Drosophila, an Integrative Model to Study the Features of Muscle Stem Cells in Development and Regeneration
Abstract
1. Introduction
2. Making a Muscle Stem Cell
2.1. Specification and Positioning of the AMPs
2.2. The Control of the AMPs Diversity
3. Role of the Microenvironment in Muscle Stem Cell Maintenance and Activation
3.1. Connecting to the Muscles; ‘Homing Behavior’
3.2. Muscle-Driven Insulin Signal Reactivates Dormant AMPs
3.3. Interplay between the AMPs and Motor Neurons
3.4. Signals from the Epithelial Tissue Maintain the Undifferentiated AMPs and Promote Their Proliferation
4. Drosophila, a New Model to Study Adult MuSCs
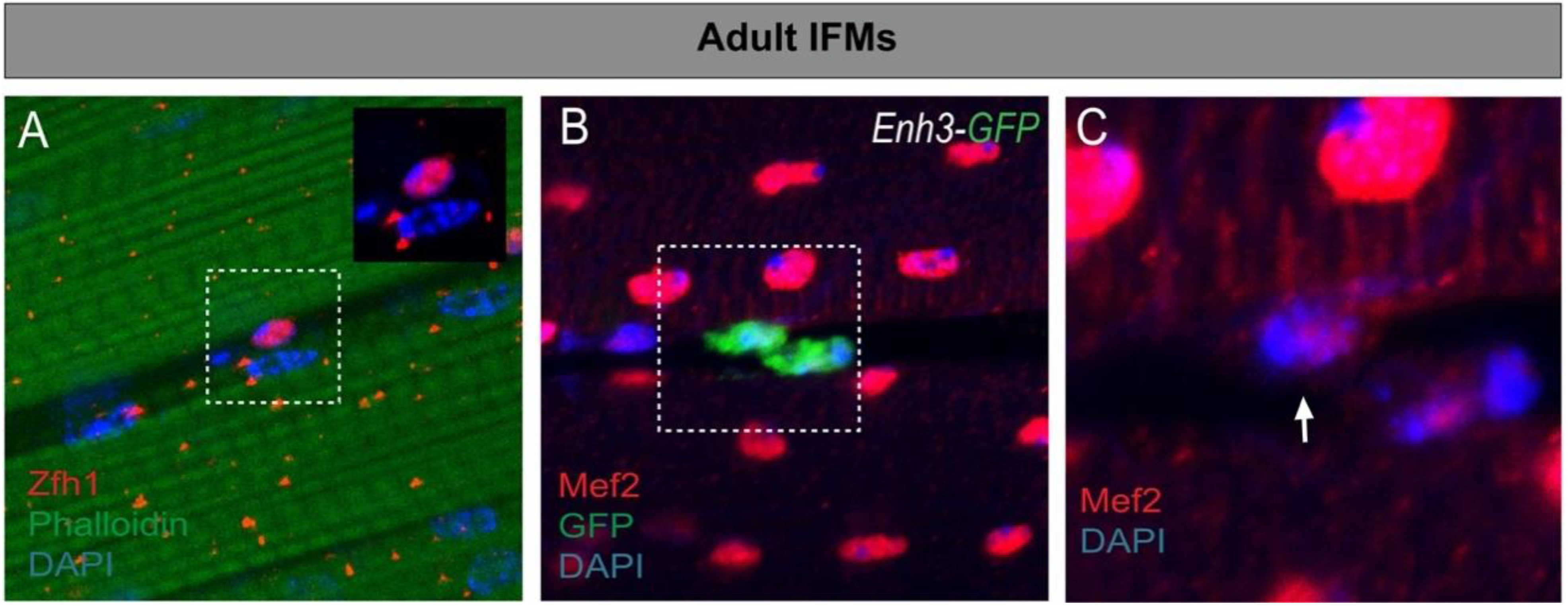
4.1. Characterization of the Drosophila Satellite Cells
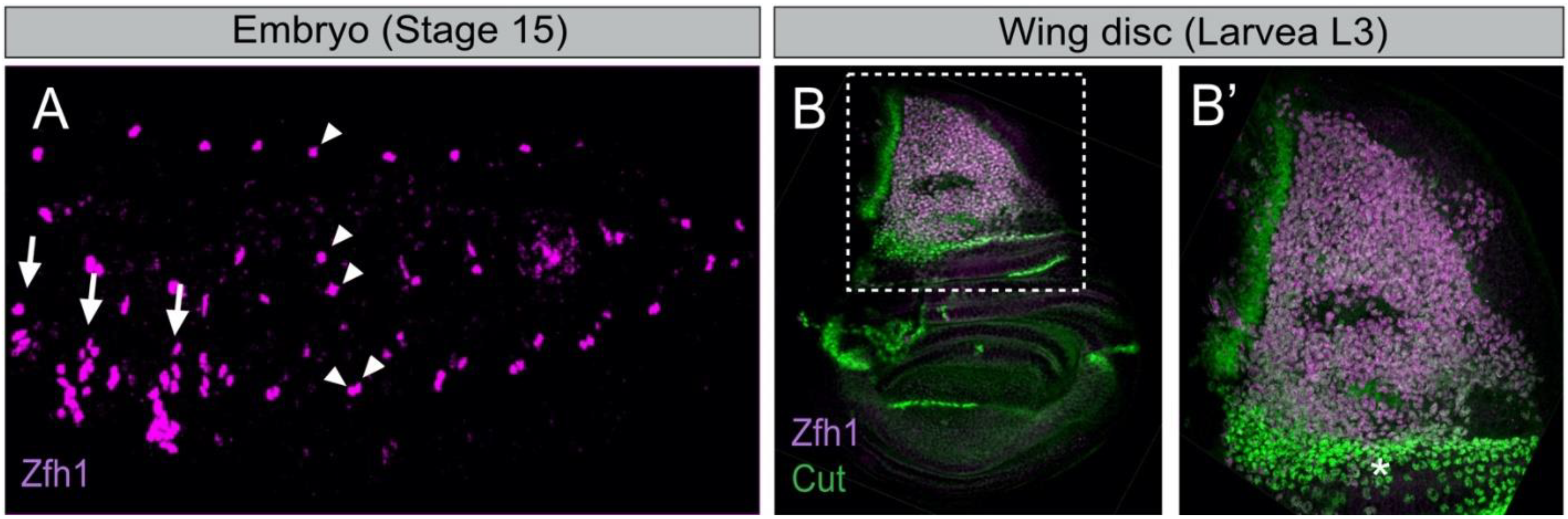
4.2. Zfh1/ZEB Maintains Undifferentiated MuSCs: An Evolutionarily Conserved Function
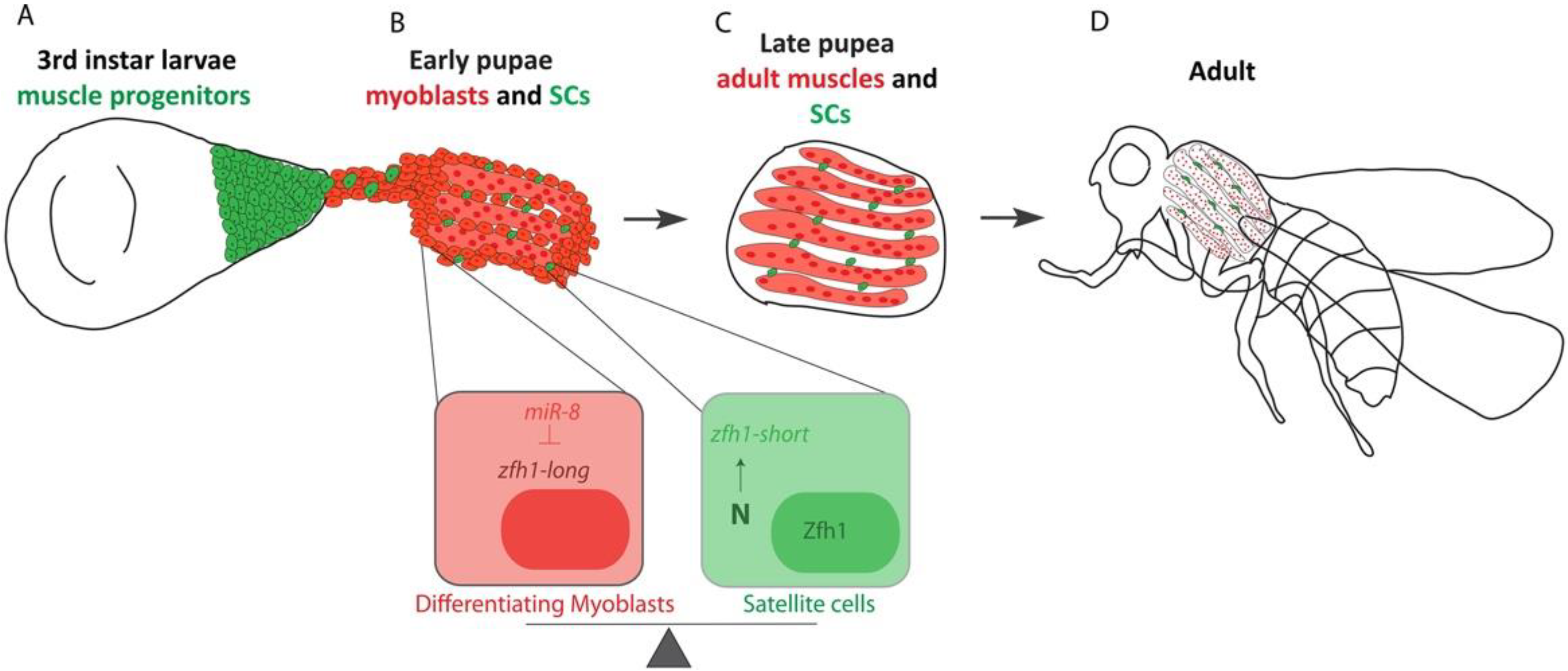
4.3. Setting Aside MuSCs during Development
5. Conclusions and Prospects
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MuSC | Muscle Stem Cell |
| AMP | Adult Muscle Precursor |
| MP | Muscle Progenitor |
| iTF | Identity Transcription Factor |
| FC | Founder Cell |
| IFM | Indirect Flight Muscle |
| DFM | Direct Flight Muscle |
References
- Chal, J.; Pourquié, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Cell Biol. 1961, 9, 493–495. [Google Scholar] [CrossRef]
- Lepper, C.; Partridge, T.A.; Fan, C.-M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 2011, 138, 3639–3646. [Google Scholar] [CrossRef]
- Fukada, S.-I. The roles of muscle stem cells in muscle injury, atrophy and hypertrophy. J. Biochem. 2018, 163, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Giordani, L.; Parisi, A.; Le Grand, F. Satellite cell self-renewal. Curr. Top. Dev. Biol. 2018, 126, 177–203. [Google Scholar] [CrossRef] [PubMed]
- Hwang, A.B.; Brack, A.S. Muscle stem cells and aging. Curr. Top. Dev. Biol. 2018, 126, 299–322. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Serra, C.; Lee, G.; Wagner, K.R. Stem cell-based therapies for Duchenne muscular dystrophy. Exp. Neurol. 2019, 323, 113086. [Google Scholar] [CrossRef]
- Chal, J.; Al Tanoury, Z.; Hestin, M.; Gobert, B.; Aivio, S.; Hick, A.; Cherrier, T.; Nesmith, A.P.; Parker, K.K.; Pourquie, O. Generation of human muscle fibers and satellite-like cells from human pluripotent stem cells in vitro. Nat. Protoc. 2016, 11, 1833–1850. [Google Scholar] [CrossRef] [PubMed]
- Gattazzo, F.; Laurent, B.; Relaix, F.; Rouard, H.; Didier, N. Distinct phases of postnatal skeletal muscle growth govern the progressive establishment of muscle stem cell quiescence. Stem Cell Rep. 2020, 15, 597–611. [Google Scholar] [CrossRef]
- Machado, L.; Geara, P.; Camps, J.; Dos Santos, M.; Teixeira-Clerc, F.; Van Herck, J.; Varet, H.; Legendre, R.; Pawlotsky, J.-M.; Sampaolesi, M.; et al. Tissue damage induces a conserved stress response that initiates quiescent muscle stem cell activation. Cell Stem Cell 2021, 28, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Ratnayake, D.; Nguyen, P.D.; Rossello, F.J.; Wimmer, V.C.; Tan, J.L.; Galvis, L.A.; Julier, Z.; Wood, A.J.; Boudier, T.; Isiaku, A.I.; et al. Macrophages provide a transient muscle stem cell niche via NAMPT secretion. Nat. Cell Biol. 2021, 591, 281–287. [Google Scholar] [CrossRef]
- Gurevich, D.B.; Nguyen, P.D.; Siegel, A.L.; Ehrlich, O.V.; Sonntag, C.; Phan, J.M.N.; Berger, S.; Ratnayake, D.; Hersey, L.; Berger, J.; et al. Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science 2016, 353, aad9969. [Google Scholar] [CrossRef] [PubMed]
- Konstantinides, N.; Averof, M. A common cellular basis for muscle regeneration in arthropods and vertebrates. Science 2014, 343, 788–791. [Google Scholar] [CrossRef]
- Fox, D.T.; Cohen, E.; Smith-Bolton, R. Model systems for regeneration: Drosophila. Development 2020, 147. [Google Scholar] [CrossRef] [PubMed]
- Khodabukus, A. Tissue-engineered skeletal muscle models to study muscle function, plasticity, and disease. Front. Physiol. 2021, 12. [Google Scholar] [CrossRef]
- Boukhatmi, H.; Bray, S. A population of adult satellite-like cells in Drosophila is maintained through a switch in RNA-isoforms. eLife 2018, 7, e35954. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, D.; Reichert, H.; Gunage, R.D.; VijayRaghavan, K. Identification and functional characterization of muscle satellite cells in Drosophila. eLife 2017, 6, e30107. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, G.; Soler, C.; Zmojdzian, M.; Jagla, K. Characterization of Drosophila muscle stem cell-like adult muscle precursors. In Muscle Stem Cells; Humana: New York, NY, USA, 2017; Volume 1556, pp. 103–116. [Google Scholar]
- Laurichesse, Q.; Soler, C. Muscle development: A view from adult myogenesis in Drosophila. Semin. Cell Dev. Biol. 2020, 104, 39–50. [Google Scholar] [CrossRef]
- Gunage, R.D.; Dhanyasi, N.; Reichert, H.; VijayRaghavan, K. Drosophila adult muscle development and regeneration. Semin. Cell Dev. Biol. 2017, 72, 56–66. [Google Scholar] [CrossRef]
- Schnorrer, F.; Schönbauer, C.; Langer, C.C.H.; Dietzl, G.; Novatchkova, M.; Schernhuber, K.; Fellner, M.; Azaryan, A.; Radolf, M.; Stark, A.; et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nat. Cell Biol. 2010, 464, 287–291. [Google Scholar] [CrossRef]
- Gunage, R.D.; Reichert, H.; VijayRaghavan, K. Identification of a new stem cell population that generates Drosophila flight muscles. eLife 2014, 3, e03126. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.; Bate, M.; Vijayraghavan, K. Development of the indirect flight muscles of Drosophila. Development 1991, 113, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Figeac, N.; Daczewska, M.; Marcelle, C.; Jagla, K. Muscle stem cells and model systems for their investigation. Dev. Dyn. 2007, 236, 3332–3342. [Google Scholar] [CrossRef] [PubMed]
- Baylies, M.K.; Bate, M. Twist: A myogenic switch in Drosophila. Science 1996, 272, 1481–1484. [Google Scholar] [CrossRef]
- Bate, M.; Rushton, E.; Currie, D. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development 1991, 113, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Currie, D.A.; Bate, M. The development of adult abdominal muscles in Drosophila: Myoblasts express twist and are associated with nerves. Development 1991, 113, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Boukhatmi, H.; Frendo, J.-L.; Enriquez, J.; Crozatier, M.; Dubois, L.; Vincent, A. Tup/Islet1 integrates time and position to specify muscle identity in Drosophila. Development 2012, 139, 3572–3582. [Google Scholar] [CrossRef]
- Roy, S.; Shashidhara, L.; VijayRaghavan, K. Muscles in the Drosophila second thoracic segment are patterned independently of autonomous homeotic gene function. Curr. Biol. 1997, 7, 222–227. [Google Scholar] [CrossRef]
- Roy, S.; VijayRaghavan, K. Homeotic genes and the regulation of myoblast migration, fusion, and fibre-specific gene ex-pression during adult myogenesis in Drosophila. Development 1997, 124, 3333–3341. [Google Scholar] [CrossRef]
- Greig, S.; Akam, M. Homeotic genes autonomously specify one aspect of pattern in the Drosophlla mesoderm. Nat. Cell Biol. 1993, 362, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Zmojdzian, M.; Jagla, K. The relationship between muscle stem cells and motor neurons. Cell. Mol. Life Sci. 2021, 78, 5043–5049. [Google Scholar] [CrossRef]
- Figeac, N.; Jagla, T.; Aradhya, R.; Da Ponte, J.P.; Jagla, K. Drosophila adult muscle precursors form a network of interconnected cells and are specified by the rhomboid-triggered EGF pathway. Development 2010, 137, 1965–1973. [Google Scholar] [CrossRef]
- Sudarsan, V.; Anant, S.; Guptan, P.; VijayRaghavan, K.; Skaer, H. Myoblast diversification and ectodermal signaling in Drosophila. Dev. Cell 2001, 1, 829–839. [Google Scholar] [CrossRef]
- Everetts, N.J.; Worley, M.I.; Yasutomi, R.; Yosef, N.; Hariharan, I.K. Single-cell transcriptomics of the Drosophila wing disc reveals instructive epithelium-to-myoblast interactions. eLife 2021, 10, e61276. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, M.; Backer, S.; Saintpierre, B.; Izac, B.; Andrieu, M.; Letourneur, F.; Relaix, F.; Sotiropoulos, A.; Maire, P. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat. Commun. 2020, 11, 5102. [Google Scholar] [CrossRef] [PubMed]
- Petrany, M.J.; Swoboda, C.O.; Sun, C.; Chetal, K.; Chen, X.; Weirauch, M.T.; Salomonis, N.; Millay, D.P. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 2020, 11, 6374. [Google Scholar] [CrossRef]
- De Micheli, A.; Laurilliard, E.J.; Heinke, C.L.; Ravichandran, H.; Fraczek, P.; Soueid-Baumgarten, S.; De Vlaminck, I.; Elemento, O.; Cosgrove, B.D. Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration. Cell Rep. 2020, 30, 3583–3595. [Google Scholar] [CrossRef] [PubMed]
- Baylies, M.K.; Bate, M.; Gomez, M.R. The specification of muscle in Drosophila. CSH Symp. Quant. Biol. 1997, 62, 385–393. [Google Scholar]
- Poovathumkadavil, P.; Jagla, K. Genetic control of muscle diversification and homeostasis: Insights from Drosophila. Cells 2020, 9, 1543. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Azevedo, M.; Baylies, M. Acting on identity: Myoblast fusion and the formation of the syncytial muscle fiber. Semin. Cell Dev. Biol. 2017, 72, 45–55. [Google Scholar] [CrossRef]
- Bataille, L.; Boukhatmi, H.; Frendo, J.-L.; Vincent, A. Dynamics of transcriptional (re)-programming of syncytial nuclei in developing muscles. BMC Biol. 2017, 15, 1–19. [Google Scholar] [CrossRef]
- Maqbool, T.; Soler, C.; Jagla, T.; Daczewska, M.; Lodha, N.; Palliyil, S.; VijayRaghavan, K.; Jagla, K. Shaping leg muscles in Drosophila: Role of ladybird, a conserved regulator of appendicular myogenesis. PLoS ONE 2006, 1, e122. [Google Scholar] [CrossRef]
- Schönbauer, C.; Distler, J.; Jährling, N.; Radolf, M.; Dodt, H.-U.; Frasch, M.; Schnorrer, F. Spalt mediates an evolutionarily conserved switch to fibrillar muscle fate in insects. Nat. Cell Biol. 2011, 479, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Spletter, M.; Barz, C.; Yeroslaviz, A.; Schönbauer, C.; Ferreira, I.R.S.; Sarov, M.; Gerlach, D.; Stark, A.; Habermann, B.; Schnorrer, F. The RNA -binding protein Arrest (Bruno) regulates alternative splicing to enable myofibril maturation in Drosophila flight muscle. EMBO Rep. 2014, 16, 178–191. [Google Scholar] [CrossRef] [PubMed]
- Avellaneda, J.; Rodier, C.; Daian, F.; Brouilly, N.; Rival, T.; Luis, N.M.; Schnorrer, F. Myofibril and mitochondria morphogenesis are coordinated by a mechanical feedback mechanism in muscle. Nat. Commun. 2021, 12, 2091. [Google Scholar] [CrossRef]
- Bryantsev, A.; Duong, S.; Brunetti, T.M.; Chechenova, M.B.; Lovato, T.L.; Nelson, C.; Shaw, E.; Uhl, J.D.; Gebelein, B.; Cripps, R.M. Extradenticle and homothorax control adult muscle fiber identity in Drosophila. Dev. Cell 2012, 23, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Wang, Y.; Zhang, L.; Yang, Y.; Huang, S.; Wang, J.; Ge, H.; Ishibashi, T.; Yan, Y. Single cell transcriptomic landscapes of pattern formation, proliferation and growth in Drosophila wing imaginal discs. Development 2019, 146, 179754. [Google Scholar] [CrossRef] [PubMed]
- Bageritz, J.; Willnow, P.; Valentini, E.; Leible, S.; Boutros, M.; Teleman, A.A. Gene expression atlas of a developing tissue by single cell expression correlation analysis. Nat. Methods 2019, 16, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Zappia, M.P.; De Castro, L.; Ariss, M.M.; Jefferson, H.; Islam, A.B.; Frolov, M.V. A cell atlas of adult muscle precursors uncovers early events in fibre-type divergence in Drosophila. EMBO Rep. 2020, 21, e49555. [Google Scholar] [CrossRef] [PubMed]
- Vishal, K.; Lovato, T.L.; Bragg, C.; Chechenova, M.B.; Cripps, R.M. FGF signaling promotes myoblast proliferation through activation of wingless signaling. Dev. Biol. 2020, 464, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Vishal, K.; Brooks, D.S.; Bawa, S.; Gameros, S.; Stetsiv, M.; Geisbrecht, E.R. Adult muscle formation requires Drosophila moleskin for proliferation of wing disc-associated muscle precursors. Genetics 2017, 206, 199–213. [Google Scholar] [CrossRef]
- Aradhya, R.; Zmojdzian, M.; Da Ponte, J.P.; Jagla, K. Muscle niche-driven Insulin-Notch-Myc cascade reactivates dormant adult muscle precursors in Drosophila. eLife 2015, 4, e08497. [Google Scholar] [CrossRef]
- Bröhl, D.; Vasyutina, E.; Czajkowski, M.; Griger, J.; Rassek, C.; Rahn, H.-P.; Purfürst, B.; Wende, H.; Birchmeier, C. Colonization of the satellite cell niche by skeletal muscle progenitor cells depends on Notch signals. Dev. Cell 2012, 23, 469–481. [Google Scholar] [CrossRef]
- Aradhya, R.; Jagla, K. Insulin-dependent non-canonical activation of Notch in Drosophila: A story of Notch-induced muscle stem cell proliferation. Notch Signal. Embryol. Cancer 2020, 1227, 131–144. [Google Scholar] [CrossRef]
- Lavergne, G.; Zmojdzian, M.; Da Ponte, J.P.; Junion, G.; Jagla, K. Drosophila adult muscle precursor cells contribute to motor axon pathfinding and proper innervation of embryonic muscles. Development 2020, 147, dev183004. [Google Scholar] [CrossRef]
- Bray, S. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Boukhatmi, H.; Martins, T.; Pillidge, Z.; Kamenova, T.; Bray, S. Notch mediates inter-tissue communication to promote tumorigenesis. Curr. Biol. 2020, 30, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- Krejci, A.; Bernard, F.; Housden, B.; Collins, S.; Bray, S. Direct response to Notch activation: Signaling crosstalk and incoherent logic. Sci. Signal. 2009, 2, ra1. [Google Scholar] [CrossRef] [PubMed]
- Bernard, F.; Krejci, A.; Housden, B.; Adryan, B.; Bray, S.J. Specificity of Notch pathway activation: Twist controls the transcriptional output in adult muscle progenitors. Development 2010, 137, 2633–2642. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, M.B.; Firmino, J.; Soni, K.; Evano, B.; Di Girolamo, D.; Mourikis, P.; Castel, D.; Tajbakhsh, S. Notch-induced miR-708 antagonizes satellite cell migration and maintains quiescence. Cell Stem Cell 2018, 23, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, M.B.; Castel, D.; Machado, L.; Fukada, S.-I.; Birk, D.E.; Relaix, F.; Tajbakhsh, S.; Mourikis, P. Reciprocal signalling by Notch–collagen V–CALCR retains muscle stem cells in their niche. Nat. Cell Biol. 2018, 557, 714–718. [Google Scholar] [CrossRef]
- Fujimaki, S.; Seko, D.; Kitajima, Y.; Yoshioka, K.; Tsuchiya, Y.; Masuda, S.; Ono, Y. Notch1 and Notch2 coordinately regulate stem cell function in the quiescent and activated states of muscle satellite cells. Stem Cells 2017, 36, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Soler, C.; Taylor, M.V. The Him gene inhibits the development of Drosophila flight muscles during metamorphosis. Mech. Dev. 2009, 126, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Krejci, A.; Bray, S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007, 21, 1322–1327. [Google Scholar] [CrossRef]
- Taylor, M.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 2017, 72, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Postigo, A.; Ward, E.; Skeath, J.B.; Dean, D.C. Zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol. Cell. Biol. 1999, 19, 7255–7263. [Google Scholar] [CrossRef]
- Kornberg, T.B. Distributing signaling proteins in space and time: The province of cytonemes. Curr. Opin. Genet. Dev. 2017, 45, 22–27. [Google Scholar] [CrossRef]
- González-Méndez, L.; Gradilla, A.-C.; Guerrero, I. The cytoneme connection: Direct long-distance signal transfer during development. Development 2019, 146, dev174607. [Google Scholar] [CrossRef]
- Huang, H.; Kornberg, T.B. Myoblast cytonemes mediate Wg signaling from the wing imaginal disc and Delta-Notch signaling to the air sac primordium. eLife 2015, 4, e06114. [Google Scholar] [CrossRef]
- Gildor, B.; Schejter, E.D.; Shilo, B.-Z. Bidirectional Notch activation represses fusion competence in swarming adult Drosophila myoblasts. Development 2012, 139, 4040–4050. [Google Scholar] [CrossRef]
- Segal, D.; Dhanyasi, N.; Schejter, E.D.; Shilo, B.-Z. Adhesion and fusion of muscle cells are promoted by filopodia. Dev. Cell 2016, 38, 291–304. [Google Scholar] [CrossRef][Green Version]
- Schmidt, M.; Schüler, S.C.; Hüttner, S.S.; Von Eyss, B.; Von Maltzahn, J. Adult stem cells at work: Regenerating skeletal muscle. Cell. Mol. Life Sci. 2019, 76, 2559–2570. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Brabletz, T. The ZEB/miR-200 feedback loop—A motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Tilló, E.; Siles, L.; de Barrios, O.; Cuatrecasas, M.; Vaquero, E.C.; Castells, A.; Postigo, A. Expanding roles of ZEB factors in tumorigenesis and tumor progression. Am. J. Cancer Res. 2011, 1, 897–912. [Google Scholar] [PubMed]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Wu, W.; Kuo, T.; Kao, C.; Girardot, C.; Hung, S.; Liu, T.; Furlong, E.E.M.; Liu, Y. Expanding the mesodermal transcriptional network by genome-wide identification of Zinc finger homeodomain 1 (Zfh1) targets. FEBS Lett. 2019, 593, 1698–1710. [Google Scholar] [CrossRef]
- Antonello, Z.A.; Reiff, T.; Ballesta-Illan, E.; Dominguez, M. Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by miR-8-Escargot switch. EMBO J. 2015, 34, 2025–2041. [Google Scholar] [CrossRef]
- Siles, L.; Ninfali, C.; Cortés, M.; Darling, D.S.; Postigo, A. ZEB1 protects skeletal muscle from damage and is required for its regeneration. Nat. Commun. 2019, 10, 1364. [Google Scholar] [CrossRef]
- Almeida, C.F.; Fernandes, S.A.; Junior, A.F.R.; Okamoto, O.K.; Vainzof, M. Muscle satellite cells: Exploring the basic biology to rule them. Stem Cells Int. 2016, 2016, 1–14. [Google Scholar] [CrossRef]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nat. Cell Biol. 2005, 435, 948–953. [Google Scholar] [CrossRef]
- Manceau, M.; Marcelle, C.; Gros, J. Une source unique de progéniteurs musculaires. MS Med. Sci. 2005, 21, 915–917. [Google Scholar] [CrossRef][Green Version]
- Maqbool, M.A.; Pioger, L.; El Aabidine, A.Z.; Karasu, N.; Molitor, A.M.; Dao, L.T.; Charbonnier, G.; Van Laethem, F.; Fenouil, R.; Koch, F.; et al. Alternative enhancer usage and targeted polycomb marking hallmark promoter choice during T cell differentiation. Cell Rep. 2020, 32. [Google Scholar] [CrossRef] [PubMed]
- Lamas-Maceiras, M.; Singh, B.; Hampsey, M.; Freire-Picos, M.A. Promoter-terminator gene loops affect alternative 3′-end processing in yeast. J. Biol. Chem. 2016, 291, 8960–8968. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in colorectal cancer: A culprit in metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Fereres, S.; Hatori, R.; Hatori, M.; Kornberg, T.B. Cytoneme-mediated signaling essential for tumorigenesis. PLoS Genet. 2019, 15, e1008415. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.T.; Manor, U.; Lippincott-Schwartz, J.; Fan, C.-M. Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell 2015, 18, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Ratnayake, D.; Currie, P.D. Stem cell dynamics in muscle regeneration: Insights from live imaging in different animal models. BioEssays 2017, 39, 1700011. [Google Scholar] [CrossRef] [PubMed]
- Konagaya, Y.; Takakura, K.; Sogabe, M.; Bisaria, A.; Liu, C.; Meyer, T.; Sehara-Fujisawa, A.; Matsuda, M.; Terai, K. Intravital imaging reveals cell cycle-dependent myogenic cell migration during muscle regeneration. Cell Cycle 2020, 19, 3167–3181. [Google Scholar] [CrossRef]
- Kimmel, J.C.; Hwang, A.B.; Scaramozza, A.; Marshall, W.F.; Brack, A.S. Aging induces aberrant state transition kinetics in murine muscle stem cells. Development 2020, 147, dev183855. [Google Scholar] [CrossRef]
- Wosczyna, M.N.; Rando, T.A. A muscle stem cell support group: Coordinated cellular responses in muscle regeneration. Dev. Cell 2018, 46, 135–143. [Google Scholar] [CrossRef]
- Dumont, N.A.; Wang, Y.X.; Rudnicki, M.A. Intrinsic and extrinsic mechanisms regulating satellite cell function. Development 2015, 142, 1572–1581. [Google Scholar] [CrossRef]
- Dumont, N.A.; Bentzinger, C.F.; Sincennes, M.; Rudnicki, M.A. Satellite cells and skeletal muscle regeneration. Compr. Physiol. 2015, 5, 1027–1059. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.L.; Sanders, E.; Moreno-Roman, P.; Koyama, L.A.; Balachandra, S.; Du, X.; O’Brien, L.E. Long-term live imaging of the Drosophila adult midgut reveals real-time dynamics of division, differentiation and loss. eLife 2018, 7, e36248. [Google Scholar] [CrossRef]
- Port, F.; Strein, C.; Stricker, M.; Rauscher, B.; Heigwer, F.; Zhou, J.; Beyersdörffer, C.; Frei, J.; Hess, A.; Kern, K.; et al. A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. eLife 2020, 9, e53865. [Google Scholar] [CrossRef]
- Li, H. Single-cell RNA sequencing in Drosophila: Technologies and applications. Wiley Interdiscip. Rev. Dev. Biol. 2020, e396. [Google Scholar] [CrossRef]
- Koyama, L.A.J.; Aranda-Díaz, A.; Su, Y.-H.; Balachandra, S.; Martin, J.L.; Ludington, W.B.; Huang, K.C.; O’Brien, L.E. Bellymount enables longitudinal, intravital imaging of abdominal organs and the gut microbiota in adult Drosophila. PLoS Biol. 2020, 18, e3000567. [Google Scholar] [CrossRef] [PubMed]
- Boutet, S.C.; Cheung, T.H.; Quach, N.L.; Liu, L.; Prescott, S.L.; Edalati, A.; Iori, K.; Rando, T.A. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell 2012, 10, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Sanjuan, J.F.; Lammers, N.; Garcia, H.G.; Bray, S.J. Enhancer priming enables fast and sustained transcriptional responses to Notch signaling. Dev. Cell 2019, 50, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Garcia, H.G.; Tikhonov, M.; Lin, A.; Gregor, T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr. Biol. 2013, 23, 2140–2145. [Google Scholar] [CrossRef]
- Starosta, A.; Konieczny, P. Therapeutic aspects of cell signaling and communication in Duchenne muscular dystrophy. Cell. Mol. Life Sci. 2021, 78, 4867–4891. [Google Scholar] [CrossRef]
- Choi, S.; Ferrari, G.; Tedesco, F.S. Cellular dynamics of myogenic cell migration: Molecular mechanisms and implications for skeletal muscle cell therapies. EMBO Mol. Med. 2020, 12, e12357. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boukhatmi, H. Drosophila, an Integrative Model to Study the Features of Muscle Stem Cells in Development and Regeneration. Cells 2021, 10, 2112. https://doi.org/10.3390/cells10082112
Boukhatmi H. Drosophila, an Integrative Model to Study the Features of Muscle Stem Cells in Development and Regeneration. Cells. 2021; 10(8):2112. https://doi.org/10.3390/cells10082112
Chicago/Turabian StyleBoukhatmi, Hadi. 2021. "Drosophila, an Integrative Model to Study the Features of Muscle Stem Cells in Development and Regeneration" Cells 10, no. 8: 2112. https://doi.org/10.3390/cells10082112
APA StyleBoukhatmi, H. (2021). Drosophila, an Integrative Model to Study the Features of Muscle Stem Cells in Development and Regeneration. Cells, 10(8), 2112. https://doi.org/10.3390/cells10082112




