Modeling Drug and Radiation Resistance with Patient-Derived Organoids: Recent Progress, Unmet Needs, and Future Directions for Lung Cancer
Highlights
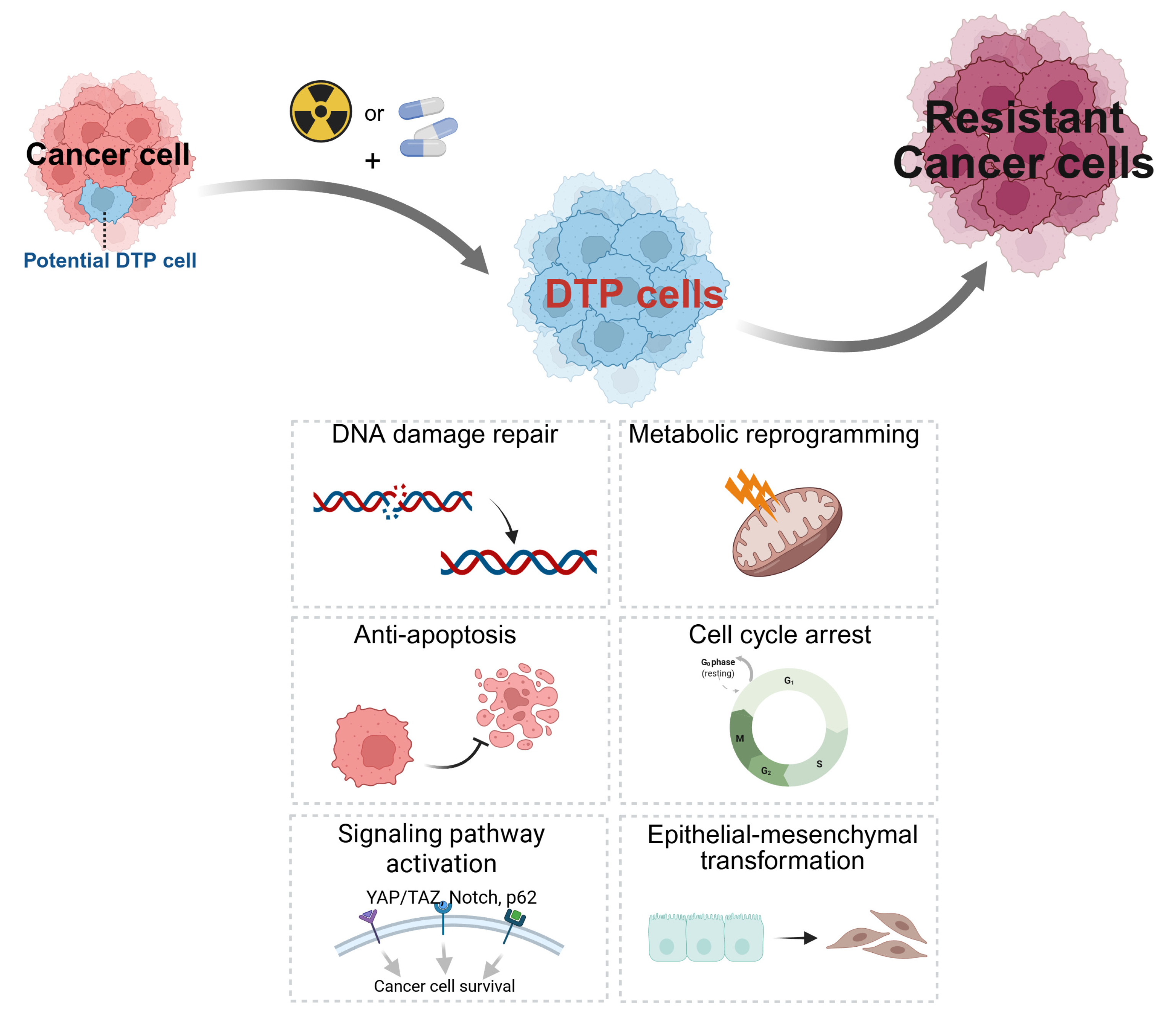
- Patient-derived organoids (PDOs) faithfully recapitulate the genetic heterogeneity and histological architecture of original tumors, serving as a superior platform for modeling intrinsic and acquired resistance mechanisms, particularly the reversible state of drug-tolerant persisters (DTPs).
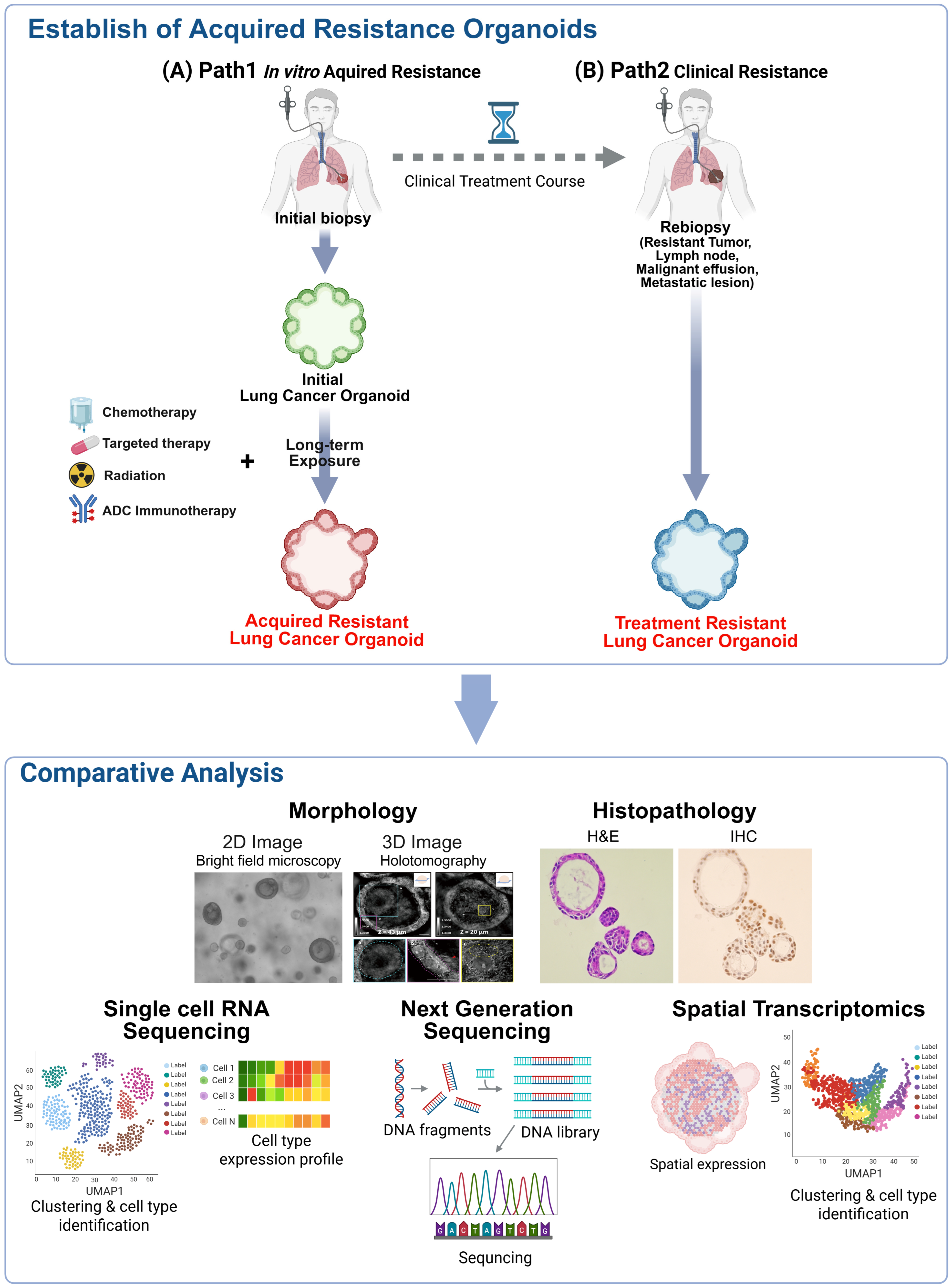
- Despite their utility, current PDO models face significant challenges, including the absence of tumor microenvironment (TME) components, the lack of standardized drug sensitivity cutoff values, and the scarcity of acquired resistance models in lung cancer due to re-biopsy limitations.
- Integrating PDOs with advanced technologies—such as single-cell RNA sequencing, spatial transcriptomics, and immune co-culture systems—enables the precise mapping of resistance evolutionary trajectories and the evaluation of novel therapeutics like antibody-drug conjugates (ADCs).
- Establishing standardized culture protocols and large-scale biobanks will transition PDOs from experimental tools to robust “pharmacotyping” platforms, facilitating functional precision medicine and improved treatment strategies for relapsed lung cancer patients.
Abstract
1. Introduction
2. Overview of Organoid Technology
3. Recent Advances in Organoid-Based Studies of Drug and Radiotherapy Resistance
3.1. Organoids for Modeling Drug Resistance
| Cancer | Drug Used | Organoid Type | Key Finding | Reference |
|---|---|---|---|---|
| Colorectal cancer | Oxaliplatin | Intrinsic/Acquired | Lnc-RP11-536K7.3/SOX2–HIF1α pathway drives Oxaliplatin resistance; knockout restores sensitivity. | [34] |
| 5-FU, Oxaliplatin, Irinotecan, Paclitaxel | Acquired | PDO drug response strongly correlated with clinical outcomes (100% sensitivity for paclitaxel). | [59] | |
| Irinotecan (monotherapy), 5-FU + Irinotecan (FI) | Intrinsic | PDO-based test successfully predicted clinical response (non-responders) to irinotecan-based therapies. | [60] | |
| Gastric cancer | 5-FU | Acquired | KHDRBS3 upregulation induced 5-FU multidrug resistance and cancer stem cell features. | [61] |
| Oxaliplatin | Acquired | Myoferlin (MYOF) upregulation drove acquired oxaliplatin resistance; knockdown restored sensitivity. | [62] | |
| 37 anticancer drugs (HTS) | Intrinsic (Biobank) | Established a large biobank capturing GC molecular subtypes. HTS identified novel drug sensitivities and linked ARID1A mutations to increased sensitivity to ATR inhibitors (VE-822). | [54] | |
| Ovarian cancer | Cisplatin | Intrinsic/Acquired | Aurora A–SOX8–FOXK1 axis promoted Cisplatin resistance via senescence/glycolysis regulation. | [57] |
| Cisplatin, others (X22) | Intrinsic/Acquired | PDOs recapitulated tumor genetics; useful for evaluating sensitivity to platinum and PARP inhibitors. | [58] | |
| Pancreatic cancer | Gemcitabine, others (X 5) | Intrinsic | Pharmacotyping of 66 PDOs correlated with clinical response; tracked resistance evolution. | [30] |
| Gemcitabine, Paclitaxel | Intrinsic | Identified BARD1, RAD50, and other genes as potential biomarkers for Gem/Paclitaxel resistance. | [31] | |
| Gemcitabine, 5-Fluorouracil, Paclitaxel | Intrinsic (PDO-CAF Co-culture) | CAF co-culture induced chemoresistance and an EMT phenotype in PDOs. | [63] | |
| Esophageal cancer | Cisplatin + 5-FU (CF) | Intrinsic/Acquired | Resistant PDOs showed NRF2 pathway hyperactivation; Fedratinib identified as potential therapy. | [64] |
| Cisplatin, others (X4) | Intrinsic | PDO drug sensitivity profiles correlated with patient clinical outcomes. | [32] | |
| Head and neck squamous cell carcinoma | Nutlin-3a, Alpelisib, PRMT5 inhibitor (EZP015566), Radiotherapy (RT) | Intrinsic (Biobank)/Gene-edited | PDO response to RT correlated with patient relapse. HTS linked CDKN2A loss to PRMT5 inhibitor sensitivity. | [51] |
| Lung cancer | EGFR-TKI, Cisplatin | Intrinsic | PDO drug response correlated with patient clinical outcomes for targeted therapies (e.g., Src activation). | [65] |
| Osimertinib, others (X7) | Acquired | PDOs effectively predicted chemotherapy and targeted therapy resistance, reflecting clinical responses. | [33] |
3.2. Organoids for Modeling Radiotherapy Resistance
3.3. Modeling Resistance to Novel Therapeutics: ICIs and ADCs
4. Clinical Trials Involving Cancer Organoids Resistant to Drug or Radiotherapy
5. Key Challenges and Unmet Needs in Lung Cancer Resistance Research
5.1. Technical Limitations and Standardization Issues
Challenges Specific to Lung Cancer
5.2. Clinical Translation Barriers
5.3. Critical Research Gaps in Lung Cancer Resistance
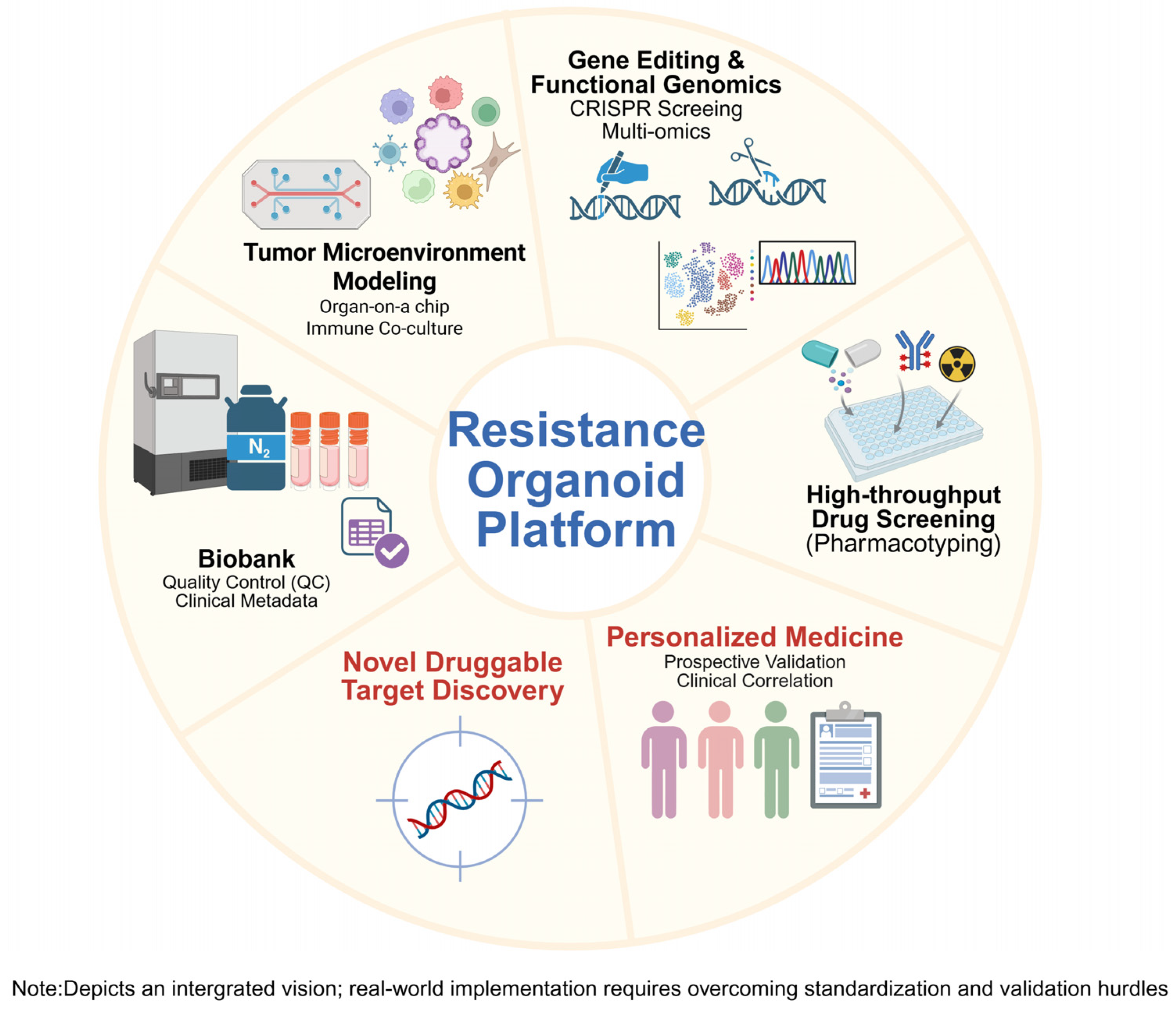
6. Overcoming Technical Limitations and Future Directions
6.1. Integrating Advanced Technologies to Overcome Limitations
6.2. Ultimate Goals and Vision for Research
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| 5-FU | 5-Fluorouracil |
| ADC | Antibody-Drug Conjugate |
| CAF | cancer-associated fibroblast |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CRT | Chemoradiation |
| CSC | Cancer Stem Cell |
| ctDNA | Circulating Tumor DNA |
| DTP | Drug-Tolerant Persister |
| EMT | Epithelial-to-mesenchymal transition |
| GLI | Gel–Liquid Interface |
| HTS | High-throughput screening |
| ICI | Immune Checkpoint Inhibitor |
| IDLD | Interstitial Lung Disease |
| iPSC | Induced Pluripotent Stem Cell |
| lncRNA | long non-coding RNA |
| MDR | Multidrug Resistance |
| MDSC | Myeloid-Derived Suppressor Cell |
| NSCLC | Non-Small Cell Lung Cancer |
| PBMC | Peripheral Blood Mononuclear Cell |
| PDO | Patient-Derived Organoid |
| PDOX | Patient-derived organoid xenograft |
| PDX | Patient-Derived Xenograft |
| scRNA-seq | single-cell RNA sequencing |
| ST | Spatial Transcriptomics |
| TAM | Tumor-Associated Macrophage |
| TIL | Tumor-Infiltrating Lymphocyte |
| TKI | Tyrosine Kinase Inhibitor |
| TME | Tumor Microenvironment |
References
- Gottesman, M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002, 53, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xing, Y.; Li, B.; Li, X.; Liu, B.; Wang, Y. Molecular pathways, resistance mechanisms and targeted interventions in non-small-cell lung cancer. Mol. Biomed. 2022, 3, 42. [Google Scholar] [CrossRef]
- Min, H.Y.; Lee, H.Y. Mechanisms of resistance to chemotherapy in non-small cell lung cancer. Arch. Pharm. Res. 2021, 44, 146–164. [Google Scholar] [CrossRef]
- Tahayneh, K.; Idkedek, M.; Abu Akar, F. NSCLC: Current Evidence on Its Pathogenesis, Integrated Treatment, and Future Perspectives. J. Clin. Med. 2025, 14, 1025. [Google Scholar] [CrossRef]
- Chhouri, H.; Alexandre, D.; Grumolato, L. Mechanisms of Acquired Resistance and Tolerance to EGFR Targeted Therapy in Non-Small Cell Lung Cancer. Cancers 2023, 15, 504. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef]
- Kochanowski, K.; Morinishi, L.; Altschuler, S.; Wu, L. Drug persistence—From antibiotics to cancer therapies. Curr. Opin. Syst. Biol. 2018, 10, 1–8. [Google Scholar] [CrossRef]
- Izumi, M.; Costa, D.B.; Kobayashi, S.S. Targeting of drug-tolerant persister cells as an approach to counter drug resistance in non-small cell lung cancer. Lung Cancer 2024, 194, 107885. [Google Scholar] [CrossRef]
- Lee, J.E.; Park, H.S.; Lee, D.; Yoo, G.; Kim, T.; Jeon, H.; Yeo, M.K.; Lee, C.S.; Moon, J.Y.; Jung, S.S.; et al. Hippo pathway effector YAP inhibition restores the sensitivity of EGFR-TKI in lung adenocarcinoma having primary or acquired EGFR-TKI resistance. Biochem. Biophys. Res. Commun. 2016, 474, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Lee, D.H.; Kang, D.H.; Yeo, M.K.; Bae, G.; Lee, D.; Yoo, G.; Kim, J.O.; Moon, E.; Huh, Y.H.; et al. Targeting YAP-p62 signaling axis suppresses the EGFR-TKI-resistant lung adenocarcinoma. Cancer Med. 2021, 10, 1405–1417. [Google Scholar] [CrossRef]
- Kamb, A. What’s wrong with our cancer models? Nat. Rev. Drug Discov. 2005, 4, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.C. Organoids: Avatars for Personalized Medicine. Keio J. Med. 2019, 68, 95. [Google Scholar] [CrossRef]
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Choi, J.; Iich, E.; Lee, J.H. Organogenesis of adult lung in a dish: Differentiation, disease and therapy. Dev. Biol. 2016, 420, 278–286. [Google Scholar] [CrossRef]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.J.; Chun, S.M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Verduin, M.; Hoeben, A.; De Ruysscher, D.; Vooijs, M. Patient-Derived Cancer Organoids as Predictors of Treatment Response. Front. Oncol. 2021, 11, 641980. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.H.; Lee, J.; Im, S.; Chung, C. Navigating the Complexity of Resistance in Lung Cancer Therapy: Mechanisms, Organoid Models, and Strategies for Overcoming Treatment Failure. Cancers 2024, 16, 3996. [Google Scholar] [CrossRef]
- Park, Y.; Kang, D.H.; Chung, C. Integrating tumor macroenvironment, microenvironment and mechanobiology with organoid and organ-on-a-chip models for lung cancer immunotherapy. Lung Cancer 2025, 207, 108726. [Google Scholar] [CrossRef]
- Lin, X.; Kang, K.; Chen, P.; Zeng, Z.; Li, G.; Xiong, W.; Yi, M.; Xiang, B. Regulatory mechanisms of PD-1/PD-L1 in cancers. Mol. Cancer 2024, 23, 108. [Google Scholar] [CrossRef]
- Taylor, C.; Patterson, K.M.; Friedman, D.; Bacot, S.M.; Feldman, G.M.; Wang, T. Mechanistic Insights into the Successful Development of Combination Therapy of Enfortumab Vedotin and Pembrolizumab for the Treatment of Locally Advanced or Metastatic Urothelial Cancer. Cancers 2024, 16, 3071. [Google Scholar] [CrossRef]
- Chang, H.L.; Schwettmann, B.; McArthur, H.L.; Chan, I.S. Antibody-drug conjugates in breast cancer: Overcoming resistance and boosting immune response. J. Clin. Investig. 2023, 133, e172156. [Google Scholar] [CrossRef]
- Khoury, R.; Saleh, K.; Khalife, N.; Saleh, M.; Chahine, C.; Ibrahim, R.; Lecesne, A. Mechanisms of Resistance to Antibody-Drug Conjugates. Int. J. Mol. Sci. 2023, 24, 9674. [Google Scholar] [CrossRef]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschênes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef]
- Oyama, S.; Matsuda, A.; Murakami, R.; Kakizaki, Y.; Ishizawa, T.; Kobayashi, T.; Nakamura, H.; Nawa, Y.; Otaki, Y.; Nagata, Y.; et al. Pancreatic cancer organoids derived from EUS-guided fine needle aspiration specimens can be used to predict chemotherapy resistance. Sci. Rep. 2025, 15, 23818. [Google Scholar] [CrossRef]
- Shen, S.; Liu, B.; Guan, W.; Liu, Z.; Han, Y.; Hu, Y.; Chen, Y.; Liu, S.; He, J.; Li, Z.; et al. Advancing precision medicine in esophageal squamous cell carcinoma using patient-derived organoids. J. Transl. Med. 2024, 22, 1168. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.M.; Zhang, C.Y.; Peng, K.C.; Chen, Z.X.; Su, J.W.; Li, Y.F.; Li, W.F.; Gao, Q.Y.; Zhang, S.L.; Chen, Y.Q.; et al. Using patient-derived organoids to predict locally advanced or metastatic lung cancer tumor response: A real-world study. Cell Rep. Med. 2023, 4, 100911. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sun, H.; Luo, D.; Gan, L.; Mo, S.; Dai, W.; Liang, L.; Yang, Y.; Xu, M.; Li, J.; et al. Lnc-RP11-536 K7.3/SOX2/HIF-1α signaling axis regulates oxaliplatin resistance in patient-derived colorectal cancer organoids. J. Exp. Clin. Cancer Res. 2021, 40, 348. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Cao, Z.; Wang, Z.; Xin, J.; Kong, B.; Xu, J.; Zhang, L.; Chen, P. Patient-Derived Organoid Model in the Prediction of Chemotherapeutic Drug Response in Colorectal Cancer. ACS Biomater. Sci. Eng. 2022, 8, 3515–3525. [Google Scholar] [CrossRef]
- Yan, H.H.N.; Chan, A.S.; Lai, F.P.; Leung, S.Y. Organoid cultures for cancer modeling. Cell Stem Cell 2023, 30, 917–937. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Translational applications of adult stem cell-derived organoids. Development 2017, 144, 968–975. [Google Scholar] [CrossRef]
- Azar, J.; Bahmad, H.F.; Daher, D.; Moubarak, M.M.; Hadadeh, O.; Monzer, A.; Al Bitar, S.; Jamal, M.; Al-Sayegh, M.; Abou-Kheir, W. The Use of Stem Cell-Derived Organoids in Disease Modeling: An Update. Int. J. Mol. Sci. 2021, 22, 7667. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A.; et al. Organoids. Nat. Rev. Methods Primers 2022, 2, 94. [Google Scholar] [CrossRef] [PubMed]
- Szepanowski, L.P.; Wruck, W.; Kapr, J.; Rossi, A.; Fritsche, E.; Krutmann, J.; Adjaye, J. Cockayne Syndrome Patient iPSC-Derived Brain Organoids and Neurospheres Show Early Transcriptional Dysregulation of Biological Processes Associated with Brain Development and Metabolism. Cells 2024, 13, 591. [Google Scholar] [CrossRef] [PubMed]
- Heinzelmann, E.; Piraino, F.; Costa, M.; Roch, A.; Norkin, M.; Garnier, V.; Homicsko, K.; Brandenberg, N. iPSC-derived and Patient-Derived Organoids: Applications and challenges in scalability and reproducibility as pre-clinical models. Curr. Res. Toxicol. 2024, 7, 100197. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Dye, B.R.; Ferrer-Torres, D.; Hill, D.R.; Overeem, A.W.; Shea, L.D.; Spence, J.R. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019, 14, 518–540. [Google Scholar] [CrossRef]
- Wang, J.; Tao, X.; Zhu, J.; Dai, Z.; Du, Y.; Xie, Y.; Chu, X.; Fu, G.; Lei, Z. Tumor organoid-immune co-culture models: Exploring a new perspective of tumor immunity. Cell Death Discov. 2025, 11, 195. [Google Scholar] [CrossRef]
- Li, K.; Liu, C.; Sui, X.; Li, C.; Zhang, T.; Zhao, T.; Zhang, D.; Wu, H.; Liu, Y.; Wang, S.; et al. An organoid co-culture model for probing systemic anti-tumor immunity in lung cancer. Cell Stem Cell 2025, 32, 1218–1234.e7. [Google Scholar] [CrossRef]
- Rodrigues, R.O.; Sousa, P.C.; Gaspar, J.; Bañobre-López, M.; Lima, R.; Minas, G. Organ-on-a-Chip: A Preclinical Microfluidic Platform for the Progress of Nanomedicine. Small 2020, 16, e2003517. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, T.; Cao, L. Use and application of organ-on-a-chip platforms in cancer research. J. Cell Commun. Signal 2023, 17, 1163–1179. [Google Scholar] [CrossRef]
- Chen, K.Y.; Srinivasan, T.; Lin, C.; Tung, K.L.; Gao, Z.; Hsu, D.S.; Lipkin, S.M.; Shen, X. Single-Cell Transcriptomics Reveals Heterogeneity and Drug Response of Human Colorectal Cancer Organoids. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; Volume 2018, pp. 2378–2381. [Google Scholar] [CrossRef]
- Wang, R.; Mao, Y.; Wang, W.; Zhou, X.; Wang, W.; Gao, S.; Li, J.; Wen, L.; Fu, W.; Tang, F. Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis. Genome Biol. 2022, 23, 106. [Google Scholar] [CrossRef]
- Millen, R.; De Kort, W.W.B.; Koomen, M.; van Son, G.J.F.; Gobits, R.; Penning de Vries, B.; Begthel, H.; Zandvliet, M.; Doornaert, P.; Raaijmakers, C.P.J.; et al. Patient-derived head and neck cancer organoids allow treatment stratification and serve as a tool for biomarker validation and identification. Med 2023, 4, 290–310.e12. [Google Scholar] [CrossRef]
- Jun, H.R.; Kang, H.J.; Ju, S.H.; Kim, J.E.; Jeon, S.Y.; Ku, B.; Lee, J.J.; Kim, M.; Kim, M.J.; Choi, J.J.; et al. High-throughput organo-on-pillar (high-TOP) array system for three-dimensional ex vivo drug testing. Biomaterials 2023, 296, 122087. [Google Scholar] [CrossRef]
- Ramakrishna, G.; Babu, P.E.; Singh, R.; Trehanpati, N. Application of CRISPR-Cas9 based gene editing to study the pathogenesis of colon and liver cancer using organoids. Hepatol. Int. 2021, 15, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.K.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.; Lam, K.O.; et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018, 23, 882–897.e11. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, S.; Wang, H.; Zhu, H.; Lu, Y.; Zhang, Y.; Guo, S.; He, J.; Li, Y.; Zhang, Y.; et al. A pancreatic cancer organoid biobank links multi-omics signatures to therapeutic response and clinical evaluation of statin combination therapy. Cell Stem Cell 2025, 32, 1369–1389.e14. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sui, X.; Song, F.; Li, Y.; Li, K.; Chen, Z.; Yang, F.; Chen, X.; Zhang, Y.; Wang, X.; et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat. Commun. 2021, 12, 2581. [Google Scholar] [CrossRef]
- Sun, H.; Wang, H.; Wang, X.; Aoki, Y.; Wang, X.; Yang, Y.; Cheng, X.; Wang, Z.; Wang, X. Aurora-A/SOX8/FOXK1 signaling axis promotes chemoresistance via suppression of cell senescence and induction of glucose metabolism in ovarian cancer organoids and cells. Theranostics 2020, 10, 6928–6945. [Google Scholar] [CrossRef]
- Nanki, Y.; Chiyoda, T.; Hirasawa, A.; Ookubo, A.; Itoh, M.; Ueno, M.; Akahane, T.; Kameyama, K.; Yamagami, W.; Kataoka, F.; et al. Patient-derived ovarian cancer organoids capture the genomic profiles of primary tumours applicable for drug sensitivity and resistance testing. Sci. Rep. 2020, 10, 12581. [Google Scholar] [CrossRef]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef]
- Ukai, S.; Honma, R.; Sakamoto, N.; Yamamoto, Y.; Pham, Q.T.; Harada, K.; Takashima, T.; Taniyama, D.; Asai, R.; Fukada, K.; et al. Molecular biological analysis of 5-FU-resistant gastric cancer organoids; KHDRBS3 contributes to the attainment of features of cancer stem cell. Oncogene 2020, 39, 7265–7278. [Google Scholar] [CrossRef]
- Harada, K.; Sakamoto, N.; Ukai, S.; Yamamoto, Y.; Pham, Q.T.; Taniyama, D.; Honma, R.; Maruyama, R.; Takashima, T.; Ota, H.; et al. Establishment of oxaliplatin-resistant gastric cancer organoids: Importance of myoferlin in the acquisition of oxaliplatin resistance. Gastric Cancer 2021, 24, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- Schuth, S.; Le Blanc, S.; Krieger, T.G.; Jabs, J.; Schenk, M.; Giese, N.A.; Büchler, M.W.; Eils, R.; Conrad, C.; Strobel, O. Patient-specific modeling of stroma-mediated chemoresistance of pancreatic cancer using a three-dimensional organoid-fibroblast co-culture system. J. Exp. Clin. Cancer Res. 2022, 41, 312. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Sato, T.; Ohashi, E.; Kajita, M.; Miya, F.; Yamamoto, K.; Yotsumata, H.; Yamaguchi, K.; Nakajima, Y.; Miura, A.; et al. An organoid library of human esophageal squamous cell carcinomas (ESCCs) uncovers the chemotherapy-resistant ESCC features. Commun. Biol. 2025, 8, 507. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, S.M.; Lim, S.; Lee, J.Y.; Choi, S.J.; Yang, S.D.; Yun, M.R.; Kim, C.G.; Gu, S.R.; Park, C.; et al. Modeling Clinical Responses to Targeted Therapies by Patient-Derived Organoids of Advanced Lung Adenocarcinoma. Clin. Cancer Res. 2021, 27, 4397–4409. [Google Scholar] [CrossRef]
- Rycaj, K.; Tang, D.G. Cancer stem cells and radioresistance. Int. J. Radiat. Biol. 2014, 90, 615–621. [Google Scholar] [CrossRef]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Szeglin, B.C.; Zheng, Y.; Sauvé, C.G.; Adileh, M.; Wasserman, I.; Marco, M.R.; Kim, A.S.; et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 2019, 25, 1607–1614. [Google Scholar] [CrossRef]
- Andel, D.; Nouwens, A.J.; Klaassen, S.; Laoukili, J.; Viergever, B.; Verheem, A.; Intven, M.P.W.; Zandvliet, M.; Hagendoorn, J.; Borel Rinkes, I.H.M.; et al. Rational design of alternative treatment options for radioresistant rectal cancer using patient-derived organoids. Br. J. Cancer 2025, 132, 973–981. [Google Scholar] [CrossRef]
- Andel, D.; Viergever, B.J.; Peters, N.A.; Elisabeth Raats, D.A.; Schenning-van Schelven, S.J.; Willem Intven, M.P.; Zandvliet, M.; Hagendoorn, J.; Max Borel Rinkes, I.H.; Kranenburg, O. Pre-existing subclones determine radioresistance in rectal cancer organoids. Cell Rep. 2024, 43, 113735. [Google Scholar] [CrossRef]
- Al Bitar, S.; Ballout, F.; Monzer, A.; Kanso, M.; Saheb, N.; Mukherji, D.; Faraj, W.; Tawil, A.; Doughan, S.; Hussein, M.; et al. Thymoquinone Radiosensitizes Human Colorectal Cancer Cells in 2D and 3D Culture Models. Cancers 2022, 14, 1363. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef]
- Lucky, S.S.; Law, M.; Lui, M.H.; Mong, J.; Shi, J.; Yu, S.; Yoon, D.K.; Djeng, S.K.; Wang, J.; Lim, C.M.; et al. Patient-Derived Nasopharyngeal Cancer Organoids for Disease Modeling and Radiation Dose Optimization. Front. Oncol. 2021, 11, 622244. [Google Scholar] [CrossRef] [PubMed]
- Issing, C.; Menche, C.; Richter, M.R.; Mosa, M.H.; von der Grün, J.; Fleischmann, M.; Thoenissen, P.; Winkelmann, R.; Darvishi, T.; Loth, A.G.; et al. Head and neck tumor organoid biobank for modelling individual responses to radiation therapy according to the TP53/HPV status. J. Exp. Clin. Cancer Res. 2025, 44, 85. [Google Scholar] [CrossRef] [PubMed]
- van Goor, I.; Raymakers, L.; Andel, D.S.H.; Brosens, L.A.A.; Kranenburg, O.; Leusen, J.H.W.; Meijer, G.J.; Molenaar, I.Q.; van Santvoort, H.C.; de Vries, J.H.W.; et al. Radiation response assessment of organoids derived from patients with pancreatic cancer. Clin. Transl. Radiat. Oncol. 2024, 48, 100829. [Google Scholar] [CrossRef] [PubMed]
- Nounsi, A.; Seitlinger, J.; Ponté, C.; Demiselle, J.; Idoux-Gillet, Y.; Pencreach, E.; Beau-Faller, M.; Lindner, V.; Balloul, J.M.; Quemeneur, E.; et al. Patient-Derived Tumoroid for the Prediction of Radiotherapy and Chemotherapy Responses in Non-Small-Cell Lung Cancer. Biomedicines 2023, 11, 1824. [Google Scholar] [CrossRef]
- Mariniello, A.; Borgeaud, M.; Weiner, M.; Frisone, D.; Kim, F.; Addeo, A. Primary and Acquired Resistance to Immunotherapy with Checkpoint Inhibitors in NSCLC: From Bedside to Bench and Back. BioDrugs 2025, 39, 215–235. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef]
- Magré, L.; Verstegen, M.M.A.; Buschow, S.; van der Laan, L.J.W.; Peppelenbosch, M.; Desai, J. Emerging organoid-immune co-culture models for cancer research: From oncoimmunology to personalized immunotherapies. J. Immunother. Cancer 2023, 11, e006290. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef]
- Fu, Z.; Li, S.; Han, S.; Shi, C.; Zhang, Y. Antibody drug conjugate: The “biological missile” for targeted cancer therapy. Signal Transduct. Target. Ther. 2022, 7, 93. [Google Scholar] [CrossRef]
- Yu, H.A.; Goto, Y.; Hayashi, H.; Felip, E.; Chih-Hsin Yang, J.; Reck, M.; Yoh, K.; Lee, S.H.; Paz-Ares, L.; Besse, B.; et al. HERTHENA-Lung01, a Phase II Trial of Patritumab Deruxtecan (HER3-DXd) in Epidermal Growth Factor Receptor-Mutated Non-Small-Cell Lung Cancer After Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Therapy and Platinum-Based Chemotherapy. J. Clin. Oncol. 2023, 41, 5363–5375. [Google Scholar] [CrossRef]
- Powell, C.A.; Modi, S.; Iwata, H.; Takahashi, S.; Smit, E.F.; Siena, S.; Chang, D.Y.; Macpherson, E.; Qin, A.; Singh, J.; et al. Pooled analysis of drug-related interstitial lung disease and/or pneumonitis in nine trastuzumab deruxtecan monotherapy studies. ESMO Open 2022, 7, 100554. [Google Scholar] [CrossRef]
- Ying, W.; Pi, H.; He, X.; Sheng, Y. Case Report: Personalized treatment in stage IV breast cancer using the patient-derived organoids. Front. Oncol. 2025, 15, 1629690. [Google Scholar] [CrossRef] [PubMed]
- Hosni, R.; Klümper, N.; Sanders, C.; Hosni, S.; Branchi, V.; Semaan, A.; Alajati, A.; Pelusi, N.; Ng, S.S.; Ralser, D.J.; et al. The Antibody-Drug Conjugate Sacituzumab Govitecan (IMMU-132) Represents a Potential Novel Therapeutic Strategy in Cholangiocarcinoma. Mol. Cancer Ther. 2025, 24, 1775–1788. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, M. Recent advances in lung cancer organoid (tumoroid) research (Review). Exp. Ther. Med. 2024, 28, 383. [Google Scholar] [CrossRef]
- Zhou, R.; Brislinger, D.; Fuchs, J.; Lyons, A.; Langthaler, S.; Hauser, C.A.E.; Baumgartner, C. Vascularised organoids: Recent advances and applications in cancer research. Clin. Transl. Med. 2025, 15, e70258. [Google Scholar] [CrossRef]
- Ehab, S.; Gaser, O.A.; Dayem, A.A. Hypoxia and Multilineage Communication in 3D Organoids for Human Disease Modeling. Biomimetics 2025, 10, 624. [Google Scholar] [CrossRef]
- Yu, J.; Wang, K.; Tang, Y.; Zheng, D. Applications and perspectives of tumor organoids in radiobiology (Review). Oncol. Rep. 2024, 52, 100. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Monkhorst, K.; Schipper, L.J.; Hartemink, K.J.; Smit, E.F.; Kaing, S.; de Groot, R.; Wolkers, M.C.; Clevers, H.; Cuppen, E.; et al. Challenges in Establishing Pure Lung Cancer Organoids Limit Their Utility for Personalized Medicine. Cell Rep. 2020, 31, 107588. [Google Scholar] [CrossRef]
- Chen, X.; Ye, L.; Wang, H.; Liu, X.; Zhao, L.; Xu, K.; Liu, Y.; He, Y. Promising preclinical models for lung cancer research-lung cancer organoids: A narrative review. Transl. Lung Cancer Res. 2024, 13, 623–634. [Google Scholar] [CrossRef]
- Choi, S.Y.; Cho, Y.H.; Kim, D.S.; Ji, W.; Choi, C.M.; Lee, J.C.; Rho, J.K.; Jeong, G.S. Establishment and Long-Term Expansion of Small Cell Lung Cancer Patient-Derived Tumor Organoids. Int. J. Mol. Sci. 2021, 22, 1349. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Kim, Y.; Chung, C. Scientific Validation and Clinical Application of Lung Cancer Organoids. Cells 2021, 10, 3012. [Google Scholar] [CrossRef] [PubMed]
- Yokota, E.; Iwai, M.; Yukawa, T.; Yoshida, M.; Naomoto, Y.; Haisa, M.; Monobe, Y.; Takigawa, N.; Guo, M.; Maeda, Y.; et al. Clinical application of a lung cancer organoid (tumoroid) culture system. NPJ Precis. Oncol. 2021, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Hocking, A.J.; Mortimer, L.A.; Farrall, A.L.; Russell, P.A.; Klebe, S. Establishing mesothelioma patient-derived organoid models from malignant pleural effusions. Lung Cancer 2024, 191, 107542. [Google Scholar] [CrossRef]
- Park, Y.; Lee, D.; Lee, J.E.; Park, H.S.; Jung, S.S.; Park, D.; Kang, D.H.; Lee, S.I.; Woo, S.D.; Chung, C. The Matrix Stiffness Coordinates the Cell Proliferation and PD-L1 Expression via YAP in Lung Adenocarcinoma. Cancers 2024, 16, 598. [Google Scholar] [CrossRef]
- Park, D. Advanced Bronchoscopic Diagnostic Techniques in Lung Cancer. Tuberc. Respir. Dis. 2024, 87, 282–291. [Google Scholar] [CrossRef]
- Park, D.; Lee, D.; Kim, Y.; Park, Y.; Lee, Y.J.; Lee, J.E.; Yeo, M.K.; Kang, M.W.; Chong, Y.; Han, S.J.; et al. Cryobiopsy: A Breakthrough Strategy for Clinical Utilization of Lung Cancer Organoids. Cells 2023, 12, 1854. [Google Scholar] [CrossRef]
- Liu, C.; Li, K.; Sui, X.; Zhao, T.; Zhang, T.; Chen, Z.; Wu, H.; Li, C.; Li, H.; Yang, F.; et al. Patient-Derived Tumor Organoids Combined with Function-Associated ScRNA-Seq for Dissecting the Local Immune Response of Lung Cancer. Adv. Sci. 2024, 11, e2400185. [Google Scholar] [CrossRef]
- Moye, A.L.; Dost, A.F.; Ietswaart, R.; Sengupta, S.; Ya, V.; Aluya, C.; Fahey, C.G.; Louie, S.M.; Paschini, M.; Kim, C.F. Early-stage lung cancer is driven by a transitional cell state dependent on a KRAS-ITGA3-SRC axis. EMBO J. 2024, 43, 2843–2861. [Google Scholar] [CrossRef]
- Ma, S.; Wang, W.; Zhou, J.; Liao, S.; Hai, C.; Hou, Y.; Zhou, Z.; Wang, Z.; Su, Y.; Zhu, Y.; et al. Lamination-based organoid spatially resolved transcriptomics technique for primary lung and liver organoid characterization. Proc. Natl. Acad. Sci. USA 2024, 121, e2408939121. [Google Scholar] [CrossRef]
- Wensink, G.E.; Elias, S.G.; Mullenders, J.; Koopman, M.; Boj, S.F.; Kranenburg, O.W.; Roodhart, J.M.L. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precis. Oncol. 2021, 5, 30. [Google Scholar] [CrossRef]
| Cancer | Radiation Exposure | Key Findings | Reference |
|---|---|---|---|
| Rectal cancer | Chemoradiation (CRT) | PDO radiosensitivity strongly correlated with patient clinical outcomes. Radioresistant PDOs showed activation of DNA damage repair pathways (ATM/ATR). | [67] |
| Irradiation | PDOs showed enhanced DNA repair pathways and antioxidant metabolism, and treatment with GCLC inhibitor and RRx-001 induced oxidative stress and improved radiotherapy sensitivity, overcoming resistance. | [68] | |
| Irradiation | In rectal cancer radiotherapy-resistant organoids, enhanced survival of subclones and reduced chromosomal instability suggest that subclonal populations play a crucial role in the development of radiotherapy resistance. | [69] | |
| Colorectal cancer | Irradiation + Thymoquinone | Thymoquinone acted as a radiosensitizer, suppressing NF-κB and Wnt/β-catenin pathways and inhibiting cancer stem cell features. | [70] |
| Glioblastoma cancer | Irradiation | Modeled radioresistant cancer stem cell (CSC) niches, revealing enhanced DNA damage repair and hypoxia-induced survival signaling as key resistance mechanisms. | [71] |
| Nasopharyngeal carcinoma (NPC) | Irradiation | Hypoxia-induced HIF-1α expression increased radioresistance. The model served as a platform for personalized radiation dose optimization. | [72] |
| Irradiation | PDOs can effectively model radiotherapy resistance, and HPV infection and TP53 mutations significantly influence the radiation response. | [73] | |
| Pancreatic cancer | Irradiation | PDOs showed varying sensitivity to radiation in the dose–response relationship, with one PDO demonstrating higher radiation sensitivity, making it useful for predicting individual responses to radiation therapy. | [74] |
| Lung cancer | Irradiation + Cytotoxic chemo (Cisplatin + Vinorelbine) | Patient-derived tumoroids exhibited varying sensitivity depending on the radiation dose, with radiation-resistant tumoroids showing reduced DNA damage and enhanced survival, making them a useful model for studying responses to radiotherapy. | [75] |
| Cancer | Treatment | Research Overview | Trial ID |
|---|---|---|---|
| Lung cancer | Chemotherapy | Establishment of NSCLC patient-derived organoids for drug sensitivity testing and prediction of treatment response and personalized therapy. | NCT06406608, NCT05669586 |
| Cervical cancer | Chemotherapy + Radiotherapy | Evaluation of organoid response to chemo-radiotherapy, exploration of treatment resistance mechanisms, and identification of new therapeutic targets. | NCT06786780 |
| Ovary cancer | Chemotherapy (Paclitaxel, Platinum–Taxane) | Development of personalized drug regimens and precision medicine approaches using ovarian cancer organoids. | NCT05813509, NCT04846933 |
| Breast cancer | Chemotherapy | Establishment of organoid-based drug sensitivity testing and investigation of drug resistance mechanisms. | NCT03925233 |
| Prostate cancer | Chemotherapy | Organoid-based drug sensitivity screening for castration-resistant prostate cancer with bone metastasis. | NCT06529549 |
| Head and neck cancer | Chemotherapy | Characterization of p53-mutant tumor organoids to understand recurrence mechanisms and identify therapeutic strategies. | NCT0671941 |
| Glioma | Chemotherapy (Temozolomide) + Radiotherapy | Use of high-grade glioma organoids for optimal drug combination screening and ctDNA/proteomics-based biomarker identification. | NCT05532397, NCT04868396 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.; Kim, Y.; Kang, D.H.; Chung, C. Modeling Drug and Radiation Resistance with Patient-Derived Organoids: Recent Progress, Unmet Needs, and Future Directions for Lung Cancer. Cells 2025, 14, 1994. https://doi.org/10.3390/cells14241994
Lee D, Kim Y, Kang DH, Chung C. Modeling Drug and Radiation Resistance with Patient-Derived Organoids: Recent Progress, Unmet Needs, and Future Directions for Lung Cancer. Cells. 2025; 14(24):1994. https://doi.org/10.3390/cells14241994
Chicago/Turabian StyleLee, Dahye, Yoonjoo Kim, Da Hyun Kang, and Chaeuk Chung. 2025. "Modeling Drug and Radiation Resistance with Patient-Derived Organoids: Recent Progress, Unmet Needs, and Future Directions for Lung Cancer" Cells 14, no. 24: 1994. https://doi.org/10.3390/cells14241994
APA StyleLee, D., Kim, Y., Kang, D. H., & Chung, C. (2025). Modeling Drug and Radiation Resistance with Patient-Derived Organoids: Recent Progress, Unmet Needs, and Future Directions for Lung Cancer. Cells, 14(24), 1994. https://doi.org/10.3390/cells14241994






