Deep Learning-Based Computational Cytopathologic Diagnosis of Metastatic Breast Carcinoma in Pleural Fluid
Abstract
1. Introduction
2. Materials and Methods
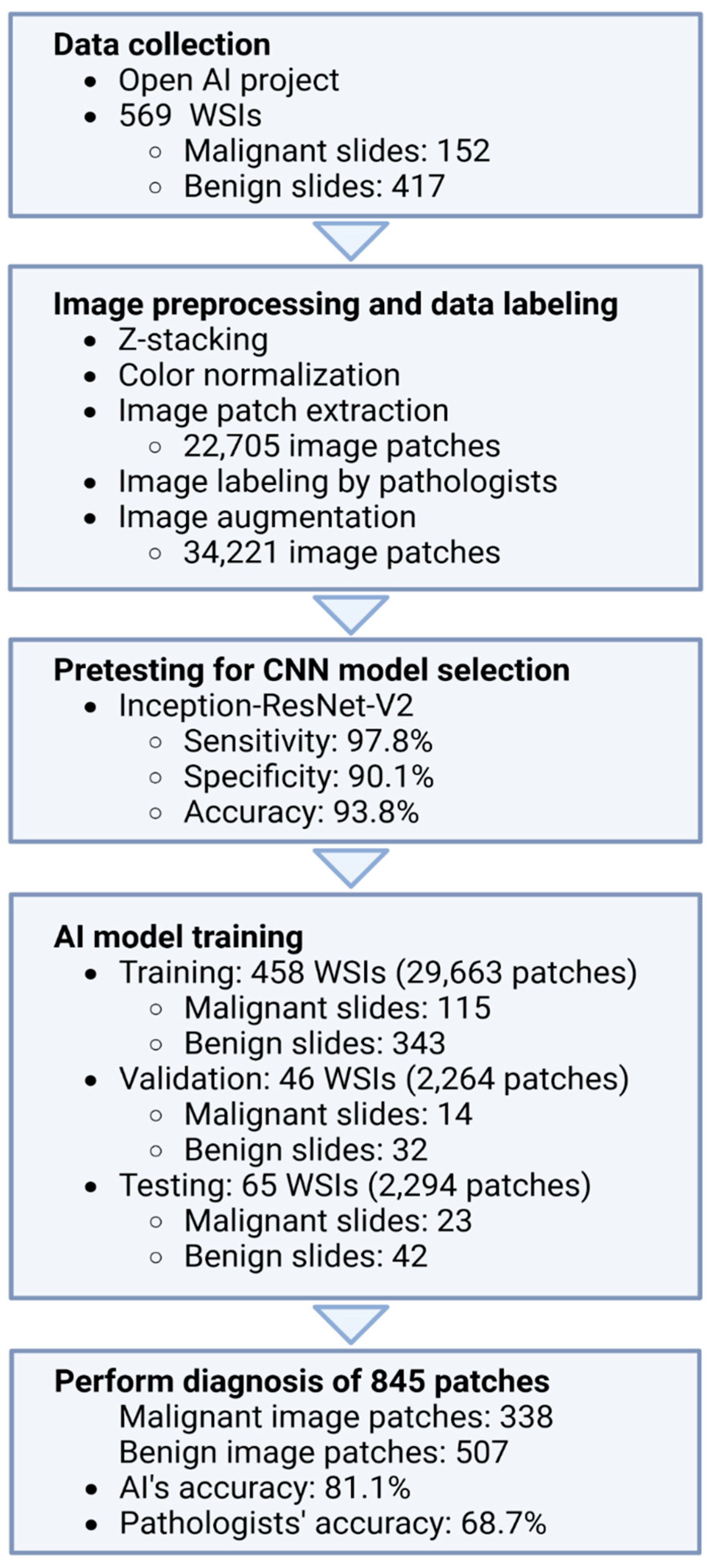
2.1. Data Collection
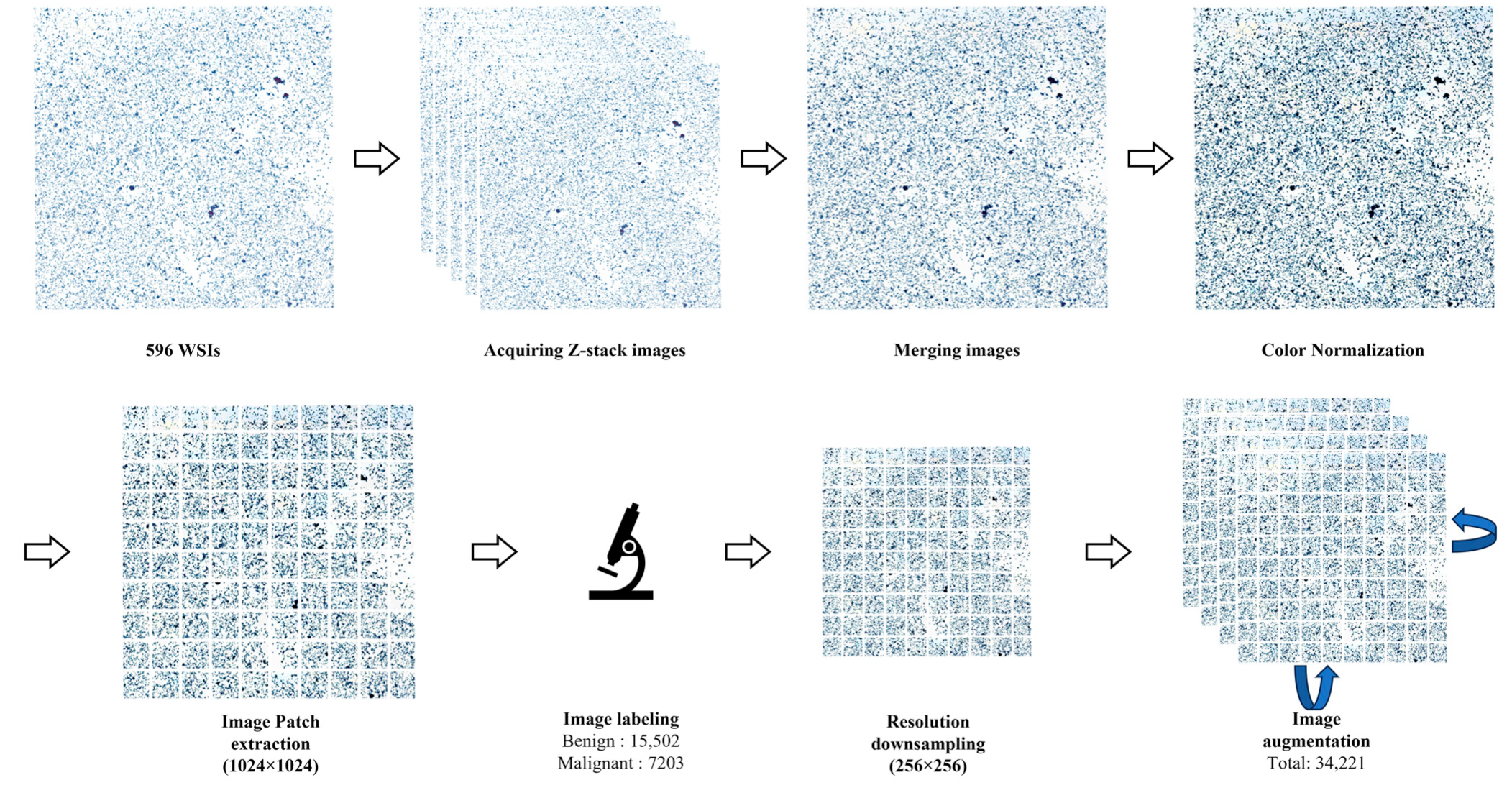
2.2. Image Preprocessing
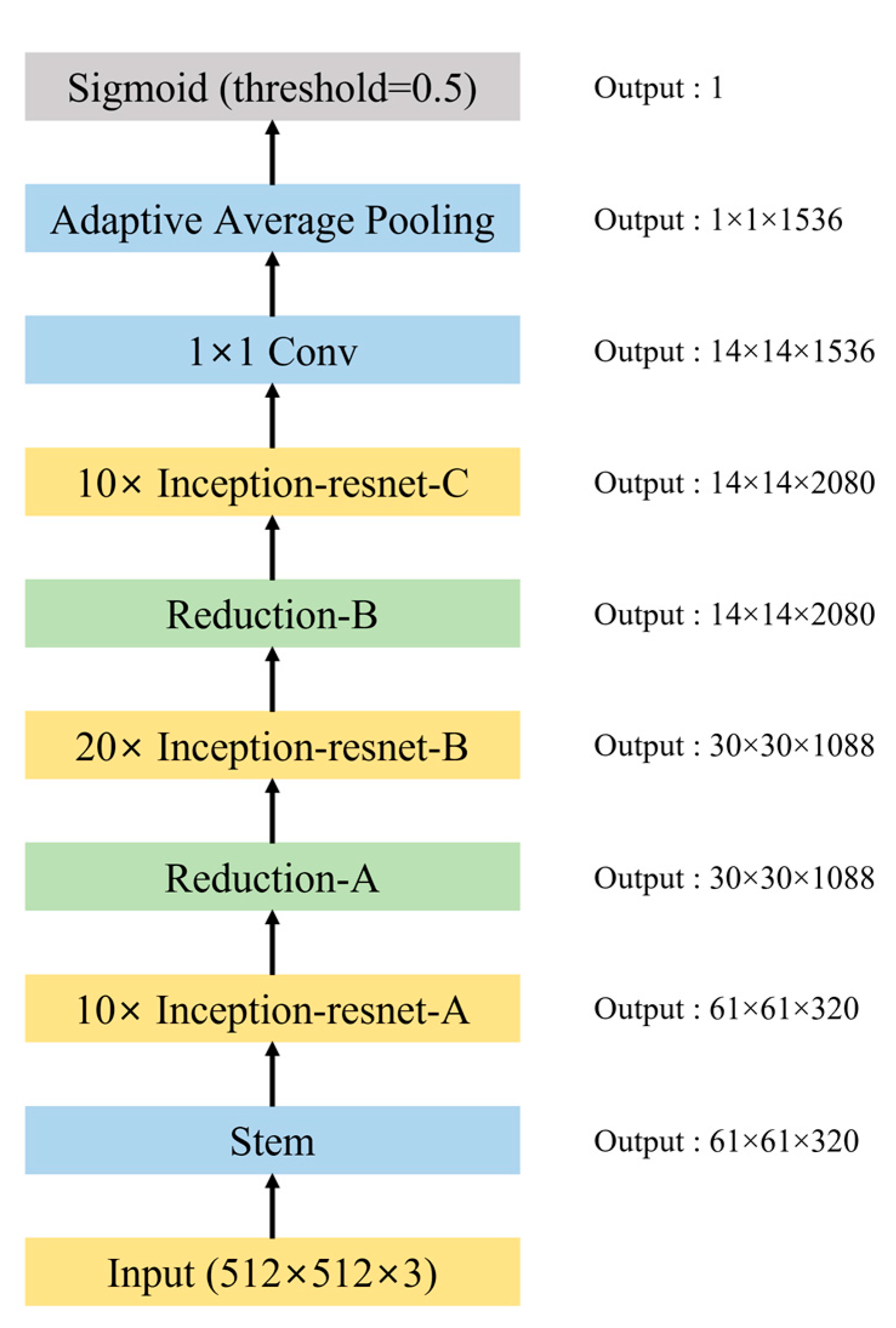
2.3. Pretesting for DCNN Model Selection
2.4. AI Model Training
2.5. Comparison of the Performance of the AI Model and the Pathologists
3. Results
3.1. Data Characteristics
3.2. Pretesting for DCNN Model Selection
3.3. AI Model Training
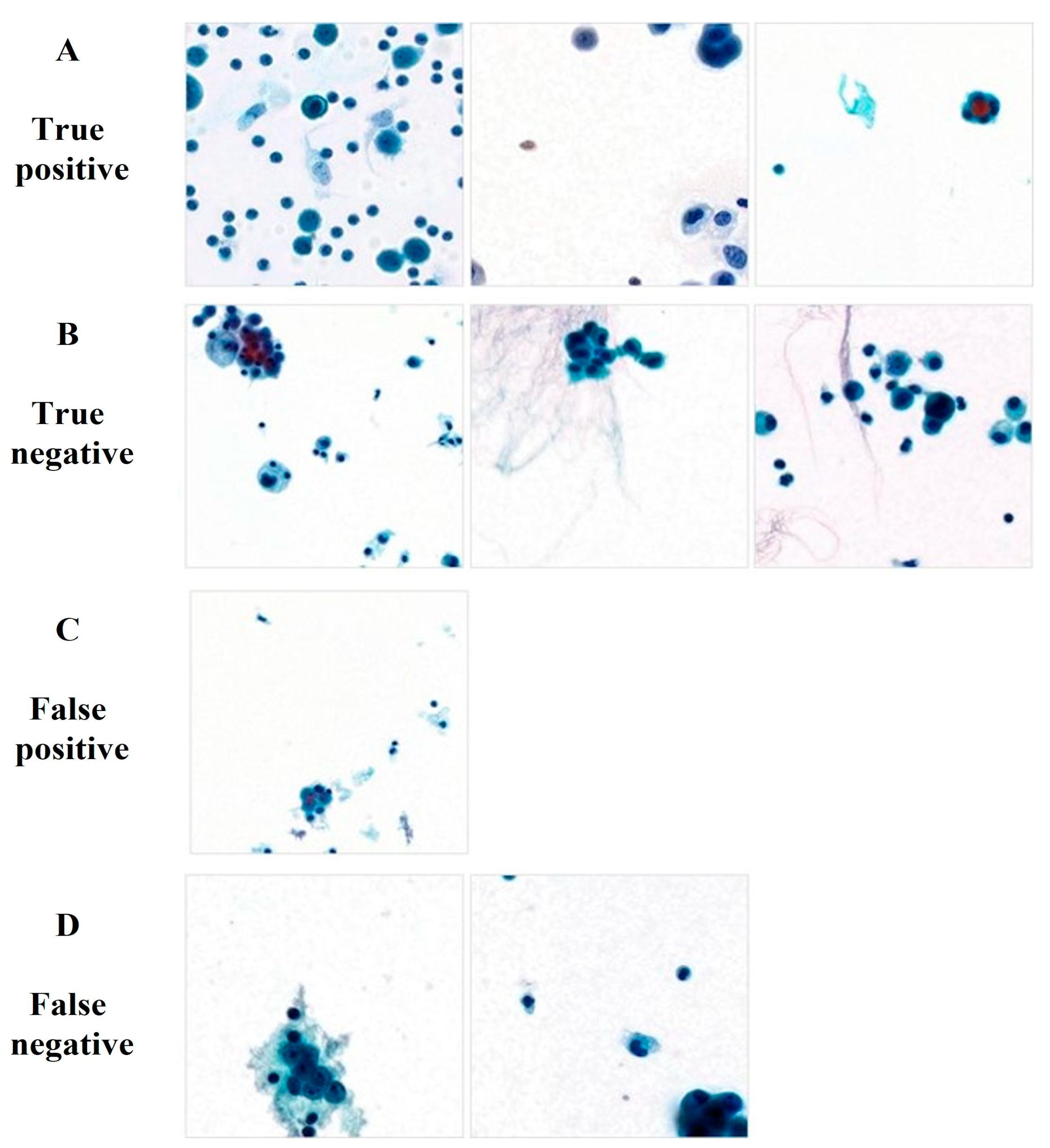
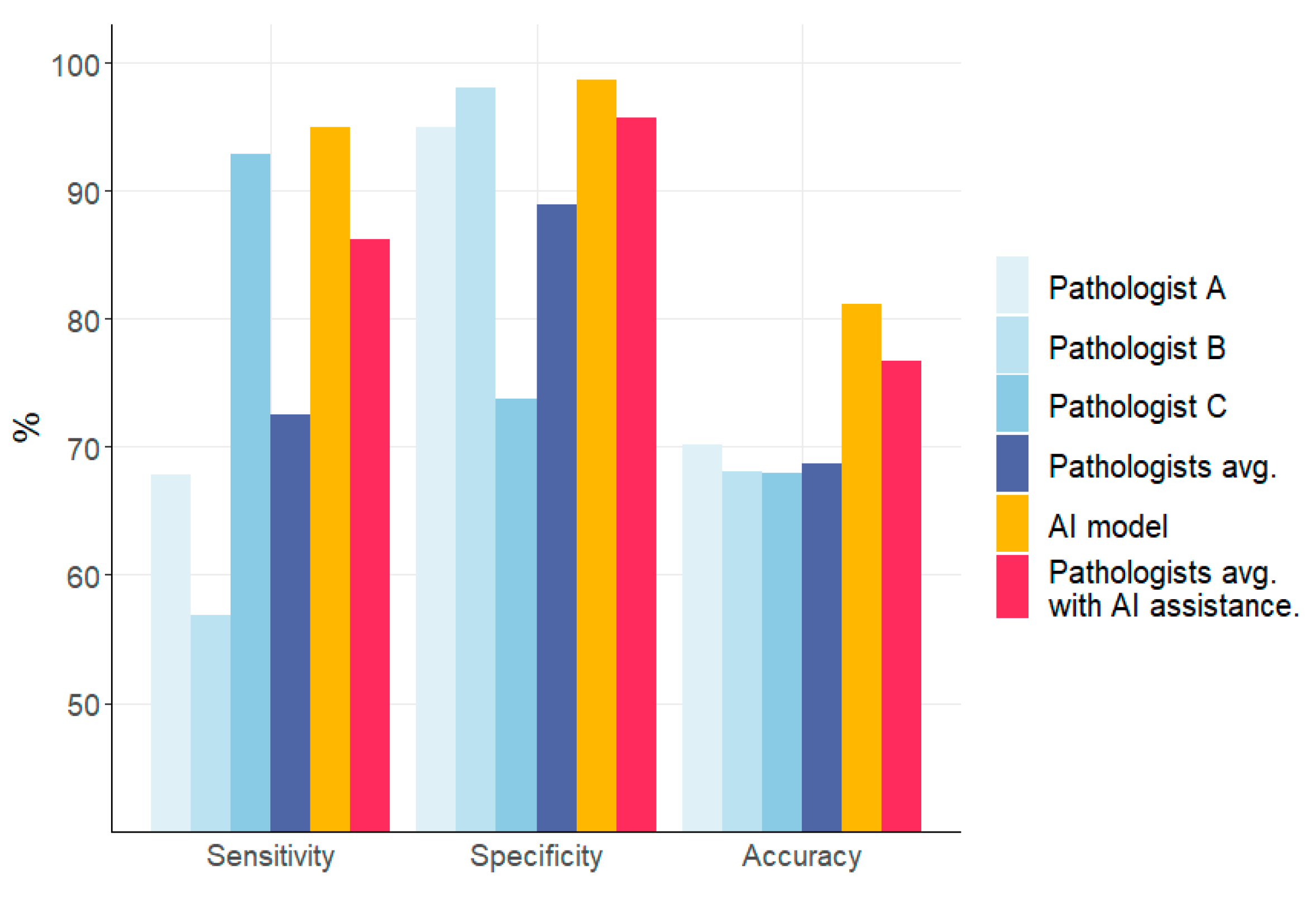
3.4. Comparison of the Performances of the AI Model and Pathologists
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Chen, M.-T.; Sun, H.-F.; Zhao, Y.; Fu, W.-Y.; Yang, L.-P.; Gao, S.-P.; Li, L.-D.; Jiang, H.-l.; Jin, W. Comparison of patterns and prognosis among distant metastatic breast cancer patients by age groups: A SEER population-based analysis. Sci. Rep. 2017, 7, 9254. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, X.; Wei, B.; Liu, N.; Li, Q.; Su, X. Mucinous breast carcinoma metastatic to thyroid gland: Report of a case diagnosed by fine-needle aspiration cytology. Diagn. Cytopathol. 2020, 48, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Cancer.Net. Breast Cancer—Metastatic: Statistics. Available online: https://www.cancer.net/cancer-types/breast-cancer-metastatic/statistics (accessed on 1 June 2023).
- Gayen, S. Malignant pleural effusion: Presentation, diagnosis, and management. Am. J. Med. 2022, 135, 1188–1192. [Google Scholar] [CrossRef]
- Shinohara, T.; Yamada, H.; Fujimori, Y.; Yamagishi, K. Malignant pleural effusion in breast cancer 12 years after mastectomy that was successfully treated with endocrine therapy. Am. J. Case Rep. 2013, 14, 184. [Google Scholar]
- Dermawan, J.K.T.; Policarpio-Nicolas, M.L. Malignancies in Pleural, Peritoneal, and Pericardial EffusionsA 17-Year Single-Institution Review From 30 085 Specimens. Arch. Pathol. Lab. Med. 2020, 144, 1086–1091. [Google Scholar] [CrossRef]
- Layfield, L.J.; Yang, Z.; Vazmitsel, M.; Zhang, T.; Esebua, M.; Schmidt, R. The international system for serous fluid cytopathology: Interobserver agreement. Diagn. Cytopathol. 2022, 50, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Kassirian, S.; Hinton, S.N.; Cuninghame, S.; Chaudhary, R.; Iansavitchene, A.; Amjadi, K.; Dhaliwal, I.; Zeman-Pocrnich, C.; Mitchell, M.A. Diagnostic sensitivity of pleural fluid cytology in malignant pleural effusions: Systematic review and meta-analysis. Thorax 2023, 78, 32–40. [Google Scholar] [CrossRef]
- Xie, X.; Fu, C.-C.; Lv, L.; Ye, Q.; Yu, Y.; Fang, Q.; Zhang, L.; Hou, L.; Wu, C. Deep convolutional neural network-based classification of cancer cells on cytological pleural effusion images. Mod. Pathol. 2022, 35, 609–614. [Google Scholar] [CrossRef]
- Bode-Lesniewska, B. Flow cytometry and effusions in lymphoproliferative processes and other hematologic neoplasias. Acta Cytol. 2016, 60, 354–364. [Google Scholar] [CrossRef]
- Thakur, N.; Alam, M.R.; Abdul-Ghafar, J.; Chong, Y. Recent Application of Artificial Intelligence in Non-Gynecological Cancer Cytopathology: A Systematic Review. Cancers 2022, 14, 3529. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Yang, B.; Wang, J.; Liu, M.; Yin, L.; Zheng, W.; Yin, Z.; Liu, C. Identifying malignant breast ultrasound images using ViT-patch. Appl. Sci. 2023, 13, 3489. [Google Scholar] [CrossRef]
- Zhuang, Y.; Chen, S.; Jiang, N.; Hu, H. An Effective WSSENet-Based Similarity Retrieval Method of Large Lung CT Image Databases. KSII Trans. Internet Inf. Syst. 2022, 16, 2359–2376. [Google Scholar]
- Wentzensen, N.; Lahrmann, B.; Clarke, M.A.; Kinney, W.; Tokugawa, D.; Poitras, N.; Locke, A.; Bartels, L.; Krauthoff, A.; Walker, J. Accuracy and efficiency of deep-learning–based automation of dual stain cytology in cervical Cancer screening. JNCI J. Natl. Cancer Inst. 2021, 113, 72–79. [Google Scholar] [CrossRef]
- Muralidaran, C.; Dey, P.; Nijhawan, R.; Kakkar, N. Artificial neural network in diagnosis of urothelial cell carcinoma in urine cytology. Diagn. Cytopathol. 2015, 43, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, P.; Mukherjee, T.; Barui, S.; Das, A.; Gangopadhyay, P. Artificial intelligence in cytopathology: A neural network to identify papillary carcinoma on thyroid fine-needle aspiration cytology smears. J. Pathol. Inform. 2018, 9, 43. [Google Scholar] [CrossRef]
- Gonzalez, D.; Dietz, R.L.; Pantanowitz, L. Feasibility of a deep learning algorithm to distinguish large cell neuroendocrine from small cell lung carcinoma in cytology specimens. Cytopathology 2020, 31, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Dey, P.; Logasundaram, R.; Joshi, K. Artificial neural network in diagnosis of lobular carcinoma of breast in fine-needle aspiration cytology. Diagn. Cytopathol. 2013, 41, 102–106. [Google Scholar] [CrossRef]
- Subbaiah, R.; Dey, P.; Nijhawan, R. Artificial neural network in breast lesions from fine-needle aspiration cytology smear. Diagn. Cytopathol. 2014, 42, 218–224. [Google Scholar] [CrossRef]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ailia, M.J.; Thakur, N.; Abdul-Ghafar, J.; Jung, C.K.; Yim, K.; Chong, Y. Current trend of artificial intelligence patents in digital pathology: A systematic evaluation of the patent landscape. Cancers 2022, 14, 2400. [Google Scholar] [CrossRef]
- Alam, M.R.; Abdul-Ghafar, J.; Yim, K.; Thakur, N.; Lee, S.H.; Jang, H.-J.; Jung, C.K.; Chong, Y. Recent applications of artificial intelligence from histopathologic image-based prediction of microsatellite instability in solid cancers: A systematic review. Cancers 2022, 14, 2590. [Google Scholar] [CrossRef]
- Dey, P. Artificial neural network in diagnostic cytology. CytoJournal 2022, 19, 27. [Google Scholar] [CrossRef]
- Shidham, V.B. Metastatic carcinoma in effusions. CytoJournal 2022, 19, 4. [Google Scholar] [CrossRef]
- Pereira, T.C.; Saad, R.S.; Liu, Y.; Silverman, J.F. The diagnosis of malignancy in effusion cytology: A pattern recognition approach. Adv. Anat. Pathol. 2006, 13, 174–184. [Google Scholar] [CrossRef]
- Chowdhuri, S.R.; Fetsch, P.; Squires, J.; Kohn, E.; Filie, A.C. Adenocarcinoma cells in effusion cytology as a diagnostic pitfall with potential impact on clinical management: A case report with brief review of immunomarkers. Diagn. Cytopathol. 2014, 42, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Shidham, V.B. The panorama of different faces of mesothelial cells. CytoJournal 2021, 18, 31. [Google Scholar] [CrossRef]
- Ashton, P.; Hollingsworth, A., Jr.; Johnston, W. The cytopathology of metastatic breast cancer. Acta Cytol. 1975, 19, 1–6. [Google Scholar]
- Borst, M.J.; Ingold, J.A. Metastatic patterns of invasive lobular versus invasive ductal carcinoma of the breast. Surgery 1993, 114, 637–642. [Google Scholar] [PubMed]
- Martinez, V.; Azzopardi, J. Invasive lobular carcinoma of the breast: Incidence and variants. Histopathology 1979, 3, 467–488. [Google Scholar] [CrossRef] [PubMed]
- Li, C.I.; Anderson, B.O.; Porter, P.; Holt, S.K.; Daling, J.R.; Moe, R.E. Changing incidence rate of invasive lobular breast carcinoma among older women. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2000, 88, 2561–2569. [Google Scholar] [CrossRef]
- Fisher, E.R.; Gregorio, R.M.; Fisher, B.; Redmond, C.; Vellios, F.; Sommers, S.C. The pathology of invasive breast cancer. A syllabus derived from findings of the National Surgical Adjuvant Breast Project (protocol no. 4). Cancer 1975, 36, 1–85. [Google Scholar] [CrossRef]
- Su, D.; Zhang, H.; Chen, H.; Yi, J.; Chen, P.-Y.; Gao, Y. Is Robustness the Cost of Accuracy?--A Comprehensive Study on the Robustness of 18 Deep Image Classification Models. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 631–648. [Google Scholar]
- De Logu, F.; Ugolini, F.; Maio, V.; Simi, S.; Cossu, A.; Massi, D.; Group, I.A.f.C.R.S.; Nassini, R.; Laurino, M. Recognition of cutaneous melanoma on digitized histopathological slides via artificial intelligence algorithm. Front. Oncol. 2020, 10, 1559. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, R.; Biswas, M.; Sen, S.; de Souza Júnior, L.A.; Papa, J.P.; Sarkar, R. A fuzzy distance-based ensemble of deep models for cervical cancer detection. Comput. Methods Programs Biomed. 2022, 219, 106776. [Google Scholar] [CrossRef] [PubMed]
- Whitney, J.; Corredor, G.; Janowczyk, A.; Ganesan, S.; Doyle, S.; Tomaszewski, J.; Feldman, M.; Gilmore, H.; Madabhushi, A. Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer. BMC Cancer 2018, 18, 610. [Google Scholar] [CrossRef] [PubMed]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.; Hermsen, M.; Manson, Q.F.; Balkenhol, M. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Xu, H.; Xu, J.; Gilmore, H.; Mandal, M.; Madabhushi, A. Multi-pass adaptive voting for nuclei detection in histopathological images. Sci. Rep. 2016, 6, 33985. [Google Scholar] [CrossRef]
- Couture, H.D.; Williams, L.A.; Geradts, J.; Nyante, S.J.; Butler, E.N.; Marron, J.; Perou, C.M.; Troester, M.A.; Niethammer, M. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 2018, 4, 30. [Google Scholar] [CrossRef]
- Al-Kofahi, Y.; Lassoued, W.; Lee, W.; Roysam, B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans. Biomed. Eng. 2009, 57, 841–852. [Google Scholar] [CrossRef]
- Basavanhally, A.N.; Ganesan, S.; Agner, S.; Monaco, J.P.; Feldman, M.D.; Tomaszewski, J.E.; Bhanot, G.; Madabhushi, A. Computerized image-based detection and grading of lymphocytic infiltration in HER2+ breast cancer histopathology. IEEE Trans. Biomed. Eng. 2009, 57, 642–653. [Google Scholar] [CrossRef]
- Senaras, C.; Sahiner, B.; Tozbikian, G.; Lozanski, G.; Gurcan, M.N. Creating synthetic digital slides using conditional generative adversarial networks: Application to Ki67 staining. In Proceedings of the Medical Imaging 2018: Digital Pathology, Houston, TX, USA, 11–12 February 2018; pp. 15–20. [Google Scholar]
- Hossain, M.S.; Hanna, M.G.; Uraoka, N.; Nakamura, T.; Edelweiss, M.; Brogi, E.; Hameed, M.R.; Yamaguchi, M.; Ross, D.S.; Yagi, Y. Automatic quantification of HER2 gene amplification in invasive breast cancer from chromogenic in situ hybridization whole slide images. J. Med. Imaging 2019, 6, 047501. [Google Scholar] [CrossRef]
- Abdollahi, J.; Davari, N.; Panahi, Y.; Gardaneh, M. Detection of Metastatic Breast Cancer from Whole-Slide Pathology Images Using an Ensemble Deep-Learning Method: Detection of Breast Cancer using Deep-Learning. Arch. Breast Cancer 2022, 9, 364–376. [Google Scholar] [CrossRef]
- Ren, W.; Zhu, Y.; Wang, Q.; Jin, H.; Guo, Y.; Lin, D. Deep Learning-Based Classification and Targeted Gene Alteration Prediction from Pleural Effusion Cell Block Whole-Slide Images. Cancers 2023, 15, 752. [Google Scholar] [CrossRef] [PubMed]
- Barwad, A.; Dey, P.; Susheilia, S. Artificial neural network in diagnosis of metastatic carcinoma in effusion cytology. Cytom. Part B: Clin. Cytom. 2012, 82, 107–111. [Google Scholar] [CrossRef]
- Tosun, A.B.; Yergiyev, O.; Kolouri, S.; Silverman, J.F.; Rohde, G.K. Detection of malignant mesothelioma using nuclear structure of mesothelial cells in effusion cytology specimens. Cytom. Part A 2015, 87, 326–333. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Mohseni, S.; Zarei, N.; Ragan, E.D. A multidisciplinary survey and framework for design and evaluation of explainable AI systems. ACM Trans. Interact. Intell. Syst. TiiS 2021, 11, 1–45. [Google Scholar] [CrossRef]
- Tosun, A.B.; Pullara, F.; Becich, M.J.; Taylor, D.; Fine, J.L.; Chennubhotla, S.C. Explainable AI (xAI) for anatomic pathology. Adv. Anat. Pathol. 2020, 27, 241–250. [Google Scholar] [CrossRef]
- Xie, Q.; Luong, M.-T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10687–10698. [Google Scholar]
- Pham, H.; Dai, Z.; Xie, Q.; Le, Q.V. Meta pseudo labels. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021; pp. 11557–11568. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]

| No. of Whole Slide Images (Image Patches) | ||||
|---|---|---|---|---|
| Training | Validation | Testing | Total | |
| Benign | 330 (12,389) | 44 (1545) | 43 (1568) | 417 (15,502) |
| Malignant | 111 (17,274) | 18 (719) | 23 (726) | 152 (18,179) |
| Total | 441 (29,663) | 62 (2264) | 66 (2294) | 569 (34,221) |
| Characteristics | Number of Cases (n = 569) |
|---|---|
| Age (median) | 18–104 (66) |
| Cytologic diagnosis | |
| Malignant lesions | 152 (26.7%) |
| Benign lesions | 417 (73.3%) |
| Preparation method | |
| Conventional | 564 (99.1%) |
| Liquid-based preparation | 5 (0.9%) |
| Z-stack layers | |
| 3-layers | 490 (86.1%) |
| 5-layers | 79 (13.9%) |
| Scanner | |
| Pannoramic Flash 250 III (3DHISTECH) | 540 (94.9%) |
| AT2 (Leica) | 2 (0.4%) |
| NanoZoomer S360 (Hamamatsu) | 27 (4.7%) |
| Accuracy | Sensitivity | Specificity | |
|---|---|---|---|
| Inception ResNet v2 | 0.9382 | 0.9777 | 0.9009 |
| Efficientnet-b1 | 0.9267 | 0.8855 | 0.9657 |
| ResNext50 | 0.9348 | 0.9679 | 0.9036 |
| Mobilenet v2 | 0.8948 | 0.9162 | 0.8745 |
| Densenet 121 | 0.9246 | 0.9218 | 0.9273 |
| ResNet 50 | 0.9192 | 0.8980 | 0.9392 |
| Actual Diagnosis | AI Diagnosis | |
|---|---|---|
| Total (n = 845) | Malignant (328) | Benign (517) |
| Malignant (338) | True positive (321) | False negative (17) |
| Benign (507) | False positive (7) | True negative (500) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.S.; Chong, Y.; Lee, Y.; Yim, K.; Seo, K.J.; Hwang, G.; Kim, D.; Gong, G.; Cho, N.H.; Yoo, C.W.; et al. Deep Learning-Based Computational Cytopathologic Diagnosis of Metastatic Breast Carcinoma in Pleural Fluid. Cells 2023, 12, 1847. https://doi.org/10.3390/cells12141847
Park HS, Chong Y, Lee Y, Yim K, Seo KJ, Hwang G, Kim D, Gong G, Cho NH, Yoo CW, et al. Deep Learning-Based Computational Cytopathologic Diagnosis of Metastatic Breast Carcinoma in Pleural Fluid. Cells. 2023; 12(14):1847. https://doi.org/10.3390/cells12141847
Chicago/Turabian StylePark, Hong Sik, Yosep Chong, Yujin Lee, Kwangil Yim, Kyung Jin Seo, Gisu Hwang, Dahyeon Kim, Gyungyub Gong, Nam Hoon Cho, Chong Woo Yoo, and et al. 2023. "Deep Learning-Based Computational Cytopathologic Diagnosis of Metastatic Breast Carcinoma in Pleural Fluid" Cells 12, no. 14: 1847. https://doi.org/10.3390/cells12141847
APA StylePark, H. S., Chong, Y., Lee, Y., Yim, K., Seo, K. J., Hwang, G., Kim, D., Gong, G., Cho, N. H., Yoo, C. W., & Choi, H. J. (2023). Deep Learning-Based Computational Cytopathologic Diagnosis of Metastatic Breast Carcinoma in Pleural Fluid. Cells, 12(14), 1847. https://doi.org/10.3390/cells12141847








