Pre-Harvest Application of Strigolactone (GR24) Accelerates Strawberry Ripening and Improves Fruit Quality
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of the Strawberry Fruit Quality Attributes
2.3. RNA Extraction, Library Construction, Sequencing, and Data Assembly
2.4. Metabolite Extraction, Detection, and Data Analysis
2.5. Gene Expression Validation
2.6. Data Processing and Analysis
3. Results
3.1. Effects of Pre-Harvest GR24 Treatment on Appearance Characteristics of Strawberry Fruit
3.2. Effects of Pre-Harvest GR24 Treatment on Nutritional Quality Parameters of Strawberries
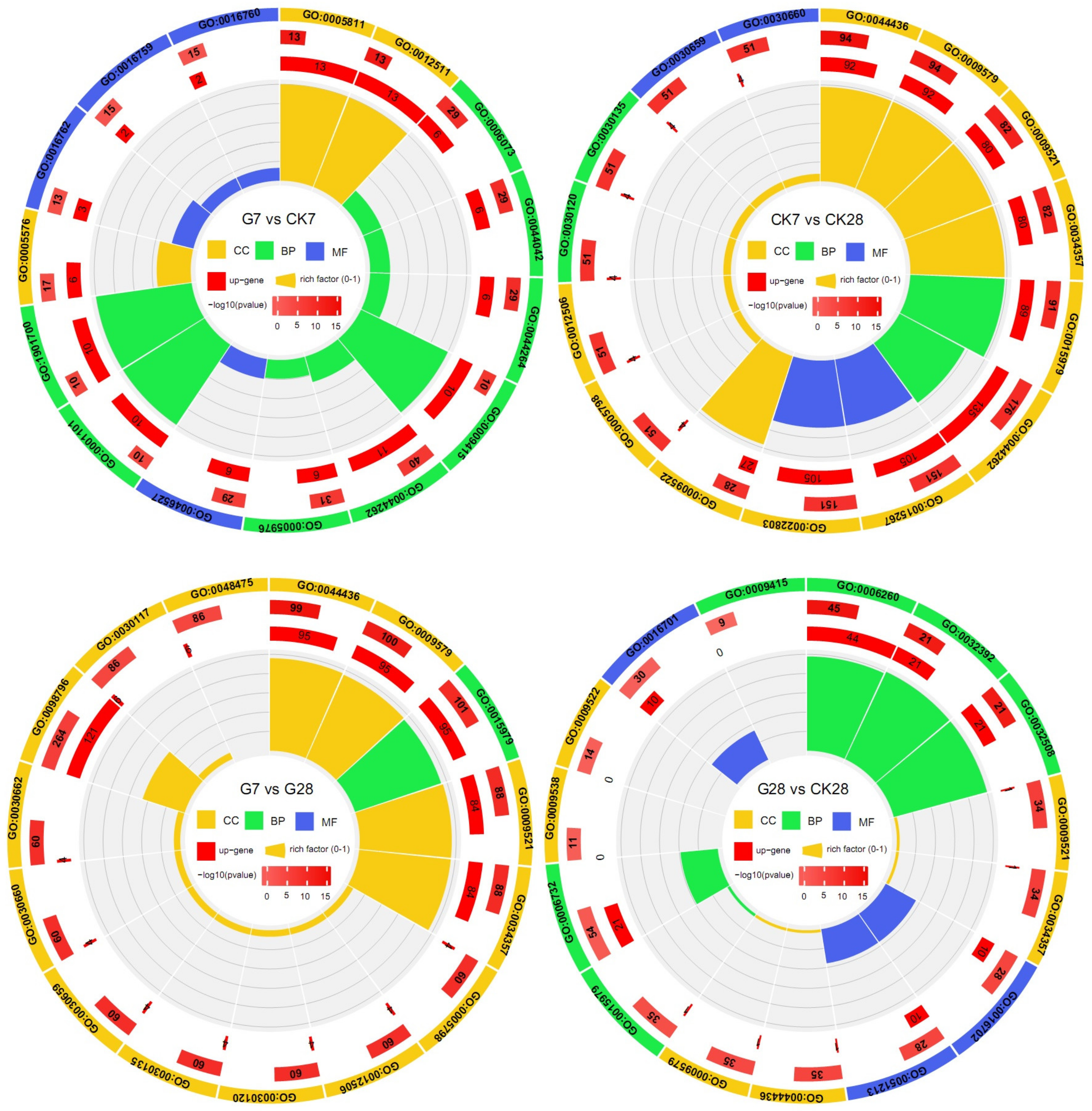
3.3. Transcriptomic Analysis of the Effect of GR24 on Fruit Quality
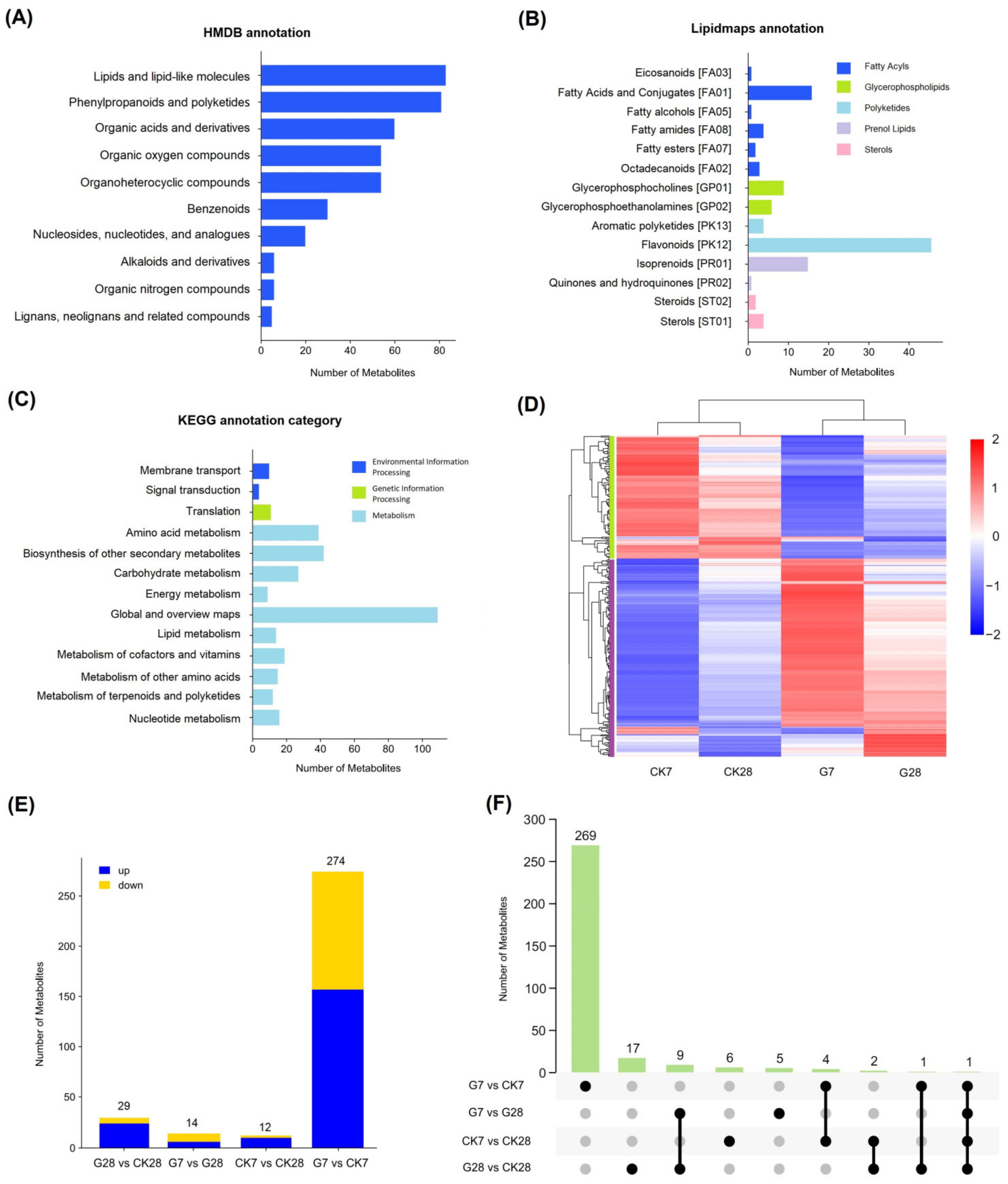
3.4. Metabolomic Analysis of the Effect of GR24 on Strawberry Fruit Quality
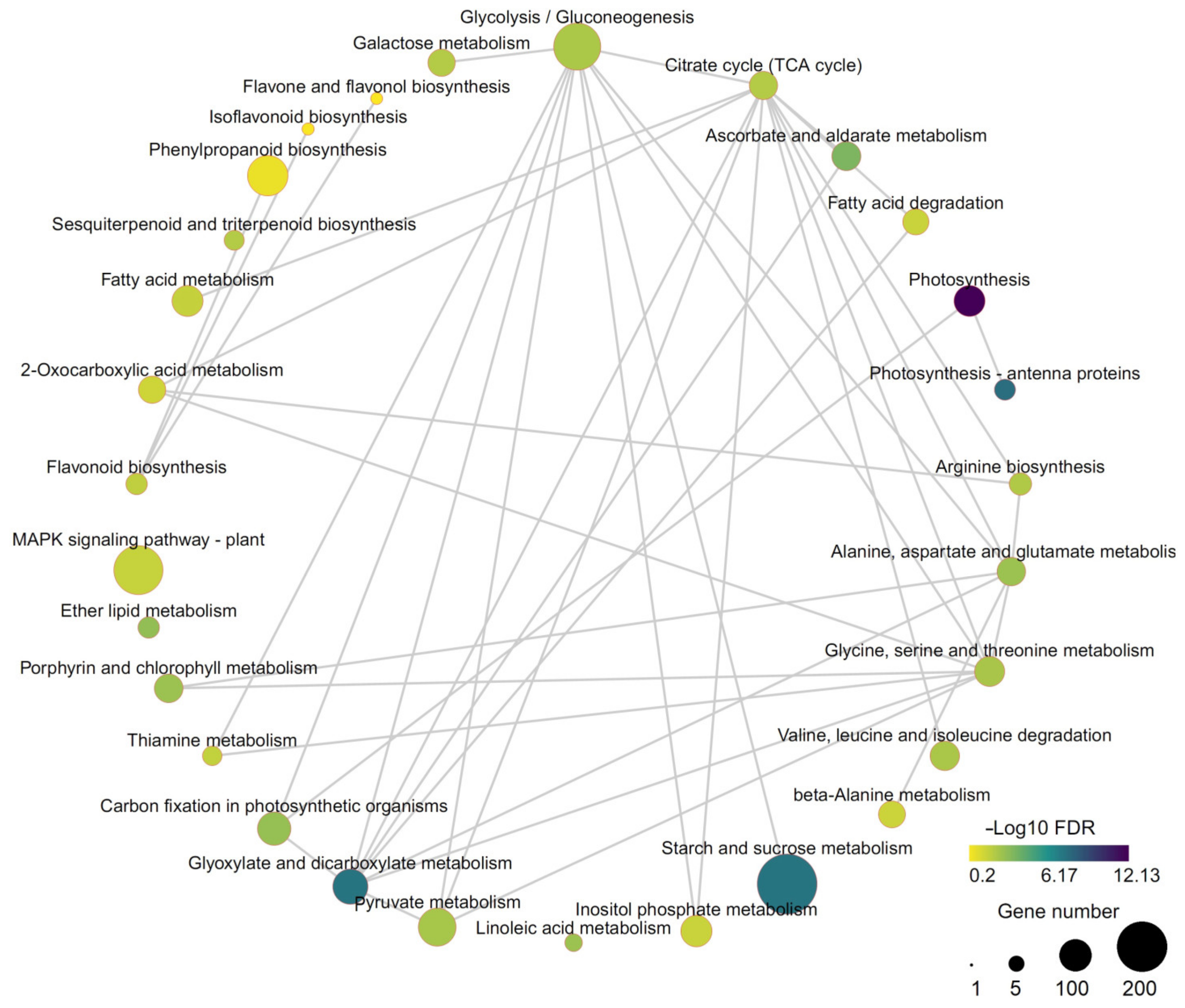
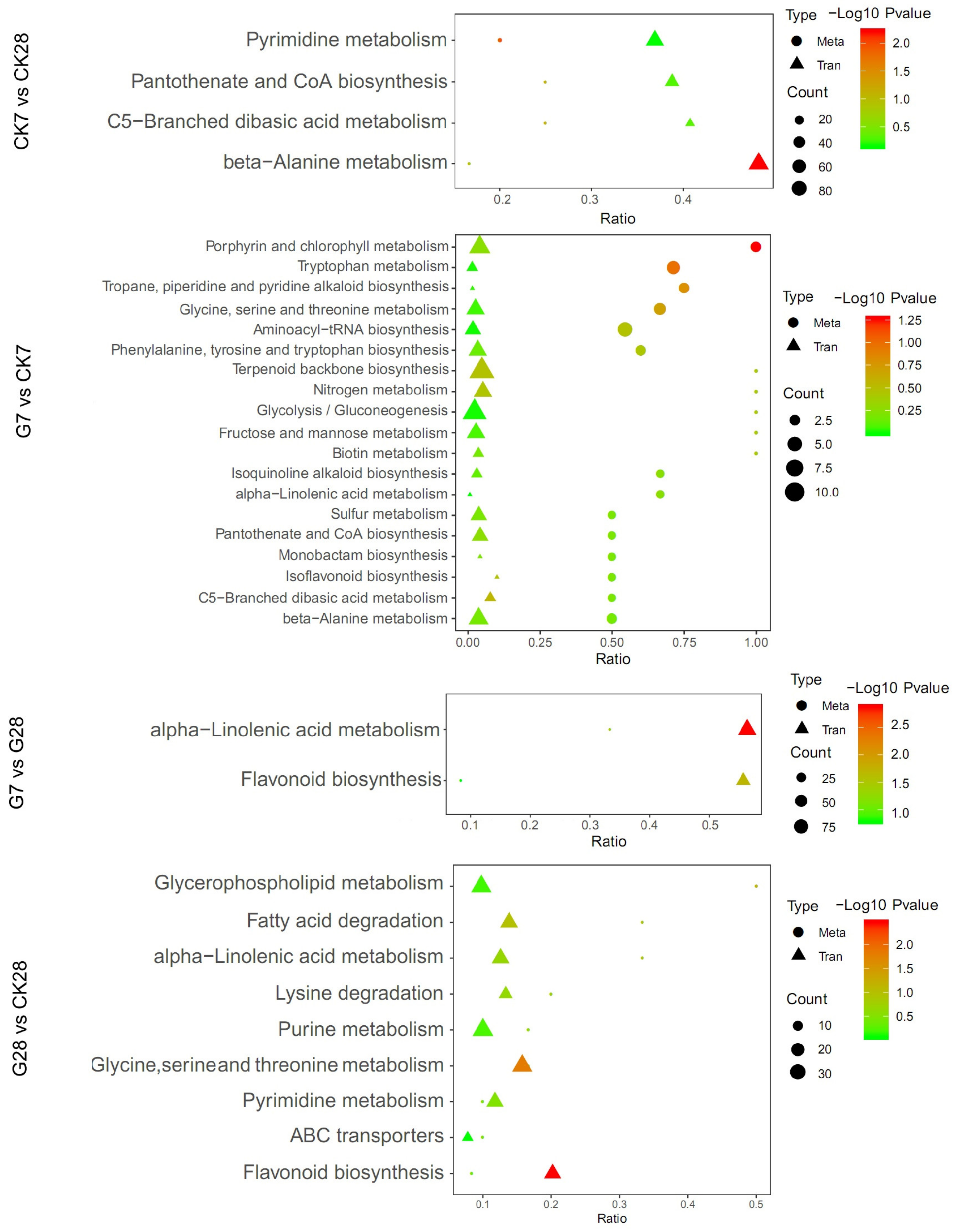
3.5. KEGG Enrichment Analysis and Transcriptome and Metabolome Correlation Analysis
3.6. Analysis of the Sugar Biosynthesis Pathway Regulated by GR24 in Strawberry Fruit Ripening
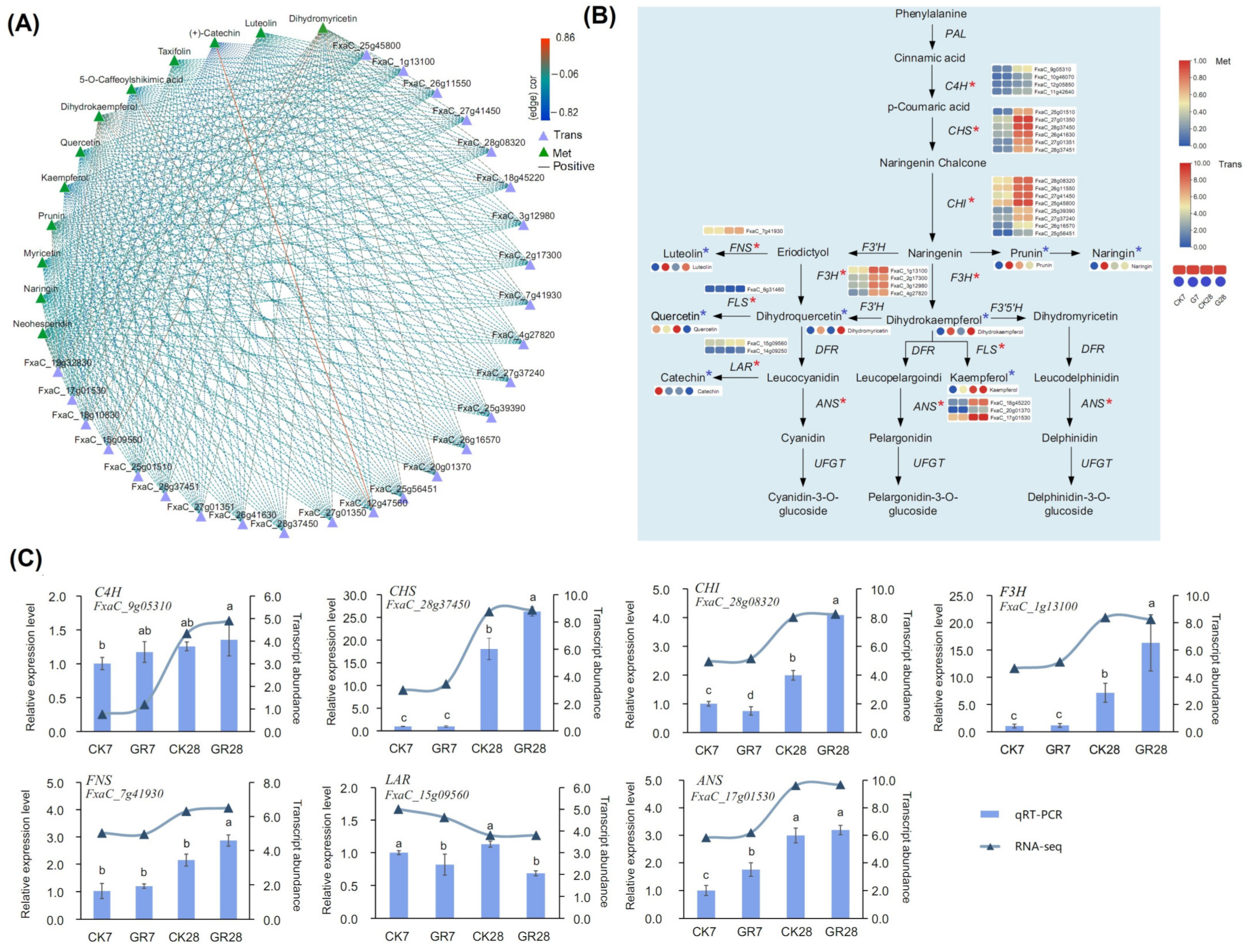
3.7. Analysis of the Anthocyanin Pathway Regulated by GR24 in Strawberry Fruit Ripening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Giampieri, F.; Alvarez-Suarez, J.M.; Battino, M. Strawberry and human health: Effects beyond antioxidant activity. J. Agric. Food Chem. 2014, 62, 3867–3876. [Google Scholar] [CrossRef] [PubMed]
- Taha, I.M.; Zaghlool, A.; Nasr, A.; Nagib, A.; El Azab, I.H.; Mersal, G.A.M.; Ibrahim, M.M.; Fahmy, A. Impact of Starch Coating Embedded with Silver Nanoparticles on Strawberry Storage Time. Polymers 2022, 14, 1439. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, Y.; Wei, X.; Wang, Z.; Wang, C.; Du, X.; Lin, Y.; Zhang, Y.; He, W.; Wang, X.; et al. Effects of pretreatment and freezing storage on the bioactive components and antioxidant activity of two kinds of celery after postharvest. Food Chem. X 2023, 18, 100655. [Google Scholar] [CrossRef]
- Li, D.; Li, L.; Xiao, G.; Limwachiranon, J.; Xu, Y.; Lu, H.; Yang, D.; Luo, Z. Effects of elevated CO2 on energy metabolism and γ-aminobutyric acid shunt pathway in postharvest strawberry fruit. Food Chem. 2018, 265, 281–289. [Google Scholar] [CrossRef]
- Silva, G.M.C.; Silva, W.B.; Medeiros, D.B.; Salvador, A.R.; Cordeiro, M.H.M.; da Silva, N.M.; Santana, D.B.; Mizobutsi, G.P. The chitosan affects severely the carbon metabolism in mango (Mangifera indica L. cv. Palmer) fruit during storage. Food Chem. 2017, 237, 372–378. [Google Scholar] [CrossRef]
- Xu, Y.; Charles, M.T.; Luo, Z.; Mimee, B.; Tong, Z.; Roussel, D.; Rolland, D.; Véronneau, P.Y. Preharvest UV-C treatment affected postharvest senescence and phytochemicals alternation of strawberry fruit with the possible involvement of abscisic acid regulation. Food Chem. 2019, 299, 125138. [Google Scholar] [CrossRef]
- Forges, M.; Bardin, M.; Urban, L.; Aarrouf, J.; Charles, F. Impact of UV-C radiation applied during plant growth on pre- and postharvest disease sensitivity and fruit quality of strawberry. Plant Dis. 2020, 104, 3239–3247. [Google Scholar] [CrossRef]
- Zuñiga, P.E.; Castañeda, Y.; Arrey-Salas, O.; Fuentes, L.; Aburto, F.; Figueroa, C.R. Methyl jasmonate applications from flowering to ripe fruit stages of strawberry (Fragaria × ananassa ‘Camarosa’) reinforce the fruit antioxidant response at post-harvest. Front. Plant Sci. 2020, 11, 538. [Google Scholar] [CrossRef]
- Feliziani, E.; Landi, L.; Romanazzi, G. Preharvest treatments with chitosan and other alternatives to conventional fungicides to control postharvest decay of strawberry. Carbohydr. Polym. 2015, 132, 111–117. [Google Scholar] [CrossRef]
- Kim, I.; Ahn, D.; Choi, J.H.; Lim, J.H.; Ok, G.; Park, K.J.; Lee, J. Changes in volatile compounds in short-term high CO2-treated ‘Seolhyang’ strawberry (Fragaria × ananassa) fruit during cold storage. Molecules 2022, 19, 6599. [Google Scholar] [CrossRef]
- Zhang, C.; Li, W.; Zhu, B.; Chen, H.; Chi, H.; Li, L.; Qin, Y.; Xue, J. The quality evaluation of postharvest strawberries stored in nano-ag packages at refrigeration temperature. Polymers 2018, 10, 894. [Google Scholar] [CrossRef]
- Humphrey, A.J.; Galster, A.M.; Beale, M.H. Strigolactones in chemical ecology: Waste products or vital allelochemicals? Nat. Prod. Rep. 2006, 23, 592–614. [Google Scholar] [CrossRef]
- Kapulnik, Y.; Koltai, H. Strigolactone involvement in root development, response to abiotic stress, and interactions with the biotic soil environment. Plant Physiol. 2014, 166, 560–569. [Google Scholar] [CrossRef]
- Koltai, H.; Prandi, C. Strigolactones: Biosynthesis, synthesis and functions in plant growth and stress responses. Phytohorm. Window Metab. Signal. Biotechnol. Appl. 2014, 265–288. [Google Scholar] [CrossRef]
- Cui, H.M.; Cao, X.W.; Wang, J.J.; Xiong, A.S.; Hou, X.L.; Li, Y. Effects of exogenous GR24 on the growth of axillary bud of non- heading Chinese cabbage. J. Nanjing Agric. Univ. 2016, 3, 7. [Google Scholar]
- Wani, K.I.; Zehra, A.; Choudhary, S.; Naeem, M.; Khan, M.M.A.; Khan, R.; Aftab, T. Exogenous strigolactone (gr24) positively regulates growth, photosynthesis, and improves glandular trichome attributes for enhanced artemisinin production in artemisia annua. J. Plant Growth Regul. 2022, 42, 4606–4615. [Google Scholar] [CrossRef]
- Ma, Q.L.; Lin, X.; Zhan, M.X.; Chen, Z.X.; Wang, H.G.; Yao, F.X.; Chen, J.Y. Effect of an exogenous strigolactone GR24 on the antioxidant capacity and quality deterioration in postharvest sweet orange fruit stored at ambient temperature. Int. J. Food Sci. Technol. 2022, 57, 619–630. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; Wang, S.; Wang, W.; Xu, X.; Wu, J.; Fang, Y.; Ju, Y. Effects of strigolactone and abscisic acid on the quality and anti-oxidant activity of grapes (Vitis vinifera L.) and wines. Food Chem. X 2022, 16, 100496. [Google Scholar] [CrossRef]
- Reis, L.; Forney, C.F.; Jordan, M.; Munro Pennell, K.; Fillmore, S.; Schemberger, M.O.; Ayub, R.A. Metabolic Profile of Strawberry Fruit Ripened on the Plant Following Treatment With an Ethylene Elicitor or Inhibitor. Front. Plant Sci. 2020, 11, 995. [Google Scholar] [CrossRef]
- Acquaviva, R.; Russo, A.; Galvano, F.; Galvano, G.; Barcellona, M.L.; Volti, G.L.; Vanella, A. Cyanidin and cyanidin 3-O-β-D-glucoside as DNA cleavage protectors and antioxidants. Cell Biol. Toxicol. 2003, 19, 243–252. [Google Scholar] [CrossRef]
- Ganhão, R.; Pinheiro, J.; Tino, C.; Faria, H.; Gil, M.M. Characterization of nutritional, physicochemical, and phytochemical composition and antioxidant capacity of three strawberry “Fragaria × ananassa Duch.” Cultivars (“primoris”, “endurance”, and “portola”) from western region of portugal. Foods 2019, 8, 682. [Google Scholar] [CrossRef] [PubMed]
- Parra-Palma, C.; Fuentes, E.; Palomo, I.; Torres, C.A.; Ramos, P. Linking the platelet antiaggregation effect of different strawberries species with antioxidants: Metabolomic and transcript profiling of polyphenols. Bol. Latinoam. Caribe Plantas Med. Aromát. 2017, 17, 36. [Google Scholar]
- Li, M.; Li, J.; Tan, H.; Luo, Y.; Zhang, Y.; Chen, Q.; Wang, Y.; Lin, Y.; Zhang, Y.; Wang, X.; et al. Comparative metabolomics provides novel insights into the basis of petiole color differences in celery (Apium graveolens L.). J. Zhejiang Univ. B 2022, 23, 300–314. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, L.; Liu, Y.; Lin, Y.; Zhang, Y.; Long, Y.; Luo, C.; Zhang, Y.; Chen, Q.; Chen, P.; et al. Characterization and regulation mechanism analysis of ubiqui-tin-conjugating family genes in strawberry reveals a potential role in fruit ripening. BMC Plant Biol. 2022, 22, 39–54. [Google Scholar] [CrossRef]
- García-Pastor, M.E.; Zapata, P.J.; Castillo, S.; Martínez-Romero, D.; Valero, D.; Serrano, M.; Guillén, F. Preharvest salicylate treatments enhance antioxidant compounds, color and crop yield in low pigmented-table grape cultivars and preserve quality traits during storage. Antioxidants 2020, 9, 832. [Google Scholar] [CrossRef]
- Erogul, D.; Sen, F. Effects of gibberellic acid tretatment on fruit thinning and fruit quality in Japanese plum (Prunus salicina Lindl). Sci. Hortic. 2015, 186, 137–142. [Google Scholar] [CrossRef]
- Lama, K.; Yadav, S.; Rosianski, Y.; Shaya, F.; Lichter, A.; Chai, L.; Dahan, Y.; Freiman, Z.; Peer, R.; Flaishman, M.A. The distinct ripening processes in the reproductive and non-reproductive parts of the fig syconium are driven by ABA. J. Exp. Bot. 2019, 70, 115–131. [Google Scholar] [CrossRef]
- Rozpądek, P.; Domka, A.M.; Nosek, M.; Ważny, R.; Jędrzejczyk, R.J.; Wiciarz, M.; Turnau, K. The role of strigolactone in the cross-talk between arabidopsis thaliana and the endophytic fungus mucorsp. Front. Microbiol. 2018, 9, 441–454. [Google Scholar] [CrossRef]
- Ruyter-Spira, C.; Kohlen, W.; Charnikhova, T.; van Zeijl, A.; van Bezouwen, L.; De Ruijter, N.; Cardoso, C.; Lopez-Raez, J.A.; Matusova, R.; Bours, R.; et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2010, 2, 721–734. [Google Scholar] [CrossRef]
- Qiu, S.; Wang, J.; Gao, L. Discrimination and characterization of strawberry juice based on electronic nose and tongue: Com-parison of different juice processing approaches by LDA, PLSR, RF, and SVM. J. Agric. Food Chem. 2014, 27, 6426–6434. [Google Scholar] [CrossRef]
- Akhatou, I.; Fernández-Recamales, A. Nutritional and nutraceutical quality of strawberries in relation to harvest time and crop conditions. J. Agric. Food Chem. 2014, 62, 5749–5760. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, H.; Zhou, X.; Zhang, J.; Xiao, H. Transcriptome reveals insights into biosynthesis of ginseng polysaccharides. BMC Plant Biol. 2022, 22, 594. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; Macrae, E. New complexities in the synthesis of sucrose. Curr. Opin. Plant Biol. 2003, 6, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Cabello, S.; Lorenz, C.; Crespo, S.; Cabrera, J.; Ludwig, R.; Escobar, C.; Hofmann, J. Altered sucrose synthase and invertase expression affects the local and systemic sugar metabolism of nematode-infected Arabidopsis thaliana plants. J. Exp. Bot. 2014, 65, 201–212. [Google Scholar] [CrossRef]
- Griesser, M.; Hoffmann, T.; Bellido, M.L.; Rosati, C.; Fink, B.; Kurtzer, R.; Aharoni, A.; Muñoz-Blanco, J.; Schwab, W. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 2008, 146, 1528–1539. [Google Scholar] [CrossRef]
- Li, Y.; Fang, J.; Qi, X.; Lin, M.; Zhong, Y.; Sun, L.; Cui, W. Combined analysis of the fruit metabolome and transcriptome reveals candidate genes involved in flavonoid biosynthesis in Actinidia arguta. Int. J. Mol. Sci. 2018, 19, 1471. [Google Scholar] [CrossRef]
- Yuan, H.; Yu, H.; Huang, T.; Shen, X.; Xia, J.; Pang, F.; Wang, J.; Zhao, M. The complexity of the Fragaria × ananassa (octoploid) tran-scriptome by single-molecule long-read sequencing. Hort. Res. 2019, 6, 46–59. [Google Scholar] [CrossRef]
- Carbone, F.; Preuss, A.; De Vos, R.C.H.; D’Amico, E.; Perrotta, G.; Bovy, A.G.; Martens, S.; Rosati, C. Developmental, genetic and environmental factors affect the expression of flavonoid genes, enzymes and metabolites in strawberry fruits. Plant Cell Environ. 2009, 32, 1117–1131. [Google Scholar] [CrossRef]
- Seymour, G.B.; Chapman, N.H.; Chew, B.L.; Rose, J.K.C. Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol. J. 2013, 11, 269–278. [Google Scholar] [CrossRef]
- Yi, G.; Shin, H.; Min, K.; Lee, E.J. Expanded transcriptomic view of strawberry fruit ripening through meta-analysis. PLoS ONE 2021, 16, e0252685. [Google Scholar] [CrossRef]
- Darwish, A.G.; Das, P.R.; Ismail, A.; Gajjar, P.; Balasubramani, S.P.; Sheikh, M.B.; Tsolova, V.; Sherif, S.M.; El-Sharkawy, I. Untargeted metabolomics and antioxidant capacities of muscadine grape genotypes during berry development. Antioxidants 2021, 10, 914. [Google Scholar] [CrossRef] [PubMed]
- Pillet, J.; Yu, H.-W.; Chambers, A.H.; Whitaker, V.M.; Folta, K.M. Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J. Exp. Bot. 2015, 66, 4455–4467. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A.C.; Acunha, T.d.S.; Perin, E.C.; Rombaldi, C.V.; Galli, V.; Chaves, F.C. Untargeted metabolomics of strawberry (Fragaria x ananassa ‘Camarosa’) fruit from plants grown under osmotic stress conditions. J. Sci. Food Agric. 2019, 99, 6973–6980. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, L.; Luo, Z.; Mou, W.; Mao, L.; Ying, T. Comparative transcriptome analysis reveals the influence of abscisic acid on the metabolism of pigments, ascorbic acid and folic acid during strawberry fruit ripening. PLoS ONE 2015, 10, e0130037. [Google Scholar] [CrossRef]
- Li, M.; Li, J.; Zhang, R.; Lin, Y.; Xiong, A.; Tan, G.; Luo, Y.; Zhang, Y.; Chen, Q.; Wang, Y.; et al. Combined analysis of the metabolome and transcriptome to explore heat stress responses and adaptation mechanisms in celery (Apium graveolens L.). Int. J. Mol. Sci. 2022, 23, 3367. [Google Scholar] [CrossRef]
- Zheng, T.; Lv, J.; Sadeghnezhad, E.; Cheng, J.; Jia, H. Transcriptomic and metabolomic profiling of strawberry during postharvest cooling and heat storage. Front. Plant Sci. 2022, 13, 1009747. [Google Scholar] [CrossRef]
- Yang, H.; Tian, C.; Ji, S.; Ni, F.; Fan, X.; Yang, Y.; Sun, C.; Gong, H.; Zhang, A. Integrative analyses of metabolome and transcriptome reveals metabolomic variations and candidate genes involved in sweet cherry (Prunus avium L.) fruit quality during development and ripening. PLoS ONE 2021, 16, e0260004. [Google Scholar] [CrossRef]
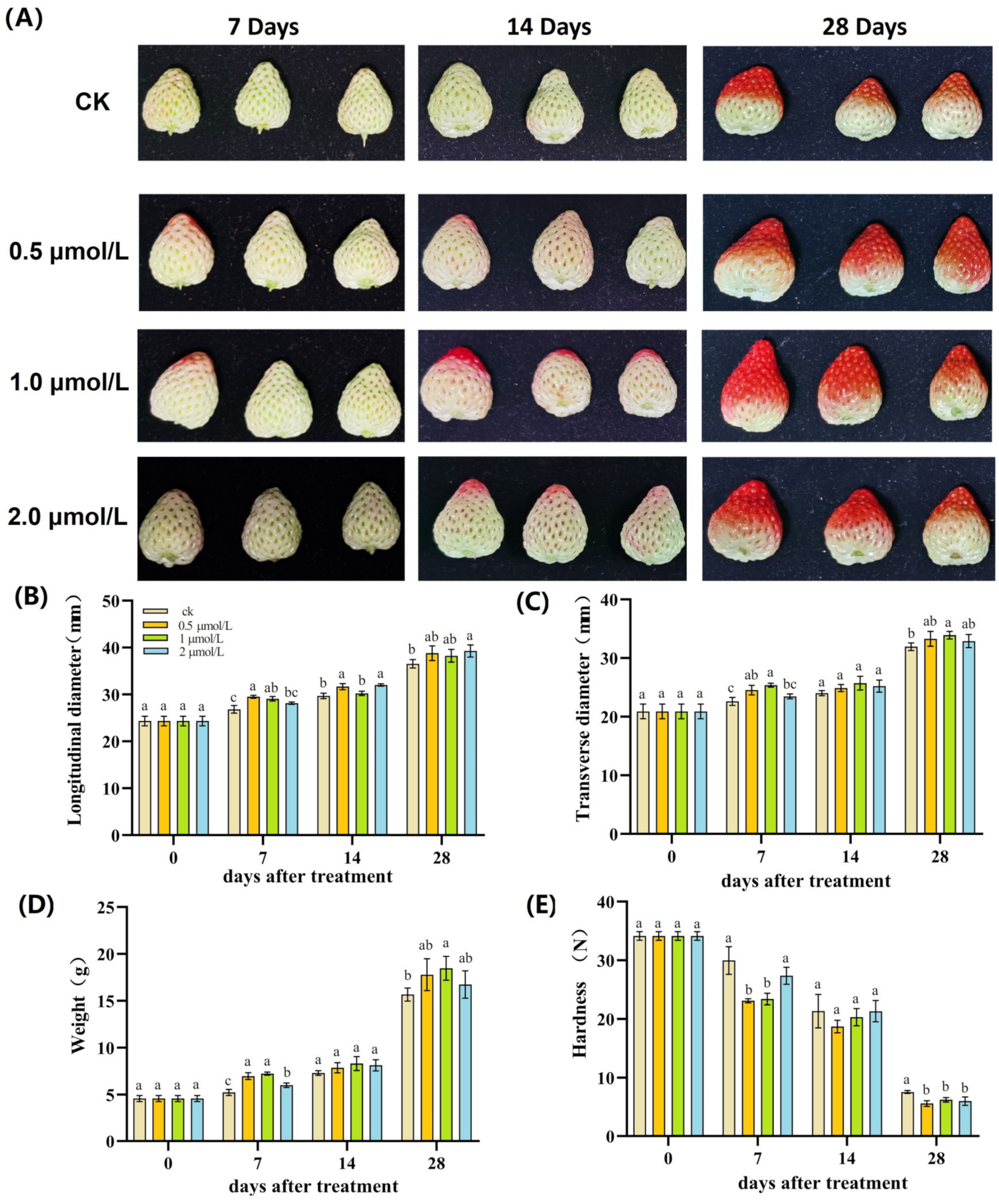
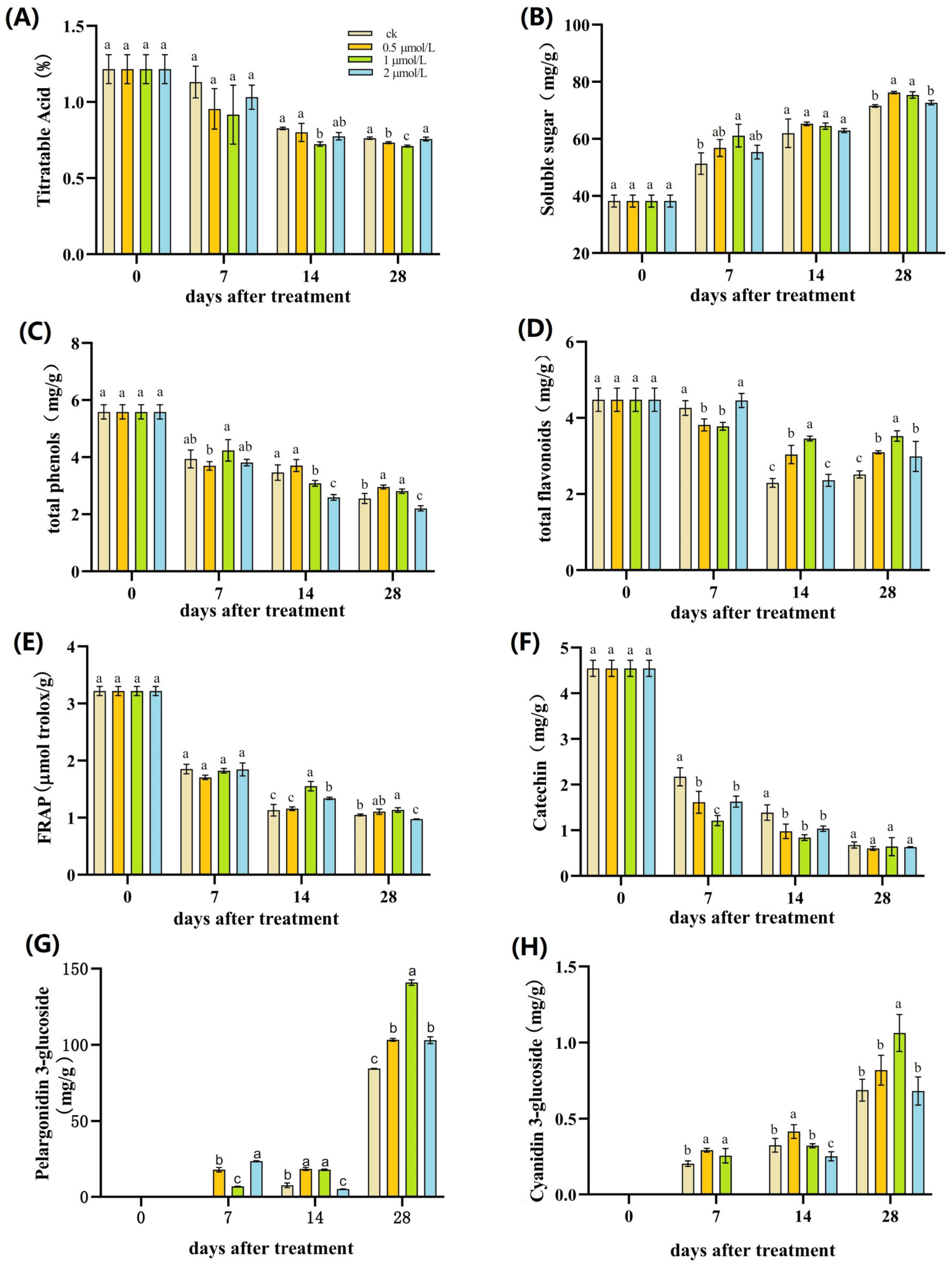


| Days | Treatments | L* | a* | b* |
|---|---|---|---|---|
| DAT 7 | CK | 74.473 ± 0.150 a | −6.851 ± 0.183 a | 46.105 ± 0.332 a |
| 0.5 µmol/L GR24 | 75.691 ± 0.312 a | −6.578 ± 0.114 a | 45.474 ± 0.151 a | |
| 1 µmol/L GR24 | 75.431 ± 1.083 a | −6.564 ± 0.116 a | 45.510 ± 0.702 a | |
| 2 µmol/L GR24 | 76.073 ± 0.286 a | −6.903 ± 0.403 a | 46.728 ± 0.148 a | |
| DAT 14 | CK | 67.427 ± 0.456 a | −7.374 ± 0.247 a | 34.056 ± 0.508 a |
| 0.5 µmol/L GR24 | 68.173 ± 0.832 a | −5.474 ± 1.367 a | 32.924 ± 1.520 ab | |
| 1 µmol/L GR24 | 67.787 ± 0.442 a | −5.454 ± 0.204 a | 31.830 ± 0.053 ab | |
| 2 µmol/L GR24 | 67.615 ± 0.730 a | −5.014 ± 0.357 a | 31.149 ± 0.235 b | |
| DAT 28 | CK | 46.256 ± 2.106 a | 33.138 ± 1.783 b | 32.372 ± 0.225 a |
| 0.5 µmol/L GR24 | 42.241 ± 1.168 a | 37.256 ± 0.826 a | 30.527 ± 1.469 a | |
| 1 µmol/L GR24 | 42.523 ± 0.347 a | 37.485 ± 1.251 a | 31.495 ± 0.445 a | |
| 2 µmol/L GR24 | 41.898 ± 1.201 a | 38.331 ± 0.228 a | 30.912 ± 0.432 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Yang, M.; Liu, X.; Hou, G.; Jiang, Y.; She, M.; He, C.; Peng, Y.; Lin, Y.; Zhang, Y.; et al. Pre-Harvest Application of Strigolactone (GR24) Accelerates Strawberry Ripening and Improves Fruit Quality. Agronomy 2023, 13, 2699. https://doi.org/10.3390/agronomy13112699
Li M, Yang M, Liu X, Hou G, Jiang Y, She M, He C, Peng Y, Lin Y, Zhang Y, et al. Pre-Harvest Application of Strigolactone (GR24) Accelerates Strawberry Ripening and Improves Fruit Quality. Agronomy. 2023; 13(11):2699. https://doi.org/10.3390/agronomy13112699
Chicago/Turabian StyleLi, Mengyao, Min Yang, Xiaoyang Liu, Guoyan Hou, Yuyan Jiang, Musha She, Caixia He, Yuting Peng, Yuanxiu Lin, Yunting Zhang, and et al. 2023. "Pre-Harvest Application of Strigolactone (GR24) Accelerates Strawberry Ripening and Improves Fruit Quality" Agronomy 13, no. 11: 2699. https://doi.org/10.3390/agronomy13112699
APA StyleLi, M., Yang, M., Liu, X., Hou, G., Jiang, Y., She, M., He, C., Peng, Y., Lin, Y., Zhang, Y., Wang, Y., He, W., Chen, Q., Zhang, Y., Wang, X., Tang, H., & Luo, Y. (2023). Pre-Harvest Application of Strigolactone (GR24) Accelerates Strawberry Ripening and Improves Fruit Quality. Agronomy, 13(11), 2699. https://doi.org/10.3390/agronomy13112699








