In Silico Evaluation of Natural Compounds for an Acidic Extracellular Environment in Human Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Processing
2.3. Acidosis-Dependent Prognosis-Related Genes
2.4. Selection Method to Identify Compounds with Therapeutic Potential against Acidosis-Dependent Prognosis-Related Genes
2.5. Evaluation of Compound Effect on 14 Different Functional States from CancerSEA Database
2.6. Single-Cell Sequencing Analysis
2.7. Cell-to-Cell Interaction Analysis with scRNA-seq
2.8. Statistical Analysis
3. Results
3.1. Identification of Acidosis-Dependent Prognosis-Related Genes
3.2. Evaluation of Therapeutic Compounds against an Acidosis-Dependent Manner in BC
3.3. Effect of BD on 14 Different Cancer-Related Functional States with the CancerSEA Database
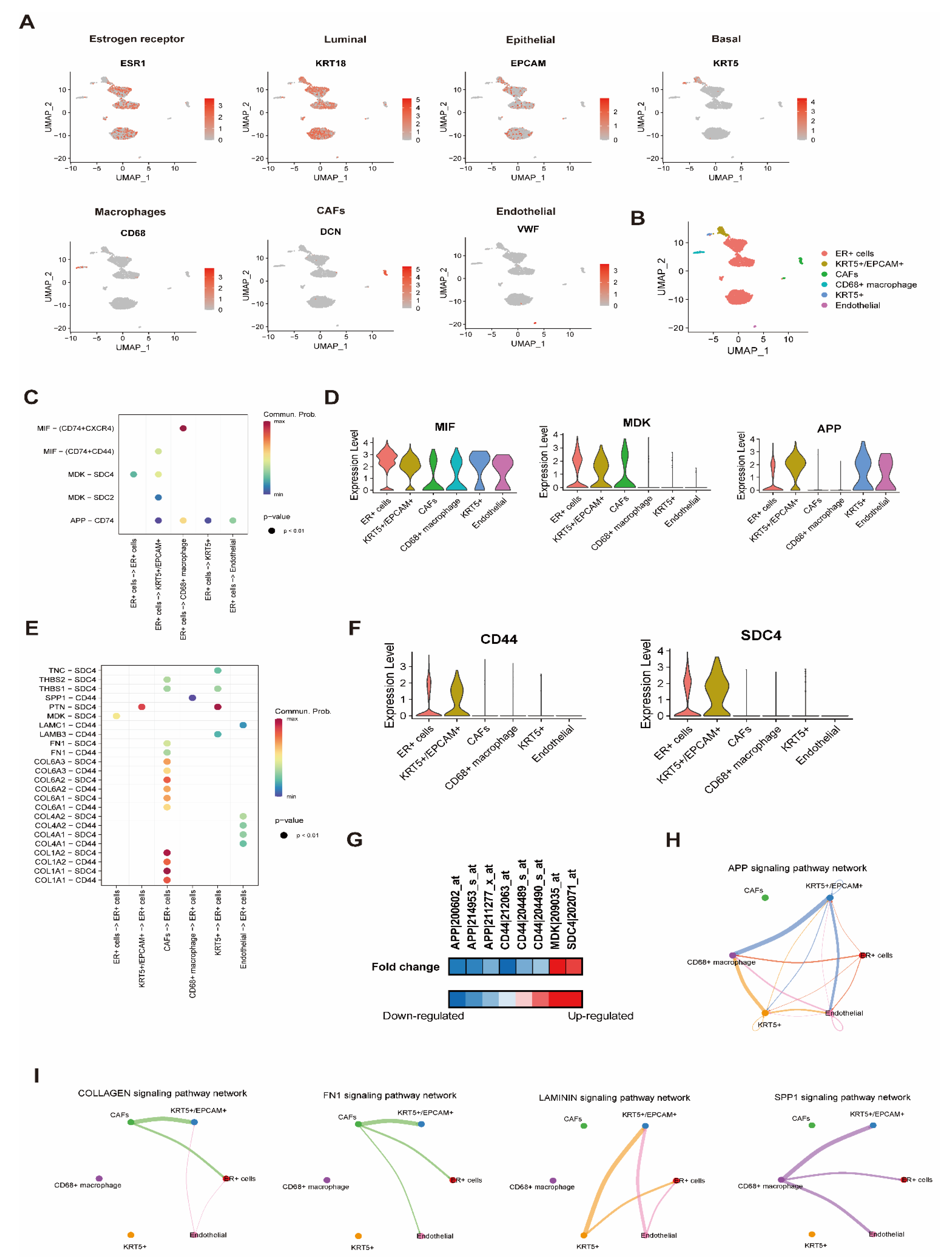
3.4. Effect of BD on Cell-to-Cell Interaction Analysis with scRNA Resolution
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Haussmann, J.; Corradini, S.; Nestle-Kraemling, C.; Bolke, E.; Njanang, F.J.D.; Tamaskovics, B.; Orth, K.; Ruckhaeberle, E.; Fehm, T.; Mohrmann, S.; et al. Recent advances in radiotherapy of breast cancer. Radiat. Oncol. 2020, 15, 71. [Google Scholar] [CrossRef]
- Gote, V.; Nookala, A.R.; Bolla, P.K.; Pal, D. Drug resistance in metastatic breast cancer: Tumor targeted nanomedicine to the rescue. Int. J. Mol. Sci. 2021, 22, 4673. [Google Scholar] [CrossRef]
- Riggio, A.I.; Varley, K.E.; Welm, A.L. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer 2021, 124, 13–26. [Google Scholar] [CrossRef]
- Hu, T.; Zhou, R.; Zhao, Y.; Wu, G. Integrin alpha6/akt/erk signaling is essential for human breast cancer resistance to radiotherapy. Sci Rep. 2016, 6, 33376. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, Y.; Kiani, M.F.; Wang, B. Classification, treatment strategy, and associated drug resistance in breast cancer. Clin. Breast Cancer 2016, 16, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Sonveaux, P.; Copetti, T.; De Saedeleer, C.J.; Vegran, F.; Verrax, J.; Kennedy, K.M.; Moon, E.J.; Dhup, S.; Danhier, P.; Frerart, F.; et al. Targeting the lactate transporter mct1 in endothelial cells inhibits lactate-induced hif-1 activation and tumor angiogenesis. PLoS ONE 2012, 7, e33418. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. A microenvironmental model of carcinogenesis. Nat. Rev. Cancer 2008, 8, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Damaghi, M.; Wojtkowiak, J.W.; Gillies, R.J. Ph sensing and regulation in cancer. Front. Physiol 2013, 4, 370. [Google Scholar] [CrossRef]
- Moellering, R.E.; Black, K.C.; Krishnamurty, C.; Baggett, B.K.; Stafford, P.; Rain, M.; Gatenby, R.A.; Gillies, R.J. Acid treatment of melanoma cells selects for invasive phenotypes. Clin. Exp. Metastasis 2008, 25, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Estrella, V.; Chen, T.; Lloyd, M.; Wojtkowiak, J.; Cornnell, H.H.; Ibrahim-Hashim, A.; Bailey, K.; Balagurunathan, Y.; Rothberg, J.M.; Sloane, B.F.; et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013, 73, 1524–1535. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gawlinski, E.T.; Gmitro, A.F.; Kaylor, B.; Gillies, R.J. Acid-mediated tumor invasion: A multidisciplinary study. Cancer Res. 2006, 66, 5216–5223. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fan, Z.; Zhang, J.; Changyi, Y.; Huang, C.; Gu, Y.; Xu, Z.; Tang, Z.; Lu, W.; Wei, X.; et al. Evaluating tumor metastatic potential by imaging intratumoral acidosis via ph-activatable near-infrared fluorescent probe. Int J. Cancer 2015, 136, E107–E116. [Google Scholar] [CrossRef]
- Thews, O.; Riemann, A. Tumor ph and metastasis: A malignant process beyond hypoxia. Cancer Metastasis Rev. 2019, 38, 113–129. [Google Scholar] [CrossRef]
- Tavares-Valente, D.; Baltazar, F.; Moreira, R.; Queiros, O. Cancer cell bioenergetics and ph regulation influence breast cancer cell resistance to paclitaxel and doxorubicin. J. Bioenerg Biomembr 2013, 45, 467–475. [Google Scholar] [CrossRef]
- Li, Z.; Zong, L.; Liu, H.; Yao, Z.; Sun, Y.; Li, Z. A solid-state sb/sb2o3 biosensor for the in situ measurement of extracellular acidification associated with the multidrug resistance phenotype in breast cancer cells. Analytical Methods 2018, 10, 4445–4453. [Google Scholar] [CrossRef]
- Anemone, A.; Consolino, L.; Conti, L.; Irrera, P.; Hsu, M.Y.; Villano, D.; Dastru, W.; Porporato, P.E.; Cavallo, F.; Longo, D.L. Tumour acidosis evaluated in vivo by mri-cest ph imaging reveals breast cancer metastatic potential. Br. J. Cancer 2021, 124, 207–216. [Google Scholar] [CrossRef]
- Luo, H.; Vong, C.T.; Chen, H.; Gao, Y.; Lyu, P.; Qiu, L.; Zhao, M.; Liu, Q.; Cheng, Z.; Zou, J.; et al. Naturally occurring anti-cancer compounds: Shining from chinese herbal medicine. Chin. Med. 2019, 14, 48. [Google Scholar] [CrossRef]
- Yao, J.; Czaplinska, D.; Ialchina, R.; Schnipper, J.; Liu, B.; Sandelin, A.; Pedersen, S.F. Cancer cell acid adaptation gene expression response is correlated to tumor-specific tissue expression profiles and patient survival. Cancers (Basel) 2020, 12, 2183. [Google Scholar] [CrossRef]
- Lv, C.; Wu, X.; Wang, X.; Su, J.; Zeng, H.; Zhao, J.; Lin, S.; Liu, R.; Li, H.; Li, X.; et al. The gene expression profiles in response to 102 traditional chinese medicine (tcm) components: A general template for research on tcms. Sci Rep. 2017, 7, 352. [Google Scholar] [CrossRef]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. Cancersea: A cancer single-cell state atlas. Nucleic Acids Res. 2019, 47, D900–D908. [Google Scholar] [CrossRef]
- Pal, B.; Chen, Y.; Vaillant, F.; Capaldo, B.D.; Joyce, R.; Song, X.; Bryant, V.L.; Penington, J.S.; Di Stefano, L.; Tubau Ribera, N.; et al. A single-cell rna expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 2021, 40, e107333. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 2015, 33, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using cellchat. Nat. Commun 2021, 12, 1088. [Google Scholar] [CrossRef]
- Swietach, P.; Vaughan-Jones, R.D.; Harris, A.L.; Hulikova, A. The chemistry, physiology and pathology of ph in cancer. Philos Trans. R Soc. Lond B Biol Sci 2014, 369, 20130099. [Google Scholar] [CrossRef]
- Andersen, A.P.; Moreira, J.M.; Pedersen, S.F. Interactions of ion transporters and channels with cancer cell metabolism and the tumour microenvironment. Philos Trans. R Soc. Lond B Biol Sci 2014, 369, 20130098. [Google Scholar] [CrossRef] [PubMed]
- Corbet, C.; Feron, O. Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer 2017, 17, 577–593. [Google Scholar] [CrossRef]
- Dong, J. The relationship between traditional chinese medicine and modern medicine. Evid Based Complement. Alternat Med. 2013, 2013, 153148. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal. 2020, 18, 59. [Google Scholar] [CrossRef]
- Xu, J.-P. Cancer inhibitors from chinese natural medicines; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Sin, Z.W.; Bhardwaj, V.; Pandey, A.K.; Garg, M. A brief overview of antitumoral actions of bruceine d. Exploration of Targeted Anti-tumor Therapy 2020, 1, 200–217. [Google Scholar] [CrossRef]
- Wang, S.; Hu, H.; Zhong, B.; Shi, D.; Qing, X.; Cheng, C.; Deng, X.; Zhang, Z.; Shao, Z. Bruceine d inhibits tumor growth and stem cell-like traits of osteosarcoma through inhibition of stat3 signaling pathway. Cancer Med. 2019, 8, 7345–7358. [Google Scholar] [CrossRef]
- Luo, C.; Wang, Y.; Wei, C.; Chen, Y.; Ji, Z. The anti-migration and anti-invasion effects of bruceine d in human triple-negative breast cancer mda-mb-231 cells. Exp. Ther Med. 2020, 19, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.Q.; Ip, S.P.; Liao, H.J.; Lu, Z.; Xie, J.H.; Su, Z.R.; Chen, Y.L.; Xian, Y.F.; Leung, P.S.; Lin, Z.X. Brucein d, a naturally occurring tetracyclic triterpene quassinoid, induces apoptosis in pancreatic cancer through ros-associated pi3k/akt signaling pathway. Front. Pharmacol 2017, 8, 936. [Google Scholar] [CrossRef]
- Wang, T.; Yuan, J.; Zhang, J.; Tian, R.; Ji, W.; Zhou, Y.; Yang, Y.; Song, W.; Zhang, F.; Niu, R. Anxa2 binds to stat3 and promotes epithelial to mesenchymal transition in breast cancer cells. Oncotarget 2015, 6, 30975–30992. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Ban, Z.; Nai, M.; Yang, L.; Chen, Y.; Xu, Y. Annexin a2 inhibition suppresses ovarian cancer progression via regulating beta-catenin/emt. Oncol Rep. 2017, 37, 3643–3650. [Google Scholar] [CrossRef] [PubMed]
- Santana-Codina, N.; Muixi, L.; Foj, R.; Sanz-Pamplona, R.; Badia-Villanueva, M.; Abramowicz, A.; Marce-Grau, A.; Cosialls, A.M.; Gil, J.; Archilla, I.; et al. Grp94 promotes brain metastasis by engaging pro-survival autophagy. Neuro Oncol 2020, 22, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhao, C.; Ye, Y.; Wang, Z.; He, Y.; Li, Y.; Mao, H. High expression of fibronectin 1 indicates poor prognosis in gastric cancer. Oncol Lett 2020, 19, 93–102. [Google Scholar] [CrossRef]
- Yang, X.; Hu, Q.; Hu, L.X.; Lin, X.R.; Liu, J.Q.; Lin, X.; Dinglin, X.X.; Zeng, J.Y.; Hu, H.; Luo, M.L.; et al. Mir-200b regulates epithelial-mesenchymal transition of chemo-resistant breast cancer cells by targeting fn1. Discov Med. 2017, 24, 75–85. [Google Scholar]
- Fico, F.; Santamaria-Martinez, A. Tgfbi modulates tumour hypoxia and promotes breast cancer metastasis. Mol. Oncol 2020, 14, 3198–3210. [Google Scholar] [CrossRef]
- Meyer, T.; Xu, L.; Chang, J.; Liu, E.T.; Craven, R.J.; Cance, W.G. Breast cancer cell line proliferation blocked by the src-related rak tyrosine kinase. Int J. Cancer 2003, 104, 139–146. [Google Scholar] [CrossRef]
- Yim, E.K.; Peng, G.; Dai, H.; Hu, R.; Li, K.; Lu, Y.; Mills, G.B.; Meric-Bernstam, F.; Hennessy, B.T.; Craven, R.J.; et al. Rak functions as a tumor suppressor by regulating pten protein stability and function. Cancer Cell 2009, 15, 304–314. [Google Scholar] [CrossRef]
- Jin, L.; Craven, R.J. The rak/frk tyrosine kinase associates with and internalizes the epidermal growth factor receptor. Oncogene 2014, 33, 326–335. [Google Scholar] [CrossRef][Green Version]
- Ogunbolude, Y.; Dai, C.; Bagu, E.T.; Goel, R.K.; Miah, S.; MacAusland-Berg, J.; Ng, C.Y.; Chibbar, R.; Napper, S.; Raptis, L.; et al. Frk inhibits breast cancer cell migration and invasion by suppressing epithelial-mesenchymal transition. Oncotarget 2017, 8, 113034–113065. [Google Scholar] [CrossRef]
- Runa, F.; Hamalian, S.; Meade, K.; Shisgal, P.; Gray, P.C.; Kelber, J.A. Tumor microenvironment heterogeneity: Challenges and opportunities. Curr Mol. Biol Rep. 2017, 3, 218–229. [Google Scholar] [CrossRef]
- O’Brien, R.J.; Wong, P.C. Amyloid precursor protein processing and alzheimer’s disease. Annu Rev. Neurosci 2011, 34, 185–204. [Google Scholar] [CrossRef]
- Matsuda, S.; Matsuda, Y.; D’Adamio, L. Cd74 interacts with app and suppresses the production of abeta. Mol. Neurodegener 2009, 4, 41. [Google Scholar] [CrossRef]
- Takagi, K.; Ito, S.; Miyazaki, T.; Miki, Y.; Shibahara, Y.; Ishida, T.; Watanabe, M.; Inoue, S.; Sasano, H.; Suzuki, T. Amyloid precursor protein in human breast cancer: An androgen-induced gene associated with cell proliferation. Cancer Sci 2013, 104, 1532–1538. [Google Scholar] [CrossRef]
- Lim, S.; Yoo, B.K.; Kim, H.S.; Gilmore, H.L.; Lee, Y.; Lee, H.P.; Kim, S.J.; Letterio, J.; Lee, H.G. Amyloid-beta precursor protein promotes cell proliferation and motility of advanced breast cancer. BMC Cancer 2014, 14, 928. [Google Scholar] [CrossRef]
- Louderbough, J.M.; Schroeder, J.A. Understanding the dual nature of cd44 in breast cancer progression. Mol. Cancer Res. 2011, 9, 1573–1586. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Stamenkovic, I. Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for cd44-mediated tumor invasion. Genes Dev. 1999, 13, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Stamenkovic, I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates tgf-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000, 14, 163–176. [Google Scholar]
- Bourguignon, L.Y.W.; Xia, W.; Wong, G. Hyaluronan-mediated cd44 interaction with p300 and sirt1 regulates beta-catenin signaling and nfkappab-specific transcription activity leading to mdr1 and bcl-xl gene expression and chemoresistance in breast tumor cells. J. Biol Chem 2009, 284, 2657–2671. [Google Scholar] [CrossRef]
- Rao, G.; Du, L.; Chen, Q. Osteopontin, a possible modulator of cancer stem cells and their malignant niche. Oncoimmunology 2013, 2, e24169. [Google Scholar] [CrossRef][Green Version]
- Pietras, A.; Katz, A.M.; Ekstrom, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-cd44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, Y.; Jeong, J.; Seong, S.; Kim, W. In Silico Evaluation of Natural Compounds for an Acidic Extracellular Environment in Human Breast Cancer. Cells 2021, 10, 2673. https://doi.org/10.3390/cells10102673
Park Y, Jeong J, Seong S, Kim W. In Silico Evaluation of Natural Compounds for an Acidic Extracellular Environment in Human Breast Cancer. Cells. 2021; 10(10):2673. https://doi.org/10.3390/cells10102673
Chicago/Turabian StylePark, YoungJoon, Jaekwang Jeong, Shin Seong, and Wonnam Kim. 2021. "In Silico Evaluation of Natural Compounds for an Acidic Extracellular Environment in Human Breast Cancer" Cells 10, no. 10: 2673. https://doi.org/10.3390/cells10102673
APA StylePark, Y., Jeong, J., Seong, S., & Kim, W. (2021). In Silico Evaluation of Natural Compounds for an Acidic Extracellular Environment in Human Breast Cancer. Cells, 10(10), 2673. https://doi.org/10.3390/cells10102673




