 9370–9374. [Google Scholar] [CrossRef]
- Diener, T.O. Viroids: “living fossils” of primordial RNAs? Biol. Direct. 2016, 11, 15. [Google Scholar] [CrossRef]
- Chela-Flores, J. Are Viroids Molecular Fossils of the RNA World? J. Theor. Biol. 1994, 166, 163–166. [Google Scholar] [CrossRef]
- Flores, R.; Gago-Zachert, S.; Serra, P.; Sanjuan, R.; Elena, S.F. Viroids: Survivors from the RNA world? Annu. Rev. Microbiol. 2014, 68, 395–414. [Google Scholar] [CrossRef]
- De la Peña, M.; García-Robles, I.; Cervera, A. The hammerhead ribozyme: A long history for a short RNA. Molecules 2017, 22, 78. [Google Scholar] [CrossRef]
- De la Peña, M.; Cervera, A. Circular RNAs with hammerhead ribozymes encoded in eukaryotic genomes: The enemy at home. RNA Biol. 2017, 14, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, P.J.; Zaug, A.J.; Cech, T.R. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of tetrahymena. Cell 1981, 23, 467–476. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Haugen, P.; Simon, D.M.; Bhattacharya, D. The natural history of group I introns. Trends Genet. 2005, 21, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.; Fiskaa, T.; Birgisdottir, Å.B.; Haugen, P.; Einvik, C.; Johansen, S. The ability to form full-length intron RNA circles is a general property of nuclear group I introns. RNA 2003, 9, 1464–1475. [Google Scholar] [CrossRef]
- Murray, H.L.; Mikheeva, S.; Coljee, V.W.; Turczyk, B.M.; Donahue, W.F.; Bar-Shalom, A.; Jarrell, K.A. Excision of Group II Introns as Circles. Mol. Cell 2001, 8, 201–211. [Google Scholar] [CrossRef]
- Molina-Sánchez, M.D.; Martinez-Abarca, F.; Toro, N. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J. Biol. Chem. 2006, 281, 28737–28744. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Daubersies, P.; Majerus, M.A.; Kerckaert, J.P.; Bailleul, B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992, 11, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Talhouarne, G.J.; Gall, J.G. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 2014, 20, 1476–1487. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Cervera, A.; Urbina, D.; de la Peña, M. Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs. Genome Biol. 2016, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Cervera, A.; de la Peña, M. Small circRNAs with self-cleaving ribozymes are highly expressed in diverse metazoan transcriptomes. Nucleic Acids Res. 2020, 48, 5054–5064. [Google Scholar] [CrossRef] [PubMed]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Gilbert, W. The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Steitz, T.A.; Moore, P.B. RNA, the first macromolecular catalyst: The ribosome is a ribozyme. Trends Biochem. Sci. 2003, 28, 411–418. [Google Scholar] [CrossRef]
- Prody, G.A.; Bakos, J.T.; Buzayan, J.M.; Schneider, I.R.; Bruening, G. Autolytic processing of dimeric plant virus satellite RNA. Science 1986, 231, 1577–1580. [Google Scholar] [CrossRef]
- Hutchins, C.J.; Rathjen, P.D.; Forster, A.C.; Symons, R.H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986, 14, 3627–3640. [Google Scholar] [CrossRef]
- Buzayan, J.M.; Gerlach, W.L.; Bruening, G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 1986, 323, 349–353. [Google Scholar] [CrossRef]
- Kuo, M.Y.; Sharmeen, L.; Dinter-Gottlieb, G.; Taylor, J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol. 1988, 62, 4439–4444. [Google Scholar] [CrossRef]
- Saville, B.J.; Collins, R.A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 1990, 61, 685–696. [Google Scholar] [CrossRef]
- Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004, 428, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.; Weinberg, Z.; Chen, A.G.; Kim, P.B.; Ames, T.D.; Breaker, R.R. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2014, 10, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, Z.; Kim, P.B.; Chen, T.H.; Li, S.; Harris, K.A.; Lünse, C.E.; Breaker, R.R. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 2015, 11, 606–610. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, M.; Gago, S.; Flores, R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003, 22, 5561–5570. [Google Scholar] [CrossRef]
- Khvorova, A.; Lescoute, A.; Westhof, E.; Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003, 10, 708–712. [Google Scholar] [CrossRef]
- Martick, M.; Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 2006, 126, 309–320. [Google Scholar] [CrossRef]
- Przybilski, R.; Graf, S.; Lescoute, A.; Nellen, W.; Westhof, E.; Steger, G.; Hammann, C. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell 2005, 17, 1877–1885. [Google Scholar] [CrossRef]
- Ferbeyre, G.; Smith, J.M.; Cedergren, R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell Biol. 1998, 18, 3880–3888. [Google Scholar] [CrossRef]
- Rojas, A.A.; Vazquez-Tello, A.; Ferbeyre, G.; Venanzetti, F.; Bachmann, L.; Paquin, B.; Sbordoni, V.; Cedergren, R. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res. 2000, 28, 4037–4043. [Google Scholar] [CrossRef]
- Epstein, L.M.; Gall, J.G. Self-cleaving transcripts of satellite DNA from the newt. Cell 1987, 48, 535–543. [Google Scholar] [CrossRef]
- Martick, M.; Horan, L.H.; Noller, H.F.; Scott, W.G. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 2008, 454, 899–902. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, M.; García-Robles, I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA 2010, 16, 1943–1950. [Google Scholar] [CrossRef]
- Jimenez, R.M.; Delwart, E.; Luptak, A. Structure-based search reveals hammerhead ribozymes in the human microbiome. J. Biol. Chem. 2011, 286, 7737–7743. [Google Scholar] [CrossRef] [PubMed]
- Perreault, J.; Weinberg, Z.; Roth, A.; Popescu, O.; Chartrand, P.; Ferbeyre, G.; Breaker, R.R. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011, 7, e1002031. [Google Scholar] [CrossRef] [PubMed]
- Seehafer, C.; Kalweit, A.; Steger, G.; Gräf, S.; Hammann, C. From alpaca to zebrafish: Hammerhead ribozymes wherever you look. RNA 2011, 17, 21–26. [Google Scholar] [CrossRef]
- De la Peña, M.; García-Robles, I. Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep. 2010, 11, 711–716. [Google Scholar] [CrossRef]
- Hammann, C.; Luptak, A.; Perreault, J.; de la Peña, M. The ubiquitous hammerhead ribozyme. RNA 2012, 18, 871–885. [Google Scholar] [CrossRef]
- García-Robles, I.; Sanchez-Navarro, J.; de la Peña, M. Intronic hammerhead ribozymes in mRNA biogenesis. Biol. Chem. 2012, 393, 1317–1326. [Google Scholar] [CrossRef]
- Webb, C.H.T.; Riccitelli, N.J.; Ruminski, D.J.; Lupták, A. Widespread occurrence of self-cleaving ribozymes. Science 2009, 326, 953. [Google Scholar] [CrossRef]
- Salehi-Ashtiani, K.; Lupták, A.; Litovchick, A.; Szostak, J.W. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 2006, 313, 1788–1792. [Google Scholar] [CrossRef]
- Eickbush, D.G.; Eickbush, T.H. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol. Cell Biol. 2010, 30, 3142–3150. [Google Scholar] [CrossRef] [PubMed]
- Ruminski, D.J.; Webb, C.H.T.; Riccitelli, N.J.; Lupták, A. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J. Biol. Chem. 2011, 286, 41286–41295. [Google Scholar] [CrossRef]
- Sánchez-Luque, F.J.; López, M.C.; Macias, F.; Alonso, C.; Thomas, M.C. Identification of an hepatitis delta virus-like ribozyme at the mRNA 5′-end of the L1Tc retrotransposon from Trypanosoma cruzi. Nucleic Acids Res. 2011, 39, 8065–8077. [Google Scholar] [CrossRef] [PubMed]
- Cervera, A.; de la Peña, M. Eukaryotic penelope-like retroelements encode hammerhead ribozyme motifs. Mol. Biol. Evol. 2014, 31, 2941–2947. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R.; Yushenova, I.A.; Rodriguez, F. Giant reverse transcriptase-encoding transposable elements at telomeres. Mol. Biol. Evol. 2017, 34, 2245–2257. [Google Scholar] [CrossRef] [PubMed]
- Gorinsek, B.; Gubensek, F.; Kordis, D. Evolutionary genomics of chromoviruses in eukaryotes. Mol. Biol. Evol. 2004, 21, 781–798. [Google Scholar] [CrossRef] [PubMed]
- Witte, C.P.; Le, Q.H.; Bureau, T.; Kumar, A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc. Natl. Acad Sci. USA 2001, 98, 13778–13783. [Google Scholar] [CrossRef]
- Gao, D.; Chen, J.; Chen, M.; Meyers, B.C.; Jackson, S. A highly conserved, small LTR retrotransposon that preferentially targets genes in grass genomes. PLoS ONE 2012, 7, e32010. [Google Scholar] [CrossRef]
- Nohales, M.A.; Molina-Serrano, D.; Flores, R.; Daròs, J.A. Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae. J. Virol. 2012, 86, 8269–8276. [Google Scholar] [CrossRef]
- Chi, Y.I.; Martick, M.; Lares, M.; Kim, R.; Scott, W.G.; Kim, S.H. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008, 6, e234. [Google Scholar] [CrossRef]
- Dufour, D.; de la Peña, M.; Gago, S.; Flores, R.; Gallego, J. Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: A loop-loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res. 2009, 37, 368–381. [Google Scholar] [CrossRef] [PubMed]
- Gago, S.; de la Peña, M.; Flores, R. A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA 2005, 11, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.C.; Davies, C.; Sheldon, C.C.; Jeffries, A.C.; Symons, R.H. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature 1988, 334, 265–267. [Google Scholar] [CrossRef] [PubMed]
- Lunse, C.E.; Weinberg, Z.; Breaker, R.R. Numerous small hammerhead ribozyme variants associated with Penelope-like retrotransposons cleave RNA as dimers. RNA Biol. 2017, 14, 1499–1507. [Google Scholar] [CrossRef]
- Koonin, E.V.; Krupovic, M. The depths of virus exaptation. Curr. Opin. Virol. 2018, 31, 1–8. [Google Scholar] [CrossRef]
- Jangam, D.; Feschotte, C.; Betran, E. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet. 2017, 33, 817–831. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de la Peña, M.; Ceprián, R.; Cervera, A. A Singular and Widespread Group of Mobile Genetic Elements: RNA Circles with Autocatalytic Ribozymes. Cells 2020, 9, 2555. https://doi.org/10.3390/cells9122555
de la Peña M, Ceprián R, Cervera A. A Singular and Widespread Group of Mobile Genetic Elements: RNA Circles with Autocatalytic Ribozymes. Cells. 2020; 9(12):2555. https://doi.org/10.3390/cells9122555
Chicago/Turabian Stylede la Peña, Marcos, Raquel Ceprián, and Amelia Cervera. 2020. "A Singular and Widespread Group of Mobile Genetic Elements: RNA Circles with Autocatalytic Ribozymes" Cells 9, no. 12: 2555. https://doi.org/10.3390/cells9122555
APA Stylede la Peña, M., Ceprián, R., & Cervera, A. (2020). A Singular and Widespread Group of Mobile Genetic Elements: RNA Circles with Autocatalytic Ribozymes. Cells, 9(12), 2555. https://doi.org/10.3390/cells9122555





