Abstract
Physical exercise (PE) is recommended for Rheumatoid Arthritis (RA), but the molecular and biological mechanisms that impact the inflammatory process and joint destruction in RA remain unknown. The objective of this study was to evaluate the effect of PE on the histological and transcriptional changes in the joints of adjuvant-induced arthritis (AIA) rat model. AIA rats were subjected to PE on a treadmill for eight weeks. The joints were subjected to histological and microarray analysis. The differentially expressed genes (DEGs) by PE in the arthritic rats were obtained from the microarray. The bioinformatic analysis allowed the association of these genes in biological processes and signaling pathways. PE induced the differential expression of 719 genes. The DEGs were significantly associated with pathogenic mechanisms in RA, including HIF-1, VEGF, PI3-Akt, and Jak-STAT signaling pathways, as well as response to oxidative stress and inflammatory response. At a histological level, PE exacerbated joint inflammatory infiltrate and tissue destruction. The PE exacerbated the stressed joint environment aggravating the inflammatory process, the hypoxia, and the oxidative stress, conditions described as detrimental in the RA joints. Research on the effect of PE on the pathogenesis process of RA is still necessary for animal models and human.
1. Introduction
Rheumatoid arthritis (RA) is one of the most common inflammatory arthritis affecting 0.5–1.0% of the population [1]. RA is a systemic autoimmune disease characterized by inflammation of the synovial membrane and periarticular structures. Joint pain, stiffness, and swelling are the primary symptoms, which eventually lead to joint deformity and functional disability. Extra-articular manifestations, including cardiovascular, pulmonary, psychological and skeletal disorders are common in RA patients [2]. The etiology and pathophysiology of RA remain unresolved; however, genetic and environmental factors have been associated with the inappropriate immunomodulation that triggers the inflammatory process in the joints and the subsequent damage to synovial structures [3]. The treatment of RA aims to reduce pain and inflammation in the joints and also to preserve their structural integrity and the patient’s functionality. Currently, treatment includes a variety of pharmacological agents, education programs, joint protection, lifestyle changes, PE and surgical intervention as a final step [4,5,6].
PE is considered in the multidisciplinary management of RA patients. The European League Against Rheumatism (EULAR) has established the recommendations for physical activity in people with inflammatory arthritis [4], and its recommendations for cardiovascular disease risk management in RA include regular exercise [7]. It has been previously shown that PE improves joint health [8], muscle strength [9], cardiovascular fitness [10], vascular function [11], and psychological well being [12]. PE also reduces the inflammation [13], rheumatoid cachexia [14], and fatigue [15] in RA patients.
Although the systemic benefits of PE in rheumatic patients are well recognized, the direct consequences of PE in active synovitis and the potential role of PE accelerating joint destruction are not clear. Some reports have shown that mechanical load influences the onset and worsens the progression of the disease in experimental animal models of arthritis [16,17]; therefore, the consequences of PE on inflamed joints remain a topic that should be addressed.
We selected adjuvant-induced arthritis (AIA) because it mimics the signs and symptoms of human RA, such as the histopathological changes of inflammation, vascular proliferation, and importantly, the progression to joint destruction [18]. The AIA model has been widely used in the development of antirheumatic drugs, including several non-steroid anti-inflammatories, methotrexate [19], and also biologic drugs, including tocilizumab [20] and Jak-inhibitors like tofacitinib [21].
To understand the effects of PE in the complexity of the inflamed joint, we selected an unbiased approach to maintain a broad perspective. We selected the bioinformatic analysis of the transcriptome from the tarsal joint through a DNA microarray. The microarray simultaneously evaluates the expression levels for a large number of genes [22]; and, identifies the most relevant mediators in complex processes such as the interplay between PE and inflammation.
A better understanding of the molecular consequences of PE on the inflamed joints may contribute to improving its prescription for RA patients for the protection of joint integrity. In this regard, the objective of this study was to determine the effect of an exercise intervention on the transcriptional expression of genes in the AIA rat model using microarray technology.
2. Materials and Methods
2.1. Animals and Study Groups
Male Wistar rats (300–350g) were used for this study. The animals were housed in an animal facility with a 12 h light/dark cycle maintained at temperatures between 22 ± 1 °C with food and water provided at libitum. The experimental protocol was approved by the Committee of Ethics of the Instituto Chihuahuense de Salud-Secretaría de Salud-Facultad de Medicina y Ciencias Biomédicas, UACH (protocol number CEI-EXP-140/15). The animals were randomly divided into two groups of seven animals each: (1) arthritis group and (2) arthritis-exercise group.
2.2. Arthritis Induction
The AIA model was conducted as previously reported [23]. Rats were injected in the footpads with 0.2 mL of Complete Freund’s Adjuvant (CFA) (Sigma Chemicals, St. Louis, MO, USA) mixed with phosphate buffer saline in the 1:1 ratio. To increase the severity of arthritis, a booster injection with 0.1 mL of the emulsion was administered in the same way on day 5–post first injection.
2.3. Familiarization
Animals belonging to the arthritis-exercise group were familiarized with the treadmill for three weeks to reduce their stress level during the PE. Each daily familiarization session included placing the rats on the treadmill switched off for 10 min (visual/olfactory adaptation) and then turning on the treadmill at the minimum speed (3 m/min) for 5 min (sound/movement adaptation). After the first week of the treadmill familiarization, arthritis induction was started and the familiarization period continued for two more weeks.
2.4. Physical Capacity Test
After the familiarization, a physical capacity test (PCT) was conducted to establish the maximum speed reached by the animals before the intervention of PE. The PCT consisted of a single treadmill session in which the rats, after 5 min of warm-up (3 m/min), ran in the band with an increase in the speed of 3 m/min every 2 min until they fatigued and stopped running. The maximal physical capacity (100%) was defined as the maximum speed reached by each animal. The average speed per group was calculated, and this was used to establish the speed of the exercise sessions.
2.5. Exercise Program
The rats were exercised three times a week for eight weeks. The exercise started with a speed of 20% of the PCT and increases of 13% in speed were applied every two weeks until reaching 60%. Each exercise session included four phases: (1) acclimatization: rats were placed on the treadmill switched off for 5 min; (2) warm-up: rats walked for 5 min at lowest speed; (3) exercise: rats run for 25 min at the corresponding speed; and (4) cool down: rats walked at lowest speed for 5 min. The exercise sessions were administered at the same time each day.
2.6. Histological Analysis
After exercise intervention, the rats were euthanized, and the tarsal joints were immediately dissected. The ankle, the subtalar and the navicular cuboid joints were extracted and cut proximally in the distal fibula and tibia; and, distally in the metatarsal bones at the diaphysis. The muscle was dissected as much as possible and the synovial and ligament structures were preserved and fixed in 10% formalin phosphate buffer for 48 h and decalcified with 5% nitric acid for 24 h [24]. The tissues were dehydrated in graded ethanol, and embedded in paraffin, sectioned, and stained with hematoxylin-eosin (H&E Merck, Darmstadt, Germany). The histological variables: (a) inflammatory infiltrate, (b) synovial hyperplasia, (c) pannus formation, (d) synovial vascularity, (e) cartilage damage and (f) bone erotion were semi-quantitatively evaluated using a scale of 0 = absent, 1 = mild, 2 = moderate and, 3 = severe on each animal. The maximum score reached per animal for each parameter was 12 when the four joints showed a severe level. The scoring average per study group was estimated. The histological variables were evaluated bu two experienced operators blinded to the different groups.
2.7. DNA Microarray and Bioinformatics Analysis
The effect of PE on transcriptome was evaluated in the DNA microarray by comparing the arthritis-exercise group (experimental group) and the arthritis group (reference group). The tarsal joints dissected were immediately placed in liquid nitrogen and disrupted with mortar and pestle. Total RNA was extracted using RNeasy® Lipid Tissue Mini Kit (QIAGEN, Germantown, MD, USA) according to the manufacturer´s instructions. The RNA quality and integrity were verified using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). For the microarray, the RNA of every group was pooled keeping equimolar quantities of every individual.
The microarray analysis was performed at the Institute of Cellular Physiology of the National Autonomous University of Mexico (México City, México). Briefly, the cDNA was synthesized and labeled for subsequent hybridization in the Rn5K microchip containing 5000 rat genes. Scanning and signal acquisition was performed using the ScanArray 4000 (Packard BioChips Technologies, Billerica, MA, USA). The GenArise software was used for the analysis of the microarray scan, and the lists of DEGs [Z-score ≥ 1.5 standard deviations (SD)] were obtained. To assess the biological relevance of the DEGs, DAVID Bioinformatics Resources 6.8 platform (https://david.ncifcrf.gov/) was used. Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway mapper with significant associations (p ≤ 0.5) were obtained [25]. Also, the STRING 10.5 database (http://string-db.org/) was used to obtain the analysis and integration of direct and indirect protein-protein interactions (IPP) centered on the functional association [26]. The DEGs identified in the microarray were loaded, and the interactions were selected with minimal confidence (interaction score > 0.4). The obtained IPP network was analyzed more thoroughly to obtain primary clusters of sub-networks using the Cytoscape software version 3.7.0 (Bethesda, Rockville, MD, USA) with the Molecular Complex Detection (MCODE) complement [27,28].
2.8. Statistical Analysis
The statistical analysis was made in SPSS statistics v22 software (SPSS Science Inc., Chicago, IL, USA). Measures of central tendency were estimated for each variable. T-test was used to compare the effect of PE on histological parameters. Differences were considered significant when p < 0.05.
3. Results
3.1. Maximal Physical Capacity
The maximal physical capacity obtained in the PCT for the arthritis-exercise group was 21.28 ± 5.96 m/min. The initial speed and its increments along the exercise intervention were based on this value.
3.2. Histological Analysis
The effect of exercise in the joints was histologically evaluated using H&E staining. The scores of inflammatory infiltrate, synovial hyperplasia, pannus formation, synovial vascularity, and cartilage and erosions obtained for each study group are shown in Figure 1. The highest scores of all evaluated parameters were reached in the arthritis exercise group. However, only hyperplasia, cartilage damage and bone erosion were statistically different between the groups.
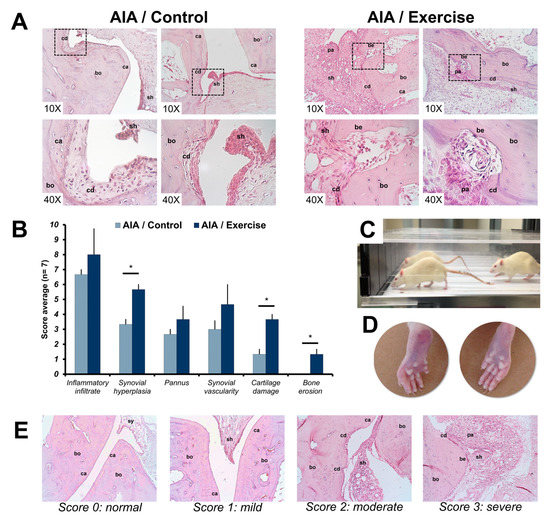
Figure 1.
Effects of exercise on tarsal bone histological parameters in adjuvant-induced arthritis rats. (A) Representative images of histological findings in the tarsal joints of the study groups at the end of the exercise intervention using H&E staining. (B) Joint involvement was scored by the semi-quantitative scale (showed in E) to describe inflammatory changes and structural remodeling in the tarsal joints (7 rats per group). The t-student test was used to compare histological measurements between groups. * p < 0.01. (C) Exercised rats on a treadmill. (D) Representative images of clinical changes on hind paws of adjuvant-induced arthritis (AIA) rats exercised (left) and non-exercised (right). (E) Representative images of the inflammation and structural joint damage scores in the tarsal joints of AIA rats. The 0 (normal) score was established in healthy rats, where the bone (bo), cartilage (ca) and synovium (sy) did not show alterations. The arthritis scores 1 (mild), 2 (moderate), and 3 (severe) were based on the inflammatory changes: the presence of synovial hyperplasia (sh) and pannus (pa) and structural remodeling: cartilage damage (cd) and bone erosion (be). The images were acquired with a 10× and 40× amplification. AIA: adjuvant-induced arthritis.
3.3. Microarray and Bioinformatic Analysis
The transcriptome-wide microarray analysis identified a pool of DEGs in the joints from arthritic exercised rats in comparison to arthritic non-exercised rats. A total of 719 genes were differentially expressed (Z-score ≥ 1.5 SD), 361 up-regulated (Appendix A), and 358 down-regulated (Appendix B). AB000928 (Zp1) and X68101 (Trg) were the most significantly up- and down-expressed genes by PE, respectively.
To understand the related biological functions of the 719 DEGs, we used the DAVID database for the enrichment analysis of GO and KEGG pathways. The top 10 enriched terms of GO and KEGG pathways are shown in Figure 2. Response to organic cyclic compound, response to drug, and response to ethanol were among the top enriched biological processes. The DEGs were enriched in molecular functions like protein binding. The most enriched KEGG pathways included hypoxia-induced factor-1 (HIF-1), cyclic adenosine monophosphate (cAMP) and mitogen-activated protein kinase (MAPK) signaling pathways. Subsequently, we selected the biological processes and KEGG signaling pathways known to be relevant in the pathogenesis of RA, which included: response to hypoxia, response to oxidative stress, angiogenesis and inflammatory/immune response (Table 1).
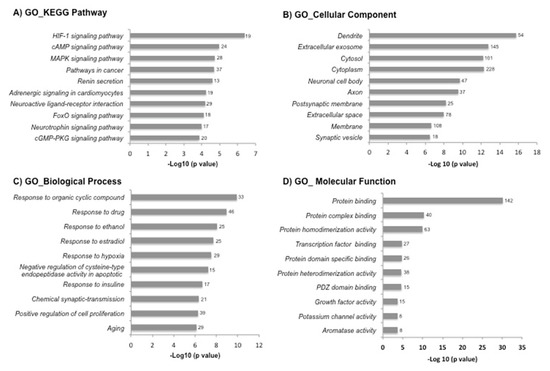
Figure 2.
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes in the DAVID database. The 719 differentially expressed genes by exercise were uploaded into the DAVID database for enrichment analysis. The top 10 GO analysis results of these dysregulated genes were displayed in the bar chart: (A) KEGG pathway, (B) cellular component, (C) biological process and (D) molecular function. The bars indicated the -Log10 (p value) of each GO and KEGG term. The number of genes involved in each term is shown on the right side of each bar.

Table 1.
Differentially expressed genes by physical exercise associated with pathogenic processes in Rheumatoid Arthritis.
After the analysis on DAVID, the list of the 719 DEGs was analyzed on the STRING and Cytoscape-MCODE platforms. The first three clusters obtained are shown in Figure 3, Figure 4 and Figure 5. The genes from the first three clusters were loaded in STRING to identify the associated KEGG signaling pathways; those relevant in RA were selected and marked with different colors. The pathways considered as relevant included the HIF-1 signaling pathway. The analysis of the protein networks unveiled relevant genes dysregulated by PE. Those presented multiple functional connections included up-regulated genes such as serine-threonine protein kinases Akt1, Akt2, and Akt3; the vascular endothelial growth factor A (Vefga), the mitogen-activated protein kinase kinase 1 (Mapk1), phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform (Pik3cb), glucokinase (Gck), fructose-biphosphatase2 (Fbp2); and also down-regulated genes such as interleukin 6 (Il6).
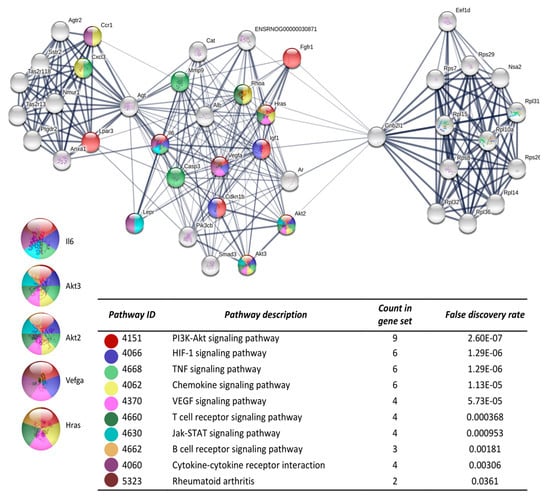
Figure 3.
The protein-protein interaction network construction of the cluster number one obtained with the differentially expressed genes. The lists of the differentially expressed genes (Z-score ≥ 1.5 SD) were analyzed on the STRING and Cytoscape platforms. The primary clusters of sub-networks were obtained using the Molecular Complex Detection (MCODE) complement (cutoff = 0.2). Line thickness indicates the strength of data support; colored nodes indicate query proteins and first shell of interactors; white nodes indicate the second shell of interactors.
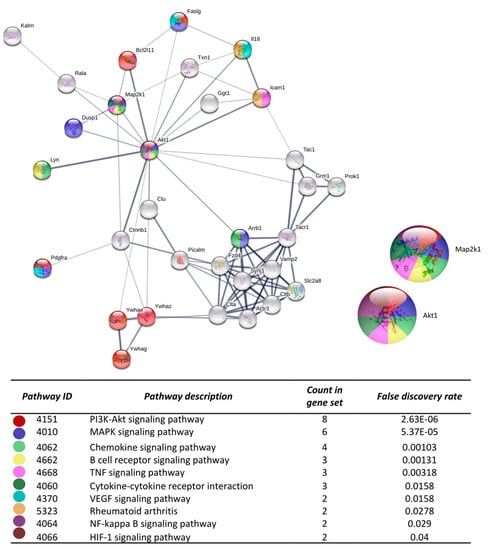
Figure 4.
The protein-protein interaction network construction of the cluster number two obtained with the differentially expressed genes. The lists of the differentially expressed genes (Z-score ≥ 1.5 SD) were analyzed on the STRING and Cytoscape platforms. The primary clusters of sub-networks were obtained using the Molecular Complex Detection (MCODE) complement (cutoff = 0.2). Line thickness indicates the strength of data support; colored nodes indicate query proteins and first shell of interactors; white nodes indicate the second shell of interactors.
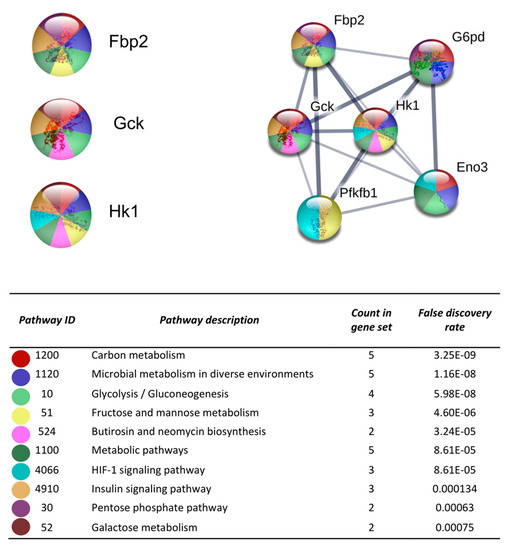
Figure 5.
The protein-protein interaction network construction of the cluster number three obtained with the differentially expressed genes. The lists of the differentially expressed genes (Z-score ≥ 1.5 SD) were analyzed on the STRING and Cytoscape platforms. The primary clusters of sub-networks were obtained using the Molecular Complex Detection (MCODE) complement (cutoff = 0.2). Line thickness indicates the strength of data support; colored nodes indicate query proteins and first shell of interactors; white nodes indicate the second shell of interactors.
4. Discussion
Current evidence describing the influence of PE on the pathogenic mechanisms of the arthritides is scarce and limited to studies in animal models. Most of these studies have focused on animal models of osteoarthritis and, in most cases, only clinical and histological parameters have been evaluated, limiting our understanding of the PE in the pathogenesis of active synovitis. To our knowledge, no previous study has assessed the effects of PE on the transcriptome profile in the AIA model.
Our study shows that PE induced several genes linked to RA pathogenesis. Specifically, PE exacerbated hypoxia, oxidative stress, and the immune/inflammatory response in rats with AIA. The histology confirms our transcriptome results; in the joints of the exercised animals, we observed an increase in the inflammatory infiltrate and more severe scores for bone and cartilage destruction.
In our opinion, the connection between PE and its potential detrimental role at active stages of synovitis should be considered as a relevant event in patients with inflammatory arthritides. Current recommendations for physical activity and PE in people with inflammatory and non-inflammatory arthritis [4,5,7,29] have been established mostly on clinical studies in humans, which the intimate effect of PE in the arthritic joints at a molecular level has not been considered. These recommendations have not been specified according to the disease activity; however, as our study shows, likely at very active stages of inflammatory arthritides, PE could increase the inflammatory process. It is unlikely that PE will be prescribed in patients with active arthritis; however, a wide prescription of PE could also include the large proportion of patients whom remain with some degree of inflammatory activity despite therapy. Probably, the PE benefits may not be as clear in those patients with partial disease control. Therefore, the disease activity should be considered as an element for PE prescription.
RA pathogenesis remains a primary research field; we now understand that the rheumatoid pannus is an altered microenvironment that carries a significant array of abnormal immune processes and also several metabolic alterations. The increase in hypoxia and oxidative stress are recognized as relevant inflammation drivers within the rheumatic synovium. Their pathological implications are now sustained by experimental results both: in animal models [30,31] and human disease [32,33]. Synovial hypoxia promotes inflammation, angiogenesis, production of chemokines, and cartilage and bone destruction, mediated in part, through HIF-1 that is a potent pleiotropic mediator [32,34].
Our study confirms that PE up-regulates genes linked to hypoxia and oxidative stress in the arthritic joints; indeed, the HIF-1 signaling was the first enriched pathway in our bioinformatic analysis, suggesting that this pathway was the strongest influenced by the PE. Downstream HIF-linked genes as the vascular endothelial growth factor (VEGF) are also up-regulated. VEGF is critical for the onset and perpetuation of vascular proliferation in synovitis [35,36,37]. Also, it was previously reported the VEGF increase in the serum and synovial fluid of RA patients [35,38] as well as in experimental animal models [39].
Hypoxia could activate the genes encoding nearly every step of glycolysis [32]. Abnormal glucose metabolism was observed in the synovial fluid and synovium of RA, which is evidenced by increased glycolytic enzyme activity [40]. A recent study proposes the inhibition of hexokinases as a potential therapeutic strategy in the treatment of patients with RA [41]. According to this, the glucokinase (Gck) and the hexokinase 1 (Hk1) were up-regulated by PE in our study. Additionally, genes involved in the oxidative stress process were also dysregulated by PE. It is recognized that in an inflamed joint, hypoxia and reperfusion cycles occur, producing an excessive amount of reactive oxygen species (ROS) known as oxidative stress [33,42].
In addition to hypoxia and oxidative stress conditions induced by the effect of PE, other KEGG pathways were dysregulated (Table 1) such as rheumatoid arthritis, T-cell and B-cell receptor signaling, chemokine, Jak-Stat, TNF, Toll-like-receptor, WNT and osteoclast differentiation signaling pathways. This array of dysregulated pathways suggests a widespread effect of PE both in the adaptive and innate immune response.
Other genes participating in several KEGG pathways in our study include Akt1, Akt2, Akt3, Map2k1 and PIk3cb which belong to PI3K/AKT/mTOR and MAPK pathways that are considered central in RA pathogenesis [43,44]. They have been implicated in fibroblast-like synoviocyte proliferation and activation. They stimulate the production of pro-inflammatory and osteoclastogenic cytokines; besides, the inhibition of either pathway has been correlated with the improvement of disease activity parameters. Therefore, the use of agents with the potential to regulate either PI3K/AKT/mTOR or MAPK pathways has been considered as a potential therapeutic strategy [45].
The molecular results of the PE effect are not easily comparable with previous studies due to the low number of reports, which had objectives aimed at particular aspects of the disease. However, we can distinguish that the negative effect of PE has also been reported in the two most recent and complete studies in this field [16,17]. The results of these studies suggest a direct link between the degree of mechanical stress to inflammation and tissue localization, which strengthens previous studies that propose biomechanical factors as potential determinants for the topographic pattern of joint disorders in arthritis [46]. Cambré et al., suggest that the link between mechanical stress and the onset of arthritis is explained by local recruitment of Ly6high inflammatory cells elicited by mechanostress-induced chemokine induction in resident mesenchyme cells [17]. They also provide evidence of the local participation of the complement activators to maintain the progression and chronicity of arthritis through an impaired resolution [16].
On the other hand, it has been shown that in healthy rats, PE has a positive effect in the joints it induces extracellular matrix biosynthesis, cartilage strengthening, and attenuation of inflammatory pathways [47]. This suggests that the gene induction pattern of mechanosensitive genes seems to be distinct between different arthritis models and healthy animals, and likely also in regarding to the degree of arthritis activity.
Consistent with the genetic analysis, our histological findings showed that PE had an adverse effect on increasing the inflammatory infiltrate and the joint destruction. In experimental models of arthritis, it has been confirmed that mechanical stress increase joint inflammation, stimulate the progression and chronicity of arthritis and structural damage [16,17,48]; besides, the mechanical stress decrease the bone quality [16]. In contrast to our findings, other studies in the AIA model have shown an anti-inflammatory effect of PE in some parameters such as synovial hyperplasia [49,50], and joint destruction [51].
Mode, intensity, frequency, timing, and duration of the PE protocol are decisive aspects to the physiological responses and different outcomes [52]. Moreover, the severity of arthritis at the starting time of PE could be definitory. Studies in the AIA model that started the PE at the pre-arthritic stage [49,50] showed beneficial results. However, we started the PE at an arthritic stage because we think this scenario is more representative of RA patients.
5. Conclusions
The present study explored the influence of PE in the genetic expression and the histology of the actively swollen joints in the AIA model. Our results describe an exploratory and preliminary scenario in which PE increased the expression of genes with a known pathogenic role in RA; these findings were consistent with the histological findings. These results suggest that the PE exacerbated the stressed joint environment, where the inflammation, hypoxia, and the oxidative stress prevail, rendering clear parallels to the RA joints. The molecular effect of exercise on active stages of inflammatory arthropathies requires further studies in animal models and humans, which could contribute with the development of adequate programs for RA patients that can ensure a beneficial effect without negative implications at the tissue level.
Author Contributions
Conceptualization: C.M.Q.-F.; methodology: S.A.G.-C., G.P.E.-S. and J.Á.V.-C.; writing original draft preparation, C.P.-T. and S.A.G.-C.; writing-review and editing, C.M.Q.-F., and C.P.-T.
Funding
This research received no external funding.
Acknowledgments
The authors thank Lorena Chávez, José Luis Santillán, Simón Guzmán and Jorge Ramírez for the microarray analysis. Likewise, the authors thank Rogelio González and Adrián Chávez for their support in the design and construction of the software used in this research.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. List of Genes Up-Regulated by PE Effect
| Gene Id | Gene | Z-Score |
| AA818786 | No data | 1.770223 |
| AA963507 | No data | 1.500387 |
| AB000928 | zona pellucida glycoprotein 1(Zp1) | 3.330347 |
| AB003753 | No data | 2.348897 |
| AB012139 | bone morphogenetic protein 1(Bmp1) | 1.717532 |
| AB022014 | proteasome 26S subunit, non-ATPase 10(Psmd10) | 1.800734 |
| AB024333 | barrier to autointegration factor 1(Banf1) | 2.409521 |
| AB027143 | septin 5(Sept5) | 2.332071 |
| AB037248 | ATPase H+ transporting V0 subunit e1(Atp6v0e1) | 1.790973 |
| AB041723 | apoptosis inducing factor, mitochondria associated 1(Aifm1) | 2.20147 |
| AB043959 | ubiquitin C-terminal hydrolase L3(Uchl3) | 2.083474 |
| AB070355 | kinesin family member 1B(Kif1b) | 2.560659 |
| AB073318 | brain and acute leukemia, cytoplasmic(Baalc) | 1.825273 |
| AF000139 | cytochrome P450, family 27, subfamily b, polypeptide 1(Cyp27b1) | 3.142163 |
| AF016183 | vomeronasal 2 receptor, pseudogene 45(Vom2r-ps45) | 2.284991 |
| AF026554 | solute carrier family 5 member 6(Slc5a6) | 1.552609 |
| AF030243 | interleukin 3 receptor subunit alpha(Il3ra) | 1.890042 |
| AF041374 | phosphatidylinositol binding clathrin assembly protein(Picalm) | 2.019124 |
| AF053989 | vomeronasal 2 receptor 44(Vom2r44) | 1.534172 |
| AF059678 | replication factor C subunit 1(Rfc1) | 1.961506 |
| AF063103 | adhesion G protein-coupled receptor L3(Adgrl3) | 1.510015 |
| AF063890 | protein tyrosine kinase 2 beta(Ptk2b) | 2.392998 |
| AF065161 | cytokine inducible SH2-containing protein(Cish) | 2.074343 |
| AF102262 | beta-1,4-galactosyltransferase 1(B4galt1) | 1.819501 |
| AF106657 | ubiquitin specific peptidase 15(Usp15) | 2.061666 |
| AF115768 | defensin alpha 24(Defa24) | 2.430658 |
| AF121265 | catenin beta 1(Ctnnb1) | 1.995608 |
| AF121893 | single stranded DNA binding protein 3(Ssbp3) | 1.660423 |
| AF145050 | eukaryotic translation elongation factor 1 delta(Eef1d) | 1.902054 |
| AF146738 | centrosomal protein 19(Cep19) | 1.815362 |
| AF155196 | No data | 1.575313 |
| AF169390 | phosphodiesterase 6H(Pde6h) | 1.774068 |
| AF169636 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3-like(Lilrb3l) | 3.098234 |
| AF173834 | calpain 3(Capn3) | 1.680331 |
| AF176023 | PR/SET domain 4(Prdm4) | 2.242755 |
| AF178689 | carbohydrate sulfotransferase 3(Chst3) | 1.70415 |
| AF187323 | cathepsin Q(Ctsq) | 2.106614 |
| AF190256 | phosphate cytidylyltransferase 1, choline, beta(Pcyt1b) | 2.363304 |
| AF201901 | interferon gamma receptor 1(Ifngr1) | 1.542342 |
| AF208125 | acyl-CoA synthetase bubblegum family member 1(Acsbg1) | 2.403616 |
| AF214647 | N-acylsphingosine amidohydrolase 1(Asah1) | 1.539499 |
| AF218575 | nibrin(Nbn) | 1.875441 |
| AF228043 | nuclear receptor coactivator 6(Ncoa6) | 1.721794 |
| AF228917 | zinc finger, DHHC-type containing 2(Zdhhc2) | 1.609594 |
| AF237778 | calcium/calmodulin-dependent protein kinase II alpha(Camk2a) | 1.638852 |
| AF273025 | solute carrier family 38, member 3(Slc38a3) | 2.373408 |
| AF276940 | ectonucleoside triphosphate diphosphohydrolase 2(Entpd2) | 2.0331 |
| AF281304 | potassium channel, two pore domain subfamily K, member 6(Kcnk6) | 2.029054 |
| AF288611 | RUN domain containing 3A(Rundc3a) | 2.840557 |
| AF303035 | PARP1 binding protein(Parpbp) | 2.933101 |
| AF308818 | KH-type splicing regulatory protein(Khsrp) | 1.694262 |
| AF311055 | thioredoxin 1(Txn1) | 2.848762 |
| AF333325 | HPS1, biogenesis of lysosomal organelles complex 3 subunit 1(Hps1) | 1.833169 |
| AF333986 | oxidation resistance 1(Oxr1) | 2.393664 |
| AF336113 | actin-binding Rho activating protein(Abra) | 2.216764 |
| AF345444 | potassium voltage-gated channel interacting protein 4(Kcnip4) | 2.743564 |
| AF368269 | cytochrome P450, family 2, subfamily t, polypeptide 1(Cyp2t1) | 1.620044 |
| AF393750 | BPI fold containing family A, member 1(Bpifa1) | 2.365618 |
| AF398465 | dihydropyrimidinase-like 3(Dpysl3) | 1.872371 |
| AF419333 | gamma-aminobutyric acid type A receptor theta subunit(Gabrq) | 1.792286 |
| AI059116 | No data | 1.626247 |
| AI102932 | No data | 1.930485 |
| AI104638 | No data | 2.595373 |
| AI105022 | No data | 1.646591 |
| AI113337 | No data | 1.994172 |
| AI230498 | No data | 1.568518 |
| AI230682 | No data | 2.036962 |
| AI231775 | No data | 1.560267 |
| AI385377 | No data | 3.220256 |
| AI406694 | No data | 2.313044 |
| AI500802 | No data | 2.504556 |
| AI535093 | No data | 1.900459 |
| AI599423 | No data | 1.514733 |
| AI602844 | No data | 1.652924 |
| AI717432 | No data | 1.677302 |
| AJ012482 | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta(Pik3cb) | 2.039374 |
| AT005481 | No data | 1.678314 |
| AW141757 | No data | 1.778821 |
| AW520335 | No data | 2.449793 |
| AW533345 | No data | 1.75804 |
| AW914284 | No data | 1.704328 |
| AW915815 | No data | 1.91521 |
| AW916410 | No data | 1.858779 |
| AW918157 | No data | 1.600146 |
| AW918457 | No data | 1.504258 |
| AW918850 | No data | 1.940086 |
| AW921253 | No data | 1.848394 |
| AY026068 | ras homolog family member A(Rhoa) | 2.033377 |
| AY030278 | Wnt inhibitory factor 1(Wif1) | 1.764736 |
| AY034383 | dynein light chain LC8-type 2(Dynll2) | 1.527297 |
| AY035403 | kinesin family member 6(Kif6) | 1.9322 |
| AY122322 | protein phosphatase 1, regulatory (inhibitor) subunit 14D(Ppp1r14d) | 2.374861 |
| BE100543 | No data | 2.356497 |
| BE113282 | No data | 2.311542 |
| BE113420 | No data | 1.660364 |
| BE118455 | No data | 1.925824 |
| BF390636 | No data | 1.846075 |
| BF405195 | No data | 1.559474 |
| BF406553 | No data | 1.625267 |
| BF411273 | No data | 1.537562 |
| BF418537 | No data | 1.791187 |
| BF420605 | No data | 1.918056 |
| BF522262 | No data | 2.816838 |
| BF522647 | No data | 1.689307 |
| BF525282 | No data | 2.164385 |
| BF543281 | No data | 1.772391 |
| BF543321 | No data | 1.81299 |
| BF546465 | No data | 2.021733 |
| BF553424 | No data | 1.799168 |
| BF561374 | No data | 1.753152 |
| BF563877 | No data | 2.224055 |
| BF564018 | No data | 1.608414 |
| BF565044 | No data | 1.715644 |
| BF565565 | No data | 1.534809 |
| BG374035 | No data | 1.848573 |
| BG664137 | No data | 1.654493 |
| BG665039 | No data | 1.996679 |
| BG666176 | No data | 2.409512 |
| BG668317 | No data | 1.503399 |
| BG668660 | No data | 1.902199 |
| BG671436 | No data | 2.520046 |
| BG671873 | No data | 2.019116 |
| BG673258 | No data | 2.563218 |
| BI282584 | No data | 2.135856 |
| BM383757 | No data | 1.584869 |
| BQ207374 | No data | 1.532423 |
| BQ211469 | No data | 1.62545 |
| BU671396 | No data | 1.554288 |
| BU671701 | No data | 2.802387 |
| D00729 | enoyl-CoA delta isomerase 1(Eci1) | 2.814065 |
| D14480 | calpain 8(Capn8) | 1.674437 |
| D21095 | chemokine (C-X-C motif) ligand 3(Cxcl3) | 1.92518 |
| D30040 | AKT serine/threonine kinase 1(Akt1) | 1.680116 |
| D38261 | protein phosphatase 2, regulatory subunit B, gamma(Ppp2r2c) | 1.79365 |
| D85435 | protein kinase C, delta binding protein(Prkcdbp) | 1.78163 |
| D87950 | ubiquilin 1(Ubqln1) | 1.763773 |
| J00744 | No data | 1.516659 |
| L04760 | tripartite motif-containing 23(Trim23) | 2.018875 |
| L07315 | dipeptidase 1 (renal)(Dpep1) | 1.601127 |
| L13606 | myosin heavy chain 2(Myh2) | 1.787821 |
| L15618 | casein kinase 2 alpha 1(Csnk2a1) | 1.7187 |
| L28801 | general transcription factor IIIC subunit 1(Gtf3c1) | 1.516161 |
| L29427 | No data | 2.009211 |
| L35921 | G protein subunit gamma 8(Gng8) | 1.713815 |
| M13011 | No data | 1.614206 |
| M15883 | clathrin, light chain B(Cltb) | 1.806758 |
| M21759 | filaggrin(Flg) | 1.813956 |
| M22030 | No data | 1.780357 |
| M32062 | Fc fragment of IgG, low affinity IIa, receptor(Fcgr2a) | 1.60304 |
| M33821 | gamma-glutamyltransferase 1(Ggt1) | 2.263792 |
| M91450 | No data | 1.564184 |
| M96626 | ATPase plasma membrane Ca2+ transporting 3(Atp2b3) | 2.172804 |
| M97255 | trefoil factor 2(Tff2) | 1.647276 |
| NM_012519 | calcium/calmodulin-dependent protein kinase II delta(Camk2d) | 1.505653 |
| NM_012520 | catalase(Cat) | 2.274713 |
| NM_012526 | chromogranin B(Chgb) | 2.018038 |
| NM_012528 | cholinergic receptor nicotinic beta 1 subunit(Chrnb1) | 1.680012 |
| NM_012531 | catechol-O-methyltransferase(Comt) | 2.231328 |
| NM_012543 | D-box binding PAR bZIP transcription factor(Dbp) | 1.532421 |
| NM_012549 | endothelin 2(Edn2) | 1.830311 |
| NM_012565 | glucokinase(Gck) | 3.003472 |
| NM_012572 | glutamate ionotropic receptor kainate type subunit 4(Grik4) | 1.960676 |
| NM_012594 | lactalbumin, alpha(Lalba) | 2.35724 |
| NM_012596 | leptin receptor(Lepr) | 1.572461 |
| NM_012600 | malic enzyme 1(Me1) | 1.705429 |
| NM_012612 | natriuretic peptide A(Nppa) | 1.752513 |
| NM_012619 | phenylalanine hydroxylase(Pah) | 1.861168 |
| NM_012653 | solute carrier family 9 member A2(Slc9a2) | 1.680892 |
| NM_012663 | vesicle-associated membrane protein 2(Vamp2) | 1.751666 |
| NM_012667 | tachykinin receptor 1(Tacr1) | 2.182398 |
| NM_012695 | sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 6(Sult2a6) | 1.605457 |
| NM_012730 | cytochrome P450, family 2, subfamily d, polypeptide 2(Cyp2d2) | 1.829295 |
| NM_012734 | hexokinase 1(Hk1) | 1.709348 |
| NM_012788 | discs large MAGUK scaffold protein 1(Dlg1) | 1.700899 |
| NM_012850 | growth hormone releasing hormone receptor(Ghrhr) | 1.557048 |
| NM_012876 | ribosomal protein S29(Rps29) | 2.638145 |
| NM_012904 | annexin A1(Anxa1) | 1.502437 |
| NM_012908 | Fas ligand(Faslg) | 2.142956 |
| NM_012910 | arrestin, beta 1(Arrb1) | 1.672619 |
| NM_012941 | cytochrome P450, family 51(Cyp51) | 1.657199 |
| NM_012963 | high mobility group box 1(Hmgb1) | 1.666837 |
| NM_012978 | luteinizing hormone/choriogonadotropin receptor(Lhcgr) | 3.050579 |
| NM_012980 | matrix metallopeptidase 11(Mmp11) | 2.630092 |
| NM_012993 | nardilysin convertase(Nrdc) | 2.298098 |
| NM_013005 | phosphoinositide-3-kinase regulatory subunit 1(Pik3r1) | 2.802073 |
| NM_013008 | POU class 1 homeobox 1(Pou1f1) | 1.552156 |
| NM_013030 | solute carrier family 34 member 1(Slc34a1) | 1.569844 |
| NM_013183 | meprin A subunit beta(Mep1b) | 1.901411 |
| NM_013224 | ribosomal protein S26(Rps26) | 1.620632 |
| NM_013226 | ribosomal protein L32(Rpl32) | 2.284438 |
| NM_013413 | relaxin 1(Rln1) | 1.971464 |
| NM_017015 | glucuronidase, beta(Gusb) | 1.8737 |
| NM_017042 | protein phosphatase 3 catalytic subunit beta(Ppp3cb) | 1.989406 |
| NM_017137 | chloride channel, voltage-sensitive 2(Clcn2) | 1.867684 |
| NM_017144 | troponin I3, cardiac type(Tnni3) | 1.571609 |
| NM_017169 | peroxiredoxin 2(Prdx2) | 2.126629 |
| NM_017193 | aminoadipate aminotransferase(Aadat) | 2.633207 |
| NM_017251 | gap junction protein, beta 1(Gjb1) | 1.77218 |
| NM_017256 | transforming growth factor beta receptor 3(Tgfbr3) | 1.864647 |
| NM_017264 | proteasome activator subunit 1(Psme1) | 1.819425 |
| NM_017279 | proteasome subunit alpha 2(Psma2) | 1.725385 |
| NM_017348 | solute carrier family 6 member 8(Slc6a8) | 1.628447 |
| NM_019137 | early growth response 4(Egr4) | 1.894886 |
| NM_019151 | myostatin(Mstn) | 2.351448 |
| NM_019161 | cadherin 22(Cdh22) | 2.206278 |
| NM_019165 | interleukin 18(Il18) | 1.52836 |
| NM_019170 | carbonyl reductase 1(Cbr1) | 1.714897 |
| NM_019179 | thymidylate synthetase(Tyms) | 1.837275 |
| NM_019198 | fibroblast growth factor 17(Fgf17) | 2.35561 |
| NM_019216 | growth differentiation factor 15(Gdf15) | 1.772315 |
| NM_019266 | sodium voltage-gated channel alpha subunit 8(Scn8a) | 2.082371 |
| NM_019272 | ssemaphorin 4F(Sema4f) | 1.685348 |
| NM_019277 | exocyst complex component 6(Exoc6) | 1.957294 |
| NM_019291 | carbonic anhydrase 2(Car2) | 2.185707 |
| NM_019323 | mast cell protease 9(Mcpt9) | 1.647125 |
| NM_019340 | regulator of G-protein signaling 3(Rgs3) | 1.639602 |
| NM_019353 | thyroid peroxidase(Tpo) | 1.757899 |
| NM_019376 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma(Ywhag) | 1.753748 |
| NM_019904 | galectin 1(Lgals1) | 1.747295 |
| NM_020082 | ribonuclease A family member 4(Rnase4) | 1.5499 |
| NM_021581 | prolyl 3-hydroxylase family member 4(P3h4) | 1.874377 |
| NM_021585 | mast cell immunoglobulin-like receptor 1(Milr1) | 1.878356 |
| NM_021598 | mast cell protease 8(Mcpt8) | 2.72646 |
| NM_021671 | transmembrane protein 33(Tmem33) | 1.613433 |
| NM_021684 | adenylate cyclase 10 (soluble)(Adcy10) | 1.83554 |
| NM_021697 | potassium voltage-gated channel modifier subfamily V member 1(Kcnv1) | 1.756715 |
| NM_021701 | protein phosphatase 3, regulatory subunit B, beta(Ppp3r2) | 1.764523 |
| NM_021748 | N-ethylmaleimide sensitive factor, vesicle fusing ATPase(Nsf) | 1.720353 |
| NM_021772 | cyclin-dependent kinase-like 3(Cdkl3) | 1.589505 |
| NM_021849 | RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase(Rfng) | 1.75421 |
| NM_021859 | megakaryocyte-associated tyrosine kinase(Matk) | 1.846828 |
| NM_021997 | CAP-GLY domain containing linker protein 2(Clip2) | 1.829356 |
| NM_022203 | No data | 1.914261 |
| NM_022210 | MYC associated factor X(Max) | 1.814325 |
| NM_022254 | G protein-coupled receptor 85(Gpr85) | 1.800299 |
| NM_022282 | discs large MAGUK scaffold protein 2(Dlg2) | 1.768177 |
| NM_022284 | guanylate cyclase activator 2B(Guca2b) | 2.039829 |
| NM_022296 | xylosyltransferase 2(Xylt2) | 2.198775 |
| NM_022297 | dimethylarginine dimethylaminohydrolase 1(Ddah1) | 1.685947 |
| NM_022509 | survival of motor neuron 1, telomeric(Smn1) | 1.906114 |
| NM_022538 | phospholipid phosphatase 1(Plpp1) | 1.909921 |
| NM_022600 | adenylate cyclase 5(Adcy5) | 1.520435 |
| NM_022613 | No data | 1.929265 |
| NM_022623 | frizzled class receptor 4(Fzd4) | 2.230565 |
| NM_022625 | tropic 1808(Tpc1808) | 1.653187 |
| NM_022638 | transient receptor potential cation channel, subfamily C, member 2, pseudogene(Trpc2) | 1.663797 |
| NM_022639 | cholinergic receptor nicotinic alpha 10 subunit(Chrna10) | 1.732239 |
| NM_022686 | histone cluster 1, H4b(Hist1h4b) | 2.369333 |
| NM_022704 | mannose-binding lectin (protein C) 2(Mbl2) | 1.568571 |
| NM_022850 | dipeptidyl peptidase like 6(Dpp6) | 2.262092 |
| NM_022859 | cysteine-rich secretory protein 1(Crisp1) | 1.609319 |
| NM_022929 | potassium voltage-gated channel interacting protein 1(Kcnip1) | 1.663328 |
| NM_022947 | ClpB homolog, mitochondrial AAA ATPase chaperonin(Clpb) | 1.811601 |
| NM_022954 | FAT atypical cadherin 2(Fat2) | 2.727616 |
| NM_023102 | casein kinase 1, gamma 2(Csnk1g2) | 1.702925 |
| NM_023963 | caudal type homeo box 2(Cdx2) | 1.683684 |
| NM_023969 | lysophosphatidic acid receptor 3(Lpar3) | 1.63442 |
| NM_023973 | indoleamine 2,3-dioxygenase 1(Ido1) | 1.519611 |
| NM_023981 | colony stimulating factor 1(Csf1) | 1.538487 |
| NM_023994 | taste receptor, type 2, member 118(Tas2r118) | 1.695958 |
| NM_023995 | taste receptor, type 2, member 107(Tas2r107) | 2.24126 |
| NM_024127 | growth arrest and DNA-damage-inducible, alpha(Gadd45a) | 2.152067 |
| NM_024381 | glycerol kinase(Gk) | 1.811678 |
| NM_024487 | GrpE-like 1, mitochondrial(Grpel1) | 1.943116 |
| NM_024489 | zinc finger and BTB domain containing 10(Zbtb10) | 2.056393 |
| NM_030826 | glutathione peroxidase 1(Gpx1) | 1.652839 |
| NM_030837 | kidney specific organic anion transporter(Slc21a4) | 2.998452 |
| NM_030843 | syntaxin binding protein 5(Stxbp5) | 2.170744 |
| NM_030860 | myocyte enhancer factor 2D(Mef2d) | 2.048593 |
| NM_030868 | nephroblastoma overexpressed(Nov) | 1.600397 |
| NM_030875 | sodium voltage-gated channel alpha subunit 1(Scn1a) | 1.622929 |
| NM_031000 | aldo-keto reductase family 1 member A1(Akr1a1) | 1.567229 |
| NM_031021 | casein kinase 2 beta(Csnk2b) | 1.917293 |
| NM_031036 | G protein subunit alpha q(Gnaq) | 1.918023 |
| NM_031059 | msh homeobox 1(Msx1) | 1.99165 |
| NM_031065 | ribosomal protein L10A(Rpl10a) | 2.300643 |
| NM_031085 | protein kinase C, eta(Prkch) | 2.372214 |
| NM_031093 | RAS like proto-oncogene A(Rala) | 2.39697 |
| NM_031120 | signal sequence receptor, gamma(Ssr3) | 1.674383 |
| NM_031129 | transcription elongation factor B subunit 2(Tceb2) | 2.331149 |
| NM_031130 | nuclear receptor subfamily 2, group F, member 1(Nr2f1) | 1.84239 |
| NM_031133 | thrombopoietin(Thpo) | 1.757085 |
| NM_031136 | thymosin beta 4, X-linked(Tmsb4x) | 2.204979 |
| NM_031142 | double C2 domain beta(Doc2b) | 2.438734 |
| NM_031240 | cysteine-rich secretory protein 2(Crisp2) | 2.969829 |
| NM_031352 | drebrin-like(Dbnl) | 1.689691 |
| NM_031360 | sphingomyelin phosphodiesterase 2(Smpd2) | 1.509688 |
| NM_031537 | RoBo-1(LOC24906) | 2.382064 |
| NM_031570 | ribosomal protein S7(Rps7) | 1.720451 |
| NM_031575 | AKT serine/threonine kinase 3(Akt3) | 1.827906 |
| NM_031601 | calcium voltage-gated channel subunit alpha1 G(Cacna1g) | 2.076481 |
| NM_031603 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon(Ywhae) | 1.516651 |
| NM_031604 | ATPase H+ transporting V0 subunit a1(Atp6v0a1) | 2.734505 |
| NM_031621 | SH2B adaptor protein 3(Sh2b3) | 1.87789 |
| NM_031624 | immunoglobulin (CD79A) binding protein 1(Igbp1) | 1.647906 |
| NM_031627 | nuclear receptor subfamily 1, group H, member 3(Nr1h3) | 1.513038 |
| NM_031643 | mitogen activated protein kinase kinase 1(Map2k1) | 2.033606 |
| NM_031728 | synaptosomal-associated protein 91(Snap91) | 1.989374 |
| NM_031729 | protein phosphatase 5, catalytic subunit(Ppp5c) | 1.7209 |
| NM_031762 | cyclin-dependent kinase inhibitor 1B(Cdkn1b) | 1.807938 |
| NM_031781 | amyloid beta precursor protein binding family A member 3(Apba3) | 1.770669 |
| NM_031789 | nuclear factor, erythroid 2-like 2(Nfe2l2) | 2.362231 |
| NM_031821 | polo-like kinase 2(Plk2) | 2.022837 |
| NM_031823 | wolframin ER transmembrane glycoprotein(Wfs1) | 2.469074 |
| NM_031832 | galectin 3(Lgals3) | 1.597594 |
| NM_031836 | vascular endothelial growth factor A(Vegfa) | 1.578976 |
| NM_031974 | clathrin, light chain A(Clta) | 1.664435 |
| NM_032079 | DnaJ heat shock protein family (Hsp40) member A2(Dnaja2) | 1.580883 |
| NM_032082 | hydroxyacid oxidase 2(Hao2) | 1.813424 |
| NM_033376 | potassium two pore domain channel subfamily K member 3(Kcnk3) | 1.540317 |
| NM_053323 | delta(4)-desaturase, sphingolipid 1(Degs1) | 1.581652 |
| NM_053716 | fructose-bisphosphatase 2(Fbp2) | 1.901898 |
| NM_057098 | transcription elongation factor A2(Tcea2) | 2.570171 |
| NM_057099 | proteasome subunit beta 6(Psmb6) | 1.767274 |
| NM_078622 | phosphate cytidylyltransferase 1, choline, alpha(Pcyt1a) | 1.676003 |
| NM_080900 | actin filament associated protein 1(Afap1) | 1.565316 |
| NM_130420 | tripartite motif-containing 9(Trim9) | 2.025582 |
| NM_130421 | lymphocyte cytosolic protein 2(Lcp2) | 1.912839 |
| NM_130753 | fibroblast growth factor 19(Fgf19) | 1.524216 |
| NM_133309 | calpain 8(Capn8) | 2.126877 |
| NM_133321 | potassium voltage-gated channel subfamily J member 15(Kcnj15) | 2.331074 |
| NM_133383 | serine carboxypeptidase 1(Scpep1) | 1.565457 |
| NM_133392 | serine/threonine kinase 17b(Stk17b) | 2.265261 |
| NM_133406 | 1-acylglycerol-3-phosphate O-acyltransferase 4(Agpat4) | 1.599979 |
| NM_133612 | No data | 1.653442 |
| NM_134376 | calsyntenin 3(Clstn3) | 1.500037 |
| NM_134462 | ATPase secretory pathway Ca2+ transporting 2(Atp2c2) | 2.49921 |
| NM_134465 | cytokine receptor-like factor 2(Crlf2) | 2.519908 |
| NM_138710 | DAB2 interacting protein(Dab2ip) | 1.61244 |
| NM_139193 | prolactin releasing hormone receptor(Prlhr) | 1.542231 |
| NM_144741 | resistin(Retn) | 1.849312 |
| NM_144742 | ATPase phospholipid transporting 11A(Atp11a) | 2.002968 |
| NM_147207 | ischemia related factor vof-16(Vof16) | 1.623283 |
| U03390 | receptor for activated C kinase 1(Rack1) | 1.944424 |
| U05593 | Cd80 molecule(Cd80) | 2.654472 |
| U06434 | C-C motif chemokine ligand 4(Ccl4) | 1.819756 |
| U14746 | von Hippel-Lindau tumor suppressor(Vhl) | 1.543271 |
| U17604 | reticulon 1(Rtn1) | 1.825625 |
| U18771 | RAB26, member RAS oncogene family(Rab26) | 1.820437 |
| U39208 | cytochrome P450, family 4, subfamily f, polypeptide 6(Cyp4f6) | 1.645854 |
| U48828 | No data | 1.727898 |
| U51583 | zinc finger E-box binding homeobox 1(Zeb1) | 1.51051 |
| U53475 | RAB8B, member RAS oncogene family(Rab8b) | 1.913337 |
| U57063 | granzyme F(Gzmf) | 1.980644 |
| U57391 | SH2B adaptor protein 1(Sh2b1) | 2.074809 |
| U72353 | lamin B1(Lmnb1) | 2.074554 |
| U92564 | zinc finger protein 423(Zfp423) | 2.090788 |
| U93851 | cyclic nucleotide gated channel alpha 1(Cnga1) | 1.506566 |
| U94856 | paraoxonase 1(Pon1) | 1.546413 |
| X51992 | gamma-aminobutyric acid type A receptor alpha 5 subunit(Gabra5) | 1.648049 |
| X52952 | Moloney sarcoma oncogene(Mos) | 1.892727 |
| X56190 | No data | 1.501611 |
| X63281 | No data | 2.264276 |
| X69029 | No data | 1.553785 |
| X80671 | olfactory receptor 1271(Olr1271) | 1.74883 |
| X81193 | cysteine and glycine rich protein 3(Csrp3) | 1.681766 |
| X89603 | metallothionein 3(Mt3) | 2.362925 |
| X89962 | cold shock domain containing C2(Csdc2) | 2.244 |
| X96790 | glutamate metabotropic receptor 7(Grm7) | 1.589366 |
| X98490 | replication protein A2(Rpa2) | 1.518939 |
| Z75029 | heat shock 70kD protein 1B (mapped)(Hspa1b) | 1.844737 |
Appendix B. List of Genes Down-Regulated by PE Effect
| Gene Id | Gene | Z-Score |
| AA944170 | S/D | −1.506199 |
| AA944489 | No data | −1.530233 |
| AA996993 | No data | −2.983268 |
| AB011529 | cadherin, EGF LAG seven-pass G-type receptor 2(Celsr2) | −1.74635 |
| AB011679 | tubulin, beta 5 class I(Tubb5) | −2.325114 |
| AB020504 | No data | −2.211124 |
| AB020757 | chymotrypsin-like(Ctrl) | −2.059928 |
| AB025017 | No data | −2.29894 |
| AB033418 | No data | −2.907947 |
| AB047540 | isocitrate dehydrogenase 3 (NAD+) beta(Idh3B) | −1.980831 |
| AB067445 | integrin alpha 2(Itga2) | −1.977614 |
| AF000944 | general transcription factor IIA, 2(Gtf2a2) | −2.614055 |
| AF006664 | NK2 homeobox 5(Nkx2-5) | −1.571779 |
| AF012714 | multiple inositol-polyphosphate phosphatase 1(Minpp1) | −1.517925 |
| AF015953 | aryl hydrocarbon receptor nuclear translocator-like(Arntl) | −1.67005 |
| AF016387 | retinoid X receptor gamma(Rxrg) | −1.847981 |
| AF021936 | CDC42 binding protein kinase beta(Cdc42bpb) | −2.024961 |
| AF026505 | sorbin and SH3 domain containing 2(Sorbs2) | −1.84203 |
| AF032120 | GIPC PDZ domain containing family, member 1(Gipc1) | −1.832343 |
| AF037071 | nitric oxide synthase 1 adaptor protein(Nos1ap) | −2.242322 |
| AF039584 | CD55 molecule, decay accelerating factor for complement(Cd55) | −1.707524 |
| AF053093 | No data | −1.976489 |
| AF053097 | No data | −1.603483 |
| AF084544 | versican(Vcan) | −1.711944 |
| AF092090 | polyamine modulated factor 1 binding protein 1(Pmfbp1) | −1.629489 |
| AF123651 | spermatogenesis associated 2(Spata2) | −1.791841 |
| AF151710 | No data | −1.874231 |
| AF170284 | gap junction protein, beta 6(Gjb6) | −2.501274 |
| AF193757 | N-terminal EF-hand calcium binding protein 2(Necab2) | −1.515331 |
| AF200359 | UDP-glucose glycoprotein glucosyltransferase 1(Uggt1) | −2.250208 |
| AF243515 | BCL2/adenovirus E1B interacting protein 3(Bnip3) | −1.627503 |
| AF247450 | hyperpolarization-activated cyclic nucleotide-gated potassium channel 1(Hcn1) | −1.51455 |
| AF277901 | zinc finger protein 483(Zfp483) | −1.798414 |
| AF302047 | CXADR-like membrane protein(Clmp) | −2.140061 |
| AF304429 | mitochondrial pyruvate carrier 1(Mpc1) | −2.257792 |
| AF307852 | ARP3 actin related protein 3 homolog(Actr3) | −1.95466 |
| AF323615 | phospholipase C, epsilon 1(Plce1) | −4.175461 |
| AF329856 | phosphodiesterase 1C(Pde1c) | −1.504374 |
| AF347935 | interleukin 11(Il11) | −1.674463 |
| AF361476 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2(Cited2) | −1.710103 |
| AF367467 | selenoprotein S(Selenos) | −2.382484 |
| AF379608 | doublesex and mab-3 related transcription factor 1(Dmrt1) | −1.683129 |
| AF400662 | calcium voltage-gated channel auxiliary subunit alpha2delta 1(Cacna2d1) | −1.543195 |
| AF537333 | aurora kinase A(Aurka) | −1.661243 |
| AI059699 | No data | −1.842174 |
| AI136886 | No data | −1.650746 |
| AI229387 | No data | −1.511607 |
| AI598392 | No data | −1.725522 |
| AJ006855 | synaptojanin 1(Synj1) | −1.727807 |
| AJ131196 | No data | −1.769069 |
| AJ250280 | potassium voltage-gated channel subfamily H member 5(Kcnh5) | −1.520944 |
| AJ303374 | ATP binding cassette subfamily G member 1(Abcg1) | −1.609867 |
| AJ409332 | TIMP metallopeptidase inhibitor 2(Timp2) | −2.3002 |
| AW141281 | No data | −2.051067 |
| AW141994 | No data | −1.692865 |
| AW434257 | No data | −1.789087 |
| AW915590 | No data | −1.557293 |
| AW916146 | No data | −1.959788 |
| AW916635 | No data | −1.746735 |
| AW917632 | No data | −1.549931 |
| AW918748 | No data | −2.166549 |
| AW918775 | No data | −1.649553 |
| AW919008 | No data | −1.692303 |
| AY011335 | No data | −1.711045 |
| AY014898 | inositol polyphosphate multikinase(Ipmk) | −2.342213 |
| AY026512 | dynein light chain roadblock-type 1(Dynlrb1) | −2.119183 |
| BF284301 | No data | −2.121369 |
| BF287132 | No data | −2.068833 |
| BF392368 | No data | −2.212956 |
| BF395339 | No data | −1.928218 |
| BF399588 | No data | −2.70933 |
| BF414052 | No data | −2.155561 |
| BF416262 | No data | −1.546722 |
| BF524417 | No data | −2.049708 |
| BF549771 | No data | −1.690628 |
| BF555199 | No data | −1.523037 |
| BF559446 | No data | −2.697068 |
| BF560218 | No data | −2.252126 |
| BF566173 | No data | −2.261156 |
| BF567456 | No data | −1.898923 |
| BG663098 | No data | −1.5361 |
| BG664103 | No data | −2.448298 |
| BG666505 | No data | −1.572982 |
| BG667467 | No data | −2.508352 |
| BG671325 | No data | −1.655659 |
| BI278738 | No data | −2.62235 |
| BI295378 | No data | −2.278025 |
| BI296499 | No data | −2.01441 |
| BM386847 | No data | −2.077829 |
| BQ194714 | No data | −1.88522 |
| BQ205274 | NADH:ubiquinone oxidoreductase subunit A12(Ndufa12) | −2.38962 |
| BQ208291 | No data | −1.636124 |
| BU670896 | No data | −3.513474 |
| BU671010 | No data | −2.556612 |
| BU671095 | No data | −2.578096 |
| BU671151 | No data | −1.744558 |
| D00920 | seminal vesicle secretory protein 3A(Svs3a) | −2.000729 |
| D14048 | heterogeneous nuclear ribonucleoprotein U(Hnrnpu) | −2.261341 |
| D16465 | adenylate cyclase activating polypeptide 1 receptor type 1(Adcyap1r1) | −1.966142 |
| D16479 | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit(Hadhb) | −1.944151 |
| D26178 | intestinal cell kinase(Ick) | −2.540983 |
| D63772 | solute carrier family 1 member 1(Slc1a1) | −1.665296 |
| J02585 | stearoyl-CoA desaturase(Scd) | −2.299181 |
| J02868 | cytochrome P450, family 2, subfamily d, polypeptide 3(Cyp2d3) | −1.688779 |
| J04628 | 3-hydroxyisobutyrate dehydrogenase(Hibadh) | −1.717486 |
| L02121 | cyclin-dependent kinase 5(Cdk5) | −2.181096 |
| L02315 | calcium voltage-gated channel auxiliary subunit beta 4(Cacnb4) | −1.528497 |
| L10362 | synaptic vesicle glycoprotein 2b(Sv2b) | −1.569646 |
| L12407 | No data | −1.867835 |
| L22654 | gamma-2a immunoglobulin heavy chain(IgG-2a) | −1.709121 |
| L27487 | calcitonin receptor-like(Calcrl) | −1.970351 |
| M11794 | No data | −1.536746 |
| M15202 | No data | −1.947036 |
| M15327 | alcohol dehydrogenase 1 (class I)(Adh1) | −2.079242 |
| M15481 | insulin-like growth factor 1(Igf1) | −2.298139 |
| M16409 | cholinergic receptor, muscarinic 4(Chrm4) | −1.743017 |
| M18841 | No data | −1.998377 |
| M29295 | small nuclear ribonucleoprotein polypeptides B and B1(Snrpb) | −1.675114 |
| M30692 | No data | −2.027642 |
| M31322 | amyloid beta precursor like protein 2(Aplp2) | −1.814887 |
| M57705 | thyroid peroxidase(Tpo) | −1.513088 |
| M61142 | thimet oligopeptidase 1(Thop1) | −1.696997 |
| M63837 | platelet derived growth factor receptor alpha(Pdgfra) | −1.822324 |
| M64381 | olfactory receptor 1082(Olr1082) | −1.761637 |
| M76733 | glutathione peroxidase 6(Gpx6) | −1.971279 |
| M83143 | ST6 beta-galactoside alpha-2,6-sialyltransferase 1(St6gal1) | −1.602127 |
| M83209 | BPI fold containing family A, member 2(Bpifa2) | −2.386439 |
| M83210 | BPI fold containing family A, member 2F(Bpifa2f) | −1.615422 |
| M83745 | proprotein convertase subtilisin/kexin type 1(Pcsk1) | −1.961893 |
| M94043 | RAB38, member RAS oncogene family(Rab38) | −1.660598 |
| NM_012490 | acrosin(Acr) | −1.761651 |
| NM_012502 | androgen receptor(Ar) | −2.367953 |
| NM_012542 | cytochrome P450, family 2, subfamily a, polypeptide 3(Cyp2a3) | −1.563632 |
| NM_012589 | interleukin 6(Il6) | −2.279895 |
| NM_012595 | lactate dehydrogenase B(Ldhb) | −1.56024 |
| NM_012621 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1(Pfkfb1) | −2.130949 |
| NM_012635 | protease, serine 1(Prss1) | −1.938556 |
| NM_012657 | serine (or cysteine) proteinase inhibitor, clade A, member 3C(Serpina3c) | −1.73983 |
| NM_012679 | No data | −1.660105 |
| NM_012681 | transthyretin(Ttr) | −1.632171 |
| NM_012682 | uncoupling protein 1(Ucp1) | −1.956669 |
| NM_012703 | thyroid hormone responsive(Thrsp) | −2.132157 |
| NM_012706 | gastrin releasing peptide receptor(Grpr) | −2.32707 |
| NM_012740 | tyrosine hydroxylase(Th) | −1.687967 |
| NM_012777 | apolipoprotein D(Apod) | −2.293886 |
| NM_012805 | retinoid X receptor alpha(Rxra) | −1.788029 |
| NM_012825 | aquaporin 4(Aqp4) | −1.738027 |
| NM_012826 | alpha-2-glycoprotein 1, zinc-binding(Azgp1) | −1.917567 |
| NM_012841 | DCC netrin 1 receptor(Dcc) | −2.060894 |
| NM_012843 | epithelial membrane protein 1(Emp1) | −1.524748 |
| NM_012922 | caspase 3(Casp3) | −1.730399 |
| NM_012940 | cytochrome P450, family 1, subfamily b, polypeptide 1(Cyp1b1) | −1.825438 |
| NM_012949 | enolase 3(Eno3) | −2.250452 |
| NM_012951 | fibroblast growth factor 10(Fgf10) | −1.581501 |
| NM_012967 | intercellular adhesion molecule 1(Icam1) | −1.527629 |
| NM_012971 | potassium voltage-gated channel subfamily A member 4(Kcna4) | −2.126831 |
| NM_013023 | S-antigen visual arrestin(Sag) | −1.921779 |
| NM_013054 | No data | −1.571264 |
| NM_013095 | SMAD family member 3(Smad3) | −1.508985 |
| NM_013159 | insulin degrading enzyme(Ide) | −1.663445 |
| NM_013161 | pancreatic lipase(Pnlip) | −2.292426 |
| NM_013167 | uncoupling protein 3(Ucp3) | −2.046956 |
| NM_013168 | hydroxymethylbilane synthase(Hmbs) | −4.18491 |
| NM_013192 | potassium voltage-gated channel subfamily J member 6(Kcnj6) | −1.544981 |
| NM_016998 | carboxypeptidase A1(Cpa1) | −1.72664 |
| NM_017006 | glucose-6-phosphate dehydrogenase(G6pd) | −2.105296 |
| NM_017011 | glutamate receptor, metabotropic 1(Grm1) | −1.945417 |
| NM_017014 | glutathione S-transferase mu 1(Gstm1) | −2.433956 |
| NM_017019 | interleukin 1 alpha(Il1a) | −1.572188 |
| NM_017069 | gamma-aminobutyric acid type A receptor alpha3 subunit(Gabra3) | −3.176886 |
| NM_017075 | acetyl-CoA acetyltransferase 1(Acat1) | −1.693916 |
| NM_017084 | glycine N-methyltransferase(Gnmt) | −1.587023 |
| NM_017085 | cytochrome P450, family 19, subfamily a, polypeptide 1(Cyp19a1) | −3.202593 |
| NM_017093 | AKT serine/threonine kinase 2(Akt2) | −1.708931 |
| NM_017101 | peptidylprolyl isomerase A (cyclophilin A)(Ppia) | −1.962416 |
| NM_017106 | chloride voltage-gated channel 5(Clcn5) | −2.303484 |
| NM_017120 | casein beta(Csn2) | −1.753558 |
| NM_017156 | cytochrome P450, family 2, subfamily b, polypeptide 12(Cyp2b12) | −1.675282 |
| NM_017158 | cytochrome P450, family 2, subfamily c, polypeptide 7(Cyp2c7) | −1.9999 |
| NM_017177 | choline kinase beta(Chkb) | −1.598296 |
| NM_017179 | UNC homeobox(Uncx) | −1.985105 |
| NM_017190 | myelin-associated glycoprotein(Mag) | −1.957495 |
| NM_017197 | CUGBP, Elav-like family member 2(Celf2) | −2.358172 |
| NM_017228 | atrophin 1(Atn1) | −3.12621 |
| NM_017265 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 6(Hsd3b6) | −1.512438 |
| NM_017285 | proteasome subunit beta 3(Psmb3) | −2.192897 |
| NM_017291 | gamma-aminobutyric acid type A receptor rho 1 subunit(Gabrr1) | −3.620853 |
| NM_017292 | gamma-aminobutyric acid type A receptor rho 2 subunit(Gabrr2) | −1.576548 |
| NM_017326 | calmodulin 2(Calm2) | −2.141575 |
| NM_017337 | phosphodiesterase 3A(Pde3a) | −1.72263 |
| NM_017339 | ISL LIM homeobox 1(Isl1) | −2.983281 |
| NM_017343 | myosin light chain 12B(Myl12b) | −1.748003 |
| NM_019182 | ring finger protein 4(Rnf4) | −1.852677 |
| NM_019190 | CD46 molecule(Cd46) | −1.945158 |
| NM_019211 | RAS guanyl releasing protein 1(Rasgrp1) | −1.760087 |
| NM_019241 | gap junction protein, beta 5(Gjb5) | −1.748274 |
| NM_019248 | neurotrophic receptor tyrosine kinase 3(Ntrk3) | −1.880549 |
| NM_019255 | calcium voltage-gated channel auxiliary subunit gamma 1(Cacng1) | −1.569839 |
| NM_019261 | killer cell lectin-like receptor subfamily C, member 2(Klrc2) | −2.129231 |
| NM_019270 | potassium voltage-gated channel subfamily A member 3(Kcna3) | −2.943964 |
| NM_019278 | regulated endocrine-specific protein 18(Resp18) | −2.015828 |
| NM_019285 | adenylate cyclase 4(Adcy4) | −1.51557 |
| NM_019299 | clathrin heavy chain(Cltc) | −1.670579 |
| NM_019348 | somatostatin receptor 2(Sstr2) | −1.921952 |
| NM_019354 | uncoupling protein 2(Ucp2) | −2.271891 |
| NM_019363 | aldehyde oxidase 1(Aox1) | −1.50887 |
| NM_019620 | zinc finger protein 386 (Kruppel-like)(Zfp386) | −2.6169 |
| NM_019621 | discs large MAGUK scaffold protein 4(Dlg4) | −1.768061 |
| NM_019907 | CXXC repeat containing interactor of PDZ3 domain(Cript) | −1.965978 |
| NM_020074 | serglycin(Srgn) | −1.792037 |
| NM_020076 | 3-hydroxyanthranilate 3,4-dioxygenase(Haao) | −1.647168 |
| NM_020096 | interferon-induced protein with tetratricopeptide repeats 1(Ifit1) | −1.569029 |
| NM_020301 | ADAM metallopeptidase domain 7(Adam7) | −1.648235 |
| NM_020542 | C-C motif chemokine receptor 1(Ccr1) | −1.713195 |
| NM_021580 | prolactin family 8, subfamily a, member 4(Prl8a4) | −2.141944 |
| NM_021659 | synaptotagmin 7(Syt7) | −1.795637 |
| NM_021672 | growth differentiation factor 9(Gdf9) | −1.566303 |
| NM_021676 | SH3 and multiple ankyrin repeat domains 3(Shank3) | −2.237337 |
| NM_021703 | A-kinase anchoring protein 14(Akap14) | −2.557232 |
| NM_021742 | nuclear receptor subfamily 5, group A, member 2(Nr5a2) | −1.603742 |
| NM_021769 | sulfotransferase family 1D, member 1(Sult1d1) | −1.883171 |
| NM_022178 | myosin VA(Myo5a) | −2.420753 |
| NM_022196 | leukemia inhibitory factor(Lif) | −1.875411 |
| NM_022202 | glutamate metabotropic receptor 8(Grm8) | −1.511956 |
| NM_022204 | exocyst complex component 5(Exoc5) | −1.572194 |
| NM_022214 | C-X-C motif chemokine ligand 6(Cxcl6) | −2.525867 |
| NM_022241 | prostaglandin D2 receptor(Ptgdrl) | −1.557255 |
| NM_022261 | B-box and SPRY domain containing(Bspry) | −1.56046 |
| NM_022278 | glutaredoxin(Glrx) | −2.208115 |
| NM_022283 | No data | −2.543935 |
| NM_022386 | MAF bZIP transcription factor G(Mafg) | −2.383068 |
| NM_022388 | FXYD domain-containing ion transport regulator 4(Fxyd4) | −2.897865 |
| NM_022394 | scaffold attachment factor B(Safb) | −2.659302 |
| NM_022502 | palmitoyl-protein thioesterase 1(Ppt1) | −1.90058 |
| NM_022503 | cytochrome c oxidase subunit VIIa polypeptide 2(Cox7a2) | −1.523241 |
| NM_022504 | ribosomal protein L36(Rpl36) | −1.906632 |
| NM_022506 | ribosomal protein L31(Rpl31) | −2.087805 |
| NM_022530 | prolactin family 7, subfamily a, member 3(Prl7a3) | −1.576727 |
| NM_022595 | PDGFA associated protein 1(Pdap1) | −2.240738 |
| NM_022597 | cathepsin B(Ctsb) | −2.37573 |
| NM_022612 | BCL2 like 11(Bcl2l11) | −1.597404 |
| NM_022634 | leukocyte specific transcript 1(Lst1) | −1.756651 |
| NM_022694 | staphylococcal nuclease and tudor domain containing 1(Snd1) | −2.119255 |
| NM_022711 | steroid 5 alpha-reductase 2(Srd5a2) | −1.557239 |
| NM_022797 | glutamate ionotropic receptor NMDA type subunit 2D(Grin2d) | −2.63892 |
| NM_022863 | iron responsive element binding protein 2(Ireb2) | −1.923661 |
| NM_022867 | microtubule-associated protein 1 light chain 3 beta(Map1lc3b) | −1.780024 |
| NM_022949 | ribosomal protein L14(Rpl14) | −1.533009 |
| NM_023095 | mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase(Mgat5) | −2.000325 |
| NM_023100 | neuromedin U receptor 1(Nmur1) | −1.627031 |
| NM_023974 | synaptoporin(Synpr) | −1.803122 |
| NM_023998 | taste receptor, type 2, member 13(Tas2r13) | −1.675877 |
| NM_024156 | annexin A6(Anxa6) | −1.687137 |
| NM_024346 | stathmin 3(Stmn3) | −1.575717 |
| NM_024360 | hes family bHLH transcription factor 1(Hes1) | −2.146919 |
| NM_024385 | hematopoietically expressed homeobox(Hhex) | −1.692521 |
| NM_024388 | nuclear receptor subfamily 4, group A, member 1(Nr4a1) | −2.520533 |
| NM_024397 | abl-interactor 1(Abi1) | −1.624808 |
| NM_024404 | heterogeneous nuclear ribonucleoprotein D(Hnrnpd) | −3.042042 |
| NM_024483 | adrenoceptor alpha 1D(Adra1d) | −1.736564 |
| NM_024484 | 5′-aminolevulinate synthase 1(Alas1) | −1.714727 |
| NM_030857 | LYN proto-oncogene, Src family tyrosine kinase(Lyn) | −2.18526 |
| NM_031008 | adaptor-related protein complex 2, alpha 2 subunit(Ap2a2) | −1.552738 |
| NM_031055 | matrix metallopeptidase 9(Mmp9) | −2.852522 |
| NM_031057 | aldehyde dehydrogenase 6 family, member A1(Aldh6a1) | −1.519431 |
| NM_031088 | prostaglandin E receptor 2(Ptger2) | −2.221709 |
| NM_031127 | sulfite oxidase(Suox) | −2.397507 |
| NM_031236 | fucosyltransferase 1(Fut1) | −1.985466 |
| NM_031327 | cysteine-rich, angiogenic inducer, 61(Cyr61) | −1.638485 |
| NM_031346 | polypyrimidine tract binding protein 3(Ptbp3) | −1.590357 |
| NM_031531 | serine (or cysteine) peptidase inhibitor, clade A, member 3N(Serpina3n) | −2.119708 |
| NM_031549 | transgelin(Tagln) | −2.37869 |
| NM_031563 | Y box binding protein 1(Ybx1) | −1.584841 |
| NM_031590 | WNT1 inducible signaling pathway protein 2(Wisp2) | −1.877269 |
| NM_031678 | period circadian clock 2(Per2) | −1.587107 |
| NM_031706 | ribosomal protein S8(Rps8) | −2.051886 |
| NM_031719 | chloride nucleotide-sensitive channel 1A(Clns1a) | −1.549741 |
| NM_031720 | deiodinase, iodothyronine, type II(Dio2) | −1.958028 |
| NM_031786 | tripartite motif-containing 3(Trim3) | −1.668096 |
| NM_031792 | sperm associated antigen 4(Spag4) | −1.634627 |
| NM_031800 | death effector domain-containing(Dedd) | −1.641957 |
| NM_031808 | calpain 6(Capn6) | −1.820196 |
| NM_031810 | defensin beta 1(Defb1) | −2.054222 |
| NM_031828 | potassium calcium-activated channel subfamily M alpha 1(Kcnma1) | −1.606975 |
| NM_032056 | transporter 2, ATP binding cassette subfamily B member(Tap2) | −2.753854 |
| NM_032062 | kalirin, RhoGEF kinase(Kalrn) | −1.810885 |
| NM_033021 | SEC31 homolog A, COPII coat complex component(Sec31a) | −1.750599 |
| NM_033095 | crystallin, gamma D(Crygd) | −1.569465 |
| NM_053649 | kringle containing transmembrane protein 1(Kremen1) | −1.821446 |
| NM_053674 | phytanoyl-CoA 2-hydroxylase(Phyh) | −1.905711 |
| NM_053701 | calcium voltage-gated channel subunit alpha1 F(Cacna1f) | −1.62128 |
| NM_080689 | dynamin 1(Dnm1) | −1.617324 |
| NM_080783 | UDP-galactose-4-epimerase(Gale) | −2.373817 |
| NM_130405 | KH RNA binding domain containing, signal transduction associated 1(Khdrbs1) | −1.779596 |
| NM_131914 | caveolin 2(Cav2) | −1.636241 |
| NM_133403 | E1A binding protein p300(Ep300) | −2.567324 |
| NM_133594 | small ubiquitin-like modifier 2(Sumo2) | −1.569797 |
| NM_134375 | NLR family, pyrin domain containing 6(Nlrp6) | −2.192127 |
| NM_134387 | dicarbonyl and L-xylulose reductase(Dcxr) | −2.210838 |
| NM_134432 | angiotensinogen(Agt) | −1.737263 |
| NM_134457 | siah E3 ubiquitin protein ligase 2(Siah2) | −1.625673 |
| NM_138851 | prokineticin 1(Prok1) | −1.918202 |
| NM_138890 | EH-domain containing 3(Ehd3) | −1.507669 |
| NM_138976 | mitofusin 1(Mfn1) | −1.671108 |
| NM_139082 | BMP and activin membrane-bound inhibitor(Bambi) | −2.450906 |
| NM_139097 | sodium voltage-gated channel beta subunit 3(Scn3b) | −3.059727 |
| NM_139258 | Bcl2 modifying factor(Bmf) | −2.168055 |
| NM_139336 | UDP-glucuronate decarboxylase 1(Uxs1) | −1.54412 |
| NM_145673 | MAF bZIP transcription factor K(Mafk) | −1.536959 |
| NM_147137 | cystatin SC(LOC257643) | −3.032693 |
| NM_152849 | homeobox and leucine zipper encoding(Homez) | −1.725448 |
| S75437 | No data | −1.965821 |
| U03407 | proline rich, lacrimal 1(Prol1) | −2.285635 |
| U03417 | olfactomedin 1(Olfm1) | −1.797439 |
| U03630 | store-operated calcium entry-associated regulatory factor(Saraf) | −2.016451 |
| U20286 | torsin 1A interacting protein 1(Tor1aip1) | −1.727547 |
| U21954 | Eph receptor A7(Epha7) | −1.630146 |
| U22663 | No data | −1.993711 |
| U24489 | tenascin XA, pseudogene 1(Tnxa-ps1) | −2.172308 |
| U28356 | protein tyrosine phosphatase, non-receptor type 7(Ptpn7) | −2.374227 |
| U35371 | contactin 4(Cntn4) | −1.667741 |
| U40652 | protein tyrosine phosphatase, receptor type, N(Ptprn) | −1.867797 |
| U41663 | neuroligin 3(Nlgn3) | −1.591787 |
| U41803 | mitofusin 2(Mfn2) | −1.574964 |
| U41853 | hypoxia up-regulated 1(Hyou1) | −1.657766 |
| U44129 | lectin, mannose-binding, 1(Lman1) | −2.451062 |
| U49694 | acyl-CoA thioesterase 7(Acot7) | −1.924287 |
| U50948 | No data | −1.881066 |
| U53420 | No data | −4.016245 |
| U56241 | MAF bZIP transcription factor B(Mafb) | −2.421294 |
| U56936 | killer cell lectin-like receptor subfamily B member 1B(Klrb1b) | −1.830188 |
| U57362 | collagen type XII alpha 1 chain(Col12a1) | −2.0687 |
| U62316 | solute carrier family 16 member 7(Slc16a7) | −2.847701 |
| U69702 | activin A receptor type 1C(Acvr1c) | −1.705311 |
| U76206 | purinergic receptor P2Y14(P2ry14) | −2.320753 |
| U78304 | cartilage acidic protein 1(Crtac1) | −1.778874 |
| U81037 | neuronal cell adhesion molecule(Nrcam) | −1.502983 |
| U92010 | similar to RIKEN cDNA D230025D16Rik(RGD621098) | −1.975117 |
| V01222 | albumin(Alb) | −2.265804 |
| X01153 | No data | −1.843423 |
| X05034 | No data | −1.718179 |
| X13309 | WAP four-disulfide core domain 18(Wfdc18) | −2.535182 |
| X53003 | acetyl-CoA carboxylase alpha(Acaca) | −1.756607 |
| X54549 | transcription factor 3(Tcf3) | −2.43995 |
| X56328 | No data | −2.636174 |
| X60822 | methionine adenosyltransferase 1A(Mat1a) | −1.586548 |
| X64411 | ubiquitin protein ligase E3 component n-recognin 5(Ubr5) | −2.174631 |
| X68101 | dedicator of cytokinesis 9(Dock9) | −4.49773 |
| X71463 | No data | −1.940264 |
| X73579 | Fc fragment of IgE receptor II(Fcer2) | −1.640805 |
| X74815 | myosin IE(Myo1e) | −1.973621 |
| X78167 | ribosomal protein L15(Rpl15) | −2.28316 |
| X78604 | ADP-ribosylation factor like GTPase 5A(Arl5a) | −1.523338 |
| X84004 | dual specificity phosphatase 1(Dusp1) | −2.325959 |
| Y12178 | ceruloplasmin(Cp) | −1.709408 |
| Y17319 | No data | −1.766901 |
| Y17325 | NSA2 ribosome biogenesis homolog(Nsa2) | −1.702431 |
| Y17328 | crystallin, mu(Crym) | −1.788589 |
References
- Potempa, J.; Mydel, P.; Koziel, J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat. Rev. Rheumatol. 2017, 13, 606–620. [Google Scholar] [CrossRef] [PubMed]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Firestein, G.S.; McInnes, I.B. Immunopathogenesis of Rheumatoid Arthritis. Immunity 2017, 46, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Rausch, A.K.; Osthoff, K.; Niedermann, J.; Braun, J.; Adams, N.; Brodin, H.; Dagfinrud, T.; Duruoz, B.A.; Esbensen, K.P.; Günther, E.; et al. 2018 EULAR recommendations for physical activity in people with inflammatory arthritis and osteoarthritis. Ann. Rheum. Dis. 2018, 77, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Landewé, R.; Bijlsma, J.; Burmester, G.; Chatzidionysiou, K.; Dougados, M.; Nam, J.; Ramiro, S.; Voshaar, M.; van Vollenhoven, R.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2016 update. Ann. Rheum. Dis. 2017, 76, 960–977. [Google Scholar] [CrossRef] [PubMed]
- Anvar, N.; Matlabi, H.; Safaiyan, A.; Allahverdipour, H.; Kolahi, S. Effectiveness of self-management program on arthritis symptoms among older women: A randomized controlled trial study. Health Care Women Int. 2018, 39, 1326–1339. [Google Scholar] [CrossRef] [PubMed]
- Agca, R.; Heslinga, S.C.; Rollefstad, S.; Heslinga, M.; McInnes, I.B.; Peters, M.J.L.; Kvien, T.K.; Dougados, M.; Radner, H.; Atzeni, F.; et al. EULAR recommendations for cardiovascular disease risk management in patients with rheumatoid arthritis and other forms of inflammatory joint disorders: 2015/2016 update. Ann. Rheum. Dis. 2017, 76, 17–28. [Google Scholar] [CrossRef]
- Law, R.J.; Saynor, Z.L.; Gabbitas, J.; Jones, J.; Kraus, A.; Breslin, A.; Maddison, P.J.; Thom, J.M. The Effects of Aerobic and Resistance Exercise on Markers of Large Joint Health in Stable Rheumatoid Arthritis Patients: A Pilot Study. Musculoskeletal Care 2015, 13, 222–235. [Google Scholar] [CrossRef]
- Bergstra, S.A.; Murgia, A.; Te Velde, A.F.; Caljouw, S.R. A systematic review into the effectiveness of hand exercise therapy in the treatment of rheumatoid arthritis. Clin. Rheumatol. 2014, 33, 1539–1548. [Google Scholar] [CrossRef]
- van Rensburg, D.C.J.; Ker, J.A.; Grant, C.C.; Fletcher, L. Effect of exercise on cardiac autonomic function in females with rheumatoid arthritis. Clin. Rheumatol. 2012, 31, 1155–1162. [Google Scholar] [CrossRef]
- Stavropoulos-Kalinoglou, A.; Metsios, G.S.; van Zanten, J.J.J.C.S.V.; Nightingale, P.; Kitas, G.D.; Koutedakis, Y. Individualised aerobic and resistance exercise training improves cardiorespiratory fitness and reduces cardiovascular risk in patients with rheumatoid arthritis. Ann. Rheum. Dis. 2013, 72, 1819–1825. [Google Scholar] [CrossRef] [PubMed]
- Waite-Jones, J.M.; Hale, C.A.; Lee, H.Y. Psychosocial effects of Tai Chi exercise on people with rheumatoid arthritis. J. Clin. Nurs. 2013, 22, 3053–3061. [Google Scholar] [CrossRef]
- Perandini, L.A.; de Sá-Pinto, A.L.; Roschel, H.; Benatti, F.B.; Lima, F.R.; Bonfá, E.; Gualano, B. Exercise as a therapeutic tool to counteract inflammation and clinical symptoms in autoimmune rheumatic diseases. Autoimmun. Rev. 2012, 12, 218–224. [Google Scholar] [CrossRef]
- Sharif, S.; Thomas, J.M.; Donley, D.A.; Gilleland, D.L.; Bonner, D.E.; McCrory, J.L.; Hornsby, W.G.; Zhao, H.; Lively, M.W.; Hornsby, J.A.A.; et al. Resistance exercise reduces skeletal muscle cachexia and improves muscle function in rheumatoid arthritis. Case Rep. Med. 2011, 2011, 205691. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Feldthusen, C.; Dean, E.; Forsblad-d’Elia, H.; Mannerkorpi, K. Effects of Person-Centered Physical Therapy on Fatigue-Related Variables in Persons With Rheumatoid Arthritis: A Randomized Controlled Trial. Arch. Phys. Med. Rehabil. 2016, 97, 26–36. [Google Scholar] [CrossRef]
- Cambré, I.; Gaublomme, D.; Schryvers, N.; Lambrecht, S.; Lories, R.; Venken, K.; Elewaut, D. Running promotes chronicity of arthritis by local modulation of complement activators and impairing T regulatory feedback loops. Ann. Rheum. Dis. 2019, 6, 787–795. [Google Scholar]
- Cambré, I.; Gaublomme, D.; Burssens, A.; Jacques, P.; Schryvers, N.; De Muynck, A.; Meuris, L.; Lambrecht, S.; Carter, S.; de Bleser, P.; et al. Mechanical strain determines the site-specific localization of inflammation and tissue damage in arthritis. Nat. Commun. 2018, 9, 4613. [Google Scholar] [CrossRef]
- Bevaart, L.; Vervoordeldonk, M.J.; Tak, P.P. Evaluation of therapeutic targets in animal models of arthritis: How does it relate to rheumatoid arthritis? Arthritis Rheum. 2010, 62, 2192–2205. [Google Scholar] [CrossRef]
- Bordy, R.; Verhoeven, F.; Tournier-Nappey, M.; Wendling, D.; Demougeot, C.; Totoson, P. Methotrexate did not improve endothelial function in rheumatoid arthritis: A study in rats with adjuvant-induced arthritis. Clin. Exp. Rheumatol. 2018, 1, 81–88. [Google Scholar]
- Abdel-Maged, A.E.S.; Gad, A.M.; Abdel-Aziz, A.K.; Aboulwafa, M.M.; Azab, S.S. Comparative study of anti-VEGF Ranibizumab and Interleukin-6 receptor antagonist Tocilizumab in Adjuvant-induced Arthritis. Toxicol. Appl. Pharmacol. 2018, 356, 65–75. [Google Scholar] [CrossRef]
- Vidal, B.; Cascão, R.; Finnilä, M.A.J.; Lopes, I.P.; da Glória, V.G.; Saarakkala, S.; Zioupos, P.; Canhão, H.; Fonseca, J.E. Effects of tofacitinib in early arthritis-induced bone loss in an adjuvant-induced arthritis rat model. Rheumatology 2018, 57, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Kaliyappan, K.; Palanisamy, M.; Govindarajan, R.; Duraiyan, J. Microarray and its applications. J. Pharm. Bioallied Sci. 2012, 4, 310. [Google Scholar] [CrossRef] [PubMed]
- Snekhalatha, U.; Anburajan, M.; Venkatraman, B.; Menaka, M. Evaluation of complete Freund’s adjuvant-induced arthritis in a Wistar rat model. Comparison of thermography and histopathology. Z. Für Rheumatol. 2013, 72, 375–382. [Google Scholar] [CrossRef]
- González-Chávez, S.A.; Pacheco-Tena, C.; Macías-Vázquez, C.E.; Luévano-Flores, E. Assessment of different decalcifying protocols on Osteopontin and Osteocalcin immunostaining in whole bone specimens of arthritis rat model by confocal immunofluorescence. Int. J. Clin. Exp. Pathol. 2013, 6, 1972–1983. [Google Scholar] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Isserlin, R.; Merico, D.; Voisin, V.; Bader, G.D. Enrichment Map—A Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000 Res. 2014, 3, 141. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Geenen, R.; Overman, C.L.; Christensen, R.; Åsenlöf, P.; Capela, S.; Huisinga, K.L.; Husebø, M.E.P.; Köke, A.J.A.; Paskins, Z.; Pitsillidou, I.A.; et al. EULAR recommendations for the health professional’s approach to pain management in inflammatory arthritis and osteoarthritis. Ann. Rheum. Dis. 2018, 77, 797–807. [Google Scholar] [CrossRef]
- Peters, C.L.; Morris, C.J.; Mapp, P.I.; Blake, D.R.; Lewis, C.E.; Winrow, V.R. The transcription factors hypoxia-inducible factor 1alpha and Ets-1 colocalize in the hypoxic synovium of inflamed joints in adjuvant-induced arthritis. Arthritis Rheum. 2004, 50, 291–296. [Google Scholar] [CrossRef]
- Jeon, C.H.; Ahn, J.K.; Chai, J.Y.; Kim, H.J.; Bae, E.K.; Park, S.H.; Cho, E.Y.; Cha, H.S.; Ahn, K.S.; Koh, E.M. Hypoxia appears at pre-arthritic stage and shows co-localization with early synovial inflammation in collagen induced arthritis. Clin. Exp. Rheumatol. 2008, 26, 646–648. [Google Scholar] [PubMed]
- Quiñonez-Flores, C.M.; González-Chávez, S.A.; Pacheco-Tena, C. Hypoxia and its implications in rheumatoid arthritis. J. Biomed. Sci. 2016, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Quiñonez-Flores, C.M.; González-Chávez, S.A.; Del Río Nájera, D.; Pacheco-Tena, C. Oxidative Stress Relevance in the Pathogenesis of the Rheumatoid Arthritis: A Systematic Review. BioMed Res. Int. 2016, 2016, 6097417. [Google Scholar] [CrossRef] [PubMed]
- Fearon, U.; Canavan, M.; Biniecka, M.; Veale Hypoxia, D.J. mitochondrial dysfunction and synovial invasiveness in rheumatoid arthritis. Nat. Rev. Rheumatol. 2016, 12, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.E.; Harlow, L.A.; Haines, G.K.; Amento, E.P.; Unemori, E.N.; Wong, W.L.; Pope, R.M.; Ferrara, N. Vascular endothelial growth factor. A cytokine modulating endothelial function in rheumatoid arthritis. J. Immunol. Baltim. Md 1950 1994, 152, 4149–4156. [Google Scholar]
- Asahara, T.; Chen, D.; Takahashi, T.; Fujikawa, K.; Kearney, M.; Magner, M.; Yancopoulos, G.D.; Isner, J.M. Tie2 receptor ligands, angiopoietin-1 and angiopoietin-2, modulate VEGF-induced postnatal neovascularization. Circ. Res. 1998, 83, 233–240. [Google Scholar] [CrossRef]
- Holash, J.; Maisonpierre, P.C.; Compton, D.; Boland, P.; Alexander, C.R.; Zagzag, D.; Yancopoulos, G.D.; Wiegand, S.J. Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF. Science 1999, 284, 1994–1998. [Google Scholar] [CrossRef]
- Paleolog, E.M.; Young, S.; Stark, A.C.; McCloskey, R.V.; Feldmann, M.; Maini, R.N. Modulation of angiogenic vascular endothelial growth factor by tumor necrosis factor alpha and interleukin-1 in rheumatoid arthritis. Arthritis Rheum. 1998, 41, 1258–1265. [Google Scholar] [CrossRef]
- Etherington, P.J.; Winlove, P.; Taylor, P.; Paleolog, E.; Miotla, J.M. VEGF release is associated with reduced oxygen tensions in experimental inflammatory arthritis. Clin. Exp. Rheumatol. 2002, 20, 799–805. [Google Scholar]
- Chang, X.; Wei, C. Glycolysis and rheumatoid arthritis. Int. J. Rheum. Dis. 2011, 14, 217–222. [Google Scholar] [CrossRef]
- Song, G.; Lu, Q.; Fan, H.; Zhang, X.; Ge, L.; Tian, R.; Wang, S.; Feng, T.; Pan, J.; Feng, J.; et al. Inhibition of hexokinases holds potential as treatment strategy for rheumatoid arthritis. Arthritis Res. Ther. 2019, 21, 87. [Google Scholar] [CrossRef] [PubMed]
- Hitchon, C.A.; El-Gabalawy, H.S. Oxidation in rheumatoid arthritis. Arthritis Res. Ther. 2004, 6, 265–278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mountz, J.D.; Zhang, H.G.; Wang, Y.; Xie, J.F.; Liang, X.; Hsu, H.C.; Curiel, D.T. AKT regulates TNF-alpha-mediated apoptosis of rheumatoid arthritis synovial fibrblasts. Arthritis Res. 2001, 3, 8. [Google Scholar] [CrossRef]
- Clark, A.R.; Dean, J.L. The p38 MAPK Pathway in Rheumatoid Arthritis: A Sideways Look. Open Rheumatol. J. 2001, 6, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, G.; Cooles, F.A.; Isaacs, J.D.; Hilkens, C.M. Emerging immunotherapies for rheumatoid arthritis. Hum. Vaccines Immunother. 2014, 10, 822–837. [Google Scholar] [CrossRef] [PubMed]
- McGonagle, D.; Gibbon, W.; O’Connor, P.; Green, M.; Pease, C.; Emery, P. Characteristic magnetic resonance imaging entheseal changes of knee synovitis in spondylarthropathy. Arthritis Rheum. 1998, 41, 694–700. [Google Scholar] [CrossRef]
- Blazek, A.D.; Nam, J.; Gupta, R.; Pradhan, M.; Perera, P.; Weisleder, N.L.; Hewett, T.E.; Chaudhari, A.M.; Lee, B.S.; Leblebicioglu, B.; et al. Exercise-driven metabolic pathways in healthy cartilage. Osteoarthr. Cartil. 2016, 24, 1210–1222. [Google Scholar] [CrossRef]
- Shay, A.K.; Bliven, M.L.; Scampoli, D.N.; Otterness, I.G.; Milici, A.J. Effects of exercise on synovium and cartilage from normal and inflamed knees. Rheumatol. Int. 1995, 14, 183–189. [Google Scholar] [CrossRef]
- Gomes, R.P.; Bressan, E.; da Silva, T.M.; da Silva Gevaerd, M.; Tonussi, C.R.; Domenech, S.C. Effects of one minute and ten minutes of walking activity in rats with arthritis induced by complete Freund’s adjuvant on pain and edema symptoms. Rev. Bras. Reumatol. 2014, 54, 83–89. [Google Scholar] [CrossRef]
- Gomes, R.P.; Bressan, E.; da Silva, T.M.; Domenech, S.C.; Tonussi, C.R. Evidências de que um protocolo de atividade física pode reduzir a contagem de leucócitos sinoviais de ratos artríticos. Rev. Bras. Med. Esporte 2013, 19, 70–73. [Google Scholar] [CrossRef][Green Version]
- Shimomura, S.; Inoue, H.; Arai, Y.; Nakagawa, S.; Fujii, Y.; Kishida, T.; Ichimaru, S.; Tsuchida, S.; Shirai, T.; Ikoma, K.; et al. Treadmill Running Ameliorates Destruction of Articular Cartilage and Subchondral Bone, Not Only Synovitis, in a Rheumatoid Arthritis Rat Model. Int. J. Mol. Sci. 2018, 19, 6. [Google Scholar] [CrossRef] [PubMed]
- Seo, D.Y.; Lee, S.R.; Kim, N.; Ko, K.S.; Rhee, B.D.; Han, J. Humanized animal exercise model for clinical implication. Pflugers Arch. 2014, 466, 1673–1687. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).