Circadian Organelles: Rhythms at All Scales
Abstract
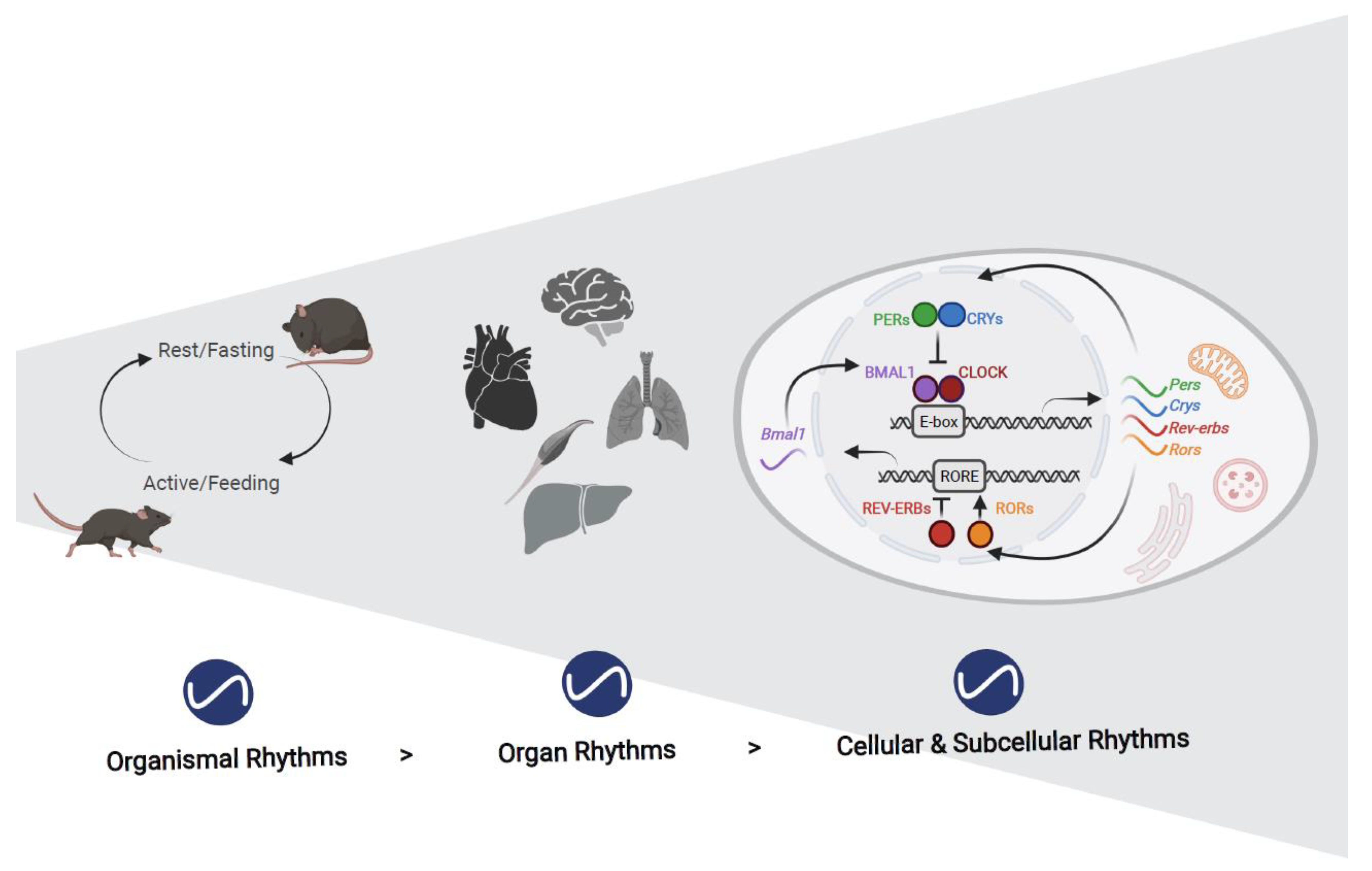
:1. Introduction
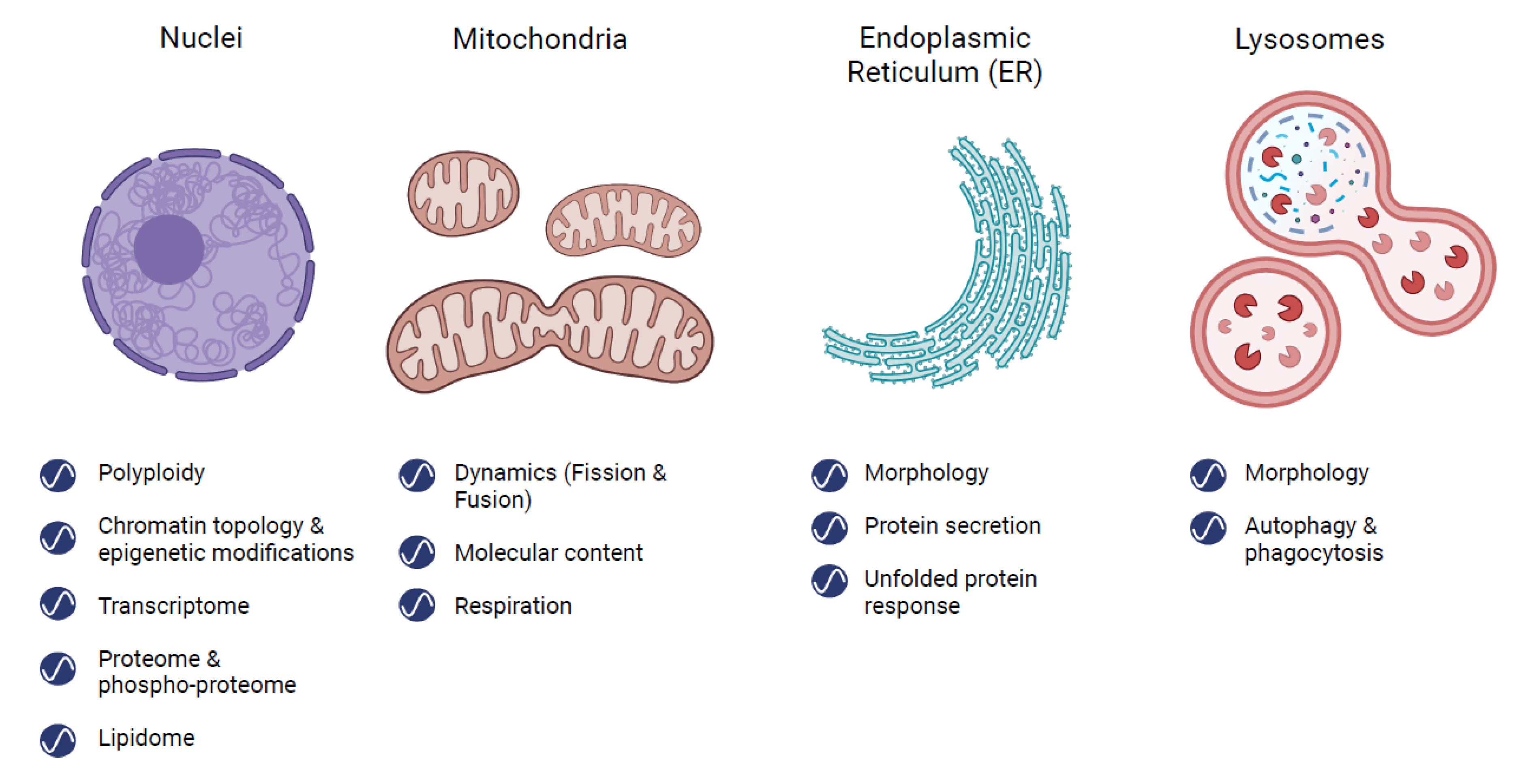
2. Rhythms in Organelles
2.1. Nucleus
2.2. Mitochondria
2.3. Endoplasmic Reticulum (ER)
2.4. Lysosomes
3. Discussion
3.1. The Importance of Circadian Organelle Research
3.2. The Challenges of Circadian Organelle Research
3.3. Potential and Speculative Implications of Circadian Organelle Research
4. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pittendrigh, C.S. On temperature independence in the clock system controlling emergence time in drosophila. Proc. Natl. Acad. Sci. USA 1954, 40, 1018–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittendrigh, C.S. Temporal organization: Reflections of a darwinian clock-watcher. Annu. Rev. Physiol. 1993, 55, 16–54. [Google Scholar] [CrossRef] [PubMed]
- Aviram, R.; Manella, G. A metaphor that keeps on ticking: The ‘clock’ as a driving force in the history of chronobiology research. PTPBio 2020, 12. [Google Scholar] [CrossRef]
- Moore, R.Y.; Eichler, V.B. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972, 42, 201–206. [Google Scholar] [CrossRef]
- Stephan, F.K.; Zucker, I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. USA 1972, 69, 1583–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagita, K.; Tamanini, F.; Van der Horst, G.T.; Okamura, H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 2001, 292, 278–281. [Google Scholar] [CrossRef]
- Yoo, S.H.; Yamazaki, S.; Lowrey, P.L.; Shimomura, K.; Ko, C.H.; Buhr, E.D.; Siepka, S.M.; Hong, H.K.; Oh, W.J.; Yoo, O.J.; et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 5339–5346. [Google Scholar] [CrossRef] [Green Version]
- Nagoshi, E.; Saini, C.; Bauer, C.; Laroche, T.; Naef, F.; Schibler, U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 2004, 119, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Marcacci, L.; Schibler, U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 2000, 10, 1291–1294. [Google Scholar] [CrossRef] [Green Version]
- Swan, J.A.; Golden, S.S.; LiWang, A.; Partch, C.L. Structure, function, and mechanism of the core circadian clock in cyanobacteria. J. Biol. Chem. 2018, 293, 5026–5034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konopka, R.J.; Benzer, S. Clock mutants of drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1971, 68, 2112–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 1990, 343, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Bargiello, T.A.; Young, M.W. Molecular genetics of a biological Clock in Drosophila. Proc. Natl. Acad. Sci. USA 1984, 81, 2142–2146. [Google Scholar] [CrossRef] [Green Version]
- Bargiello, T.A.; Jackson, F.R.; Young, M.W. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 1984, 312, 752–754. [Google Scholar] [CrossRef]
- Brown, S.A.; Kowalska, E.; Dallmann, R. (Re)inventing the circadian feedback loop. Dev. Cell 2012, 22, 477–487. [Google Scholar] [CrossRef] [Green Version]
- Debruyne, J.P.; Noton, E.; Lambert, C.M.; Maywood, E.S.; Weaver, D.R.; Reppert, S.M. A clock shock: Mouse clock is not required for circadian oscillator function. Neuron 2006, 50, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Vitaterna, M.H.; King, D.P.; Chang, A.M.; Kornhauser, J.M.; Lowrey, P.L.; McDonald, J.D.; Dove, W.F.; Pinto, L.H.; Turek, F.W.; Takahashi, J.S. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 1994, 264, 719–725. [Google Scholar] [CrossRef] [Green Version]
- Buhr, E.D.; Takahashi, J.S. Molecular Components of the Mammalian Circadian Clock. In Circadian Clocks. Handbook of Experi-mental Pharmacology; Kramer, A., Merrow, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 217, pp. 2–27. [Google Scholar]
- Dibner, C.; Schibler, U.; Albrecht, U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010, 72, 517–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honma, S.; Kawamoto, T.; Takagi, Y.; Fujimoto, K.; Sato, F.; Noshiro, M.; Kato, Y.; Honma, K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 2002, 419, 841–844. [Google Scholar] [CrossRef]
- Kato, Y.; Kawamoto, T.; Fujimoto, K.; Noshiro, M. Dec1/Stra13/Sharp2 and Dec2/Sharp1 coordinate physiological processes, including circadian rhythms in response to environmental stimuli. Curr. Top. Dev. Biol. 2014, 110, 339–372. [Google Scholar]
- Gachon, F. Physiological Function of PARbZip circadian clock-controlled transcription factors. Ann. Med. 2007, 39, 562–571. [Google Scholar] [CrossRef]
- Edgar, R.S.; Green, E.W.; Zhao, Y.; Van Ooijen, G.; Olmedo, M.; Qin, X.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, J.S.; Reddy, A.B. Circadian clocks in human red blood cells. Nature 2011, 469, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Halberg, F. Physiologic 24-hour periodicity; general and procedural considerations with reference to the adrenal cycle. Int. Z Vitam. Beih. 1959, 10, 225–296. [Google Scholar]
- Vitaterna, M.H.; Takahashi, J.S.; Turek, F.W. Overview of circadian rhythms. Alcohol Res. Health 2001, 25, 85–93. [Google Scholar] [PubMed]
- Dibner, C. The importance of being rhythmic: Living in harmony with your body clocks. Acta Physiol. 2020, 228, e13281. [Google Scholar] [CrossRef] [PubMed]
- Schibler, U.; Gotic, I.; Saini, C.; Gos, P.; Curie, T.; Emmenegger, Y.; Sinturel, F.; Gosselin, P.; Gerber, A.; Fleury-Olela, F.; et al. Clock-talk: Interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb. Symp. Quant. Biol. 2015, 80, 223–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balsalobre, A.; Brown, S.A.; Marcacci, L.; Tronche, F.; Kellendonk, C.; Reichardt, H.M.; Schutz, G.; Schibler, U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2000, 289, 2344–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirota, T.; Okano, T.; Kokame, K.; Shirotani-Ikejima, H.; Miyata, T.; Fukada, Y. Glucose down-regulates Per1 and Per2mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J. Biol. Chem. 2002, 277, 44244–44251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamajuku, D.; Inagaki, T.; Haruma, T.; Okubo, S.; Kataoka, Y.; Kobayashi, S.; Ikegami, K.; Laurent, T.; Kojima, T.; Noutomi, K.; et al. Real-time monitoring in three-dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci. Rep. 2012, 2, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Adamovich, Y.; Ladeuix, B.; Golik, M.; Koeners, M.P.; Asher, G. Rhythmic oxygen levels reset circadian clocks through Hif1α. Cell Metab. 2017, 25, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Adamovich, Y.; Ladeuix, B.; Sobel, J.; Manella, G.; Neufeld-Cohen, A.; Assadi, M.H.; Golik, M.; Kuperman, Y.; Tarasiuk, A.; Koeners, M.P.; et al. Oxygen and carbon dioxide rhythms are circadian clock controlled and differentially directed by behavioral signals. Cell Metab. 2019, 29, 1092–1103. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, Y.; Saito, K. A morphometric study of 24-hour variations in subcellular structures of the rat pancreatic acinar cell. Cell Tissue Res. 1982, 226, 609–620. [Google Scholar] [CrossRef]
- Uchiyama, Y.; Asari, A. A morphometric study of the variations in subcellular structures of rat hepatocytes during 24 hours. Cell Tissue Res. 1984, 236, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Aviram, R.; Manella, G.; Asher, G. The liver by day and by night. J. Hepatol. 2021, 74, 1240–1242. [Google Scholar] [CrossRef]
- Gentric, G.; Celton-Morizur, S.; Desdouets, C. Polyploidy and liver proliferation. Clin. Res. Hepatol. Gastroenterol. 2012, 36, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Duncan, A.W. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin. Cell Dev. Biol. 2013, 24, 347–356. [Google Scholar] [CrossRef]
- Wang, J.; Mauvoisin, D.; Martin, E.; Atger, F.; Galindo, A.N.; Dayon, L.; Sizzano, F.; Palini, A.; Kussmann, M.; Waridel, P.; et al. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 2017, 25, 102–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, H.W.; Doi, M.; Fustin, J.M.; Chen, H.; Murase, K.; Maeda, Y.; Hayashi, H.; Tanaka, R.; Sugawa, M.; Mizukuchi, N.; et al. Circadian clock regulates hepatic polyploidy by modulating Mkp1-Erk1/2 signaling pathway. Nat. Commun. 2017, 8, 2238. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lazar, M.A. Transcriptional control of circadian rhythms and metabolism: A matter of time and space. Endocr. Rev. 2020, 41, 707–732. [Google Scholar] [CrossRef]
- Yeung, J.; Naef, F. Rhythms of the genome: Circadian dynamics from chromatin topology, tissue-specific gene expression, to behavior. Trends Genet. 2018, 34, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Arnal, L.; Hakim, O.; Patel, V.R.; Baldi, P.; Hager, G.L.; Sassone-Corsi, P. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat. Struct. Mol. Biol. 2013, 20, 1206–1213. [Google Scholar] [CrossRef]
- Zhao, H.; Sifakis, E.G.; Sumida, N.; Millan-Arino, L.; Scholz, B.A.; Svensson, J.P.; Chen, X.; Ronnegren, A.L.; Mallet de Lima, C.D.; Varnoosfaderani, F.S.; et al. PARP1-and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol. Cell 2015, 59, 984–997. [Google Scholar] [CrossRef] [Green Version]
- Asher, G.; Reinke, H.; Altmeyer, M.; Gutierrez-Arcelus, M.; Hottiger, M.O.; Schibler, U. Poly (ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 2010, 142, 943–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.T.; Zhang, L.; Lin, X.; Zhang, L.C.; Garcia, V.E.; Tsai, C.W.; Ptacek, L.; Fu, Y.H. Nuclear envelope protein man1 regulates clock through Bmal1. eLife 2014, 3, e02981. [Google Scholar] [CrossRef]
- Bu, B.; He, W.; Song, L.; Zhang, L. Nuclear envelope protein Man1 regulates the drosophila circadian clock via period. Neurosci. Bull. 2019, 35, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Furlan-Magaril, M.; Ando-Kuri, M.; Arzate-Mejia, R.G.; Morf, J.; Cairns, J.; Roman-Figueroa, A.; Tenorio-Hernandez, L.; Poot-Hernandez, A.C.; Andrews, S.; Varnai, C.; et al. The global and promoter-centric 3d genome organization temporally resolved during a circadian cycle. Genome Biol. 2021, 22, 162. [Google Scholar] [CrossRef]
- Brunet, A.; Forsberg, F.; Fan, Q.; Saether, T.; Collas, P. Nuclear lamin B1 interactions with chromatin during the circadian cycle are uncoupled from periodic gene expression. Front. Genet. 2019, 10, 917. [Google Scholar] [CrossRef] [Green Version]
- Pacheco-Bernal, I.; Becerril-Perez, F.; Aguilar-Arnal, L. Circadian rhythms in the three-dimensional genome: Implications of chromatin interactions for cyclic transcription. Clin. Epigenetics 2019, 11, 79. [Google Scholar] [CrossRef]
- Kim, Y.H.; Marhon, S.A.; Zhang, Y.; Steger, D.J.; Won, K.J.; Lazar, M.A. Rev-erbα dynamically modulates chromatin looping to control circadian gene transcription. Science 2018, 359, 1274–1277. [Google Scholar] [CrossRef] [Green Version]
- Mermet, J.; Yeung, J.; Hurni, C.; Mauvoisin, D.; Gustafson, K.; Jouffe, C.; Nicolas, D.; Emmenegger, Y.; Gobet, C.; Franken, P.; et al. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev. 2018, 32, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Mermet, J.; Yeung, J.; Naef, F. Oscillating and stable genome topologies underlie hepatic physiological rhythms during the circadian cycle. PLoS Genet. 2021, 17, e1009350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [Green Version]
- Doherty, C.J.; Kay, S.A. Circadian control of global gene expression patterns. Annu. Rev. Genet. 2010, 44, 419–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, M.; Becquet, D.; Franc, J.L.; Francois-Bellan, A.M. Circadian processes in the RNA life cycle. Wiley Interdiscip. Rev. RNA 2018, 9, e1467. [Google Scholar] [CrossRef]
- Mauvoisin, D.; Wang, J.; Jouffe, C.; Martin, E.; Atger, F.; Waridel, P.; Quadroni, M.; Gachon, F.; Naef, F. Circadian clock-dependent and-independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. USA 2014, 111, 167–172. [Google Scholar] [CrossRef] [Green Version]
- Robles, M.S.; Cox, J.; Mann, M. In-Vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014, 10, e1004047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robles, M.S.; Humphrey, S.J.; Mann, M. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 2017, 25, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Song, L.; Liu, M.; Ge, R.; Zhou, Q.; Liu, W.; Li, R.; Qie, J.; Zhen, B.; Wang, Y.; et al. A proteomics landscape of circadian clock in mouse liver. Nat. Commun. 2018, 9, 1553. [Google Scholar] [CrossRef] [PubMed]
- Korge, S.; Maier, B.; Bruning, F.; Ehrhardt, L.; Korte, T.; Mann, M.; Herrmann, A.; Robles, M.S.; Kramer, A. The non-classical nuclear import carrier transportin 1 modulates circadian rhythms through its effect on PER1 nuclear localization. PLoS Genet. 2018, 14, e1007189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Shen, Y.; Francey, L.J.; Ramanathan, C.; Sehgal, A.; Liu, A.C.; Hogenesch, J.B. The NRON complex controls circadian clock function through regulated per and CRY nuclear translocation. Sci. Rep. 2019, 9, 11883. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jang, A.R.; Francey, L.J.; Sehgal, A.; Hogenesch, J.B. KPNB1 mediates PER/CRY nuclear translocation and circadian clock function. eLife 2015, 4, e08647. [Google Scholar] [CrossRef]
- Chua, E.C.; Shui, G.; Lee, I.T.; Lau, P.; Tan, L.C.; Yeo, S.C.; Lam, B.D.; Bulchand, S.; Summers, S.A.; Puvanendran, K.; et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 14468–14473. [Google Scholar] [CrossRef] [Green Version]
- Kasukawa, T.; Sugimoto, M.; Hida, A.; Minami, Y.; Mori, M.; Honma, S.; Honma, K.; Mishima, K.; Soga, T.; Ueda, H.R. Human blood metabolite timetable indicates internal body time. Proc. Natl. Acad. Sci. USA 2012, 109, 15036–15041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loizides-Mangold, U.; Perrin, L.; Vandereycken, B.; Betts, J.A.; Walhin, J.P.; Templeman, I.; Chanon, S.; Weger, B.D.; Durand, C.; Robert, M.; et al. Lipidomics reveals diurnal lipid oscillations in human skeletal muscle persisting in cellular myotubes cultured in vitro. Proc. Natl. Acad. Sci. USA 2017, 114, E8565–E8574. [Google Scholar] [CrossRef] [Green Version]
- Adamovich, Y.; Rousso-Noori, L.; Zwighaft, Z.; Neufeld-Cohen, A.; Golik, M.; Kraut-Cohen, J.; Wang, M.; Han, X.; Asher, G. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014, 19, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Aviram, R.; Manella, G.; Kopelman, N.; Neufeld-Cohen, A.; Zwighaft, Z.; Elimelech, M.; Adamovich, Y.; Golik, M.; Wang, C.; Han, X.; et al. Lipidomics analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol. Cell 2016, 62, 636–648. [Google Scholar] [CrossRef] [PubMed]
- Manella, G.; Asher, G. The circadian nature of mitochondrial biology. Front. Endocrinol. 2016, 7, 162. [Google Scholar] [CrossRef] [Green Version]
- Sardon Puig, L.; Valera-Alberni, M.; Canto, C.; Pillon, N.J. Circadian rhythms and mitochondria: Connecting the dots. Front. Genet. 2018, 9, 452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wai, T.; Langer, T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 2016, 27, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Liesa, M.; Shirihai, O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013, 17, 491–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchiyama, Y. Circadian alterations in tubular structures on the outer mitochondrial membrane of rat hepatocytes. Cell Tissue Res. 1981, 214, 519–527. [Google Scholar] [CrossRef]
- Jacobi, D.; Liu, S.; Burkewitz, K.; Kory, N.; Knudsen, N.H.; Alexander, R.K.; Unluturk, U.; Li, X.; Kong, X.; Hyde, A.L.; et al. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 2015, 22, 709–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitt, K.; Grimm, A.; Dallmann, R.; Oettinghaus, B.; Restelli, L.M.; Witzig, M.; Ishihara, N.; Mihara, K.; Ripperger, J.A.; Albrecht, U.; et al. Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab. 2018, 27, 657–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Cheng, Q.; Hua, B.; Cai, T.; Lin, J.; Yuan, G.; Yan, Z.; Li, X.; Sun, N.; Lu, C.; et al. Circadian gene clock regulates mitochondrial morphology and functions by posttranscriptional way. bioRxiv 2018, 365452. [Google Scholar] [CrossRef] [Green Version]
- Andrews, J.L.; Zhang, X.; McCarthy, J.J.; McDearmon, E.L.; Hornberger, T.A.; Russell, B.; Campbell, K.S.; Arbogast, S.; Reid, M.B.; Walker, J.R.; et al. Clock and BMAL1 Regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. USA 2010, 107, 19090–19095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohsaka, A.; Das, P.; Hashimoto, I.; Nakao, T.; Deguchi, Y.; Gouraud, S.S.; Waki, H.; Muragaki, Y.; Maeda, M. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS ONE 2014, 9, e112811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thul, P.J.; Akesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Blal, H.A.; Alm, T.; Asplund, A.; Bjork, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356. [Google Scholar] [CrossRef]
- Roger, A.J.; Munoz-Gomez, S.A.; Kamikawa, R. The origin and diversification of mitochondria. Curr Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Li, C.; Qi, X.; Song, Z.; Wu, J.; Hughes, M.E.; Li, X. The daily rhythms of mitochondrial gene expression and oxidative stress regulation are altered by aging in the mouse liver. Chronobiol. Int. 2015, 32, 1254–1263. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rey, G.; Cesbron, F.; Rougemont, J.; Reinke, H.; Brunner, M.; Naef, F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011, 9, e1000595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neufeld-Cohen, A.; Robles, M.S.; Aviram, R.; Manella, G.; Adamovich, Y.; Ladeuix, B.; Nir, D.; Rousso-Noori, L.; Kuperman, Y.; Golik, M.; et al. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by period proteins. Proc. Natl. Acad. Sci. USA 2016, 113, E1673–E1682. [Google Scholar] [CrossRef] [Green Version]
- Atger, F.; Gobet, C.; Marquis, J.; Martin, E.; Wang, J.; Weger, B.; Lefebvre, G.; Descombes, P.; Naef, F.; Gachon, F. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. USA 2015, 112, E6579–E6588. [Google Scholar] [CrossRef] [Green Version]
- Janich, P.; Arpat, A.B.; Castelo-Szekely, V.; Lopes, M.; Gatfield, D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 2015, 25, 1848–1859. [Google Scholar] [CrossRef] [Green Version]
- Peek, C.B.; Affinati, A.H.; Ramsey, K.M.; Kuo, H.Y.; Yu, W.; Sena, L.A.; Ilkayeva, O.; Marcheva, B.; Kobayashi, Y.; Omura, C.; et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 2013, 342, 1243417. [Google Scholar] [CrossRef] [Green Version]
- Mauvoisin, D.; Atger, F.; Dayon, L.; Nunez Galindo, A.; Wang, J.; Martin, E.; Da Silva, L.; Montoliu, I.; Collino, S.; Martin, F.P.; et al. Circadian and feeding rhythms orchestrate the diurnal liver acetylome. Cell Rep. 2017, 20, 1729–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masri, S.; Patel, V.R.; Eckel-Mahan, K.L.; Peleg, S.; Forne, I.; Ladurner, A.G.; Baldi, P.; Imhof, A.; Sassone-Corsi, P. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc. Natl. Acad. Sci. USA 2013, 110, 3339–3344. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef]
- Reinke, H.; Asher, G. Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 2019, 20, 227–241. [Google Scholar] [CrossRef]
- Asher, G.; Sassone-Corsi, P. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell 2015, 161, 84–92. [Google Scholar] [CrossRef] [Green Version]
- Cela, O.; Scrima, R.; Pazienza, V.; Merla, G.; Benegiamo, G.; Augello, B.; Fugetto, S.; Menga, M.; Rubino, R.; Fuhr, L.; et al. Clock genes-dependent acetylation of complex I sets rhythmic activity of mitochondrial OxPhos. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Scrima, R.; Cela, O.; Merla, G.; Augello, B.; Rubino, R.; Quarato, G.; Fugetto, S.; Menga, M.; Fuhr, L.; Relogio, A.; et al. Clock-genes and mitochondrial respiratory activity: Evidence of a reciprocal interplay. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1857, 1344–1351. [Google Scholar] [CrossRef]
- Pacelli, C.; Rotundo, G.; Lecce, L.; Menga, M.; Bidollari, E.; Scrima, R.; Cela, O.; Piccoli, C.; Cocco, T.; Vescovi, A.L.; et al. Parkin mutation affects clock gene-dependent energy metabolism. Int. J. Mol. Sci. 2019, 20, 2772. [Google Scholar] [CrossRef] [Green Version]
- Chedid, A.; Nair, V. diurnal rhythm in endoplasmic reticulum of rat liver: Electron microscopic study. Science 1972, 175, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, Y. Rhythms in morphology and function of hepatocytes. J. Gastroenterol. Hepatol. 1990, 5, 321–333. [Google Scholar] [CrossRef]
- Braakman, I.; Bulleid, N.J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 2011, 80, 71–99. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Garva, R.; Pickard, A.; Yeung, C.C.; Mallikarjun, V.; Swift, J.; Holmes, D.F.; Calverley, B.; Lu, Y.; Adamson, A.; et al. Circadian control of the secretory pathway maintains collagen homeostasis. Nat. Cell Biol. 2020, 22, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Heinemeier, K.M.; Schjerling, P.; Heinemeier, J.; Magnusson, S.P.; Kjaer, M. Lack of tissue renewal in human adult achilles tendon is revealed by nuclear bomb (14) C. FASEB J. 2013, 27, 2074–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hetz, C.; Papa, F.R. The unfolded protein response and cell fate control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Cretenet, G.; Le Clech, M.; Gachon, F. Circadian clock-coordinated 12 hr period rhythmic activation of the IRE1α pathway controls lipid metabolism in mouse liver. Cell Metab. 2010, 11, 47–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Ballance, H.; Meng, H.; Gonzalez, N.; Kim, S.M.; Abdurehman, L.; York, B.; Chen, X.; Schnytzer, Y.; Levy, O.; et al. 12-h clock regulation of genetic information flow by XBP1s. PLoS Biol. 2020, 18, e3000580. [Google Scholar] [CrossRef]
- Meng, H.; Gonzales, N.M.; Lonard, D.M.; Putluri, N.; Zhu, B.; Dacso, C.C.; York, B.; O’Malley, B.W. XBP1 links the 12-hour clock to nafld and regulation of membrane fluidity and lipid homeostasis. Nat. Commun. 2020, 11, 6215. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Zhang, Q.; Pan, Y.; Mace, E.M.; York, B.; Antoulas, A.C.; Dacso, C.C.; O’Malley, B.W. A cell-autonomous mammalian 12 hr clock coordinates metabolic and stress rhythms. Cell Metab. 2017, 25, 1305–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bu, Y.; Yoshida, A.; Chitnis, N.; Altman, B.J.; Tameire, F.; Oran, A.; Gennaro, V.; Armeson, K.E.; McMahon, S.B.; Wertheim, G.B.; et al. A PERK-miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat. Cell Biol. 2018, 20, 104–115. [Google Scholar] [CrossRef]
- Koyanagi, S.; Hamdan, A.M.; Horiguchi, M.; Kusunose, N.; Okamoto, A.; Matsunaga, N.; Ohdo, S. cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J. Biol. Chem. 2011, 286, 32416–32423. [Google Scholar] [CrossRef] [Green Version]
- De Duve, C.; Pressman, B.C.; Gianetto, R.; Wattiaux, R.; Appelmans, F. Tissue fractionation studies. 6. intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 1955, 60, 604–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Duve, C. The lysosome turns fifty. Nat. Cell Biol. 2005, 7, 847–849. [Google Scholar] [CrossRef]
- Novikoff, A.B.; Beaufay, H.; De Duve, C. Electron microscopy of lysosomerich fractions from rat liver. J. Biophys. Biochem. Cytol. 1956, 2, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Ohsumi, Y. Historical Landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, U.; Strauss, P. Autophagic vacuoles in heart muscle and liver. a comparative morphometric study including circadian variations in meal-fed rats. J. Mol. Cell Cardiol. 1981, 13, 37–49. [Google Scholar] [CrossRef]
- Ma, D.; Panda, S.; Lin, J.D. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 2011, 30, 4642–4651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryzhikov, M.; Ehlers, A.; Steinberg, D.; Xie, W.; Oberlander, E.; Brown, S.; Gilmore, P.E.; Townsend, R.R.; Lane, W.S.; Dolinay, T.; et al. Diurnal rhythms spatially and temporally organize autophagy. Cell Rep. 2019, 26, 1880–1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabinovich-Nikitin, I.; Rasouli, M.; Reitz, C.J.; Posen, I.; Margulets, V.; Dhingra, R.; Khatua, T.N.; Thliveris, J.A.; Martino, T.A.; Kirshenbaum, L.A. Mitochondrial autophagy and cell survival is regulated by the circadian clock gene in cardiac myocytes during ischemic stress. Autophagy 2021, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, S.; Molusky, M.M.; Lin, J.D. Circadian autophagy rhythm: A link between clock and metabolism? Trends Endocrinol. Metab. 2012, 23, 319–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Xu, Z.; Cai, Y.; Zeng, S.; Peng, B.; Ren, X.; Yan, Y.; Gong, Z. Rheostatic balance of circadian rhythm and autophagy in metabolism and disease. Front. Cell Dev. Biol. 2020, 8, 616434. [Google Scholar] [CrossRef]
- Milicevic, N.; Mazzaro, N.; De Bruin, I.; Wils, E.; Ten Brink, J.; Asbroek, A.T.; Mendoza, J.; Bergen, A.; Felder-Schmittbuhl, M.P. Rev-Erbα and photoreceptor outer segments modulate the circadian clock in retinal pigment epithelial cells. Sci. Rep. 2019, 9, 11790. [Google Scholar] [CrossRef] [Green Version]
- Rosbash, M. A 50-year personal journey: Location, gene expression, and circadian rhythms. Cold Spring Harb. Perspect. Biol. 2017, 9, a032516. [Google Scholar] [CrossRef] [Green Version]
- Aviram, R.; Wang, C.; Han, X.; Asher, G. A lipidomics view of circadian biology. Methods Mol. Biol. 2021, 2130, 157–168. [Google Scholar] [PubMed]
- Radhakrishnan, A.; Goldstein, J.L.; McDonald, J.G.; Brown, M.S. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab. 2008, 8, 512–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, M.P.; Attardi, G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 1989, 246, 500–503. [Google Scholar] [CrossRef]
- Eelderink-Chen, Z.; Bosman, J.; Sartor, F.; Dodd, A.N.; Kovacs, A.T.; Merrow, M. A circadian clock in a nonphotosynthetic prokaryote. Sci. Adv. 2021, 7, eabe2086. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.E.; Golden, S.S. Circadian rhythms in cyanobacteria. Microbiol. Mol. Biol. Rev. 2015, 79, 373–385. [Google Scholar] [CrossRef] [Green Version]
- Manella, G.; Sabath, E.; Aviram, R.; Dandavate, V.; Ezagouri, S.; Golik, M.; Adamovich, Y.; Asher, G. The liver-clock coordinates rhythmicity of peripheral tissues in response to feeding. Nat. Metab. 2021, 3, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.A. Retrograde signalling as an informant of circadian timing. New Phytol. 2019, 221, 1749–1753. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aviram, R.; Adamovich, Y.; Asher, G. Circadian Organelles: Rhythms at All Scales. Cells 2021, 10, 2447. https://doi.org/10.3390/cells10092447
Aviram R, Adamovich Y, Asher G. Circadian Organelles: Rhythms at All Scales. Cells. 2021; 10(9):2447. https://doi.org/10.3390/cells10092447
Chicago/Turabian StyleAviram, Rona, Yaarit Adamovich, and Gad Asher. 2021. "Circadian Organelles: Rhythms at All Scales" Cells 10, no. 9: 2447. https://doi.org/10.3390/cells10092447
APA StyleAviram, R., Adamovich, Y., & Asher, G. (2021). Circadian Organelles: Rhythms at All Scales. Cells, 10(9), 2447. https://doi.org/10.3390/cells10092447





