Abstract
The proteasome is a multi-catalytic protease complex that is involved in protein quality control via three proteolytic activities (i.e., caspase-, trypsin-, and chymotrypsin-like activities). Most cellular proteins are selectively degraded by the proteasome via ubiquitination. Moreover, the ubiquitin–proteasome system is a critical process for maintaining protein homeostasis. Here, we briefly summarize the structure of the proteasome, its regulatory mechanisms, proteins that regulate proteasome activity, and alterations to proteasome activity found in diverse diseases, chemoresistant cells, and cancer stem cells. Finally, we describe potential therapeutic modalities that use the ubiquitin–proteasome system.
1. Introduction
Reactive oxygen species (ROS), by-products of aerobic metabolism, are mostly produced in the mitochondria [1]. Low concentrations of ROS are involved as signaling molecules in various pathways, and high concentrations of ROS are removed by antioxidant proteins before they damage cells [2,3]. Nevertheless, excessive production of ROS causes imbalances between the production of free radicals, and their elimination and can lead to misfolding of native proteins [4]. To prevent the accumulation of misfolded proteins, cells have developed protein quality control machinery such as molecular chaperones [5]. Chaperone proteins facilitate the refolding of misfolded proteins; however, limitations exist since some proteins cannot be refolded to their native state by chaperones; examples include damaged or misfolded soluble proteins and obsolete proteins [6,7]. Moreover, when dysregulation of protein homeostasis (proteostasis) continues, it causes diseases such as cancer, amyotrophic lateral sclerosis, metabolic disorders, and spinal and bulbar muscular atrophy [8,9,10]. Therefore, the timely removal of non-native proteins is essential for cells to maintain protein homeostasis.
The main method of eliminating non-native proteins from cells is protein degradation (proteolysis). There are two main pathways involved in proteolysis in cells: the ubiquitin–proteasome system (UPS) and the autophagy–lysosome system [11]. Of these two, UPS prevents the abnormal accumulation of proteins by directly breaking down more than 80% of cellular proteins [12]. In contrast, the autophagy–lysosome system is involved in the decomposition of proteins that cannot be degraded even by the proteasome, such as insoluble or aggregated proteins, cellular organelles, etc. [13,14]. Thus, the proteasome is essentially involved in protein quality control against the accumulation of aggregated proteins.
Recently, various treatment methods such as proteolysis targeting chimera (PROTAC) and molecular glues have been developed for targeted protein degradation (TPD). Preclinical results from 2019 show distinct promise of this technique in the future [15]. In such treatment methods, the UPS is mainly involved in selectively degrading target proteins. Moreover, unlike existing treatments (i.e., small molecule inhibitors, siRNAs, and monoclonal antibodies, among others), this method is a new approach that can directly target and remove disease-causing proteins [16]. Thus, the development of treatments using the proteasome is expected to increase in the future. In this review, we briefly describe the structure of the proteasome, the mechanistic basis of the UPS, and the regulatory proteins involved in proteasome activity. Finally, we conclude by discussing the possibility of various therapeutic modalities using the proteasome.
2. Ubiquitin
Ubiquitin (originally called “ubiquitous immunopoietic polypeptide”) was first discovered by Gideon Goldstein in 1975 [17]. Five years after its discovery, Keith D. Wilkinson, Michael K. Urban, and Arthur L. Haas confirmed that a small heat-stable polypeptide called adenosine triphosphate (ATP)-dependent proteolysis factor1 (APF1) was actually ubiquitin [18]. In 1977, Alfred Goldberg first demonstrated ATP-dependent protein degradation in reticulocytes [19]. Three years later, Aaron Ciechanover, Avram Hershko, and Irwin A. Rose suggested that the ATP-dependent covalent binding of ubiquitin was required for targeting proteins for degradation; they were awarded the Nobel Prize in Chemistry in 2004 for the discovery of ubiquitin-mediated protein degradation. After that, extensive research on ubiquitin was steadily conducted in the following decades [20,21,22].
2.1. Type and Function of Ubiquitin Chains
Ubiquitin is a small globular protein composed of 76 amino acid polypeptides (8.6 kDa) [23]. As its name suggests, ubiquitin is ubiquitously present and is highly conserved in a wide range of eukaryotes from yeast to humans [24]. Ubiquitination (also referred to as ubiquitylation) is one of the most prevalent reversible post-translational modifications. It involves the C-terminal glycine (Gly76) of ubiquitin forming an isopeptide linkage with the internal lysine, serine, threonine, and cysteine of a target protein, followed by the subsequent formation of a mono- or polyubiquitinated chain [25,26] (Figure 1). Therefore, the complexity of the ubiquitin chain with respect to type and length constitutes its ubiquitin code [27]. Ubiquitin N-terminal methionine (i.e., Met1/linear) and seven internal lysine (K) residues (i.e., K6, K11, K27, K29, K33, K48, and K63) are involved in the formation of eight different types of ubiquitin linkages [28]. Moreover, ubiquitin is involved in diverse biological processes including protein trafficking, protein degradation, DNA repair, NF-κB activation, endocytosis, and cell cycle progression, depending on the linkage position [29,30,31].
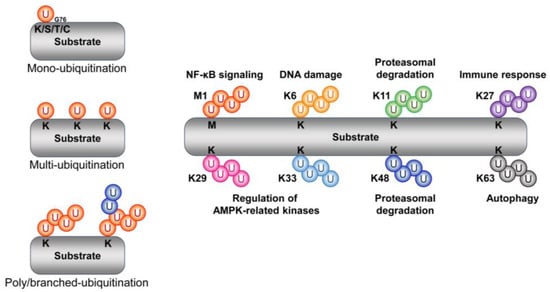
Figure 1.
Schematic representation of the type and function of ubiquitin chains. Ubiquitin forms an isopeptide bond with substrate side-chain lysine (K), serine (S), threonine (T), and cysteine (C) residues through its C-terminal glycine residue 76 (G76). These chains, through mono-, multi-, and polyubiquitination, participate in various cellular signaling pathways. The representative sites and functions of the polyubiquitination chain are as follows (right).
2.1.1. Methionine 1
The sequential binding of ubiquitin to methionine 1 (Met1) of ubiquitin causes the formation of linear polyubiquitination chains (i.e., Met1-linked or linear ubiquitin chains). At the same time, the linear ubiquitin chain assembly complex (LUBAC), a ubiquitin ligase complex consisting of SHARPIN, HOIP, and HOIL-1L, is involved in Met1/linear-linked ubiquitin chain formation. LUBACs are known to stimulate the NF-κB activation pathway via LUBAC-mediated linear polyubiquitin chains [32,33].
2.1.2. Lysine 6 (K6)
The tumor suppressor BRCA1 is implicated in both DNA repair and cell cycle regulation. In addition, BRCA1 has E3 ubiquitin ligase activity due to the presence of a novel gene (RING) finger domain located at its N-terminus. BRCA1 forms a heterodimeric complex with its N-terminal binding partner BARD1. The E3 ubiquitin ligase activity of BRCA1 is enhanced by BARD1, resulting in the auto-ubiquitination of BRCA1. The resulting ubiquitin chain is specifically formed at the K6 residue of ubiquitin. This K6-linked ubiquitination is believed to be associated with DNA double-strand break repair [34].
2.1.3. Lysine 11 (K11)
K11, together with K48, is involved in the proteasomal degradation of substrates. E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C), a key regulator of the cell cycle, is known to regulate mitosis and G1 phase. Substrates (e.g., cyclins) are bound to APC/C through the coactivator cell division cycle 20 (CDC20) or cadherin 1, and K11-linked ubiquitination and subsequent degradation is facilitated via E2 UbcH10 and Ube2S [35,36]. In another example, an E3 ubiquitin ligase neural precursor cell-expressed developmentally downregulated gene 4 (NEDD4) has been found to induce the K11-linked proteasomal degradation of Beclin1 [37].
2.1.4. Lysine 27 (K27)
Voltage-dependent anion channel 1 (VDAC1) is a target of the E3 ubiquitin ligase Parkin and is involved in the autophagy of damaged mitochondria (“mitophagy”), during which K27-linked ubiquitin chains are generated [38]. Upon cytokine stimulation, the E3 ubiquitin ligase Itch is activated and induces the K27-linked polyubiquitination of B-Raf. After activation, B-Raf then promotes tumorigenesis in melanoma cells by activating MEK/ERK signaling [39]. Another example of a protein with K27-linked ubiquitin chains is TIEG1, which is an essential transcription factor for TGF-b-induced regulatory T cell (Treg) development. The proinflammatory cytokine IL6 activates the tyrosine (Tyr) kinase Tyk2 to phosphorylate TIEG1 at Tyr179. Phosphorylated TIEG1 then undergoes K27-linked polyubiquitination via the E3 ubiquitin ligase Itch. K27-linked polyubiquitination in turn suppresses the nuclear translocation of TIEG1, suppresses Foxp3 expression and TGF-b-induced Treg development, and increases Th17 cell development, thereby reducing tumor growth [40]. In addition, K27 is involved in the innate immune response via various target proteins (e.g., cGAS and STING) [41,42].
2.1.5. Lysine 29 (K29)
AMP-activated protein kinase (AMPK)-related kinases, such as AMPK-related kinase NUAK family kinase1 (NUAK1) and microtubule-affinity-regulating kinase 4 (MARK4) are ubiquitinated by K29-linked ubiquitin chains. The polyubiquitination of NUAK1 and MARK4 was found to result in no alteration of its stability, but inhibited both its phosphorylation and activity [43]. Finally, atrophin-1-interacting protein 4 has been found to induce the polyubiquitination of deltex via K29-linked ubiquitin chains for lysosomal degradation [44].
2.1.6. Lysine 33 (K33)
The E3 ubiquitin ligase Parkin is phosphorylated and activated by AMPK. Activated Parkin then inhibits the pronecroptotic factor RIPK1−RIPK3 interaction by promoting the polyubiquitination of RIPK3 via K33-linked ubiquitin chains (note that the protein level of RIPK3 is not changed by Parkin). Through this pathway, the AMPK–Parkin axis inhibits necroptosis [45].
2.1.7. Lysine 48 (K48)
K48-linked ubiquitin chains are the most intensively studied ubiquitin modification related to proteasome-mediated protein degradation. Most proteins, including p53, HIF1α, HIPK2, and ASK1, are ubiquitinated via the ubiquitin K48, and K48-linked ubiquitin chains act as a degradation signal to induce the proteasomal degradation of proteins [46,47,48,49,50]. Table 1 summarizes the proteins degraded via the 20S proteasome and those degraded via the ubiquitin-dependent or -independent 26S proteasome (Table 1).

Table 1.
A list of substrate proteins degraded via 26S or 20S proteasome.
2.1.8. Lysine 63 (K63)
The E3 ubiquitin ligase TRAF6 promotes the ubiquitination of hexokinase 2 (HK2), a glycolytic enzyme that converts glucose to glucose-6-phosphate, at the K41 residue using K63-linked ubiquitin chains. Under high autophagic flux, ubiquitinated HK2 is recognized by the autophagy receptor SQSTM1, also known as p62, leading to the formation of autophagosomes and the subsequent selective autophagic degradation, thereby suppressing glycolysis. However, under conditions of low autophagic flux, the formation of autophagosomes is impaired, resulting in the absence of HK2 degradation. These findings demonstrate that autophagy modulates HK2-dependent glycolysis [82].
3. The Proteasome and Beyond
The proteasome participates in direct protein degradation as a high-molecular-weight protease complex. The two major species of proteasome are the 26S proteasome (2.5 MDa) and the 20S proteasome (700 kDa) [83]. The 26S proteasome consists of the 20S proteasome (also called 20S core particle, CP) and one or two 19S regulatory particles (RPs, also called PA700). The 20S proteasome is composed of two of outer heptameric rings, each containing seven α-subunits (α1–α7), and two inner heptameric rings, each containing seven β-subunits (β1–β7). These are then assembled as a β-ring barrel-shaped structure (α7-β7-β7-α7). The α-rings serve as a “gate” for substrate entry, while three subunits (i.e., β1, β2, and β5) of the β-rings hold distinct peptidase activities and directly degrade proteins [84]. The 26S proteasome engages in ATP, ubiquitin-dependent protein degradation, whereas the 20S proteasome engages in ATP, ubiquitin-independent protein degradation [85]. In addition, the type and the number of regulatory particles that interact with the 20S proteasome accounts for the diversity of proteasome complexes [86,87].
3.1. Assembly of the 20S Proteasome
For the 20S proteasome assembly, a proteasome-assembling chaperone 1 (PAC1)-PAC2 hetero-dimer and a PAC3-PAC4 hetero-dimer first create α-ring intermediates from the α4, α5, α6, and α7 structures within α-subunits. PAC1-PAC2 and PAC3-PAC4 prevent the off-pathway dimerization of α-subunits and the incorrect incorporation of the other α-ring, thereby facilitating stable α-ring formation. Once the α1, α2, and α3 subunits are assembled into an α-ring, the β-subunit starts to be incorporated. The β2 subunit then associates with the α-ring, which is promoted by ubiquitin-mediated proteolysis 1 (UMP1), and the subsequent binding of β3 dissociates the PAC3-PAC4 complex. Next, the remaining β-subunits (but not β7) are incorporated to form the 15S intermediate complex. Subsequent β7 incorporation into the 15S intermediate creates a complex, termed the ‘half-proteasome’, that triggers its own dimerization. Moreover, UMP1 is required for proper maturation of the 20S proteasome since it inhibits the premature dimerization of half-proteasomes. Finally, the mature 20S proteasome degrades the assembly factors such as UMP1 and the PAC1-PAC2 hetero-dimer (in yeast, PAC1-PAC2 is recycled for future assembly into new 20S proteasomes) (Figure 2) [88,89,90].
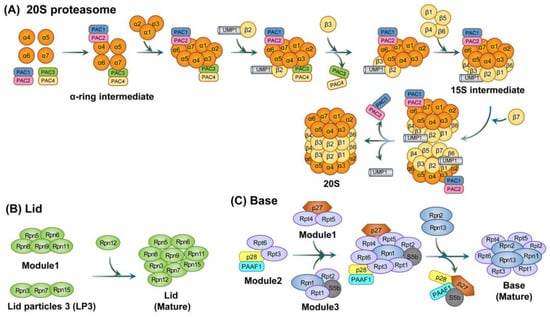
Figure 2.
The assembly of the 20S proteasome and two parts of the 19S regulatory particle, the lid and base subcomplexes. (A) Schematic diagram of the 20S proteasome. For synthesis of the 20S proteasome, the α-ring is formed first, followed by the formation of the β-subunit, thereby forming the β ring. Of the β-subunits, the catalytically active subunits that directly degrade are β1, β2, and β5. (B) Schematic diagram of the lid subcomplex. Rpn12 is an important subunit that binds to LP2 (i.e., module1 + LP3) to form the lid subcomplex. This is involved in base binding via Rpn10. (C) Schematic diagram of the base subcomplex. The basic subcomplex directly linked to 20S proteasome is complexed by various chaperone proteins. The lid subcomplex and base subcomplex are collectively referred to as 19S RPs.
3.2. Assembly of the 19S RP
The 19S RP can be subdivided into the lid subcomplex (~370 kDa) and the base subcomplex [91,92]. The lid subcomplex consists of nine regulatory particle non-ATPase (Rpn) subunits (i.e., Rpn3, Rpn5, Rpn6, Rpn7, Rpn8, Rpn9, Rpn11, Rpn12, and Rpn15). The base subcomplex consists of six regulatory particle ATPase (Rpt) subunits (i.e., Rpt1, Rpt2, Rpt3, Rpt4, Rpt5, and Rpt6) as well as three non-ATPase subunits (i.e., Rpn1, Rpn2, and Rpn13) [93]. Rpn10, which was thought to be a part of the base subcomplex, is now considered to be a factor that stabilizes the lid–base interaction and recognizes the ubiquitin chain [94,95].
The lid subcomplex assembly begins as two subcomplexes, module 1 (consisting of Rpn5, Rpn6, Rpn8, Rpn9, and Rpn11) and lid particle 3 (LP3; consisting of Rpn3, Rpn7, and Rpn15), which join to form lid intermediate LP2. Rpn12 is then incorporated into the LP2 intermediate to complete the lid subcomplex [92,96].
The base subcomplex assembly is preceded by the formation of intermediate complexes. These include a heterohexameric ATPase ring that contains three modules, i.e., module1 (p27, Rpt4, and Rpt5), module2 (p28, Rpt3, Rpt6, and proteasomal ATPase-associated factor1 (PAAF1)), and module3 (S5b, Rpt1, Rpt2, and Rpn1). This formation is regulated by four RP assembling chaperones (RACs). These include p27, p28, S5b, and PAAF1 [97]. Subsequent incorporation of the Rpn2-Rpn13 hetero-dimer causes the dissociation of RACs. Moreover, the binding of the stabilizing factor Rpn10 to the lid and base subcomplexes completes the proper assembly of the 19S RP (Figure 3). For regulatory particles, in addition to 19S RP, various regulatory particles such as 11S RP (also called proteasome activator 28, PA28, or REG), PA200 (also known as Blm10 in yeast), ATPases associated with diver cellular activities (AAA+) ATPase forming ring-shaped complexes (ARC), a homologue of 19S RP in eubacteria, and proteasome-activated nucleotidase (PAN), a homologue of 19S RP in archaea, have been identified.
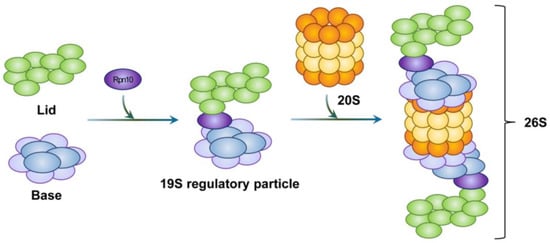
Figure 3.
Schematic representation of proteasome structure. Rpn10 acts as a hinge between the 20S proteasome and the 19S regulatory particle. Rpn10 is also involved in the formation of the 26S proteasome. As a result, one or two 19S regulatory particles associate with the 20S proteasome to form the 26S proteasome.
3.3. Assembly of the 26S Proteasome
The extracellular matrix 29 (Ecm29) proteins stabilize the 26S proteasome by tethering the 20S proteasome and the 19S RP [88,98]. Furthermore, under oxidative stress conditions, ECM29 accelerates the disassembly of the 26S proteasome [99]. Furthermore, heat shock protein 90 (Hsp90) also contributes to the association of the 26S proteasome, both in vivo and in vitro, in an ATP-dependent manner [100].
3.4. Steps Involved in Substrate Degradation via the 26S Proteasome
3.4.1. Ubiquitination
Aberrant proteins, including damaged proteins, misfolded proteins, over-expressed protein substrates, and short-lived proteins, require ATP-dependent proteolysis to be degraded by the 26S proteasome. (For some proteins, which are degraded by the 20S proteasome via an ubiquitin-independent pathway, a sequential cascade for protein degradation called ubiquitination is not required) [101] (Figure 4). Of the various types of ubiquitin chain mentioned above, the K48-linked polyubiquitin chain, which is involved in proteasomal degradation, is the most abundant and widely studied [102]. The process of ubiquitination requires three different enzymes: E1 ubiquitin-activating enzymes (of which there are 2), E2 ubiquitin conjugases (~40 in total), and E3 ubiquitin ligases (over 600 in total) [103]. These are involved in the sequential ubiquitination process as follows: Step 1: ubiquitin is activated when a thioester bond forms between the C-terminal glycine (i.e., Gly76) of ubiquitin and the active site cysteine of the E1 ubiquitin-activating enzyme in an ATP-dependent manner. Step 2: activated ubiquitin is transferred from the E1 ubiquitin-activating enzyme to E2 ubiquitin conjugase, and a thioester bond forms between the C-terminal glycine of ubiquitin and the active site cysteine of the E2 ubiquitin conjugase. Step 3: when the ubiquitin–E2 complex and substrate bind to E3 ubiquitin ligase, E3 ubiquitin ligase transfers ubiquitin to the substrate. Consequently, a covalent peptide bond is formed between the C-terminal Gly of ubiquitin and a lysine residue in the substrate protein. Step 4: in some cases, an E4 ubiquitin ligase, a polyubiquitin chain elongation factor, is involved in polyubiquitin chain assembly [104,105,106].
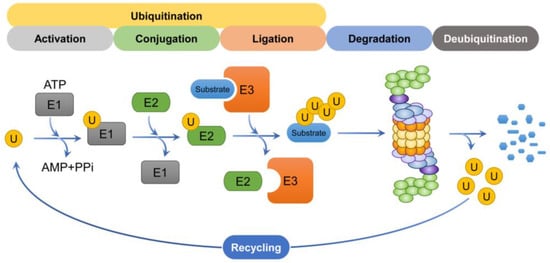
Figure 4.
Schematic representation of the degradation process. The ubiquitin mediates the K48-linked ubiquitination of substrates through a cascade involving E1, E2, and E3 enzymes. Ubiquitin chains are recognized by the 19S regulatory particle and subsequently degraded by the 26S proteasome. The ubiquitin molecules that participated in the degradation process are then recycled to participate in substrate degradation again.
The substrate specificity of E3 ubiquitin ligase is a significant feature involved in the ubiquitination process. E3 ubiquitin ligases are subdivided into four groups, including: (1) the RING-type E3 ubiquitin ligases; (2) the homology to E6-associated protein carboxyl terminus (HECT)-type E3 ubiquitin ligases; (3) the RING-between-RING (RBR) E3 ubiquitin ligases, also referred to as RING-HECT hybrids; and (4) the U-box-type E3 ubiquitin ligases [107,108]. RING-type E3 ubiquitin ligases have a RING domain and a substrate-binding domain (SBD). When ubiquitin-E2 ubiquitin conjugase binds to the RING domain of E3 ubiquitin ligases, the ubiquitin on the E2 ubiquitin conjugase is directly transferred to the substrate bound to the SBD, whereas ubiquitination via HECT type E3 ubiquitin ligases requires an additional step in which the ubiquitin is first bound to a catalytic cysteine (Cys) residue in the HECT domain within a HECT type E3 ubiquitin ligase. Here, the ubiquitin is subsequently transferred to the substrate. RBR-type E3 ubiquitin ligases have two RING domains. Once ubiquitin-E2 ubiquitin conjugase binds the RING1 domain, the ubiquitin on the E2 ubiquitin conjugase on the RING1 domain is transferred to a catalytic cysteine of the RING2 domain and is then transferred again to the substrate. U-box type E3 ubiquitin ligases were first identified in the yeast Saccharomyces cerevisiae (S. cerevisiae) [109,110]. These ligases have a conserved U-box domain that is similar to the RING domain. In these ligases, E2 ubiquitin conjugase interacts with the U-box domain and subsequently promotes the ubiquitination of the substrate. E4 ubiquitin ligase E4B, a mammalian homologue of yeast UFD2, is a U-box-containing protein that has been found to promote the elongation of polyubiquitin chains like E3 ubiquitin ligase [111,112].
3.4.2. Recognition of Ubiquitin Chains
The ubiquitin chain is recognized by three ubiquitin receptors, Rpn1, Rpn10, and Rpn13, each of which is a component of a proteasome subunit [113,114]. Rpn10 and Rpn13 are the major ubiquitin-binding subunits. In both cases, ubiquitin binds to Rpn10 and Rpn13 via a specific ubiquitin-interacting motif also known as a LALAL motif domain and a pleckstrin-like receptor for ubiquitin domain, respectively [94,115]. In addition, Rpn13 is the subunit to which the deubiquitinating enzyme UCHL5 binds [116]. Moreover, Rpn1 binds to ubiquitin through its proteasome/cyclosome repeats [117]. Rpn1 is a ubiquitin receptor and a subunit bound by the deubiquitinating enzyme USP14 [118,119].
3.4.3. Deubiquitination
Deubiquitinating enzymes (DUBs) are proteases that regulate the ubiquitin–proteasome pathway. They do so by processing ubiquitin precursors into mature ubiquitin, upon which the ubiquitin molecules are cleaved from ubiquitin-conjugated substrates. This process is therefore termed deubiquitination, since DUBs prevent protein degradation by reversing the ubiquitin process. While ubiquitinated substrates are degraded by proteasomes, DUBs prevent the degradation of substrates by separating them from substrates to recycle ubiquitin monomers for the subsequent degradation of other substrates. Human DUBs are classified into two classes: cysteine proteases and metalloproteases [120]. Cysteine proteases make up most DUB members in eukaryotic cells and can be divided into seven families: (i) ubiquitin-specific proteases, (ii) ovarian tumor proteases, (iii) monocyte chemotactic protein-induced proteases, (iv) ubiquitin C-terminal hydrolases proteases, (v) Machado–Joseph domain proteases, (vi) motif interacting with Ub-containing novel DUB family, and (vii) zinc finger (ZnF) with UFM1-specific peptidase domain protein. Interestingly, the metalloproteases have an additional family: (viii) Jab1/Pab1/MPN domain-containing metalloenzymes [121,122].
The catalytic triad of cysteine proteases is mainly composed of conserved cysteine (Cys, C), aspartate (Asp, D), and histidine (His, H) residues. These proteases hydrolyze the isopeptide bond between ubiquitin and a substrate, or between ubiquitin moieties [123]. Two DUBs (i.e., USP14 and UCHL5) of the three proteasome-associated DUBs belong to the cysteine protease family and therefore cleave the ubiquitin–ubiquitin interaction once the proteasome binds to the substrate, thereby preventing degradation by the proteasome. In contrast, Rpn11, a subunit consisting of the lid subcomplex of 26S proteasome, belongs to the metalloprotease family, and is activated by the coordination between four conserved residues (i.e., His, Asp, Glutamate, and serine) and zinc ions to catalyze isopeptide hydrolysis [120,124,125]. Once the ubiquitin chain of the substrate is recognized by Rpn1, Rpn10, and Rpn13, Rpn11—unlike USP14 and UCHL5—releases whole ubiquitin from a partially unfolded substrate before it is degraded by the proteasome [126,127]. Finally, the substrate is unfolded and translocated into the 20S proteasome.
3.4.4. Gate Opening and Translocation
The AAA+ enzymes are a superfamily of proteins and use the energy produced when ATP is hydrolyzed to ADP to drive various cellular activities [128]. Ring-shaped heterohexameric AAA+ complexes, which consist of the six ATPases (Rpt1, Rpt2, Rpt3, Rpt4, Rpt5, and Rpt6), mechanistically unfold and translocate substrates into the 20S proteasome. Rpt2, Rpt3, and Rpt5 also contain a conserved C-terminal hydrophobic-tyrosine-X (HbYX) motif, which interacts with the lysine-pocket (K-pocket) of the α-ring of the 20S proteasome. This triggers the gate opening of the α-ring pore of the 20S proteasome to convert it from a closed to an open state after ATP binding. Six ATPases change their confirmation to adopt a spiral staircase arrangement in an ATP-dependent manner, thereby permitting substrate unfolding and translocation into the 20S proteasome [129,130].
3.4.5. Proteolysis
Unfolded substrates translocated into the proteolytic chamber via the α-ring are then directly degraded by β-subunits. Of the seven β-subunits, the β3, β4, β6, and β7 subunits participate in the assembly of the structural complex of the 20S proteasome, whereas β1, β2, and β5 display caspase-like (or peptidylglutamyl-peptide-hydrolyzing), trypsin-like, and chymotrypsin-like activities, respectively [131]. β1 hydrolyzes the peptide bond on the carboxyl side of acidic or hydrophobic amino acids. Moreover, β2 cleaves the bond on the carboxyl side of basic or hydrophobic amino acids, and β5 cleaves the bond on the carboxyl side of hydrophobic amino acids. The cylindrical particle of 20S proteasomes contains two β-rings and is responsible for six substrate-degrading activities. In addition, 20S proteasomes contain highly conserved threonine (Thr) residues on the N-termini of all active β-subunits and is therefore called a Thr protease [132]. Moreover, given the proteolytic activities within the cylindrical core particle, the isopeptide bonds of the substrate are hydrolyzed, resulting in the generation of small peptides. Their average length ranges between 3 and 23 amino acids [84,85,133].
3.5. Substrate Degradation via the 20S Proteasome
In mammalian cells, approximately 20% of the proteasomes are 26S proteasomes, while the rest consist of 20S proteasomes, immunoproteasomes, thymoproteasomes, and hybrid proteasomes [134]. Despite this distribution, around 80% of the total protein is degraded by the 26S proteasome [12]. As mentioned before, the 26S proteasome recognizes substrates linked to ubiquitin chains, which are typically associated with natively folded proteins, for degradation. In contrast, the 20S proteasome is known for its role in the default degradation of proteins that exhibit certain characteristics. These include proteins containing intrinsically disordered regions (IDRs) or being intrinsically disordered proteins (IDPs) [135]. Some proteins exist in a partially or fully unfolded state without forming a defined structure. Proteins with such disordered regions, such as p21, p53, c-fos, α-synuclein, tau, and others, are degraded by the ubiquitin-independent 20S proteasome [136]. Additionally, the 20S proteasome is involved in the degradation of oxidatively damaged proteins [137]. Oxidative stress induces the dissociation of the 26S proteasome into the 20S proteasome and the 19S RP, leading to the accumulation of structurally unstable misfolded proteins. Misfolded proteins, exposing hydrophobic regions, tend to aggregate. The 20S proteasome recognizes these hydrophobic regions and facilitates the degradation of oxidized proteins [138]. However, it has recently been reported that ubiquitin-tagged proteins can also be degraded through the 20S proteasome [139]. In addition, proteins such as p53 and p21 have been shown to undergo degradation by both the 26S proteasome and the 20S proteasome [140]. This indicates that the degradation mechanisms mediated by the 26S proteasome and the 20S proteasome are not mutually exclusive but rather complementary to each other [136].
3.6. Mixed Proteasome
The mixed proteasomes, also known as intermediate proteasomes, contain a combination of immune and constitutive proteolytic subunits: β1, β2, and β5i (also known as LMP7, with chymotrypsin-like activity); β1i (also known as LMP2, with chymotrypsin-like or branched-chain-amino-acid-preferring activity), β2, and β5i. These proteasome subtypes expand the repertoire of antigens presented to CD8+ T cells [141]. In addition, proteasome subtype β1i, β2i (also known as MECL-1, with trypsin-like activity), and β5, without β5i, can be formed [142]. The high expression of proteasomes containing subunit β1i, but not β5i, is associated with the development of immunological tolerance [143]. β1i is required for the adaptation of rat ventricular cardiomyocytes to reach pressure overload [144]. Thus, the functions of proteasomes containing immune subunits in combination with constitutive ones are wider than the formation of antigenic epitopes.
3.7. Immunoproteasome
The immunoproteasome is an alternative form of the constitutive 20S proteasome that was discovered in 1994 [145]. It has distinct catalytic subunits known as β1i, β2i, and β5i. In contrast to the regular proteasome, the immunoproteasome contains these specialized subunits, which confer unique enzymatic activities. The synthesis of these subunits, β1i, β2i, and β5i, is induced by pro-inflammatory cytokines such as tumor necrosis factor-alpha (TNF-α), interferon-gamma (IFN-γ), and oxidative stress. Consequently, the immunoproteasome is generated ahead of the constitutive 20S proteasome [86,146]. Immunoproteasomes are known to be highly expressed in immune cells, such as antigen-presenting cells (APCs). They play a crucial role in generating antigenic peptides by breaking down intracellular antigens. These antigenic peptides are then transported into the endoplasmic reticulum (ER), where they form complexes with newly synthesized major histocompatibility complex (MHC) class I molecules. Subsequently, these complexes undergo processing in the Golgi apparatus and are presented on the cell surface. Once presented on the cell surface, the antigenic peptide–MHC complex can be recognized by the T cell receptors (TCRs) of CD8+ T cells, leading to the triggering of immune responses [147,148].
3.8. Thymoproteasome
The thymoproteasome, discovered in 2007, is closely related to the positive selection of T cells [149]. It is exclusively expressed in cortical thymic epithelial cells (cTECs) within the cortex and shares structural similarities with the immunoproteasome, containing unique subunits called β1i, β2i, and β5t [150]. The maturation of T cells occurs as immature T cells, formed in the bone marrow, which migrate to the thymus. The thymus can be divided into the cortex and medulla, and it is in the cortex where the initial development of T cells takes place through positive or negative selection, ultimately leading to the formation of mature T cells. Within cTECs, self-antigens are degraded by the thymoproteasome into self-peptides. These self-peptides, along with MHC class I molecules, are formed in the ER and presented on the surface of cTECs after passing through the Golgi apparatus. The T TCRs of T cells recognize these self-peptides, determining the outcome of positive selection. T cells with weak binding to self-peptides undergo positive selection, differentiating into mature CD8+ T cells that participate in adaptive immune responses. On the other hand, T cells with strong binding to self-peptides undergo negative selection, leading to apoptosis. This process prevents the development of T cells that could potentially cause autoimmune diseases [146,151].
4. Proteasome Regulatory Proteins
4.1. Proteasome-Activating Proteins
4.1.1. 11S Regulatory Particle
The 11S RP was first identified in bovine and human red blood cells in 1992 [152,153]. Due to its molecular weight of approximately 28 kDa, it is also known as proteasome activator 28 (PA28) or REG. In mammals, three homologous subunits of 11S RP (PA28α, PA28β, and PA28γ) have been identified. Among them, PA28α (28.7 kDa) and PA28β (27.1 kDa) subunits share about 47% sequence identity. PA28α and PA28β form heteroheptameric ring structures, predominantly in the form of α4β3 and α3β4 complexes [154,155]. They are primarily localized in the cytosol. On the other hand, PA28γ forms homoheptamers and is mainly localized in the nucleus [156]. 11S RP increases all three activities of the 20S proteasome, but there is no difference in the degradation rate compared to when only the 20S proteasome is present. Additionally, 11S RP facilitates the degradation of small peptides, but it is unable to degrade large proteins and ubiquitin-conjugated proteins [157,158]. Finally, 11S RP is induced by IFN-γ and binds to 20S proteasome in an ATP-independent manner, resulting in the formation of either 11S-20S proteasome-11S or 11S-20S proteasome-19S hybrid proteasomes (Table 2) [159].

Table 2.
A list of proteasome regulatory proteins.
4.1.2. PA200
PA200, another proteasome activator, is primarily localized in the nucleus and has a molecular weight of 200 kDa [160]. It binds to the 20S proteasome alone or in conjunction with the 19S RP, forming a hybrid proteasome complex (PA200-20S proteasome-19S RP), thereby enhancing the peptidase activity of the 20S proteasome [161]. PA200 stimulates the hydrolysis of small peptides and unstructured proteins, such as tau, in an ATP-dependent manner [162]. Additionally, it is involved in DNA repair and the degradation of acetylated histones [160,163].
4.1.3. Nuclear Respiratory Factor 1
Nuclear respiratory factor 1 (NRF1), also known as the endoplasmic reticulum membrane protein, is retrotranslocated to the cytosol by p97 under normal conditions and is degraded by the 26S proteasome via an ER-associated degradation pathway. However, when proteasome activity is impaired or insufficient, NRF1 is cleaved, releasing a soluble 100 kDa fragment (i.e., p110, an active form of NRF1) via aspartic protease DNA Damage Inducible 1 Homolog 2 (DDI2) to the cytosol. p110 is then translocated to the nucleus, where it dimerizes with the cofactor small musculoaponeurotic fibrosarcoma to induce proteasome gene expression, thereby recovering proteasome activity [164,165].
4.1.4. Zinc Finger AN1-Type Containing 5
Zinc finger AN1-type containing 5 (ZFAND5) is a protein that has been found to increase muscle atrophy. It directly binds to the 20S proteasome and stimulates three peptidase activities of the 26S proteasome. In particular, the AN1 domain near the C-terminus of ZFAND5 is essential for the stimulation of peptidase activity. ZFAND5 also enhances the hydrolysis of ubiquitinated dihydrofolate reductase (DHFR) by increasing ATPase activity. As a result, ZFAND5 promotes total protein degradation via the ATP- and ubiquitin-dependent 26S proteasome pathway [166].
4.1.5. Tankyrase
The 20S proteasome displays low activity when bound to a proteasome inhibitor of 31 kDa (PI31). In addition, ADP-ribotransferase tankyrase (TNKS) mediates ADP-ribosylation of PI31, which then combines with the 19S RP assembly chaperones p27 and S5b (which was previously bound to 19S RP), and the dissociated 19S RP then binds to the 20S proteasome, thereby increasing 26S proteasomal activity [167].
4.2. Proteasome-Inhibiting Proteins
4.2.1. DJ-1
DJ-1, also known as Parkinson’s disease protein 7 (PARK7), is a multifunctional protein associated with Parkinson’s disease, cancer, oxidative stress response, and mitophagy. DJ-1 binds to the 20S proteasome and inhibits its activity. Through this mechanism, α-synuclein and p53, substrates known to be degraded through the 20S proteasome, were instead protected [168].
4.2.2. NAD(P)H:Quinone-Oxidoreductase 1
NQO1 is a flavin adenine dinucleotide (FAD)-dependent flavoprotein that catalyzes the reduction of quinone to hydroquinone. NQO1, a superoxide reductase, possesses an innate antioxidant activity and can directly scavenge superoxide. Furthermore, NQO1 inhibits the degradation of p53, ornithine decarboxylase (ODC), and α-synuclein by binding to the 20Sproteasome, where it acts as a gate keeper [169,170]. Apo-NQO1, in its FAD-free form, is unstable due to a structural change caused by a partially unfolded conformation; this makes Apo-NQO1 susceptible to degradation by the 20S proteasome. Therefore, NQO1 and the 20S proteasome mutually regulate their activities via a double negative feedback loop [171].
4.2.3. PI31
PI31 is a proline-rich protein whose carboxyl-terminal proline-rich domain plays a role in inhibiting the activity of the 20S proteasome. PI31 competes with the proteasome regulatory proteins 19S RP and 11S RP when binding to the 20S proteasome, thereby blocking the activation of the 20S proteasome [172,173].
4.2.4. c-Abl
The non-receptor Tyr kinase c-Abl binds to the PSMA7 (i.e., at its α4 subunit) of the 20S proteasome and phosphorylates Tyr residue (Y106). PSMA7 phosphorylation at Y106 inhibits the ubiquitin-dependent proteasomal degradation of PSMA7 and increases its expression. This in turn increases proteasome abundance. However, proteasome activity remains suppressed, which contrasts with the cellular proteasome levels [174,175].
4.2.5. Bassoon
The presynaptic cytomatrix protein BSN is localized to the active zone of the presynaptic terminal where it interacts with the PSMB4 (i.e., at its β7 subunit) of the 20S proteasome. BSN suppresses proteasome assembly by binding to half-proteasomes at two independent regions. The reduced activity of the proteasome then leads to the inhibition of ubiquitination-dependent and independent proteolysis. Consequently, BSN reduces the degradation of presynaptic scaffolding proteins such as Rab3-interacting molecules (RIMs) and mammalian homolog of Caenorhabditis elegans unc-13 (Munc13), thus causing the accumulation of misfolded proteins [176].
5. Proteasome Activity in Diseases
5.1. Neurodegenerative Disease
A change of proteasomal activity was one of age-related dysfunction [177]. Especially, accumulation of misfolded proteins via alternative proteasome activity was induced in several neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and Huntington’s disease [178]. Proteasome activity was decreased in these diseases and leaded to the aggregation of ꞵ-amyloid, tau tangles, lewy bodies, and poly-glutamine inclusions [179]. Therefore, neurodegenerative disease observed neuronal loss and dysfunction [178].
5.2. Muscle Atrophy and Cachexia
Enhanced proteasome activity changes in the life time of a protein [180]. Muscle atrophy was observed to decrease protein synthesis; however, it was observed to increase protein degradation [178]. Cancer cachexia is also a muscle-loss-related disease [181]. Increased proteasome activity was induced in muscle protein wasting and inflammation was induced in muscle atrophy and cancer cachexia [182].
5.3. Chemoresistant Cells
A cisplatin-resistant neuroblastoma cell line has been found to show higher proteasome activity than a parental cell line [183]. Cisplatin-chemoresistant neuroblastoma cells have been found to show high expression levels of SHFM1 (i.e., 26S proteasome complex subunit SEM1) and PSMD14 (i.e., 26S proteasome non-ATPase regulatory subunit 14) using transcriptomic profiling [184]. In addition, the irreversible proteasome inhibitor TIR-199 effectively reduced the cell viability of bortezomib-related chemoresistance. This was found to suppress tumor growth in blood cancers such as multiple myeloma and mantle cell lymphoma [185].
5.4. Cancer Stem Cells
Cancer stem cells (CSCs) are characteristic of chemoresistance and tumor recurrence [186,187]. Many studies have shown that many cancer cells show high proteasomal activity. However, other studies of CSCs have reported that proteasomal activity is low [188,189]. Differences in proteasome activity have also been found among various cancers, including lung, prostate, and pancreas [189,190,191]. Moreover, the existence of CSCs has caused some solid tumors to show low proteasome activity [188]. This is notable, since low proteasome activity cells (LPACs) show increased resistance to chemotherapy and radiotherapy [187,188,192].
6. Therapeutic Modalities Targeting Proteasomes
In recent years, therapeutic modalities using TPD via the proteasome have been proposed (Table 3). These include novel strategies such as PROTAC, SNIPERs, molecular glues, and hydrophobic taggings (HyTs), and have been designed for the selective targeting of proteins associated with cancer, neurodegenerative disease, autoimmune disease, and metabolic disorders (Figure 5) [193,194].

Table 3.
Comparison of different various targeted degradation modalities.
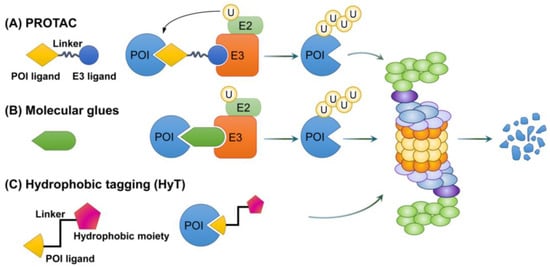
Figure 5.
Mechanisms of action of PROTAC, molecular glues, and hydrophobic tagging. A variety of degraders are being developed for TPD. (A) PROTAC involves a protein of interest (POI) ligand and an E3 ligand that are linked by a linker. When POI and E3 ubiquitin ligase bind to PROTAC, the ubiquitin of E2 is sequentially attached to POI, thereby inducing ubiquitination. Ubiquitinated POIs are eventually degraded by the proteasome. (B) Molecular glues use a mechanism similar to PROTAC. They induce molecular proximity between a POI and E3 ubiquitin ligase, resulting in the sequential attachment of ubiquitin to the POI, which triggers its ubiquitination and degradation. (C) Hydrophobic tagging combines POIs to mimic hydrophobic and partially disordered forms of POIs. Recognition of chaperones at these sites then leads to chaperone-mediated proteasomal degradation.
Therefore, a TPD-using proteasome may offer the potential to target undruggable proteins and overcome disadvantages associated with conventional small molecule inhibitor-, antibody-, or gene-based therapies (i.e., those involving small molecule inhibitors, monoclonal antibodies, or RNAi), which include: broad or smooth active sites of target proteins, low tissue penetration, high molecular weight, and low oral bioavailability (Table 4). These novel therapeutic modalities are currently under active research and are being subjected to clinical trials [15,195,196].

Table 4.
Comparison of different various therapeutic modalities.
6.1. Proteolysis-Targeting Chimera
Two decades ago, various pharmaceutical companies developed a modality called PROTAC technology. Currently, it is being developed as a protein degradation tool targeting various diseases. PROTAC involves a hetero-bifunctional small molecule in which a ligand of a POI and a ligand for E3 ubiquitin ligase-binding are typically linked by a linker. When POI and E3 ubiquitin ligase bind to each ligand, a ternary complex is formed. The POI is then ubiquitinated by the E3 ubiquitin ligase, thereby causing the POI to be directly degraded by the proteasome [15].
The first version of PROTAC was developed in 2001. In this system, which used a peptide-based PROTAC, the POI ligand was linked to the angiogenesis inhibitor ovalicin, which forms a covalent bond with methionine aminopeptidase2 (METAP2). As the E3 ligase-binding ligand, the IκBα peptide (DRHDSGLDSM) was used to target the E3 ubiquitin ligase, β-transducin repeat-containing E3 ubiquitin–protein ligase (β-TRCP). However, this peptide-based PROTAC showed problems related to low cell permeability, lipophilicity, and stability [197]. To solve these problems, small molecule-based, nucleotide-based, antibody-based, nanoparticle-based, and peptide-based PROTACs are currently being developed [198,199,200].
PROTAC therapies require the consideration of several points prior to development. Currently, most PROTACs bind to the E3 ubiquitin ligase component cereblon (CRBN) using immunomodulatory drugs (IMiDs) such as thalidomide, pomalidomide (Pom), and lenalidomide, which are used as E3 ubiquitin ligase binders. When CRBN binds to IMiDs, it induces POI ubiquitination by forming a complex (CRL4CRBN) with Cullin4-RING E3 ubiquitin ligase (CRL4), an E3 ubiquitin ligase that uses CRBN as a substrate receptor [201,202]. However, since some cells or tissues show diverse CRBN expression, new PROTAC methods are being developed that replace CRBN with other E3 ubiquitin ligases such as tumor suppressor von Hippel–Lindau, mouse double minute 2 (MDM2), and inhibitors of apoptosis (IAP) [15,203,204,205]. Moreover, when considering the ligand for the E3 ubiquitin ligase, it must be capable of high target binding specificity. Otherwise, PROTAC can generate off-target effects that affect proteins other than the POI. The POI ligand must also show a low binding affinity for the POI, since degradation via the proteasome does not occur unless the ubiquitinated POI is cleaved from PROTAC [206,207]. Furthermore, the appropriate concentration of PROTAC is also very important. PROTAC at an appropriate concentration induces the degradation of POI by forming a ternary complex structure such as POI-PROTAC-E3 ligase. However, a high concentration of PROTAC can form a binary complex as POI-PROTAC or E3 ligase-PROTAC. This phenomenon is called the “hook effect”, and results in a reduction in PROTAC activity [200]. In addition, the size of the PROTAC must also be considered. Finally, it is important to develop new PROTAC strategies, including the in-cell click-formed proteolysis-targeting chimera, which increases cell permeability by reducing molecular weight [208].
In 2019, Arvinas, Inc. conducted a Phase I trial using a PROTAC (ARV-110, NCT03888612) targeting an androgen receptor and a PROTAC (ARV-471, NCT04072952) targeting an estrogen receptor. They effectively inhibited metastatic castration-resistant prostate cancer and breast cancer, respectively, and are currently in Phase II trials. This result confirmed that PROTAC has potential as a therapeutic target. In addition, other multinational companies (e.g., Accutar Biotech, Kymera, Nurix Therapeutics, and C4 Therapeutics Inc., among others) are conducting or are planning clinical trials for various diseases using various PROTAC protocols [15].
In addition, specific non-genetic IAP-based protein eraser is another degrader molecule that works in a manner similar to that of PROTACs. This molecule features a link between a POI ligand and an antagonist (i.e., an LCL161 derivative) capable of recruiting the IAP in the presence of E3 ubiquitin ligases [209,210].
6.2. Molecular Glues
Molecular glues stabilize protein–protein interactions between proteins in homo- or hetero-dimer forms [211]. The most common molecular glues are small molecule degraders that mediate proximity-induced TPD. The simultaneous binding of E3 ubiquitin ligase and POI to molecular glues induces dimerization between E3 ubiquitin ligase and the POI. As a result, a ternary complex comprising molecular glue, E3 ubiquitin ligase, and the POI is formed, thereby leading to subsequent ubiquitination and proteasome-mediated degradation of the POI. The first molecular glues were serendipitously discovered. However, new molecular glues are currently under development via structure-based design, scalable chemical profiling, or microarray-based high-throughput screening. The most widely used molecular glues are the CRBN ligand thalidomide and its analogs, such as lenalidomide and Pom, and DCAF15 ligand sulfonamides. Both molecular glues and PROTAC induce the proteasomal degradation of a target protein; however, unlike PROTAC, molecular glues do not require a linker and binding pocket and have a lower molecular weight (i.e., ranging from 300 to 600 Da) than PROTAC (i.e., ranging from 700 to 1000 Da) [212].
6.3. Hydrophobic Tagging
Intracellular unfolded or misfolded proteins expose structurally modified hydrophobic amino acid residues or patches on their surface. These hydrophobic regions of partially denatured proteins can be recognized by the highly conserved and ubiquitous heat shock protein 70 (HSP70) chaperone. Subsequently, the co-chaperone E3 ligase C-terminus of Hsp70 interacting protein (CHIP) is recruited to induce the ubiquitination of the misfolded protein. As a result, misfolded proteins with exposed hydrophobic regions undergo quality control and degradation via the proteasome. Hydrophobic tagging (HyT) technology is a bifunctional molecule that induces the proteasomal degradation of POIs by mimicking misfolded proteins. HyT consists of a structure in which a hydrophobic moiety—e.g., an adamantyl group or Boc3Arg—and a selective ligand for a specific POI are connected by a short linker. Boc3Arg has been shown to degrade POIs by binding to the 20S proteasome involved in ATP and ubiquitin-independent degradation. However, the applications of this method are limited because it is known to inhibit Mammalian Target of Rapamycin Complex 1 signaling [213].
7. Conclusions and Future Perspectives
In this review, we summarized the basic mechanisms of proteasome-mediated degradation, its substrates, and various modalities of targeted degradation mediated by the proteasome. Decades of research on ubiquitin and proteasome have significantly broadened our understanding of the degradation mechanism and have been instrumental in elucidating signaling processes resulting from substrate degradation. Furthermore, the ongoing development of targeted degradation modalities and proteasome inhibitors is expected to position them as major therapeutic strategies and promising new therapeutic targets in the future.
For easier comprehension, we divided the degradation processes into those mediated by the 26S proteasome and the 20S proteasome. However, it is important to note that these two processes are not mutually exclusive; they are complementary to each other [140]. The 26S proteasome is not solely responsible for degrading ubiquitinated proteins, and the 20S proteasome does not exclusively target IDPs or oxidized proteins. Some studies have shown that certain proteins, such as ODC, can be degraded by the 26S proteasome without undergoing ubiquitination [67,214]. Additionally, ubiquitin-tagged proteins can also be degraded by the 20S proteasome [139]. Therefore, ubiquitin-dependent degradation via the 26S proteasome is widely recognized as the most common catalytic mechanism. However, simultaneous research on degradation through the 20S proteasome is also expected to continue steadily increasing.
Furthermore, the discovery of novel proteasome regulatory proteins is expected to greatly contribute to our understanding of the proteasome-mediated degradation process. Moreover, with the increasing research on degradation signaling pathways such as degron recognition and ubiquitin-like protein conjugation, there is a growing need for broad interest and extensive research in the field of the proteasome [26,215,216].
Author Contributions
Y.K. and E.-K.K. drafted manuscript and Y.C. and M.-J.S. summarized the literature. H.H.J. completed extensive editing and improvements to the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea grant funded by the Korean government (NRF-2021R1A2C1013988) and the Gachon University Research fund of 2020 (GCU-202002540001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- D’Autreaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Michalska, P.; Leon, R. When It Comes to an End: Oxidative Stress Crosstalk with Protein Aggregation and Neuroinflammation Induce Neurodegeneration. Antioxidants 2020, 9, 740. [Google Scholar] [CrossRef] [PubMed]
- Barral, J.M.; Broadley, S.A.; Schaffar, G.; Hartl, F.U. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell Dev. Biol. 2004, 15, 17–29. [Google Scholar] [CrossRef] [PubMed]
- De Marco, A.; Deuerling, E.; Mogk, A.; Tomoyasu, T.; Bukau, B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007, 7, 32. [Google Scholar] [CrossRef]
- Weibezahn, J.; Bukau, B.; Mogk, A. Unscrambling an egg: Protein disaggregation by AAA+ proteins. Microb. Cell Fact. 2004, 3, 1. [Google Scholar] [CrossRef]
- Mercier, R.; LaPointe, P. The role of cellular proteostasis in antitumor immunity. J. Biol. Chem. 2022, 298, 101930. [Google Scholar] [CrossRef]
- Chen, X.Q.; Shen, T.; Fang, S.J.; Sun, X.M.; Li, G.Y.; Li, Y.F. Protein homeostasis in aging and cancer. Front. Cell Dev. Biol. 2023, 11, 1143532. [Google Scholar] [CrossRef]
- Sala, A.J.; Bott, L.C.; Morimoto, R.I. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 2017, 216, 1231–1241. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wu, H. Ubiquitination-Proteasome System (UPS) and Autophagy Two Main Protein Degradation Machineries in Response to Cell Stress. Cells 2022, 11, 851. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Zheng, Q.; Li, J.; Wang, X. Interplay between the ubiquitin-proteasome system and autophagy in proteinopathies. Int. J. Physiol. Pathophysiol. Pharmacol. 2009, 1, 127–142. [Google Scholar]
- Verhoef, L.G.; Lindsten, K.; Masucci, M.G.; Dantuma, N.P. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum. Mol. Genet. 2002, 11, 2689–2700. [Google Scholar] [CrossRef]
- Bekes, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- He, M.; Lv, W.; Rao, Y. Opportunities and Challenges of Small Molecule Induced Targeted Protein Degradation. Front. Cell Dev. Biol. 2021, 9, 685106. [Google Scholar] [CrossRef]
- Goldstein, G.; Scheid, M.; Hammerling, U.; Schlesinger, D.H.; Niall, H.D.; Boyse, E.A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA 1975, 72, 11–15. [Google Scholar] [CrossRef]
- Wilkinson, K.D.; Urban, M.K.; Haas, A.L. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J. Biol. Chem. 1980, 255, 7529–7532. [Google Scholar] [CrossRef]
- Etlinger, J.D.; Goldberg, A.L. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc. Natl. Acad. Sci. USA 1977, 74, 54–58. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A.; Rose, I.A. Resolution of the ATP-dependent proteolytic system from reticulocytes: A component that interacts with ATP. Proc. Natl. Acad. Sci. USA 1979, 76, 3107–3110. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A.; Heller, H.; Haas, A.L.; Rose, I.A. Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. USA 1980, 77, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.D. The discovery of ubiquitin-dependent proteolysis. Proc. Natl. Acad. Sci. USA 2005, 102, 15280–15282. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, R.; Akasaka, K. Close identity of a pressure-stabilized intermediate with a kinetic intermediate in protein folding. Proc. Natl. Acad. Sci. USA 2003, 100, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Gatti, L.; Hoe, K.L.; Hayles, J.; Righetti, S.C.; Carenini, N.; Bo, L.D.; Kim, D.U.; Park, H.O.; Perego, P. Ubiquitin-proteasome genes as targets for modulation of cisplatin sensitivity in fission yeast. BMC Genom. 2011, 12, 44. [Google Scholar] [CrossRef]
- McClellan, A.J.; Laugesen, S.H.; Ellgaard, L. Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol. 2019, 9, 190147. [Google Scholar] [CrossRef]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef]
- Dittmar, G.; Winklhofer, K.F. Linear Ubiquitin Chains: Cellular Functions and Strategies for Detection and Quantification. Front. Chem. 2019, 7, 915. [Google Scholar] [CrossRef]
- Woelk, T.; Sigismund, S.; Penengo, L.; Polo, S. The ubiquitination code: A signalling problem. Cell Div. 2007, 2, 11. [Google Scholar] [CrossRef]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–78. [Google Scholar] [CrossRef]
- Damgaard, R.B. The ubiquitin system: From cell signalling to disease biology and new therapeutic opportunities. Cell Death Differ. 2021, 28, 423–426. [Google Scholar] [CrossRef]
- Kirisako, T.; Kamei, K.; Murata, S.; Kato, M.; Fukumoto, H.; Kanie, M.; Sano, S.; Tokunaga, F.; Tanaka, K.; Iwai, K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006, 25, 4877–4887. [Google Scholar] [CrossRef]
- Tokunaga, F.; Sakata, S.; Saeki, Y.; Satomi, Y.; Kirisako, T.; Kamei, K.; Nakagawa, T.; Kato, M.; Murata, S.; Yamaoka, S.; et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 2009, 11, 123–132. [Google Scholar] [CrossRef]
- Morris, J.R.; Solomon, E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 2004, 13, 807–817. [Google Scholar] [CrossRef]
- Zhou, Z.; He, M.; Shah, A.A.; Wan, Y. Insights into APC/C: From cellular function to diseases and therapeutics. Cell Div. 2016, 11, 9. [Google Scholar] [CrossRef]
- Schrock, M.S.; Stromberg, B.R.; Scarberry, L.; Summers, M.K. APC/C ubiquitin ligase: Functions and mechanisms in tumorigenesis. Semin. Cancer Biol. 2020, 67, 80–91. [Google Scholar] [CrossRef]
- Platta, H.W.; Abrahamsen, H.; Thoresen, S.B.; Stenmark, H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012, 441, 399–406. [Google Scholar] [CrossRef]
- Geisler, S.; Holmstrom, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef]
- Yin, Q.; Han, T.; Fang, B.; Zhang, G.; Zhang, C.; Roberts, E.R.; Izumi, V.; Zheng, M.; Jiang, S.; Yin, X.; et al. K27-linked ubiquitination of BRAF by ITCH engages cytokine response to maintain MEK-ERK signaling. Nat. Commun. 2019, 10, 1870. [Google Scholar] [CrossRef]
- Peng, D.J.; Zeng, M.; Muromoto, R.; Matsuda, T.; Shimoda, K.; Subramaniam, M.; Spelsberg, T.C.; Wei, W.Z.; Venuprasad, K. Noncanonical K27-linked polyubiquitination of TIEG1 regulates Foxp3 expression and tumor growth. J. Immunol. 2011, 186, 5638–5647. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, L.; Hong, Z.; Lv, Z.; Mao, Z.; Tang, Y.; Kong, X.; Li, S.; Cui, Y.; Liu, H.; et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 2017, 13, e1006264. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C. The emerging roles of the STING adaptor protein in immunity and diseases. Immunology 2016, 147, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Al-Hakim, A.K.; Zagorska, A.; Chapman, L.; Deak, M.; Peggie, M.; Alessi, D.R. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 2008, 411, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Chastagner, P.; Israel, A.; Brou, C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006, 7, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Kim, J.J.; Han, S.A.; Fan, Y.; Guo, L.S.; Aziz, K.; Nowsheen, S.; Kim, S.S.; Park, S.Y.; Luo, Q.; et al. The AMPK-Parkin axis negatively regulates necroptosis and tumorigenesis by inhibiting the necrosome. Nat. Cell Biol. 2019, 21, 940–951. [Google Scholar] [CrossRef]
- Kulikov, R.; Letienne, J.; Kaur, M.; Grossman, S.R.; Arts, J.; Blattner, C. Mdm2 facilitates the association of p53 with the proteasome. Proc. Natl. Acad. Sci. USA 2010, 107, 10038–10043. [Google Scholar] [CrossRef]
- Joshi, S.; Singh, A.R.; Durden, D.L. MDM2 regulates hypoxic hypoxia-inducible factor 1alpha stability in an E3 ligase, proteasome, and PTEN-phosphatidylinositol 3-kinase-AKT-dependent manner. J. Biol. Chem. 2014, 289, 22785–22797. [Google Scholar] [CrossRef]
- Abbas, T.; Sivaprasad, U.; Terai, K.; Amador, V.; Pagano, M.; Dutta, A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008, 22, 2496–2506. [Google Scholar] [CrossRef]
- Calzado, M.A.; de la Vega, L.; Moller, A.; Bowtell, D.D.; Schmitz, M.L. An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat. Cell Biol. 2009, 11, 85–91. [Google Scholar] [CrossRef]
- Maruyama, T.; Araki, T.; Kawarazaki, Y.; Naguro, I.; Heynen, S.; Aza-Blanc, P.; Ronai, Z.; Matsuzawa, A.; Ichijo, H. Roquin-2 promotes ubiquitin-mediated degradation of ASK1 to regulate stress responses. Sci. Signal. 2014, 7, ra8. [Google Scholar] [CrossRef]
- Zhu, J.; Zhao, C.; Zhuang, T.; Jonsson, P.; Sinha, I.; Williams, C.; Stromblad, S.; Dahlman-Wright, K. RING finger protein 31 promotes p53 degradation in breast cancer cells. Oncogene 2016, 35, 1955–1964. [Google Scholar] [CrossRef]
- Kim, H.; Claps, G.; Moller, A.; Bowtell, D.; Lu, X.; Ronai, Z.A. Siah2 regulates tight junction integrity and cell polarity through control of ASPP2 stability. Oncogene 2014, 33, 2004–2010. [Google Scholar] [CrossRef]
- Nakayama, K.; Frew, I.J.; Hagensen, M.; Skals, M.; Habelhah, H.; Bhoumik, A.; Kadoya, T.; Erdjument-Bromage, H.; Tempst, P.; Frappell, P.B.; et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 2004, 117, 941–952. [Google Scholar] [CrossRef]
- Garcia-Limones, C.; Lara-Chica, M.; Jimenez-Jimenez, C.; Perez, M.; Moreno, P.; Munoz, E.; Calzado, M.A. CHK2 stability is regulated by the E3 ubiquitin ligase SIAH2. Oncogene 2016, 35, 4289–4301. [Google Scholar] [CrossRef]
- Zhao, H.L.; Ueki, N.; Hayman, M.J. The Ski protein negatively regulates Siah2-mediated HDAC3 degradation. Biochem. Biophys. Res. Commun. 2010, 399, 623–628. [Google Scholar] [CrossRef]
- Zhang, C.S.; Liu, Q.; Li, M.; Lin, S.Y.; Peng, Y.; Wu, D.; Li, T.Y.; Fu, Q.; Jia, W.; Wang, X.; et al. RHOBTB3 promotes proteasomal degradation of HIFalpha through facilitating hydroxylation and suppresses the Warburg effect. Cell Res. 2015, 25, 1025–1042. [Google Scholar] [CrossRef]
- Liu, C.C.; Lin, Y.C.; Chen, Y.H.; Chen, C.M.; Pang, L.Y.; Chen, H.A.; Wu, P.R.; Lin, M.Y.; Jiang, S.T.; Tsai, T.F.; et al. Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination. Mol. Cell 2016, 61, 84–97. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Lee, D.F.; Kuo, H.P.; Liu, M.; Chou, C.K.; Xia, W.; Du, Y.; Shen, J.; Chen, C.T.; Huo, L.; Hsu, M.C.; et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol. Cell 2009, 36, 131–140. [Google Scholar] [CrossRef]
- Kravtsova-Ivantsiv, Y.; Shomer, I.; Cohen-Kaplan, V.; Snijder, B.; Superti-Furga, G.; Gonen, H.; Sommer, T.; Ziv, T.; Admon, A.; Naroditsky, I.; et al. KPC1-mediated ubiquitination and proteasomal processing of NF-kappaB1 p105 to p50 restricts tumor growth. Cell 2015, 161, 333–347. [Google Scholar] [CrossRef]
- Xia, T.; Dimitropoulou, C.; Zeng, J.; Antonova, G.N.; Snead, C.; Venema, R.C.; Fulton, D.; Qian, S.; Patterson, C.; Papapetropoulos, A.; et al. Chaperone-dependent E3 ligase CHIP ubiquitinates and mediates proteasomal degradation of soluble guanylyl cyclase. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H3080–H3087. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, H.; Xiao, N.; Wang, Y.; Wang, R.; Chen, Y.; Pan, W.; Liu, D.; Li, S.; Sun, J.; Zhang, K.; et al. Smurf1 regulates lung cancer cell growth and migration through interaction with and ubiquitination of PIPKIgamma. Oncogene 2017, 36, 5668–5680. [Google Scholar] [CrossRef] [PubMed]
- Zha, Z.; Han, X.; Smith, M.D.; Liu, Y.; Giguere, P.M.; Kopanja, D.; Raychaudhuri, P.; Siderovski, D.P.; Guan, K.L.; Lei, Q.Y.; et al. A Non-Canonical Function of Gbeta as a Subunit of E3 Ligase in Targeting GRK2 Ubiquitylation. Mol. Cell 2015, 58, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Sun, X.; Guo, P.; Dong, X.Y.; Sethi, P.; Cheng, X.; Zhou, J.; Ling, J.; Simons, J.W.; Lingrel, J.B.; et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J. Biol. Chem. 2005, 280, 41553–41561. [Google Scholar] [CrossRef]
- Azakir, B.A.; Desrochers, G.; Angers, A. The ubiquitin ligase Itch mediates the antiapoptotic activity of epidermal growth factor by promoting the ubiquitylation and degradation of the truncated C-terminal portion of Bid. FEBS J. 2010, 277, 1319–1330. [Google Scholar] [CrossRef]
- Li, D.Q.; Ohshiro, K.; Reddy, S.D.; Pakala, S.B.; Lee, M.H.; Zhang, Y.; Rayala, S.K.; Kumar, R. E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc. Natl. Acad. Sci. USA 2009, 106, 17493–17498. [Google Scholar] [CrossRef]
- Murakami, Y.; Matsufuji, S.; Kameji, T.; Hayashi, S.; Igarashi, K.; Tamura, T.; Tanaka, K.; Ichihara, A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 1992, 360, 597–599. [Google Scholar] [CrossRef]
- Pena, M.M.; Xing, Y.Y.; Koli, S.; Berger, F.G. Role of N-terminal residues in the ubiquitin-independent degradation of human thymidylate synthase. Biochem. J. 2006, 394, 355–363. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Reuven, N.; Prives, C.; Shaul, Y. Susceptibility of p53 unstructured N terminus to 20 S proteasomal degradation programs the stress response. J. Biol. Chem. 2009, 284, 26234–26242. [Google Scholar] [CrossRef]
- Huang, Y.F.; Wee, S.; Gunaratne, J.; Lane, D.P.; Bulavin, D.V. Isg15 controls p53 stability and functions. Cell Cycle 2014, 13, 2200–2210. [Google Scholar] [CrossRef]
- Takasugi, T.; Minegishi, S.; Asada, A.; Saito, T.; Kawahara, H.; Hisanaga, S. Two Degradation Pathways of the p35 Cdk5 (Cyclin-dependent Kinase) Activation Subunit, Dependent and Independent of Ubiquitination. J. Biol. Chem. 2016, 291, 4649–4657. [Google Scholar] [CrossRef]
- Wang, B.; Liu, K.; Lin, H.Y.; Bellam, N.; Ling, S.; Lin, W.C. 14-3-3Tau regulates ubiquitin-independent proteasomal degradation of p21, a novel mechanism of p21 downregulation in breast cancer. Mol. Cell Biol. 2010, 30, 1508–1527. [Google Scholar] [CrossRef]
- Ukmar-Godec, T.; Fang, P.; Ibanez de Opakua, A.; Henneberg, F.; Godec, A.; Pan, K.T.; Cima-Omori, M.S.; Chari, A.; Mandelkow, E.; Urlaub, H.; et al. Proteasomal degradation of the intrinsically disordered protein tau at single-residue resolution. Sci. Adv. 2020, 6, eaba3916. [Google Scholar] [CrossRef]
- Tofaris, G.K.; Layfield, R.; Spillantini, M.G. alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 2001, 509, 22–26. [Google Scholar] [CrossRef]
- Murai, N.; Murakami, Y.; Tajima, A.; Matsufuji, S. Novel ubiquitin-independent nucleolar c-Myc degradation pathway mediated by antizyme 2. Sci. Rep. 2018, 8, 3005. [Google Scholar] [CrossRef]
- Alvarez-Castelao, B.; Castano, J.G. Mechanism of direct degradation of IkappaBalpha by 20S proteasome. FEBS Lett. 2005, 579, 4797–4802. [Google Scholar] [CrossRef]
- Lim, S.K.; Gopalan, G. Aurora-A kinase interacting protein 1 (AURKAIP1) promotes Aurora-A degradation through an alternative ubiquitin-independent pathway. Biochem. J. 2007, 403, 119–127. [Google Scholar] [CrossRef]
- Lim, S.K.; Gopalan, G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene 2007, 26, 6593–6603. [Google Scholar] [CrossRef]
- Kalejta, R.F.; Shenk, T. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. USA 2003, 100, 3263–3268. [Google Scholar] [CrossRef]
- Sdek, P.; Ying, H.; Chang, D.L.; Qiu, W.; Zheng, H.; Touitou, R.; Allday, M.J.; Xiao, Z.X. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell 2005, 20, 699–708. [Google Scholar] [CrossRef]
- Li, Y.; Sun, D.; Ma, Z.; Yamaguchi, K.; Wang, L.; Zhong, S.; Yan, X.; Shang, B.; Nagashima, Y.; Koiwa, H.; et al. Degradation of SERRATE via ubiquitin-independent 20S proteasome to survey RNA metabolism. Nat. Plants 2020, 6, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Zhang, H.L.; Li, D.D.; Yang, K.L.; Tang, J.; Li, X.; Ji, J.; Yu, Y.; Wu, R.Y.; Ravichandran, S.; et al. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2). Autophagy 2018, 14, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Cromm, P.M.; Crews, C.M. The Proteasome in Modern Drug Discovery: Second Life of a Highly Valuable Drug Target. ACS Cent. Sci. 2017, 3, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef]
- Jang, H.H. Regulation of Protein Degradation by Proteasomes in Cancer. J. Cancer Prev. 2018, 23, 153–161. [Google Scholar] [CrossRef]
- Abi Habib, J.; Lesenfants, J.; Vigneron, N.; Van den Eynde, B.J. Functional Differences between Proteasome Subtypes. Cells 2022, 11, 421. [Google Scholar] [CrossRef]
- Morozov, A.V.; Karpov, V.L. Proteasomes and Several Aspects of Their Heterogeneity Relevant to Cancer. Front. Oncol. 2019, 9, 761. [Google Scholar] [CrossRef]
- Murata, S.; Yashiroda, H.; Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009, 10, 104–115. [Google Scholar] [CrossRef]
- Sahara, K.; Kogleck, L.; Yashiroda, H.; Murata, S. The mechanism for molecular assembly of the proteasome. Adv. Biol. Regul. 2014, 54, 51–58. [Google Scholar] [CrossRef]
- Morris, E.P.; da Fonseca, P.C.A. How to build a proteasome. Nat. Struct. Mol. Biol. 2021, 28, 409–410. [Google Scholar] [CrossRef]
- Dambacher, C.M.; Worden, E.J.; Herzik, M.A.; Martin, A.; Lander, G.C. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. Elife 2016, 5, e13027. [Google Scholar] [CrossRef]
- Tomko, R.J., Jr.; Hochstrasser, M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Mol. Cell 2011, 44, 907–917. [Google Scholar] [CrossRef]
- Kao, A.; Randall, A.; Yang, Y.; Patel, V.R.; Kandur, W.; Guan, S.; Rychnovsky, S.D.; Baldi, P.; Huang, L. Mapping the structural topology of the yeast 19S proteasomal regulatory particle using chemical cross-linking and probabilistic modeling. Mol. Cell Proteom. 2012, 11, 1566–1577. [Google Scholar] [CrossRef]
- Riedinger, C.; Boehringer, J.; Trempe, J.F.; Lowe, E.D.; Brown, N.R.; Gehring, K.; Noble, M.E.; Gordon, C.; Endicott, J.A. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J. Biol. Chem. 2010, 285, 33992–34003. [Google Scholar] [CrossRef]
- Sakata, E.; Bohn, S.; Mihalache, O.; Kiss, P.; Beck, F.; Nagy, I.; Nickell, S.; Tanaka, K.; Saeki, Y.; Forster, F.; et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 1479–1484. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef]
- Kaneko, T.; Hamazaki, J.; Iemura, S.; Sasaki, K.; Furuyama, K.; Natsume, T.; Tanaka, K.; Murata, S. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 2009, 137, 914–925. [Google Scholar] [CrossRef]
- Livneh, I.; Cohen-Kaplan, V.; Cohen-Rosenzweig, C.; Avni, N.; Ciechanover, A. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 2016, 26, 869–885. [Google Scholar] [CrossRef]
- Wang, X.; Chemmama, I.E.; Yu, C.; Huszagh, A.; Xu, Y.; Viner, R.; Block, S.A.; Cimermancic, P.; Rychnovsky, S.D.; Ye, Y.; et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Biol. Chem. 2017, 292, 16310–16320. [Google Scholar] [CrossRef]
- Imai, J.; Maruya, M.; Yashiroda, H.; Yahara, I.; Tanaka, K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003, 22, 3557–3567. [Google Scholar] [CrossRef]
- Olshina, M.A.; Arkind, G.; Kumar Deshmukh, F.; Fainer, I.; Taranavsky, M.; Hayat, D.; Ben-Dor, S.; Ben-Nissan, G.; Sharon, M. Regulation of the 20S Proteasome by a Novel Family of Inhibitory Proteins. Antioxid. Redox Signal. 2020, 32, 636–655. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tang, X.; Qi, X.; Fu, X.; Ghimire, S.; Ma, R.; Li, S.; Zhang, N.; Si, H. The Ubiquitin Conjugating Enzyme: An Important Ubiquitin Transfer Platform in Ubiquitin-Proteasome System. Int. J. Mol. Sci. 2020, 21, 2894. [Google Scholar] [CrossRef] [PubMed]
- Buetow, L.; Huang, D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016, 17, 626–642. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Qu, Z. Roles of protein ubiquitination and degradation kinetics in biological oscillations. PLoS ONE 2012, 7, e34616. [Google Scholar] [CrossRef]
- Corn, P.G. Role of the ubiquitin proteasome system in renal cell carcinoma. BMC Biochem. 2007, 8 (Suppl. 1), S4. [Google Scholar] [CrossRef]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 ubiquitin ligases: Styles, structures and functions. Mol. Biomed. 2021, 2, 23. [Google Scholar] [CrossRef]
- Garcia-Barcena, C.; Osinalde, N.; Ramirez, J.; Mayor, U. How to Inactivate Human Ubiquitin E3 Ligases by Mutation. Front. Cell Dev. Biol. 2020, 8, 39. [Google Scholar] [CrossRef]
- Tu, D.; Li, W.; Ye, Y.; Brunger, A.T. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. USA 2007, 104, 15599–15606. [Google Scholar] [CrossRef]
- Kaneko, C.; Hatakeyama, S.; Matsumoto, M.; Yada, M.; Nakayama, K.; Nakayama, K.I. Characterization of the mouse gene for the U-box-type ubiquitin ligase UFD2a. Biochem. Biophys. Res. Commun. 2003, 300, 297–304. [Google Scholar] [CrossRef]
- Benirschke, R.C.; Thompson, J.R.; Nomine, Y.; Wasielewski, E.; Juranic, N.; Macura, S.; Hatakeyama, S.; Nakayama, K.I.; Botuyan, M.V.; Mer, G. Molecular basis for the association of human E4B U box ubiquitin ligase with E2-conjugating enzymes UbcH5c and Ubc4. Structure 2010, 18, 955–965. [Google Scholar] [CrossRef]
- Antoniou, N.; Lagopati, N.; Balourdas, D.I.; Nikolaou, M.; Papalampros, A.; Vasileiou, P.V.S.; Myrianthopoulos, V.; Kotsinas, A.; Shiloh, Y.; Liontos, M.; et al. The Role of E3, E4 Ubiquitin Ligase (UBE4B) in Human Pathologies. Cancers 2019, 12, 62. [Google Scholar] [CrossRef]
- Martinez-Fonts, K.; Davis, C.; Tomita, T.; Elsasser, S.; Nager, A.R.; Shi, Y.; Finley, D.; Matouschek, A. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 2020, 11, 477. [Google Scholar] [CrossRef]
- Chen, X.; Htet, Z.M.; Lopez-Alfonzo, E.; Martin, A.; Walters, K.J. Proteasome interaction with ubiquitinated substrates: From mechanisms to therapies. FEBS J. 2021, 288, 5231–5251. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Hamazaki, J.; Iemura, S.; Natsume, T.; Yashiroda, H.; Tanaka, K.; Murata, S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006, 25, 4524–4536. [Google Scholar] [CrossRef]
- Boughton, A.J.; Liu, L.; Lavy, T.; Kleifeld, O.; Fushman, D. A novel recognition site for polyubiquitin and ubiquitin-like signals in an unexpected region of proteasomal subunit Rpn1. J. Biol. Chem. 2021, 297, 101052. [Google Scholar] [CrossRef]
- Kim, H.T.; Goldberg, A.L. The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis. J. Biol. Chem. 2017, 292, 9830–9839. [Google Scholar] [CrossRef]
- Sahu, I.; Glickman, M.H. Proteasome in action: Substrate degradation by the 26S proteasome. Biochem. Soc. Trans. 2021, 49, 629–644. [Google Scholar] [CrossRef]
- Snyder, N.A.; Silva, G.M. Deubiquitinating enzymes (DUBs): Regulation, homeostasis, and oxidative stress response. J. Biol. Chem. 2021, 297, 101077. [Google Scholar] [CrossRef]
- Zong, Z.; Zhang, Z.; Wu, L.; Zhang, L.; Zhou, F. The Functional Deubiquitinating Enzymes in Control of Innate Antiviral Immunity. Adv. Sci. 2021, 8, 2002484. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Baek, K.H. Pro-apoptotic and anti-apoptotic regulation mediated by deubiquitinating enzymes. Cell Mol. Life Sci. 2022, 79, 117. [Google Scholar] [CrossRef] [PubMed]
- Amerik, A.Y.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 2004, 1695, 189–207. [Google Scholar] [CrossRef] [PubMed]
- Chadchankar, J.; Korboukh, V.; Conway, L.C.; Wobst, H.J.; Walker, C.A.; Doig, P.; Jacobsen, S.J.; Brandon, N.J.; Moss, S.J.; Wang, Q. Inactive USP14 and inactive UCHL5 cause accumulation of distinct ubiquitinated proteins in mammalian cells. PLoS ONE 2019, 14, e0225145. [Google Scholar] [CrossRef] [PubMed]
- Maytal-Kivity, V.; Reis, N.; Hofmann, K.; Glickman, M.H. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002, 3, 28. [Google Scholar] [CrossRef]
- Lee, M.J.; Lee, B.H.; Hanna, J.; King, R.W.; Finley, D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell Proteom. 2011, 10, R110.003871. [Google Scholar] [CrossRef]
- Worden, E.J.; Dong, K.C.; Martin, A. An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Mol. Cell 2017, 67, 799–811.e8. [Google Scholar] [CrossRef]
- Miller, J.M.; Enemark, E.J. Fundamental Characteristics of AAA+ Protein Family Structure and Function. Archaea 2016, 2016, 9294307. [Google Scholar] [CrossRef]
- Yedidi, R.S.; Wendler, P.; Enenkel, C. AAA-ATPases in Protein Degradation. Front. Mol. Biosci. 2017, 4, 42. [Google Scholar] [CrossRef]
- Smith, D.M.; Chang, S.C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 2007, 27, 731–744. [Google Scholar] [CrossRef]
- Buneeva, O.A.; Medvedev, A.E. Ubiquitin-independent protein degradation in proteasomes. Biomed. Khim 2018, 64, 134–148. [Google Scholar] [CrossRef]
- Ekici, O.D.; Paetzel, M.; Dalbey, R.E. Unconventional serine proteases: Variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 2008, 17, 2023–2037. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Akopian, T.N.; Woo, K.M.; Goldberg, A.L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999, 274, 3363–3371. [Google Scholar] [CrossRef]
- Jung, T.; Grune, T. The proteasome and the degradation of oxidized proteins: Part I—Structure of proteasomes. Redox Biol. 2013, 1, 178–182. [Google Scholar] [CrossRef]
- Van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Ben-Nissan, G.; Sharon, M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 2014, 4, 862–884. [Google Scholar] [CrossRef]
- Jung, T.; Hohn, A.; Grune, T. The proteasome and the degradation of oxidized proteins: Part II—Protein oxidation and proteasomal degradation. Redox Biol. 2014, 2, 99–104. [Google Scholar] [CrossRef]
- Raynes, R.; Pomatto, L.C.; Davies, K.J. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol. Asp. Med. 2016, 50, 41–55. [Google Scholar] [CrossRef]
- Sahu, I.; Mali, S.M.; Sulkshane, P.; Xu, C.; Rozenberg, A.; Morag, R.; Sahoo, M.P.; Singh, S.K.; Ding, Z.; Wang, Y.; et al. The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag. Nat. Commun. 2021, 12, 6173. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Reuven, N.; Shaul, Y. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ. 2010, 17, 103–108. [Google Scholar] [CrossRef]
- Guillaume, B.; Chapiro, J.; Stroobant, V.; Colau, D.; Van Holle, B.; Parvizi, G.; Bousquet-Dubouch, M.P.; Theate, I.; Parmentier, N.; Van den Eynde, B.J. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 18599–18604. [Google Scholar] [CrossRef] [PubMed]
- Joeris, T.; Schmidt, N.; Ermert, D.; Krienke, P.; Visekruna, A.; Kuckelkorn, U.; Kaufmann, S.H.; Steinhoff, U. The proteasome system in infection: Impact of beta5 and LMP7 on composition, maturation and quantity of active proteasome complexes. PLoS ONE 2012, 7, e39827. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, T.M.; Bozhok, G.A.; Alabedal’karim, N.M.; Karpova, Y.D.; Lyupina, Y.V.; Ushakova, E.M.; Legach, E.I.; Bondarenko, T.P.; Sharova, N.P. Proteasome Expression in Ovarian Heterotopic Allografts of Wistar and August Rats under Induction of Donor-Specific Tolerance. Russ. J. Dev. Biol. 2019, 50, 261–267. [Google Scholar] [CrossRef]
- Petersen, A.; Kutsche, H.S.; Nippert, F.; Schreckenberg, R.; Schulz, R.; Schluter, K.D. Induction of Proteasome Subunit Low Molecular Weight Protein (LMP)-2 Is Required to Induce Active Remodeling in Adult Rat Ventricular Cardiomyocytes. Med. Sci. 2020, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Yokota, K.; Kagawa, S.; Shimbara, N.; Tamura, T.; Akioka, H.; Nothwang, H.G.; Noda, C.; Tanaka, K.; Ichihara, A. cDNA cloning and interferon gamma down-regulation of proteasomal subunits X and Y. Science 1994, 265, 1231–1234. [Google Scholar] [CrossRef]
- Murata, S.; Takahama, Y.; Kasahara, M.; Tanaka, K. The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nat. Immunol. 2018, 19, 923–931. [Google Scholar] [CrossRef]
- Kaur, G.; Batra, S. Emerging role of immunoproteasomes in pathophysiology. Immunol. Cell Biol. 2016, 94, 812–820. [Google Scholar] [CrossRef]
- Groettrup, M.; Kirk, C.J.; Basler, M. Proteasomes in immune cells: More than peptide producers? Nat. Rev. Immunol. 2010, 10, 73–78. [Google Scholar] [CrossRef]
- Murata, S.; Sasaki, K.; Kishimoto, T.; Niwa, S.; Hayashi, H.; Takahama, Y.; Tanaka, K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 2007, 316, 1349–1353. [Google Scholar] [CrossRef]
- Frantzeskakis, M.; Takahama, Y.; Ohigashi, I. The Role of Proteasomes in the Thymus. Front. Immunol. 2021, 12, 646209. [Google Scholar] [CrossRef]
- Tomaru, U.; Konno, S.; Miyajima, S.; Kimoto, R.; Onodera, M.; Kiuchi, S.; Murata, S.; Ishizu, A.; Kasahara, M. Restricted Expression of the Thymoproteasome Is Required for Thymic Selection and Peripheral Homeostasis of CD8(+) T Cells. Cell Rep. 2019, 26, 639–651.e2. [Google Scholar] [CrossRef]
- Ma, C.P.; Slaughter, C.A.; DeMartino, G.N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J. Biol. Chem. 1992, 267, 10515–10523. [Google Scholar] [CrossRef]
- Dubiel, W.; Pratt, G.; Ferrell, K.; Rechsteiner, M. Purification of an 11 S regulator of the multicatalytic protease. J. Biol. Chem. 1992, 267, 22369–22377. [Google Scholar] [CrossRef]
- Huber, E.M.; Groll, M. The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha(4)beta(3) Complex. Structure 2017, 25, 1473–1480.e3. [Google Scholar] [CrossRef]
- Ahn, K.; Erlander, M.; Leturcq, D.; Peterson, P.A.; Fruh, K.; Yang, Y. In vivo characterization of the proteasome regulator PA28. J. Biol. Chem. 1996, 271, 18237–18242. [Google Scholar] [CrossRef]
- Murata, S.; Udono, H.; Tanahashi, N.; Hamada, N.; Watanabe, K.; Adachi, K.; Yamano, T.; Yui, K.; Kobayashi, N.; Kasahara, M.; et al. Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. EMBO J. 2001, 20, 5898–5907. [Google Scholar] [CrossRef]
- Tanahashi, N.; Yokota, K.; Ahn, J.Y.; Chung, C.H.; Fujiwara, T.; Takahashi, E.; DeMartino, G.N.; Slaughter, C.A.; Toyonaga, T.; Yamamura, K.; et al. Molecular properties of the proteasome activator PA28 family proteins and gamma-interferon regulation. Genes Cells 1997, 2, 195–211. [Google Scholar] [CrossRef]
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28alphabeta reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chem. Biol. 2014, 21, 470–480. [Google Scholar] [CrossRef]
- McCarthy, M.K.; Weinberg, J.B. The immunoproteasome and viral infection: A complex regulator of inflammation. Front. Microbiol. 2015, 6, 21. [Google Scholar] [CrossRef]
- Ustrell, V.; Hoffman, L.; Pratt, G.; Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002, 21, 3516–3525. [Google Scholar] [CrossRef]
- Kocberber, Z.; Willemsen, N.; Bartelt, A. The role of proteasome activators PA28alphabeta and PA200 in brown adipocyte differentiation and function. Front. Endocrinol. 2023, 14, 1176733. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Haratake, K.; Miyahara, H.; Chiba, T. Proteasome activators, PA28gamma and PA200, play indispensable roles in male fertility. Sci. Rep. 2016, 6, 23171. [Google Scholar] [CrossRef]
- Mandemaker, I.K.; Geijer, M.E.; Kik, I.; Bezstarosti, K.; Rijkers, E.; Raams, A.; Janssens, R.C.; Lans, H.; Hoeijmakers, J.H.; Demmers, J.A.; et al. DNA damage-induced replication stress results in PA200-proteasome-mediated degradation of acetylated histones. EMBO Rep. 2018, 19, e45566. [Google Scholar] [CrossRef] [PubMed]
- Hamazaki, J.; Murata, S. ER-Resident Transcription Factor Nrf1 Regulates Proteasome Expression and Beyond. Int. J. Mol. Sci. 2020, 21, 3683. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, S.K.; den Besten, W.; Deshaies, R.J. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. Elife 2014, 3, e01856. [Google Scholar] [CrossRef]
- Lee, D.; Takayama, S.; Goldberg, A.L. ZFAND5/ZNF216 is an activator of the 26S proteasome that stimulates overall protein degradation. Proc. Natl. Acad. Sci. USA 2018, 115, E9550–E9559. [Google Scholar] [CrossRef]
- Cho-Park, P.F.; Steller, H. Proteasome regulation by ADP-ribosylation. Cell 2013, 153, 614–627. [Google Scholar] [CrossRef]
- Moscovitz, O.; Ben-Nissan, G.; Fainer, I.; Pollack, D.; Mizrachi, L.; Sharon, M. The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015, 6, 6609. [Google Scholar] [CrossRef]
- Asher, G.; Lotem, J.; Kama, R.; Sachs, L.; Shaul, Y. NQO1 stabilizes p53 through a distinct pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 3099–3104. [Google Scholar] [CrossRef]
- Asher, G.; Bercovich, Z.; Tsvetkov, P.; Shaul, Y.; Kahana, C. 20S proteasomal degradation of ornithine decarboxylase is regulated by NQO1. Mol. Cell 2005, 17, 645–655. [Google Scholar] [CrossRef]
- Moscovitz, O.; Tsvetkov, P.; Hazan, N.; Michaelevski, I.; Keisar, H.; Ben-Nissan, G.; Shaul, Y.; Sharon, M. A mutually inhibitory feedback loop between the 20S proteasome and its regulator, NQO1. Mol. Cell 2012, 47, 76–86. [Google Scholar] [CrossRef]
- Zaiss, D.M.; Standera, S.; Holzhutter, H.; Kloetzel, P.; Sijts, A.J. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett. 1999, 457, 333–338. [Google Scholar] [CrossRef]
- McCutchen-Maloney, S.L.; Matsuda, K.; Shimbara, N.; Binns, D.D.; Tanaka, K.; Slaughter, C.A.; DeMartino, G.N. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J. Biol. Chem. 2000, 275, 18557–18565. [Google Scholar] [CrossRef]
- Li, D.; Dong, Q.; Tao, Q.; Gu, J.; Cui, Y.; Jiang, X.; Yuan, J.; Li, W.; Xu, R.; Jin, Y.; et al. c-Abl regulates proteasome abundance by controlling the ubiquitin-proteasomal degradation of PSMA7 subunit. Cell Rep. 2015, 10, 484–496. [Google Scholar] [CrossRef]
- Liu, X.; Huang, W.; Li, C.; Li, P.; Yuan, J.; Li, X.; Qiu, X.B.; Ma, Q.; Cao, C. Interaction between c-Abl and Arg tyrosine kinases and proteasome subunit PSMA7 regulates proteasome degradation. Mol. Cell 2006, 22, 317–327. [Google Scholar] [CrossRef]
- Montenegro-Venegas, C.; Fienko, S.; Anni, D.; Pina-Fernandez, E.; Frischknecht, R.; Fejtova, A. Bassoon inhibits proteasome activity via interaction with PSMB4. Cell. Mol. Life Sci. 2021, 78, 1545–1563. [Google Scholar] [CrossRef]
- Saez, I.; Vilchez, D. The Mechanistic Links Between Proteasome Activity, Aging and Age-related Diseases. Curr. Genom. 2014, 15, 38–51. [Google Scholar] [CrossRef]
- Dahlmann, B. Role of proteasomes in disease. BMC Biochem. 2007, 8 (Suppl. 1), S3. [Google Scholar] [CrossRef]
- Thibaudeau, T.A.; Anderson, R.T.; Smith, D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Basler, M.; Groettrup, M. On the Role of the Immunoproteasome in Protein Homeostasis. Cells 2021, 10, 3216. [Google Scholar] [CrossRef]
- Khan, B.; Gand, L.V.; Amrute-Nayak, M.; Nayak, A. Emerging Mechanisms of Skeletal Muscle Homeostasis and Cachexia: The SUMO Perspective. Cells 2023, 12, 644. [Google Scholar] [CrossRef] [PubMed]
- Haberecht-Muller, S.; Kruger, E.; Fielitz, J. Out of Control: The Role of the Ubiquitin Proteasome System in Skeletal Muscle during Inflammation. Biomolecules 2021, 11, 1327. [Google Scholar] [CrossRef] [PubMed]
- Merlos Rodrigo, M.A.; Buchtelova, H.; de Los Rios, V.; Casal, J.I.; Eckschlager, T.; Hrabeta, J.; Belhajova, M.; Heger, Z.; Adam, V. Proteomic Signature of Neuroblastoma Cells UKF-NB-4 Reveals Key Role of Lysosomal Sequestration and the Proteasome Complex in Acquiring Chemoresistance to Cisplatin. J. Proteome Res. 2019, 18, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, M.A.M.; Buchtelova, H.; Jimenez, A.M.J.; Adam, P.; Babula, P.; Heger, Z.; Adam, V. Transcriptomic Landscape of Cisplatin-Resistant Neuroblastoma Cells. Cells 2019, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- Pierce, M.R.; Robinson, R.M.; Ibarra-Rivera, T.R.; Pirrung, M.C.; Dolloff, N.G.; Bachmann, A.S. Syrbactin proteasome inhibitor TIR-199 overcomes bortezomib chemoresistance and inhibits multiple myeloma tumor growth in vivo. Leuk. Res. 2020, 88, 106271. [Google Scholar] [CrossRef] [PubMed]
- Voutsadakis, I.A. Proteasome expression and activity in cancer and cancer stem cells. Tumour Biol. 2017, 39, 1010428317692248. [Google Scholar] [CrossRef]
- Lenos, K.J.; Vermeulen, L. Cancer stem cells don’t waste their time cleaning-low proteasome activity, a marker for cancer stem cell function. Ann. Transl. Med. 2016, 4, 519. [Google Scholar] [CrossRef]
- Vlashi, E.; Kim, K.; Lagadec, C.; Donna, L.D.; McDonald, J.T.; Eghbali, M.; Sayre, J.W.; Stefani, E.; McBride, W.; Pajonk, F. In vivo imaging, tracking, and targeting of cancer stem cells. J. Natl. Cancer Inst. 2009, 101, 350–359. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, Q.; Wang, Y.; You, M. 26S proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. PLoS ONE 2010, 5, e13298. [Google Scholar] [CrossRef]
- Della Donna, L.; Lagadec, C.; Pajonk, F. Radioresistance of prostate cancer cells with low proteasome activity. Prostate 2012, 72, 868–874. [Google Scholar] [CrossRef]
- Adikrisna, R.; Tanaka, S.; Muramatsu, S.; Aihara, A.; Ban, D.; Ochiai, T.; Irie, T.; Kudo, A.; Nakamura, N.; Yamaoka, S.; et al. Identification of pancreatic cancer stem cells and selective toxicity of chemotherapeutic agents. Gastroenterology 2012, 143, 234–245.e7. [Google Scholar] [CrossRef]
- Munakata, K.; Uemura, M.; Tanaka, S.; Kawai, K.; Kitahara, T.; Miyo, M.; Kano, Y.; Nishikawa, S.; Fukusumi, T.; Takahashi, Y.; et al. Cancer Stem-like Properties in Colorectal Cancer Cells with Low Proteasome Activity. Clin. Cancer Res. 2016, 22, 5277–5286. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, J.; Zhong, K.; Tong, A.; Jia, D. Targeted protein degradation: Mechanisms, strategies and application. Signal. Transduct. Target. Ther. 2022, 7, 113. [Google Scholar] [CrossRef]
- Zhou, Q.Q.; Xiao, H.T.; Yang, F.; Wang, Y.D.; Li, P.; Zheng, Z.G. Advancing targeted protein degradation for metabolic diseases therapy. Pharmacol. Res. 2023, 188, 106627. [Google Scholar] [CrossRef]
- Roth, S.; Fulcher, L.J.; Sapkota, G.P. Advances in targeted degradation of endogenous proteins. Cell. Mol. Life Sci. 2019, 76, 2761–2777. [Google Scholar] [CrossRef]
- Liu, Z.; Hu, M.; Yang, Y.; Du, C.; Zhou, H.; Liu, C.; Chen, Y.; Fan, L.; Ma, H.; Gong, Y.; et al. An overview of PROTACs: A promising drug discovery paradigm. Mol. Biomed. 2022, 3, 46. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef]
- Yokoo, H.; Naito, M.; Demizu, Y. Investigating the cell permeability of proteolysis-targeting chimeras (PROTACs). Expert. Opin. Drug Discov. 2023, 18, 357–361. [Google Scholar] [CrossRef]
- Au, Y.Z.; Wang, T.; Sigua, L.H.; Qi, J. Peptide-Based PROTAC: The Predator of Pathological Proteins. Cell Chem. Biol. 2020, 27, 637–639. [Google Scholar] [CrossRef]
- Cecchini, C.; Pannilunghi, S.; Tardy, S.; Scapozza, L. From Conception to Development: Investigating PROTACs Features for Improved Cell Permeability and Successful Protein Degradation. Front. Chem. 2021, 9, 672267. [Google Scholar] [CrossRef]
- Gao, S.; Wang, S.; Song, Y. Novel immunomodulatory drugs and neo-substrates. Biomark. Res. 2020, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Ito, T.; Handa, H. Cereblon-Based Small-Molecule Compounds to Control Neural Stem Cell Proliferation in Regenerative Medicine. Front. Cell Dev. Biol. 2021, 9, 629326. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, Y. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J. Hematol. Oncol. 2020, 13, 50. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xiong, H.; Gu, S.X.; Wang, M. E3 ligase ligand optimization of Clinical PROTACs. Front. Chem. 2023, 11, 1098331. [Google Scholar] [CrossRef]
- Qi, S.M.; Dong, J.; Xu, Z.Y.; Cheng, X.D.; Zhang, W.D.; Qin, J.J. PROTAC: An Effective Targeted Protein Degradation Strategy for Cancer Therapy. Front. Pharmacol. 2021, 12, 692574. [Google Scholar] [CrossRef]
- Graham, H. The mechanism of action and clinical value of PROTACs: A graphical review. Cell Signal. 2022, 99, 110446. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Ward, L.D.; Yan, Q.; Bohnuud, T.; Hernandez, R.; Lao, S.; Yuan, J.; Fan, F. A proteomic platform to identify off-target proteins associated with therapeutic modalities that induce protein degradation or gene silencing. Sci. Rep. 2021, 11, 15856. [Google Scholar] [CrossRef]
- Li, X.; Pu, W.; Zheng, Q.; Ai, M.; Chen, S.; Peng, Y. Proteolysis-targeting chimeras (PROTACs) in cancer therapy. Mol. Cancer 2022, 21, 99. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Shi, L.; Yang, S.; Chang, J.; Zhong, Y.; Li, Q.; Xing, D. Recent advances in IAP-based PROTACs (SNIPERs) as potential therapeutic agents. J. Enzym. Inhib. Med. Chem. 2022, 37, 1437–1453. [Google Scholar] [CrossRef]
- Naito, M.; Ohoka, N.; Shibata, N.; Tsukumo, Y. Targeted Protein Degradation by Chimeric Small Molecules, PROTACs and SNIPERs. Front. Chem. 2019, 7, 849. [Google Scholar] [CrossRef]
- Wu, H.; Yao, H.; He, C.; Jia, Y.; Zhu, Z.; Xu, S.; Li, D.; Xu, J. Molecular glues modulate protein functions by inducing protein aggregation: A promising therapeutic strategy of small molecules for disease treatment. Acta Pharm. Sin. B 2022, 12, 3548–3566. [Google Scholar] [CrossRef]
- Sasso, J.M.; Tenchov, R.; Wang, D.; Johnson, L.S.; Wang, X.; Zhou, Q.A. Molecular Glues: The Adhesive Connecting Targeted Protein Degradation to the Clinic. Biochemistry 2023, 62, 601–623. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; Feng, F.; Liu, W.; Sun, H. Degradation of proteins by PROTACs and other strategies. Acta Pharm. Sin. B 2020, 10, 207–238. [Google Scholar] [CrossRef]
- Schmidtke, G.; Kalveram, B.; Groettrup, M. Degradation of FAT10 by the 26S proteasome is independent of ubiquitylation but relies on NUB1L. FEBS Lett. 2009, 583, 591–594. [Google Scholar] [CrossRef]
- Heo, A.J.; Kim, S.B.; Kwon, Y.T.; Ji, C.H. The N-degron pathway: From basic science to therapeutic applications. Biochim. Biophys. Acta Gene Regul. Mech. 2023, 1866, 194934. [Google Scholar] [CrossRef]
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 2011, 10, 29–46. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).