Attenuation of Pancreatic Cancer In Vitro and In Vivo via Modulation of Nrf2 and NF-κB Signaling Pathways by Natural Compounds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
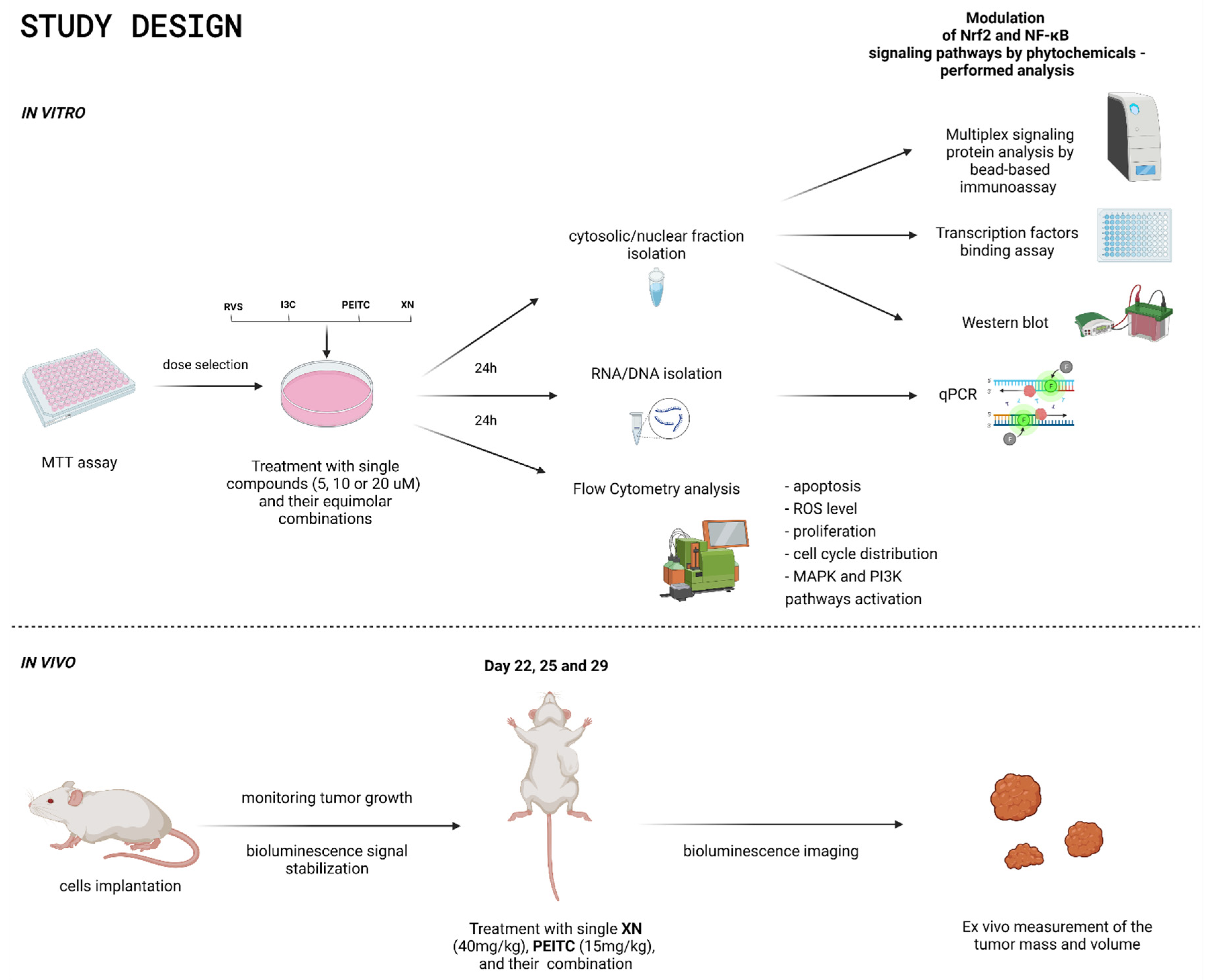
2.2. Cell Culture, Cell Viability Assay, and Treatment with Tested Compounds
2.3. Quantitative Real-Time PCR
2.4. Transcription Factors Binding Assay
2.5. Western Blot
2.6. Multiplex Signaling Protein Analysis by Bead-Based Immunoassay
2.7. Flow Cytometry Cell Analysis
2.7.1. Cell Cycle Distribution
2.7.2. Cell Proliferation
2.7.3. Apoptosis
2.7.4. The Level of ROS
2.8. In Vivo Evaluation of the Anticancer Potential of XN, PEITC, and Their Combination
2.8.1. Mice Xenograft Model
2.8.2. Treatment Protocol
2.9. Statistical Analysis
3. Results
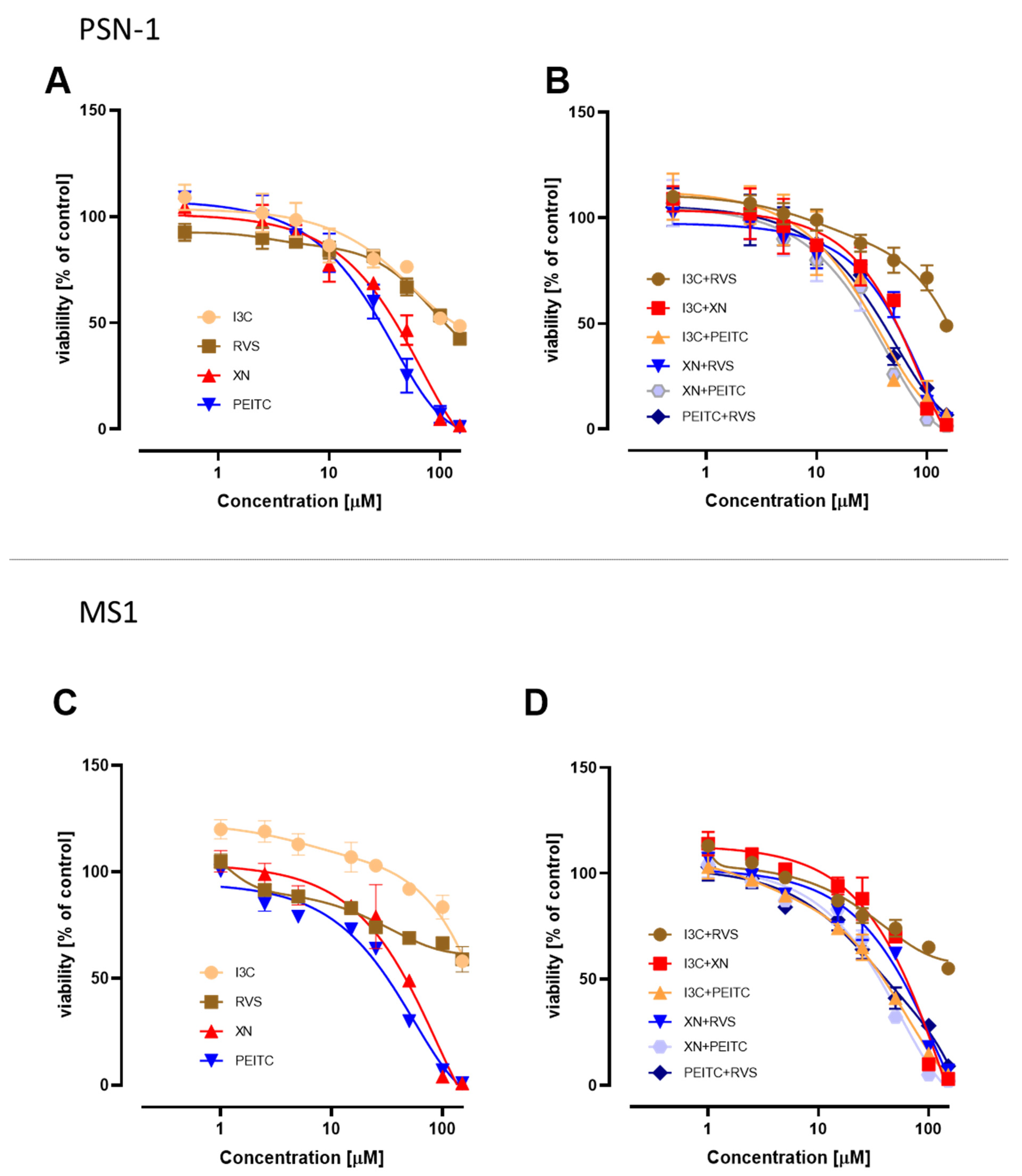
3.1. Cell Viability after Treatment with Single Phytochemicals and Their Mixtures
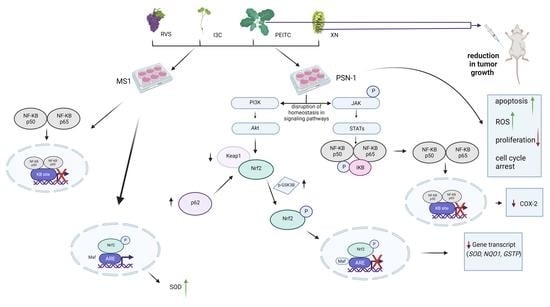
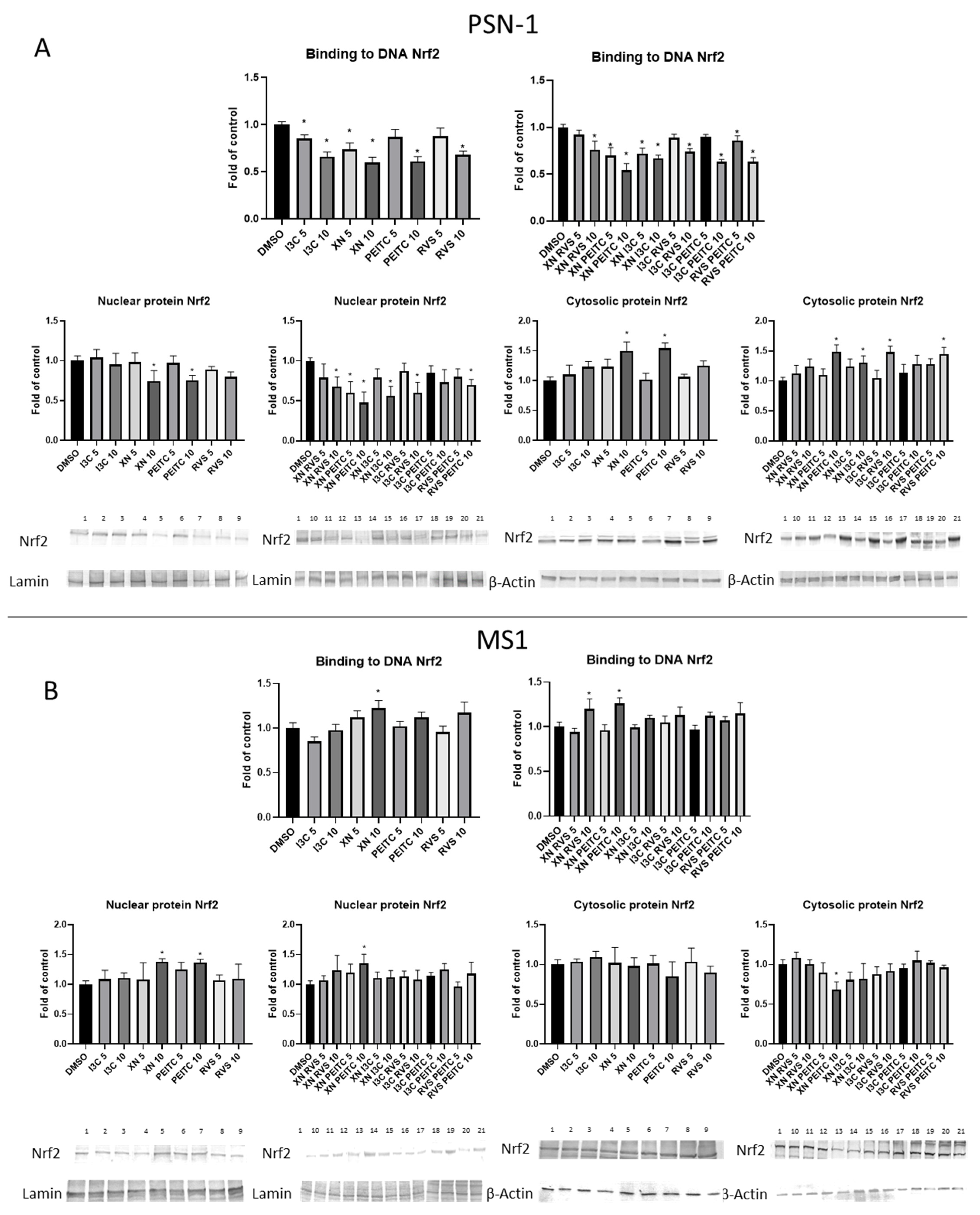
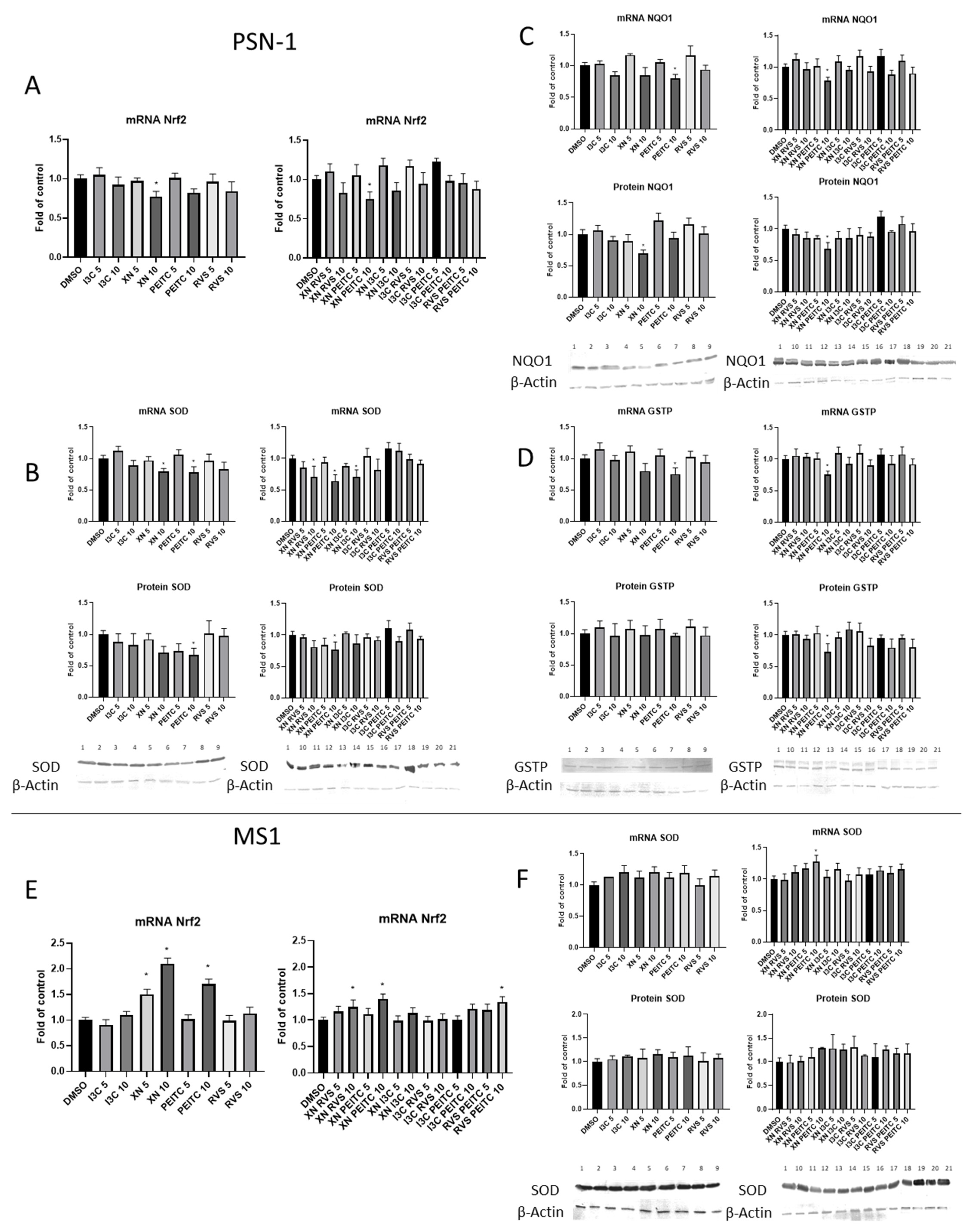
3.2. The Canonical and Non-Canonical Mechanism of Nrf2 Activation
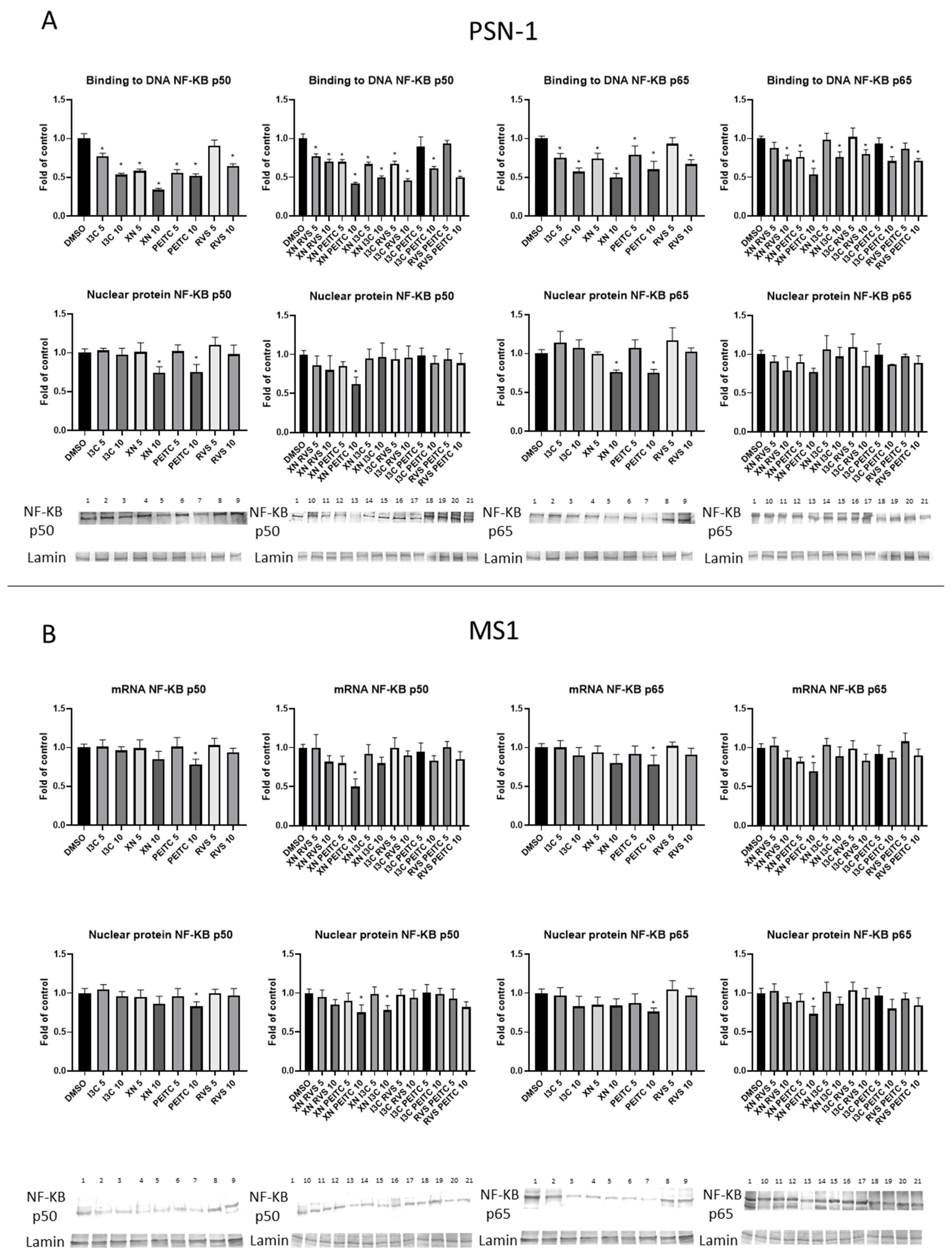
3.3. Interaction of Nrf2 with NF-κB Signaling Pathway
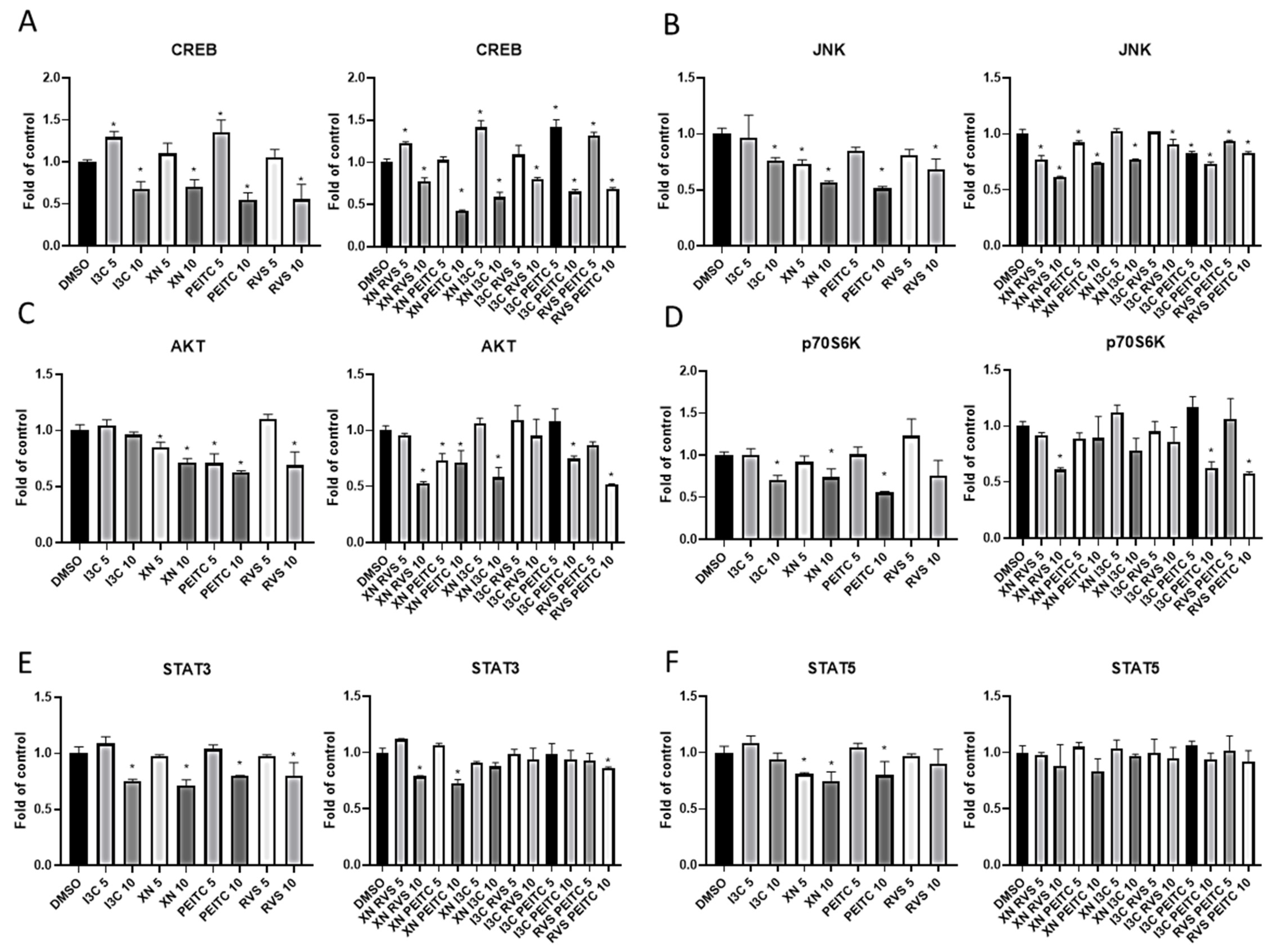
3.4. Multiplex Analysis of the Level of Proteins Regulating Several Signaling Pathways
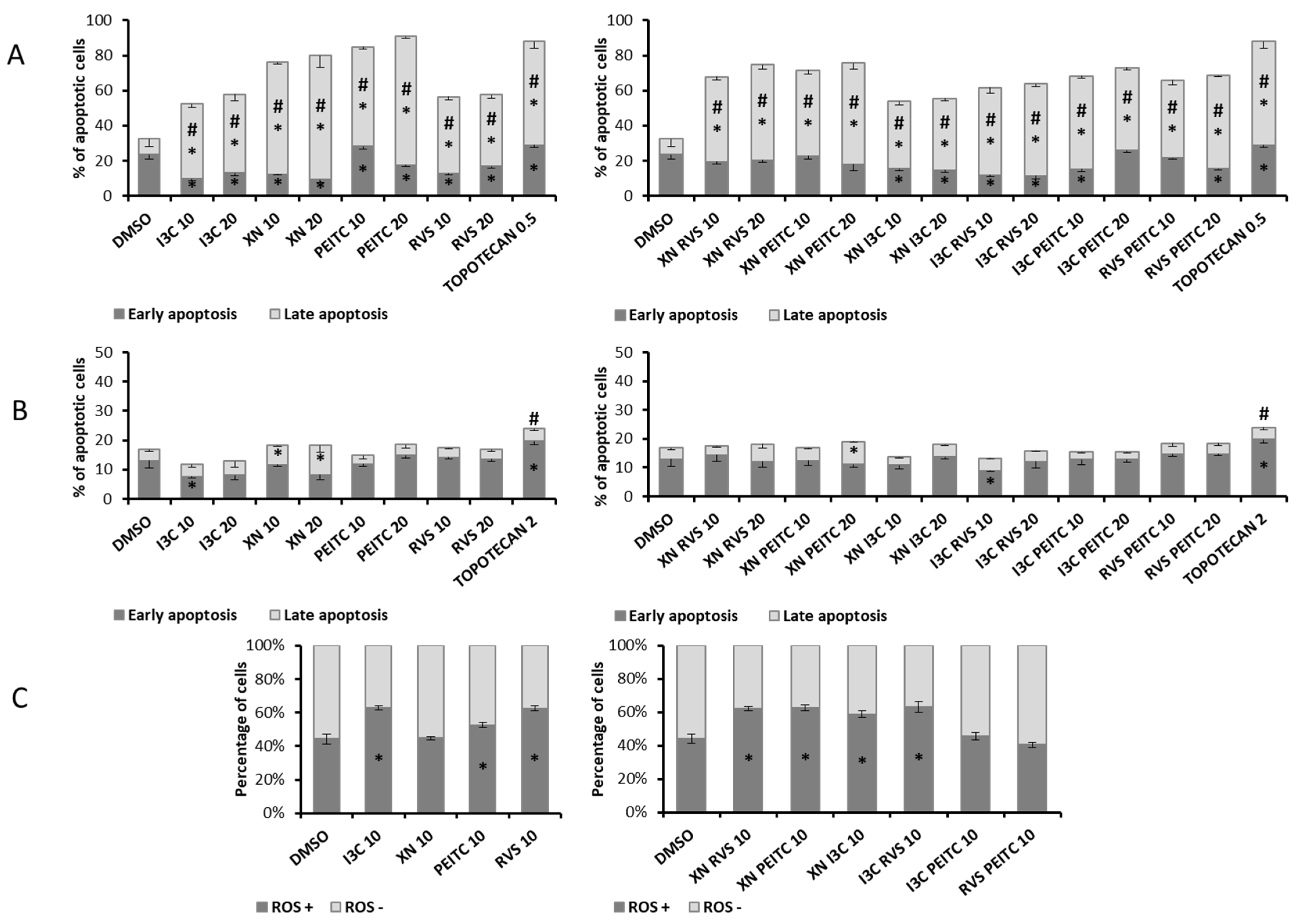
3.5. Analysis of Apoptosis and the Level of ROS
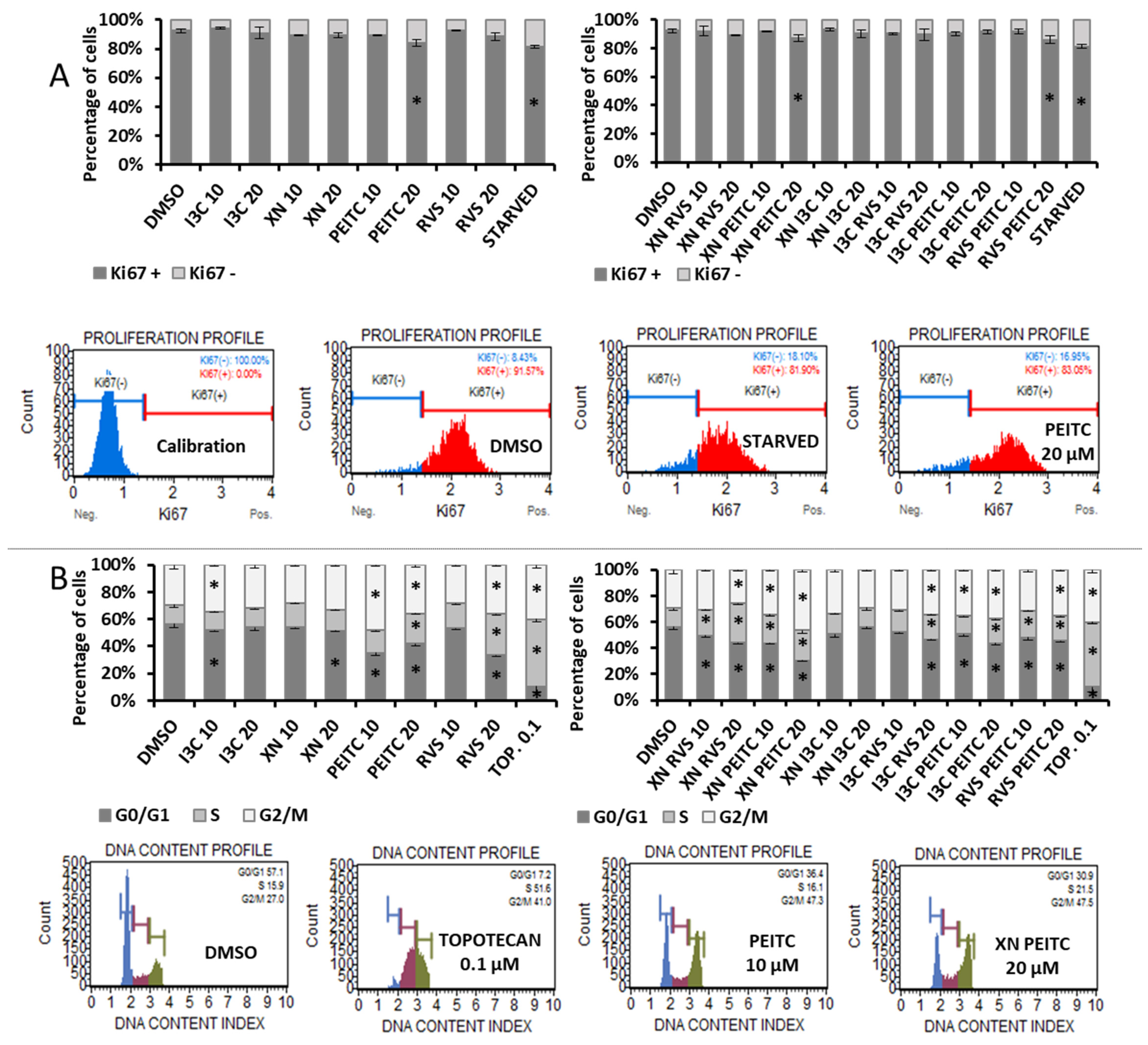
3.6. Analysis of Cell Proliferation and Cell Cycle Distribution
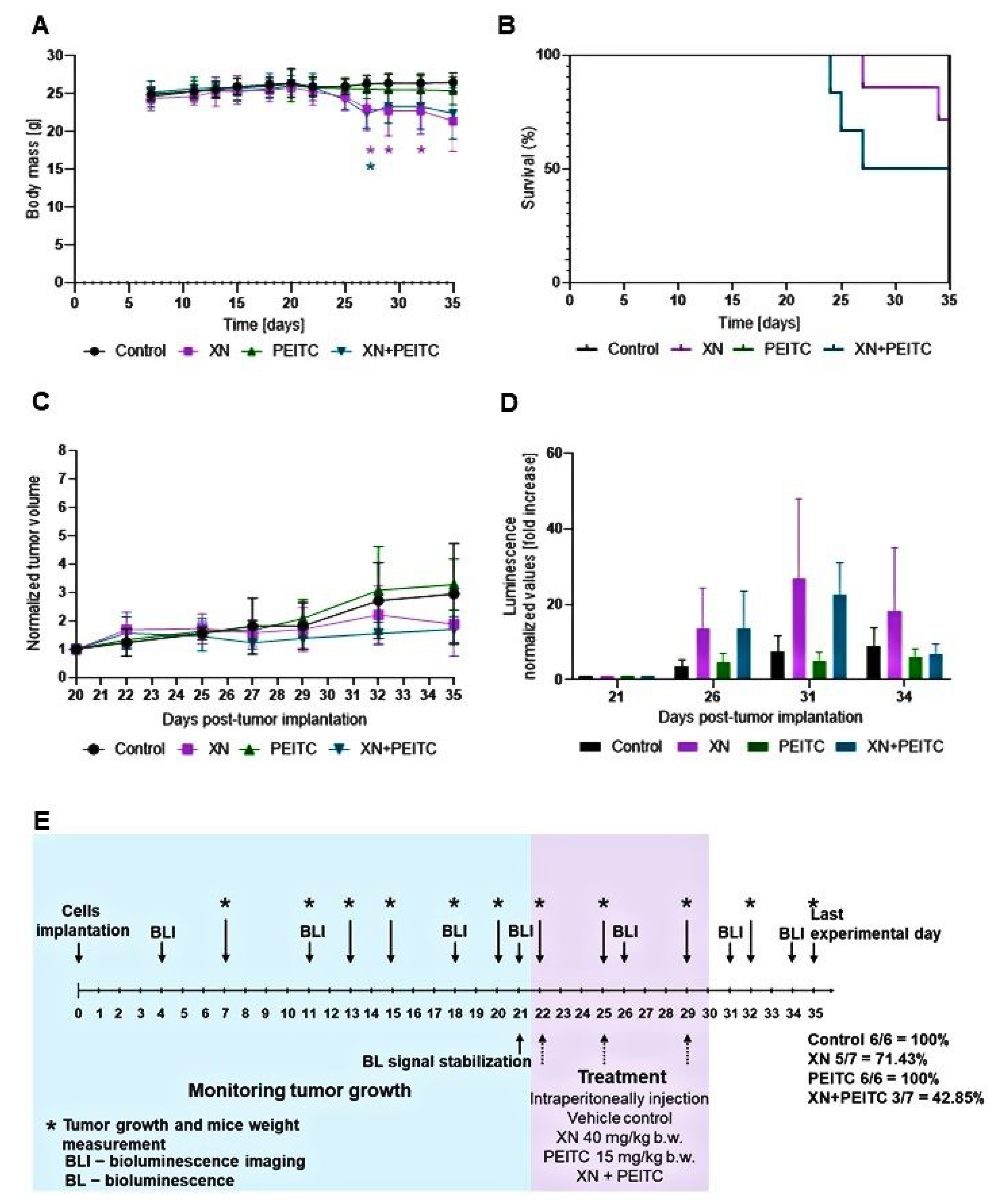
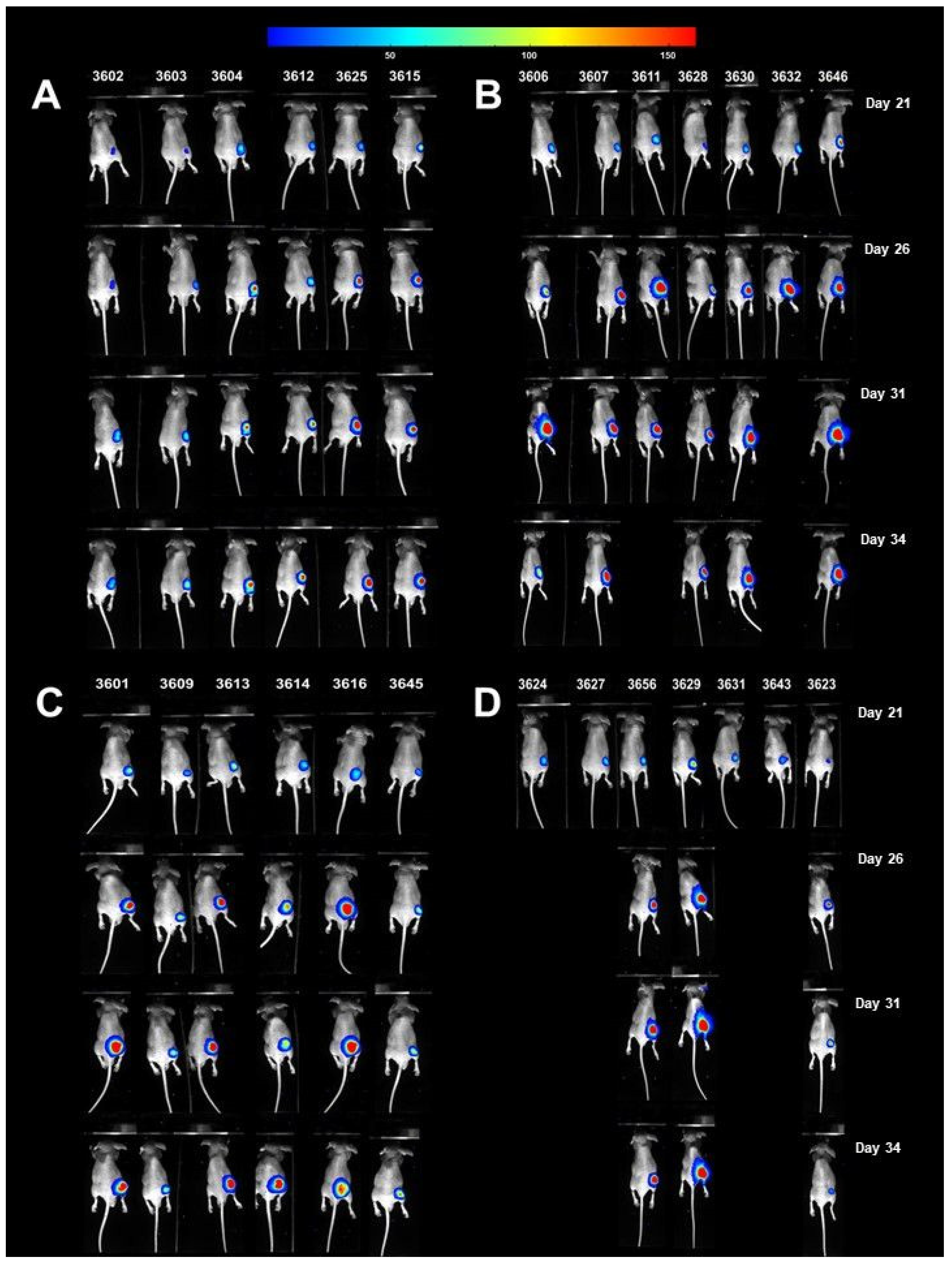
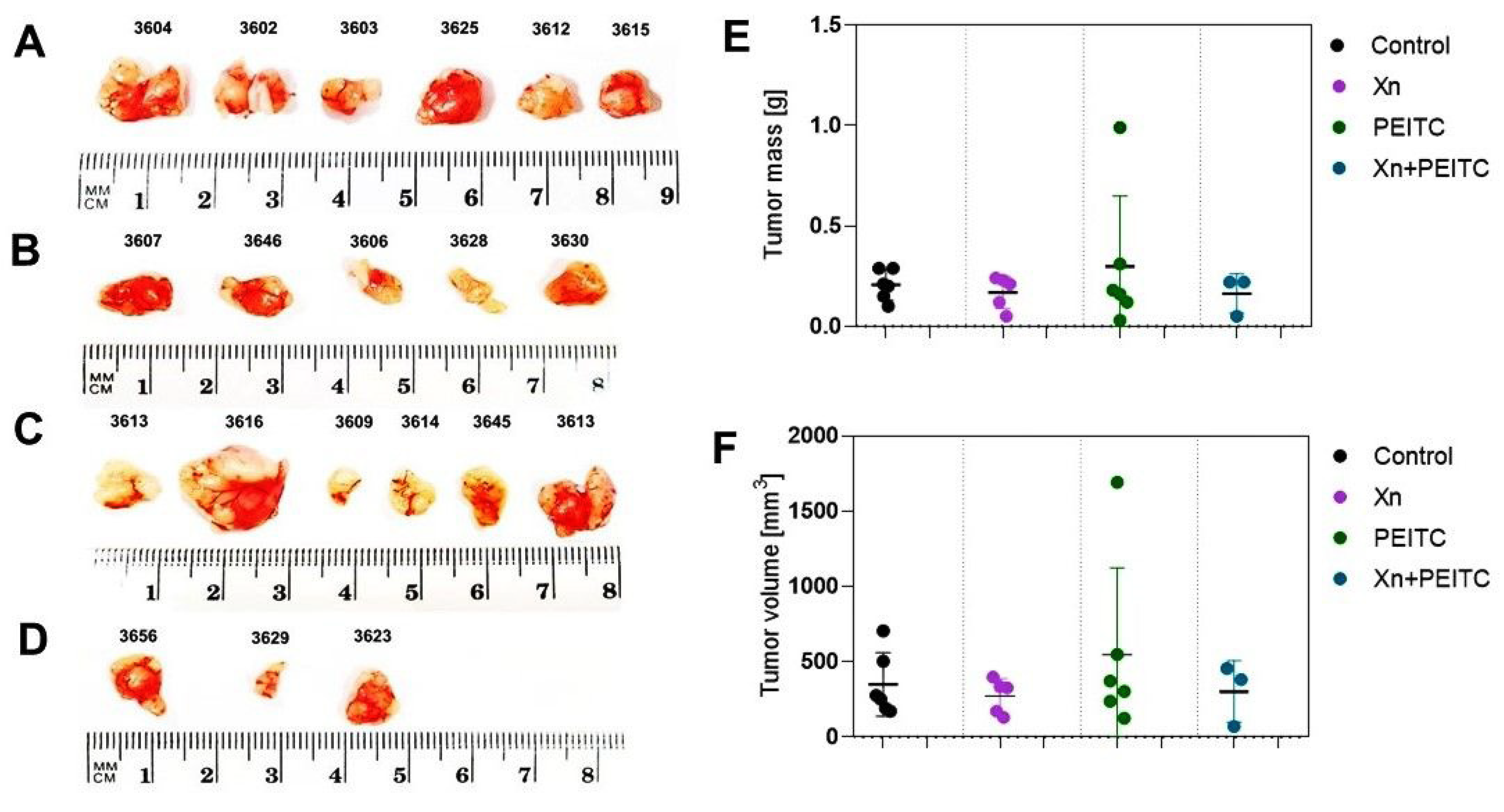
3.7. Antitumor Efficacy of Xanthohumol, Phenethyl Isothiocyanate, and Their Combination in Human Pancreatic Tumor Xenograft Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Yan, X.; Wintergerst, K.A.; Cai, L.; Keller, B.B.; Tan, Y. Nrf2: Redox and Metabolic Regulator of Stem Cell State and Function. Trends Mol. Med. 2020, 26, 185–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villavicencio Tejo, F.; Ra, Q. Contribution of the Nrf2 Pathway on Oxidative Damage and Mitochondrial Failure in Parkinson and Alzheimer’s Disease. Antioxidants 2021, 10, 1069. [Google Scholar] [CrossRef] [PubMed]
- Boo, Y.C. Natural Nrf2 Modulators for Skin Protection. Antioxidants 2020, 9, 812. [Google Scholar] [CrossRef]
- Robledinos-Antón, N.; Fernández-Ginés, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxid. Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef] [PubMed]
- Krajka-Kuźniak, V.; Paluszczak, J.; Baer-Dubowska, W. The Nrf2-ARE Signaling Pathway: An Update on Its Regulation and Possible Role in Cancer Prevention and Treatment. Pharm. Rep. 2017, 69, 393–402. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Kensler, T.W. The Role of Keap1 in Cellular Protective Responses. Chem. Res. Toxicol. 2005, 18, 1779–1791. [Google Scholar] [CrossRef]
- Tsai, T.-F.; Chen, P.-C.; Lin, Y.-C.; Chou, K.-Y.; Chen, H.-E.; Ho, C.-Y.; Lin, J.-F.; Hwang, T.I.-S. Miconazole Contributes to NRF2 Activation by Noncanonical P62-KEAP1 Pathway in Bladder Cancer Cells. Drug Des. Dev. Ther. 2020, 14, 1209–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Kern, J.T.; Walker, J.R.; Johnson, J.A.; Schultz, P.G.; Luesch, H. A Genomic Screen for Activators of the Antioxidant Response Element. Proc. Natl. Acad. Sci. USA 2007, 104, 5205–5210. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.-S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The Selective Autophagy Substrate P62 Activates the Stress Responsive Transcription Factor Nrf2 through Inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Pahl, H.L. Activators and Target Genes of Rel/NF-KappaB Transcription Factors. Oncogene 1999, 18, 6853–6866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.C.; Sundaram, C.; Reuter, S.; Aggarwal, B.B. Inhibiting NF-ΚB Activation by Small Molecules as a Therapeutic Strategy. Biochim. Biophys. Acta 2010, 1799, 775–787. [Google Scholar] [CrossRef] [Green Version]
- Li, H.Y.; Zhong, Y.F.; Wu, S.Y.; Shi, N. NF-E2 Related Factor 2 Activation and Heme Oxygenase-1 Induction by Tert-Butylhydroquinone Protect against Deltamethrin-Mediated Oxidative Stress in PC12 Cells. Chem. Res. Toxicol. 2007, 20, 1242–1251. [Google Scholar] [CrossRef]
- Sahin, K. Modulation of NF-ΚB and Nrf2 Pathways by Lycopene Supplementation in Heat-Stressed Poultry. World’s Poult. Sci. J. 2015, 71, 271–284. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Bramanti, P.; Mazzon, E. Activation of Nrf2 by Natural Bioactive Compounds: A Promising Approach for Stroke? Int. J. Mol. Sci. 2020, 21, 4875. [Google Scholar] [CrossRef] [PubMed]
- Tremmel, L.; Rho, O.; Slaga, T.J.; DiGiovanni, J. Inhibition of Skin Tumor Promotion by TPA Using a Combination of Topically Applied Ursolic Acid and Curcumin. Mol. Carcinog. 2019, 58, 185–195. [Google Scholar] [CrossRef]
- Cykowiak, M.; Krajka-Kuźniak, V.; Baer-Dubowska, W. Combinations of Phytochemicals More Efficiently than Single Components Activate Nrf2 and Induce the Expression of Antioxidant Enzymes in Pancreatic Cancer Cells. Nutr. Cancer 2021, 1–16. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Paluszczak, J.; Baer-Dubowska, W. Xanthohumol Induces Phase II Enzymes via Nrf2 in Human Hepatocytes in Vitro. Toxicol. Vitr. 2013, 27, 149–156. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Paluszczak, J.; Szaefer, H.; Baer-Dubowska, W. The Activation of the Nrf2/ARE Pathway in HepG2 Hepatoma Cells by Phytochemicals and Subsequent Modulation of Phase II and Antioxidant Enzyme Expression. J. Physiol. Biochem. 2015, 71, 227–238. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Szaefer, H.; Stefański, T.; Sobiak, S.; Cichocki, M.; Baer-Dubowska, W. The Effect of Resveratrol and Its Methylthio-Derivatives on the Nrf2-ARE Pathway in Mouse Epidermis and HaCaT Keratinocytes. Cell. Mol. Biol. Lett. 2014, 19, 500–516. [Google Scholar] [CrossRef]
- Krajka-Kuźniak, V.; Cykowiak, M.; Szaefer, H.; Kleszcz, R.; Baer-Dubowska, W. Combination of Xanthohumol and Phenethyl Isothiocyanate Inhibits NF-ΚB and Activates Nrf2 in Pancreatic Cancer Cells. Toxicol. Vitr. 2020, 65, 104799. [Google Scholar] [CrossRef] [PubMed]
- Gharibi, A.; Adamian, Y.; Kelber, J.A. Cellular and Molecular Aspects of Pancreatic Cancer. Acta Histochem. 2016, 118, 305–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paluszczak, J.; Krajka-Kuźniak, V.; Baer-Dubowska, W. The Effect of Dietary Polyphenols on the Epigenetic Regulation of Gene Expression in MCF7 Breast Cancer Cells. Toxicol. Lett. 2010, 192, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Pei, N.; Chen, X.; Chen, H.; Yan, R.; Bai, N.; Li, A.; Li, J.; Zhang, Y.; Du, H.; et al. Angiotensin 1-7 Overexpression Mediated by a Capsid-Optimized AAV8 Vector Leads to Significant Growth Inhibition of Hepatocellular Carcinoma In Vivo. Int. J. Biol. Sci. 2018, 14, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Nagai, N.; Ogura, K.; Tsuneyama, K.; Saiki, I.; Irimura, T.; Hayakawa, Y. Mammary Tissue Microenvironment Determines T Cell-Dependent Breast Cancer-Associated Inflammation. Cancer Sci. 2015, 106, 867–874. [Google Scholar] [CrossRef]
- Li, F.; Yao, Y.; Huang, H.; Hao, H.; Ying, M. Xanthohumol Attenuates Cisplatin-Induced Nephrotoxicity through Inhibiting NF-ΚB and Activating Nrf2 Signaling Pathways. Int. Immunopharmacol. 2018, 61, 277–282. [Google Scholar] [CrossRef]
- Lv, H.; Liu, Q.; Wen, Z.; Feng, H.; Deng, X.; Ci, X. Xanthohumol Ameliorates Lipopolysaccharide (LPS)-Induced Acute Lung Injury via Induction of AMPK/GSK3β-Nrf2 Signal Axis. Redox Biol. 2017, 12, 311–324. [Google Scholar] [CrossRef]
- Salazar, M.; Rojo, A.I.; Velasco, D.; de Sagarra, R.M.; Cuadrado, A. Glycogen Synthase Kinase-3beta Inhibits the Xenobiotic and Antioxidant Cell Response by Direct Phosphorylation and Nuclear Exclusion of the Transcription Factor Nrf2. J. Biol. Chem. 2006, 281, 14841–14851. [Google Scholar] [CrossRef] [Green Version]
- Jope, R.S.; Yuskaitis, C.J.; Beurel, E. Glycogen Synthase Kinase-3 (GSK3): Inflammation, Diseases, and Therapeutics. Neurochem. Res. 2007, 32, 577–595. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Lee, D.-Y.; Yun, S.-M.; Song, M.-Y.; Jung, K.; Kim, E.-H. Cyanidin Chloride Induces Apoptosis by Inhibiting NF-ΚB Signaling through Activation of Nrf2 in Colorectal Cancer Cells. Antioxidants 2020, 9, 285. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Zhou, X.; Liang, C.; Li, X.; Ge, M.; Chen, Y.; Yin, J.; Zhu, J.; Zhong, C. Apatinib Triggers Autophagic and Apoptotic Cell Death via VEGFR2/STAT3/PD-L1 and ROS/Nrf2/P62 Signaling in Lung Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 266. [Google Scholar] [CrossRef]
- Silva-Islas, C.A.; Maldonado, P.D. Canonical and Non-Canonical Mechanisms of Nrf2 Activation. Pharm. Res. 2018, 134, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; McMillan-Ward, E.; Kong, J.; Israels, S.J.; Gibson, S.B. Oxidative Stress Induces Autophagic Cell Death Independent of Apoptosis in Transformed and Cancer Cells. Cell Death Differ. 2008, 15, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Trachootham, D.; Zhou, Y.; Zhang, H.; Demizu, Y.; Chen, Z.; Pelicano, H.; Chiao, P.J.; Achanta, G.; Arlinghaus, R.B.; Liu, J.; et al. Selective Killing of Oncogenically Transformed Cells through a ROS-Mediated Mechanism by Beta-Phenylethyl Isothiocyanate. Cancer Cell 2006, 10, 241–252. [Google Scholar] [CrossRef] [Green Version]
- Schumacker, P.T. Reactive Oxygen Species in Cancer Cells: Live by the Sword, Die by the Sword. Cancer Cell 2006, 10, 175–176. [Google Scholar] [CrossRef] [Green Version]
- Waris, G.; Ahsan, H. Reactive Oxygen Species: Role in the Development of Cancer and Various Chronic Conditions. J. Carcinog. 2006, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Brücher, B.L.D.M.; Jamall, I.S. Synopsis: Special Issue on “Disruption of Signaling Homeostasis Induced Crosstalk in the Carcinogenesis Paradigm Epistemology of the Origin of Cancer”. 4open 2019, 2, 28. [Google Scholar] [CrossRef]
- Jung, B.-J.; Yoo, H.-S.; Shin, S.; Park, Y.-J.; Jeon, S.-M. Dysregulation of NRF2 in Cancer: From Molecular Mechanisms to Therapeutic Opportunities. Biomol. Ther. 2018, 26, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Jang, D.E.; Song, J.; Park, J.-W.; Yoon, S.-H.; Bae, Y.-S. Protein Kinase CK2 Activates Nrf2 via Autophagic Degradation of Keap1 and Activation of AMPK in Human Cancer Cells. BMB Rep. 2020, 53, 272–277. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Dashwood, R.H. Epigenetic Regulation of NRF2/KEAP1 by Phytochemicals. Antioxidants 2020, 9, 865. [Google Scholar] [CrossRef]
- Panieri, E.; Saso, L. Potential Applications of NRF2 Inhibitors in Cancer Therapy. Oxid. Med. Cell. Longev. 2019, 2019, 8592348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pramanik, K.C.; Makena, M.R.; Bhowmick, K.; Pandey, M.K. Advancement of NF-ΚB Signaling Pathway: A Novel Target in Pancreatic Cancer. Int. J. Mol. Sci. 2018, 19, 3890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondal, D.; Narwani, D.; Notta, S.; Ghaffar, D.; Mardhekar, N.; Quadri, S.S.A. Oxidative Stress and Redox Signaling in CRPC Progression: Therapeutic Potential of Clinically-Tested Nrf2-Activators. Cancer Drug Resist. 2021, 4, 96–124. [Google Scholar] [CrossRef]
- Pires, B.R.B.; Silva, R.C.M.C.; Ferreira, G.M.; Abdelhay, E. NF-KappaB: Two Sides of the Same Coin. Genes 2018, 9, 24. [Google Scholar] [CrossRef] [Green Version]
- Sheng, J.; Sun, H.; Yu, F.-B.; Li, B.; Zhang, Y.; Zhu, Y.-T. The Role of Cyclooxygenase-2 in Colorectal Cancer. Int. J. Med. Sci. 2020, 17, 1095–1101. [Google Scholar] [CrossRef]
- Majchrzak-Celińska, A.; Misiorek, J.O.; Kruhlenia, N.; Przybyl, L.; Kleszcz, R.; Rolle, K.; Krajka-Kuźniak, V. COXIBs and 2,5-Dimethylcelecoxib Counteract the Hyperactivated Wnt/β-Catenin Pathway and COX-2/PGE2/EP4 Signaling in Glioblastoma Cells. BMC Cancer 2021, 21, 493. [Google Scholar] [CrossRef]
- Duda, P.; Akula, S.M.; Abrams, S.L.; Steelman, L.S.; Martelli, A.M.; Cocco, L.; Ratti, S.; Candido, S.; Libra, M.; Montalto, G.; et al. Targeting GSK3 and Associated Signaling Pathways Involved in Cancer. Cells 2020, 9, 1110. [Google Scholar] [CrossRef] [PubMed]
- Domoto, T.; Uehara, M.; Bolidong, D.; Minamoto, T. Glycogen Synthase Kinase 3β in Cancer Biology and Treatment. Cells 2020, 9, 1388. [Google Scholar] [CrossRef]
- Augello, G.; Emma, M.R.; Cusimano, A.; Azzolina, A.; Montalto, G.; McCubrey, J.A.; Cervello, M. The Role of GSK-3 in Cancer Immunotherapy: GSK-3 Inhibitors as a New Frontier in Cancer Treatment. Cells 2020, 9, 1427. [Google Scholar] [CrossRef]
- Gameiro, I.; Michalska, P.; Tenti, G.; Cores, Á.; Buendia, I.; Rojo, A.I.; Georgakopoulos, N.D.; Hernández-Guijo, J.M.; Teresa Ramos, M.; Wells, G.; et al. Discovery of the First Dual GSK3β Inhibitor/Nrf2 Inducer. A New Multitarget Therapeutic Strategy for Alzheimer’s Disease. Sci. Rep. 2017, 7, 45701. [Google Scholar] [CrossRef]
- Wang, Y.-Q.; Chen, C.; Chen, Z.; Xu, Y.; Wang, Y.; Xiao, B.-K.; Chen, S.-M.; Tao, Z.-Z. Indole-3-Carbinol Inhibits Cell Proliferation and Induces Apoptosis in Hep-2 Laryngeal Cancer Cells. Oncol. Rep. 2013, 30, 227–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandberg, M.; Patil, J.; D’Angelo, B.; Weber, S.G.; Mallard, C. NRF2-Regulation in Brain Health and Disease: Implication of Cerebral Inflammation. Neuropharmacology 2014, 79, 298–306. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.-H.; Qu, J.; Shen, X. NF-KappaB/P65 Antagonizes Nrf2-ARE Pathway by Depriving CBP from Nrf2 and Facilitating Recruitment of HDAC3 to MafK. Biochim. Biophys. Acta 2008, 1783, 713–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, R.M.; Suzuki, Y.J. Cell Proliferation, Reactive Oxygen and Cellular Glutathione. Dose Response 2006, 3, 425–442. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, N.; Das, A.; Chaffee, S.; Roy, S.; Sen, C.K. Chapter 4—Reactive Oxygen Species, Oxidative Damage and Cell Death. In Immunity and Inflammation in Health and Disease; Chatterjee, S., Jungraithmayr, W., Bagchi, D., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 45–55. ISBN 978-0-12-805417-8. [Google Scholar]
- Ryter, S.W.; Kim, H.P.; Hoetzel, A.; Park, J.W.; Nakahira, K.; Wang, X.; Choi, A.M.K. Mechanisms of Cell Death in Oxidative Stress. Antioxid. Redox Signal. 2007, 9, 49–89. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, V.; Park, Y.; Chen, C.-C.; Xu, P.-Z.; Chen, M.-L.; Tonic, I.; Unterman, T.; Hay, N. Akt Determines Replicative Senescence and Oxidative or Oncogenic Premature Senescence and Sensitizes Cells to Oxidative Apoptosis. Cancer Cell 2008, 14, 458–470. [Google Scholar] [CrossRef] [Green Version]
- Raghunath, A.; Sundarraj, K.; Arfuso, F.; Sethi, G.; Perumal, E. Dysregulation of Nrf2 in Hepatocellular Carcinoma: Role in Cancer Progression and Chemoresistance. Cancers 2018, 10, 481. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, M.; Yoshida, R.; Shimada, N.; Yamazaki, H.; Yokoi, T. Inhibition and Inactivation of Human Cytochrome P450 Isoforms by Phenethyl Isothiocyanate. Drug Metab. Dispos. 2001, 29, 1110–1113. [Google Scholar]
- Liu, M.; Hansen, P.E.; Wang, G.; Qiu, L.; Dong, J.; Yin, H.; Qian, Z.; Yang, M.; Miao, J. Pharmacological Profile of Xanthohumol, a Prenylated Flavonoid from Hops (Humulus Lupulus). Molecules 2015, 20, 754–779. [Google Scholar] [CrossRef]
- Morciano, G.; Sarti, A.C.; Marchi, S.; Missiroli, S.; Falzoni, S.; Raffaghello, L.; Pistoia, V.; Giorgi, C.; Di Virgilio, F.; Pinton, P. Use of Luciferase Probes to Measure ATP in Living Cells and Animals. Nat. Protoc. 2017, 12, 1542–1562. [Google Scholar] [CrossRef]
- Auld, D.S.; Thorne, N.; Maguire, W.F.; Inglese, J. Mechanism of PTC124 Activity in Cell-Based Luciferase Assays of Nonsense Codon Suppression. Proc. Natl. Acad. Sci. USA 2009, 106, 3585–3590. [Google Scholar] [CrossRef] [Green Version]
- Braeuning, A.; Vetter, S. The Nuclear Factor ΚB Inhibitor (E)-2-Fluoro-4′-Methoxystilbene Inhibits Firefly Luciferase. Biosci. Rep. 2012, 32, 531–537. [Google Scholar] [CrossRef] [Green Version]
- Auld, D.S.; Inglese, J. Interferences with Luciferase Reporter Enzymes. In Assay Guidance Manual; Markossian, S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C.P., Baell, J., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Auld, D.S.; Southall, N.T.; Jadhav, A.; Johnson, R.L.; Diller, D.J.; Simeonov, A.; Austin, C.P.; Inglese, J. Characterization of Chemical Libraries for Luciferase Inhibitory Activity. J. Med. Chem. 2008, 51, 2372–2386. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S.; Thorne, N.; Nguyen, D.-T.; Inglese, J. A Specific Mechanism for Nonspecific Activation in Reporter-Gene Assays. ACS Chem. Biol. 2008, 3, 463–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, J.B.; Stowers, C.C.; Boczko, E.; Johnson, C.H. Real-Time Luminescence Monitoring of Cell-Cycle and Respiratory Oscillations in Yeast. Proc. Natl. Acad. Sci. USA 2008, 105, 17988–17993. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Forward Primer | Reverse Primer | |
|---|---|---|
| Human | ||
| PBGD | 5′ TCAGATAGCATACAAGAGACC | 5′ TGGAATGTTACGAGCAGTG |
| TBP | 5′ GGCACCACTCCACTGTATC | 5′ GGGATTATATTCGGCGTTTCG |
| Nrf2 | 5′ ATTGCTACTAATCAGGCTCAG | 5′ GTTTGGCTTCTGGACTTGG |
| SOD | 5′ CGACAGAAGGAAAGTAATG | 5′ TGGATAGAGGATTAAAGTGAGG |
| GSTP | 5′ GCAAATACATCTCCCTCATC | 5′AGGTTGTAGTCAGCGAAG |
| NQO1 | 5′ CAATTCAGAGTGGCATTC | 5′ GAAGTTTAGGTCAAAGAGG |
| Keap1 | 5′ ATGGGCGAGAAGTGTGTC | 5′ TCTGCTCAGCGAAGTTGG |
| p62 | 5′ TCTGGGCATTGAAGTTGA | 5′ CTCTGTGCTGGAACTCTC |
| GSK3β | 5′ ACCCAAATGTAAACTACCAAATG | 5′ TCCACGGTCTCCAGTATTAGC |
| NF-κB p50 | 5′ ATCATCCACCTTCATTCTCAA | 5′ AATCCTCCACCACATCTTCC |
| NF-κB p65 | 5′ CGCCTGTCCTTTCTCATC | 5′ ACCTCAATGTCCTCTTTCTG |
| COX-2 | 5′ CCTGTGCCTGATGATTGC | 5′ CAGCCCGTTGGTGAAAGC |
| Mouse | ||
| PBGD | 5′ GCCTACCATACTACCTCCT | 5′ AAGACAACAGCATCACAAG |
| TBP | 5′ TATTGTATCTACCGTGAAT | 5′ TAGTCTGGATTGTTCTTC |
| Nrf2 | 5′ CAGCATAGAGCAGGACAT | 5′ TTCGGTATTAAGACACTTAATTC |
| SOD | 5′ GGACAAATTACAGGATTA | 5′ TTCTTAGAGTGAGGATTA |
| NF-κB p50 | 5′ CCTCTAGTGAGAAGAACAA | 5′ TGACCAACTGAACGATAA |
| NF-κB p65 | 5′ TACTTGCCAGACACAGATG | 5′ GATACTCTTGAAGGTCTCATAGG |
| Compound/Mixture | PSN-1 | MS1 | ||
|---|---|---|---|---|
| IC50 | ±SD | IC50 | ±SD | |
| I3C | 130.0 | 6.9 | >150 | |
| XN | 46.0 | 3.0 | 49.0 | 3.0 |
| PEITC | 32.0 | 2.5 | 35.5 | 2.4 |
| RVS | 112.0 | 4.2 | >150 | |
| XN+I3C | 61.0 | 4.5 | 67.0 | 4.5 |
| XN+PEITC | 34.5 | 2.6 | 38.0 | 2.2 |
| XN+RVS | 59.0 | 6.0 | 63.0 | 4.1 |
| I3C+PEITC | 36.0 | 4.0 | 41.0 | 1.5 |
| I3C+RVS | 148.0 | 6.5 | >150 | |
| RVS+PEITC | 40.0 | 4.1 | 41.0 | 5.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cykowiak, M.; Kleszcz, R.; Kucińska, M.; Paluszczak, J.; Szaefer, H.; Plewiński, A.; Piotrowska-Kempisty, H.; Murias, M.; Krajka-Kuźniak, V. Attenuation of Pancreatic Cancer In Vitro and In Vivo via Modulation of Nrf2 and NF-κB Signaling Pathways by Natural Compounds. Cells 2021, 10, 3556. https://doi.org/10.3390/cells10123556
Cykowiak M, Kleszcz R, Kucińska M, Paluszczak J, Szaefer H, Plewiński A, Piotrowska-Kempisty H, Murias M, Krajka-Kuźniak V. Attenuation of Pancreatic Cancer In Vitro and In Vivo via Modulation of Nrf2 and NF-κB Signaling Pathways by Natural Compounds. Cells. 2021; 10(12):3556. https://doi.org/10.3390/cells10123556
Chicago/Turabian StyleCykowiak, Marta, Robert Kleszcz, Małgorzata Kucińska, Jarosław Paluszczak, Hanna Szaefer, Adam Plewiński, Hanna Piotrowska-Kempisty, Marek Murias, and Violetta Krajka-Kuźniak. 2021. "Attenuation of Pancreatic Cancer In Vitro and In Vivo via Modulation of Nrf2 and NF-κB Signaling Pathways by Natural Compounds" Cells 10, no. 12: 3556. https://doi.org/10.3390/cells10123556
APA StyleCykowiak, M., Kleszcz, R., Kucińska, M., Paluszczak, J., Szaefer, H., Plewiński, A., Piotrowska-Kempisty, H., Murias, M., & Krajka-Kuźniak, V. (2021). Attenuation of Pancreatic Cancer In Vitro and In Vivo via Modulation of Nrf2 and NF-κB Signaling Pathways by Natural Compounds. Cells, 10(12), 3556. https://doi.org/10.3390/cells10123556