Abstract
Metabolomics is a technology that generates large amounts of data and contributes to obtaining wide and integral explanations of the biochemical state of a living organism. Plants are continuously affected by abiotic stresses such as water scarcity, high temperatures and high salinity, and metabolomics has the potential for elucidating the response-to-stress mechanisms and develop resistance strategies in affected cultivars. This review describes the characteristics of each of the stages of metabolomic studies in plants and the role of metabolomics in the characterization of the response of various plant species to abiotic stresses.
1. Introduction
Global agriculture is threatened by climate change as variations in rainfall, heat waves and global CO2 levels are responsible for several types of abiotic stresses causing a negative impact on food production [1]. Production losses of about 50% have occurred due to abiotic stress [2] and the study of crops capable of withstanding abiotic stress is considered a priority [3,4]. The characterization of the effects and biochemical responses caused by abiotic stresses in different crops can contribute to understand the mechanisms of plant resistance to stress and favor the development of appropriate stress mitigation strategies including the development of abiotic stress resistant crops [4,5]. Among several alternatives, metabolomics—the study of the metabolites present in a biological system—is considered a key tool for assessing biochemical changes occurring in plants affected by abiotic stress [6].
Metabolomics is considered a fundamental branch of systems biology [7] and a powerful tool to investigate the response of organisms to external factors at the metabolite level [8,9,10,11]. Metabolomics is essential for understanding chemical signals as plants grow and develop [12]. However, the full importance of metabolomics for assessing plant responses to stress is difficult to estimate because, unlike the transcriptome and proteome, the metabolome is not necessarily associated with the plant genome [11].
Metabolites, analyzed by metabolomics, are defined as small molecules developed or modified during cellular metabolism [8]. The orderly identification and quantification of metabolites can provide the chemical fingerprint of a phenotype [13] and the biochemical response of an organism to specific conditions [14].
Plants process a wide variety of primary and secondary metabolites with diverse chemical structures. Primary metabolites are essential for plant growth and development, but secondary metabolites have more specific purposes and both types of metabolites are part of plant stress response mechanisms [15]. Primary metabolites, including amino acids, sugars, and lipids, have highly conserved structures, but secondary metabolites, such as alkaloids, polyphenols, and terpenoids, are more diverse and can vary greatly among plant species [15,16]. With current metabolomics tools, about 14,000 plant metabolites can be identified, although more than 200,000 molecules are expected in plant samples [11].
Large-scale metabolomic studies have made it possible for researchers to obtain a wealth of global data on metabolites and relevant metabolic pathways in an unprecedented manner [17]. Metabolomics has evolved into a powerful tool in many research areas, such as molecular epidemiology, toxicity assessment, biomarker discovery and identification [14,15,16,17,18] and plant research [12,17]. Currently, metabolomics has ventured into broader studies such as functional metabolomics, which is responsible for investigating the functions of specific metabolites [19]. Metabolomics has also been responsible for improving crop yield and quality [11,17,20]; and the fusion of metabolomics with other technologies related to genetic modification, transcriptomics, proteomics and quantitative genetics have boosted plant breeding [17]. Additionally, metabolomics has forayed into the observation of morphological, phenotypic and physiological responses of plants to environmental perturbations and interactions with other organisms [21].
The main objective of this review was to describe current advances in metabolomic analysis and the contributions of metabolomic tools to understanding the plant response-to-abiotic-stress mechanisms. Special emphasis is placed on state-of-the-art technologies applied to separate, identify and quantify metabolites and asses the metabolic changes suffered by plants under abiotic stress.
2. Metabolomics Analyses
Metabolomic analyses are commonly classified as directed or undirected. Directed metabolomics consists of targeted analyses focusing on the identification and quantification of as many metabolites as possible within a specific chemical group, whereas undirected metabolomics is aimed at detecting as many metabolite groups as possible to achieve patterns or fingerprints, with or without identification of the detected molecules.
The study of metabolites requires the use of various processes that incorporate aspects of chemistry, biochemistry, bioinformatics and statistics [13]. Depending on the research objective, both directed and undirected metabolomic studies involve various steps including experimental design and sample preparation as well as metabolite extraction, derivation, separation and detection followed by data analysis [22].
2.1. Design of Experiments
Experimental design (DoE) is an essential part in metabolomic studies. The goal of DoE is to obtain reliable data that lead us to appropriate conclusions while minimizing errors [23]. The objective of DoE is to allow researchers to understand the impact of independent factors on the studied variables [23].
Several sources of variation in metabolomics and related studies can be found during sample collection, storage and preparation, analysis, and data pre-treatment [24]. Therefore, the essence of planning metabolomic-based research is to account for all the potential sources of variation that could alter the assessment of a biological hypothesis, such as plant development stage, treatments, sample preparation stage (e.g., harvesting, transport and extraction protocol), analytical platform (e.g., LC-MS, GC- MS, nuclear magnetic resonance) or statistical analysis [25].
The application of design of experiments (DoE) can take into account the combined effect of several external factors and has been suggested as a strategy for the design of new protocols for metabolomic studies [23]. Therefore, DoE strategies have been used to assess the effect on the metabolome of the external factors involved in sample preparation (sample grinding and temperature) as well as metabolite extraction (extraction solvent and temperature) [23], separation and detection (mobile phases, temperature, and other instrumental conditions) [26]. As a result, the experimental conditions that maximize the number and concentration of metabolites while minimizing the errors can be selected. The most-used DoE in protocol development for metabolomics include the Box–Behnken Design, Central Composite Design, D-optimal Design, Factorial Designs, and Plackett–Burma Design, among others. A comprehensive review on DoEs applied to metabolomics can be found in the literature [23]. The best known DOE is factorial design. This design has been used to assess reducing sugars, lipids and chlorogenic acids in Coffea arabica [27]. If the number of experiments is very high a fractional factorial design can be used. Similarly, the Plackett–Burman design may be more appropriate in the absence of interactions between the independent variables [23] and has allowed the improvement of the extraction protocol (particle size of the ground tissue, the extraction time and the ethanol content in the solvents) of metabolites from Ginkgo biloba L.
Most metabolomic studies focusing on plant abiotic stress have followed a balanced one-way design as only one stress variable has been assessed in each study [28,29,30,31]. In the one-way designs, the stress variable has been evaluated at two (control vs. stress) or more (control and various levels of the stress factor) levels. For instance, the effect of drought stress in the host’s metabolome has usually been evaluated using a control vs. stress design [32,33,34], whereas various salt concentrations have been used to assess the effect of salinity in the plant’s metabolome [35,36]. In both cases, the studies have been balanced as the same number of individuals has been evaluated at each level of stress. Special care should be taken to assure that all individuals are maintained under the exact same environmental conditions during the study.
2.2. Replication and Randomization
Choosing the right number of technical and biological replicates is important for performing a powerful statistical test and executing an adequate biological interpretation of the results [22]. The technical replicates manage to assess protocol or instrumental variations, while biological replicates provide information about the natural variation of the individuals tested and are crucial for inferring significant differences among treatments [37]. Biological replicates must be specific to the study population and research conditions [22]. The minimum suggested number of replicates for metabolomic analysis is six, but the statistical power should be estimated before conducting metabolomic research in order to establish an adequate sample size [8,22]. In general, research under controlled conditions usually generates lower variability and requires a smaller sample size than field studies. However, power analysis is often excluded and sample size is usually restricted by the number of available individuals or greenhouse space. Statistical power estimation can be carried out through preliminary tests using a small number of samples or comparable literature data [38,39]. Methods for power analysis and sample size estimation are discussed in the literature [38] and various packages such as MetSizeR have been developed for sample size estimation [40]. One common strategy to reduce the biological variability and avoid outlier effects is sample compositing [37]. Typically, three or more samples can be combined before analysis, avoiding interferences resulting from individual plants while focusing on the general effect caused by the treatment or independent variables [41].
Randomization is crucial to decreasing experimental error and biological versatility, as well as preserving sample homogeneity [22,37]. If working under supervised environmental circumstances, such as in a growth chamber, plants must be rotated during the duration of the investigation in order to balance any alteration in the magnitude of light or ventilation that could modify their metabolism and the data progression [37]. For plants grown in a greenhouse or field, randomization should account for environmental diversity such as differences in landscape, soil fertility or drainage [37]. Therefore, it is necessary to record all the changes perceived in the study area and incorporate them as metadata for analysis [42]. When complete randomization is not possible, a common strategy is to arrange the plants in the form of homogeneous blocks and assess the independent variables within each block [22].
2.3. Sampling and Metabolite Quenching
Sampling steps, such as harvesting and rapid freezing, are a critical prior step to obtaining metabolomic data [22]. Depending on the magnitude of the experiment, it is important to train a large team of operators in order to decrease the harvest time. The training should take into account the number and location of samples as well as how and when to carry out the harvest, taking into account that both primary and secondary metabolites may change obeying natural cycles [43]. After harvesting, expedited metabolite quenching is of utmost importance, particularly for the evaluation of metabolites with high regeneration rates such as glycolytic intermediates and sugar phosphates [44,45]. Temperature and water-content reduction are the most common metabolite-quenching strategies. Temperature reduction can be achieved by flash-freezing the samples under liquid nitrogen immediately after the harvest. Alternatively, dry ice can be used during transport prior to the use of liquid nitrogen. Then, samples can be stored frozen at −80 °C until analysis [8]. Another metabolite-quenching strategy is moisture reduction by lyophilization [46]]. Lyophilization—aka freeze-drying—eliminates most of the water needed for the development of biochemical reactions while maintaining the sample under freezing temperatures. Additionally, sample drying reduces the variability caused by differences in the water content of plants [47] and might be a mandatory step prior to assessing metabolome changes in plants under water-affecting conditions such as various abiotic stresses [47]. Other drying strategies, such as the use of silica gel, for sample transportation from areas where both liquid nitrogen and dry ice are not accessible have been implemented [48]. However, sample drying is not recommended in experiments in which little or no water-content variability is expected as volatile metabolites can be lost and the extraction efficiency can be reduced due to metabolite adsorption to cell walls during dying [47].
In general, most metabolomic studies in plants have carried out metabolite quenching by means of liquid nitrogen for short term sample transport and freezing at −80 °C for extended storage. Samples for a flavonoid-targeted profiling through liquid chromatography photodiode array mass spectrometry (LC-PDA-MS) in Arabidopsis thalia were immediately frozen with liquid nitrogen after collection and then kept at −80 °C, until use [49]. Similarly, leaf samples of Brachystegia boehmii (Miombo) and Colophospermum mopane (Mopane) were frozen in liquid nitrogen and stored at −80 °C until freeze-drying, then analyzed using GC-TOF-M [48]. In bananas, the metabolic profiles of dwarf plants and plants infected with cucumber mosaic virus were compared by GC-MS, using leaves collected into liquid nitrogen and analyzed immediately upon arrival [50]. Similarly, rice samples were frozen with liquid nitrogen prior to metabolic profiling by gas chromatography coupled to electron impact ionization time-of-flight ionization mass spectrometry to assess plant repair mechanisms following drought and heat stress.
2.4. Metabolite Extraction
The metabolite extraction aims to maximize the number and abundance of the compounds in a sample [51]. The extraction should be achieved without altering the chemical structure of the molecules while maintaining the metabolite levels [52]. As the number of metabolites in plants is very large and encompasses a diversity of compounds, there is no single protocol capable of extracting the metabolome of vegetable samples and various solvents and combinations should be assessed. Hydrophilic metabolites are extracted with polar solvents such as methanol or a combination of methanol and water whereas nonpolar solvents such as chloroform are used to extract lipophilic metabolites [53,54]. The combination of chloroform, methanol and water is the most-used option as this combination of solvents allows to separate both hydrophilic and lipophilic metabolites in a single extraction [26,55,56]. It is valuable to carry out prior studies on the characteristics of the solvents used in the extraction of metabolites. Particularities to take into account are the solvent/sample ratio, duration and temperature of extraction [44,57].
Metabolite extraction of grapevine leaves was carried out using a solution of methanol, chloroform and water in a 2.5:1:1 ratio prior to metabolic profiling using liquid chromatography/mass spectrometry [58]. Isopropanol was used as an extraction solvent for woody tree species in an untargeted metabolite profiling study on LC-MS and high-performance liquid chromatography [59]. Similarly, 90% methanol was used to study other tree using liquid chromatography/tandem mass spectrometry and ultra-performance liquid chromatography [59].
2.5. Derivatization
Derivatization is a method in which a chemical compound is transformed at one or more of its functional groups by reaction with a derivative reagent to produce a chemically related product [60]. In plant metabolomics, derivatization is usually used before the separation of metabolites by gas chromatography (GC) [51]. In this case, the purpose of derivatization is to decrease polarity, improve thermal balance and volatility, weaken adsorption of analytes in the GC system and enrich the detector response, thereby getting a better definition of the peaks and greater detection efficiency when compared to underivatized metabolites [8]. One of the disadvantages of this process is the formation of artifacts associated with the derivatization agent used [61,62].
Derivatization is a two-step operation: (1) change of the carbonyl group (aldehyde or ketone) in an oxime or methoxyamine with the purpose of restricting monosaccharide tautomerism [26]; (2) silylation, to increase volatility, consisting in the reaction of functional groups with active hydrogens (such as H, COOH, NH and SH) with C3H10Si (trimethylsilane) products, thus obtaining minimally polar and more-thermostable trimethylsilyl (TMS) groups [63]. The most-used reagents for the silylation process with plant samples are, N-methyl-trimethylsilyltrifluoroacetamide (MSTFA) and N, O-bis-trimethylsilyltrifluoroacetamide (BSTFA).
MSTFA has been the most popular choice for derivatization. MSTFA was used to assess the metabolic profiles of maize leaves, subjected to drought, heat and combined stress, on GC-MS [64], In another culture the influence of respiration pathways using branched-chain amino acids after water shortage in Arabidopsis thaliana was determined using MSTF prior to by GC-MS analysis [65]. Similarly, derivatization of Triticeae and Citrus species was performed at 37 °C for two hours in methoxyamine hydrochloride followed by trimethylsilylation with MSTFA [66].
2.6. Separation and Detection
After metabolite extraction, techniques such as liquid chromatography (LC), gas chromatography (GC) or capillary electrophoresis (CE) are applied to separate each metabolite present in the sample matrix [67]. Chromatography-based techniques such as LC and GC are the most used for plant metabolomics and can achieve high separation efficiency of metabolites with different degrees of polarities and volatility. On the other hand, CE has been recommended to separate metabolites according to their mass to charge ratio (m/z). Among LC techniques, high performance thin layer chromatography (HPTLC) has been widely used due to its simplicity and low cost [67]. Similar techniques are high performance liquid chromatography (HPLC) and ultra-performance liquid chromatography (UHPLC), both used for the analysis of semipolar, polar and hydrophilic analytes [67].
After metabolites are separated, detection, quantification and identification are the usual follow up steps. Nuclear magnetic resonance (NMR) [68] and mass spectrometry (MS) are the most used detection techniques in metabolomics [69]. Particularly, MS devices can be coupled to LC, GC or CE separation equipment into one single system, allowing metabolite separation and detection in a single run. The advantages and disadvantages of the combined platforms are summarized in Table 1. The detailed fundamentals and applications of each technology were described in previous reviews [70,71].
The GC-MS combination has been used the most for plant metabolomics [8,9] and has shown a highly efficient separation of metabolites as well as high resolution, sensitivity and reproducibility [72], partially achieved thanks to the electron ionization (EI) of metabolites in the MS section [9,61]. Although GC-MS can mostly be applied for the analysis of volatile and thermostable compounds, these kinds of metabolites are highly abundant in plant samples and can provide relevant information about the physiological state of plants. Thus, GC-MS has been used to characterize the response of various plant species to drought [65] and heat stress [6], among other abiotic conditions. Similarly, plant metabolomics based on Gas Chromatography-Time of Flight Mass Spectrometry (GC-TOF-MS) has been used in the metabolic analysis of forest species under abiotic and biotic stress [73] providing impressive precision and mass resolution [74]. GC-MS has also been used to estimate the metabolite fluxes in plants [75,76].
The LC-MS combination can be used to assess the compounds not detectable by GC-MS, including thermolabile, polar and high molecular weight plant metabolites without the need for derivatization [9,77]. Metabolite separation is carried out thanks to a stationary phase incorporated into a column; the generally preferred types of columns are reversed phase, porous graphite carbon (PGC), anion exchange (AEC) and hydrophilic interaction chromatography (HILIC). These columns enable the separation of plant metabolites based on their interaction with the column matrix [24,63]. PGC, AEC and HILIC options are intended for the analysis of polar metabolites characteristic of the plant metabolome [9,10]. After separation, the metabolites are ionized by various techniques. Electrospray ionization (ESI) is the most widely applied [78,79] and has been used for a wide range of metabolites such as amino acids, organic acids, sugars and phospholipids [45,80]. Additionally, chemical ionization at atmospheric pressure (APCI) has also been used for nonpolar metabolites such as carotenoids and carotenoid esters [81]. Ionized metabolites are then detected by the MS. A targeted metabolic study in Epimedium pubescens using LC-MS was carried out to investigate the composition of leaf metabolites [82]. Similarly, a non-targeted metabolite profiling study used LC-MS to assess plant and root samples of six gentians (G. gelida, G. septemfida, Gentiana asclepiadea, G. cruciata, G. paradoxa, G. pneumonanthe) grown in the Caucasus [83]. The metabolic profile of various types of medical cannabis was assessed by LC-MS to identify phytocannabinoids [84].
CE is a non-chromatographic platform that has allowed the detection of an increased variety of metabolites, with shorter run times, less sample volume and minimal-to-no use of solvents when compared to traditional LC strategies [85,86]. Additionally, CE is capable of detecting metabolites that usually dissipate in the derivatization steps required by GC-MS [9]. During a CE run, metabolite separation is achieved by their m/z ratio and detection can be performed by MS. Plant diseases [51] have been characterized by CE. CE-MS has been applied to the study of changes in the primary metabolism of tobacco leaves according to their geographical location [84] and to quantifying organic acids in coffee seeds [85]. Similarly, the most abundant sugars in mature and young leaves, fruits and roots of grapevine were assessed using CE [87]. However, the low sensitivity [88] and poor reproducibility [89] of CE has minimized the use of this platform in plant research.
NMR is a non-disintegrating fingerprint technology that manages to reveal different classes of metabolites regardless of their size, charge, volatility or stability [50]. Additionally, NMR provides structural information for an unquestionable identification of metabolites [9,90]. Sample preparation for NMR is fast and easy, while the instrument shows high reproducibility [91,92]. However, due to its limited sensitivity [17,93], NMR-supported plant metabolomics have only achieved the detection and quantification of highly abundant metabolites [92,94] and potentially valuable metabolites present in reduced concentrations have not be detected by NMR [95].

Table 1.
Advantages and disadvantages of techniques used in metabolomics studies.
Table 1.
Advantages and disadvantages of techniques used in metabolomics studies.
| Technique | Advantage | Disadvantage | References |
|---|---|---|---|
| Gas chromatography coupled with mass spectrometry (GC-MS) | Efficient separation of metabolites. High resolution. Detection and quantification of metabolites, including low concentrations. Highly reproducible. | Analysis of volatile and thermostable metabolites. It is necessary to derivatize the samples. | [72,96] |
| Gas chromatography coupled to time-of-flight mass spectrometry (GC-TOF-MS) | Greater accuracy in mass measurement. High homework periods. Fast operating time. Better sample profit. | High cost. | [97] |
| Liquid chromatography coupled with mass spectrometry (LC-MS) | Remarkable additional technology for GC-MS. Identifies specific metabolites. Distinguishes thermolabile, polar metabolites and high molecular weight compounds without derivatization. Can choose column and retention mechanism. Identifies amino acids, organic acids, sugars, and phospholipids. | Each study group establishes its own library. | [98,99] |
| Ultra high performance liquid chromatography coupled with mass spectrometry (UHPLC-MS) | Determines complex samples thanks to its speed, efficiency, sensitivity and selectivity. Improved high resolution UHPLC, higher peak efficiency, fast separations and lower solvent consumption when compared to common HPLC. | Long analysis time. | [100] |
| Liquid chromatography-quadrupole-mass spectrometry (LC-Q-MS) | Performs qualitative and quantitative analysis By performing qualitative analysis, allows for obtaining information on the structure of the metabolite Quantify compounds. | Restricted only to ions present within an exclusive nominal mass unit range. | [101,102,103] |
| Triple quadrupole liquid chromatography coupled to mass spectrometry (LC-QQQ-MS) | Selects ions to collide and analyze the fragments. Detects the primary and secondary ion. Accurate, sensitive and comprehensive ion detection. Features high quantitative reproducibility compared to tandem mass spectrometry | Approves the passage of a specific mass ion category | [103] |
| Liquid chromatography-quadrupole-time of flight-mass spectrometry (LC-Q-TOF-MS) | Superior mass resolution and detection sensitivity. Accurately measures the mass. Higher qualitative capacity for fragment ions. | Low abundance ions can be difficult to identify. Difficulty setting up large mass spectrum libraries. | [99,104] |
| Matrix-assisted laser-desorption and ionization time-of-flight mass spectroscopy (MALDI-TOF-MS) | Flexible technique for the analysis of biological samples containing proteins. Fast and particularly economical in terms of reagent use and time required for sample processing Used for plant breeding | Difficulty setting up large mass spectrum libraries. Each study group establishes its own library. | [105] |
| Capillary electrophoresis coupled to mass spectrometry (CE-MS) | Does not require the derivatization step as in GC-MS. Analyzes the important metabolites from the physiological point of view. Analysis is very fast, efficient, low consumption of sample and reagents. Requires minimal sample preparation. | Quantitative analysis by CE is critical, it depends on several factors. | [106,107] |
| Nuclear magnetic resonance spectroscopy (NMR) | Relative ease of sample preparation. Quantify metabolites High reproducibility Non-destructive technique Select and detect isotopes | limited sensitivity. Detects and quantifies large metabolites. | [91] |
There is no single ideal technique that can span the entire metabolome of a biological system. Nevertheless, taking into account the objective of each investigation, a combination of techniques would provide significant data to understand the behavior of an organism and/or cell. The study [108] related the uses of GC-MS, LC-MS and GC-QTOF-MS to assess the metabolic pathways associated to the synthesis of fatty acids in Physaria fendleri. The combination of LC and GC has been the preferred choice, as both techniques complement each other by characterizing non-volatiles and volatile metabolites, respectively. LC-MS and GC-MS were used to study the contents of metabolites and hormones in rice plants subjected to drought stress [109]. Moreover, the quantitative profile of phytohormones and other metabolites was studied in barley roots subjected to salinity stress using GC-MS and LC-MS [110]. Likewise, the combination GC-TOF-MS and LC-q-TOF-MS was used to determine the metabolic profile of Arabidopsis thaliana affected by the combination of glutathione under mild oxidative stress and low phosphorus content [110]. Table 2 summarizes the technologies and tools commonly used to study abiotic stress in plants.
3. Data Treatment
The data generated during metabolomic studies usually integrate retention or migration times, m/z ratios and intensity values for each detected peak. Background noise can also be present due to column carry-overs or instrumental electronic noise. Similarly, a metabolite’s retention times and intensities can vary from run to run and need to be corrected prior to statistical analysis. Therefore, metabolomic data analysis involves a series of steps including data conversion, noise reduction, feature detection, retention time alignment, intensity normalization, metabolite identification and quality control [111,112,113,114].
Data transformation consists of the conversion of the instrument’s file format to more general formats that allow the use of standalone data analysis software. NetCDF and mzXML are the most widely applied file configurations for storing hyphenated MS data and the software associated to most MS instruments can transform their generated data into either NetCDF or mzXML [115]. After conversion, noise elimination, metabolite detection, alignment and quantification are usually carried out by ad-hoc software such as Thermo Scientific’s SIEVE [13] as well as free software packages like XCMS [116] or MZmine 2 [117]. Finally, metabolite intensities need to be normalized. The purpose of normalization is to decrease the variability caused by the inequality in the concentrations of the sample as well as analytical errors affecting the intensity of the detected peaks [118]. The most-applied normalization method is the use of an internal standard added to the sample prior to metabolite extraction. Metabolite data is then mathematically normalized to assure homogenous levels of the internal standard across the sample [11]. For this, the internal standard used should be compatible with the analysis platform and must not interfere with the metabolite profile of the samples. If this is not possible, other normalization methods can be applied, including the use of the total area [119] normalization to a uniformly detected metabolite, or the application of probabilistic methods [120], among others. A detailed review on normalization strategies for metabolomics can be found elsewhere [121]. Normalization to the total area allowed the development of predictive metabolite-based models for the detection of banana somaclonal variants [50].
4. Statistical Analysis
A wide variety of statistical procedures or techniques have been used in metabolomics [13]. The classical approach involves both multivariate and univariate analyses to assess a treatment effect on the overall metabolite profile and individual metabolites, respectively.
Multivariate analyses aims to reduce the dimensionality associated to the number of variables (metabolites) and assess the differences in the metabolite profile between the research groups. In this way, the metabolites contributing the most to the group’s separation can be obtained [122]. The most-applied multivariate techniques in plant metabolomics are Principal Components Analysis (PCA), Partial Least Squares (PLS) and Partial Least Squares with Discriminant Analysis (PLS-DA).
PCA reduces the data dimensionality into individual components explaining part of the total samples variance. Sample grouping can be observed in a two or three axis score plot while the contribution of individual metabolites to the grouping is usually shown in a two-axis loading plot. Alternatively, both the score and loading plots can be combined into a single biplot [123]. PCA has been used to evaluate potential metabolites and biological pathways involved in the Pb and Cd stress response of radish roots [124]. PCA was also used for the analysis of the metabolism of Arabidopsis thaliana, under the effect of the combination of glutathione, mild oxidative stress and low phosphorus content [125]. Similarly, PCA was used to study the combined protective responses of drought and heat activation in Eucalyptus globulus that are not activated when subjected to drought or heat stress alone [125].
PLS aims to reduce the dimensionality of the data while maximizing the relationship between the metabolite levels and the treatments [126]. For this, PLS estimates the covariance between the independent variables X (metabolomic data) and the dependent variable Y (treatments) and produces a predictive model [126,127]. The PLS-DA is an extension of PLS applied when the treatment variable is categorical instead of continuous. PLS-DA has been used to observe metabolic differences between invasive alien plants in native and invaded habitats, enabling statistical separation of the species under study [128]. In another research, PLS-DA was used for biomarker recognition of natural groups of fermented aqueous extracts of Viscum album L. [129]. In the same aspect, PLS-DA was applied to recognize a distinct pattern of 51 species, based on the leaves, stems and relative phylogeny in the metabolome of three plant families Rosaceae, Asteraceae and Fabaceae [130].
Univariate techniques have also commonly been used in metabolomic studies to assess the significant accumulation of metabolites after the treatment [131]. Additionally, univariate techniques have allowed the reduction of a large number of analytes while keeping only the significantly accumulated metabolites at a given p value [131]. Analysis of variance (ANOVA) and t-test are the most-used univariate techniques in metabolomics. Metabolic and mineral changes in response to salt stress were analyzed in genotypes of Triticum turgidum ssp. Durum using two-way ANOVA in order to estimate the effects of genotype, salt, and genotype × salt interaction in the metabolite response to salt stress [38]. Another statistical test commonly used is the Student’s t-test. The Student’s t-test has been used to determine significant differences in the levels of phytohormones and other metabolites between barley roots subjected to salinity stress [109] and normal roots. ANOVA and Student’s t-test were used to assess the influence of alternative respiration pathways using branched-chain amino acids after water stress in Arabidopsis thaliana [64]. Nevertheless, metabolomic analyses usually generate a large number of compounds, which increases the number of individual parametric tests. An increased number of individual t-tests or ANOVAS can yield false positive results [90]. To correct this, a multiple-test rectification method such as the Bonferroni and Benjamini–Hochberg corrections, aka the false discovery rate (FDR), has been applied [13]. The p-values obtained from ANOVA and Student’s t-test were corrected by Bonferroni’s test to assess the metabolome of corn hybrids [132]. Similarly, the response to drought of wheat leaf tissues was assessed using Student’s t-test in conjunction with the Bonferroni false discovery rate [90]. Likewise, the Student t-test, p-values were corrected with Bonferroni’s method to evaluate the metabolite profile of maize leaves subjected to heat, drought and combined stress [6]. However, the normality and homoscedasticity (equal variances) assumptions should be tested before using parametric tests such as ANOVA and t-tests, especially in small datasets. If assumptions are not met, non-parametric tests such as Mann–Whitney and Kruskal–Wallis should be used. Thus, the differential metabolite profile of Camellia sinensis subjected to abiotic stress was assessed using the Kruskal–Wallis nonparametric test [133].
5. Pathway Analysis
Differentially accumulated metabolites can be used for assessing significantly affected pathways. A complete metabolic pathway analysis usually includes the observation of interactions between genes, proteins, and metabolites within cells, tissues, or organs under a biological treatment [134]. Thus, the metabolomic data acquired is usually added to transcriptomic and proteomic data to obtain a complete perception of the biological processes [135]. However, metabolites alone can also provide pathway-related information thanks to the development of databases such as the Kyoto Encyclopedia of Genes and Genome (KEGG) which is able to map metabolites into pathways [136]. Metabolic pathways can be assessed from metabolomic data by overrepresentation analysis (OA). In an OA, detected metabolites are mapped in potential pathways and the statistical significance of each pathway is determined by incorporating the number of differentially accumulated metabolites in a Fisher’s exact test or hypergeometric test [137]. Metabolite enrichment and pathway studies are done using ad-hoc software to connect significant metabolites to biochemical pathways using KEGG or other public databases [138]. The various methods and software used for metabolite-based pathway analysis were reviewed elsewhere [137]. Some examples of KEGG usage are cited below [132], analyzed the metabolomic profile of two cytotypes of Solidago canadensis, once the differentiated metabolites were obtained, they used KEGG to find the significant metabolic pathways such as “diterpenoid, phenylpropanoid biosynthesis” as well as “flavone and flavonol and flavonoid biosynthesis”. In the same field [139], they employed KEGG to map the metabolic pathways involved in the biosynthesis of rotenoids from Mirabilis himalaic; the main metabolic pathway was that of amino acids, especially phenylalanine, methionine and cysteine [65], used the KEGG platform to identify the metabolic pathways related, to differential metabolites obtained from roots and leaves, generated by drought in seven species of Triticeae, the metabolic pathway identified was TCA, especially succinate. Currently, metabolomic studies follow a sequence of steps (Figure 1) to provide broad and deep explanations of the biochemistry of an organism.
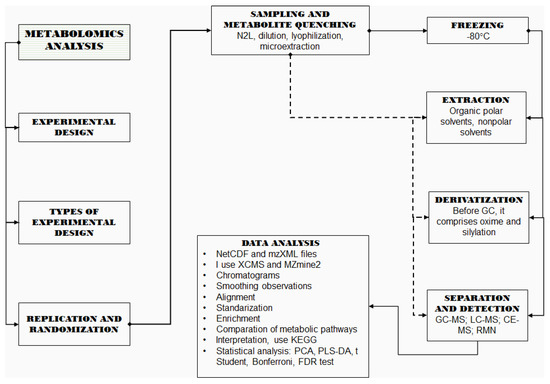
Figure 1.
Synthesis of metabolic analysis.

Table 2.
Contributions to metabolomics for understanding abiotic stress in plants.
Table 2.
Contributions to metabolomics for understanding abiotic stress in plants.
| Species Name | Type of Stress | Solution Used | Separation and Detection | Data Treatment | Tool for Data Analysis | Database | Reference |
|---|---|---|---|---|---|---|---|
| Musa spp. | No stress | Methanol-chloroform-water | GC-MS | PCA, PLS-DA | KEGG | NIST, Wiley | [50] |
| Eucaliptus globulus | Drought and Heat Stress | Methanol-chloroform-water | GC-MS | ANOVA, PCA, sPLS | R v3.1.2 | VSN., United Kingdom | [125] |
| Triticum spp. | Drought stress | Methanol-chloroform | GC-TOF-MS | PCA, OPLS-DA | KEGG | EI-MS library | [140] |
| Triticum turgidum ssp. Durum | Salt stress | --------- | GC-MS, HPLC | ANOVA, PCA, t Student | JMP software | NIST 2011 | [35] |
| Arabidopsis thaliana | Drought stress | Water, acetone | GC-TOF-MS | Welch’s t-test and FDR | R | GEO | [141] |
| Oriza sativa
cv. Nipponbare | Drought stress | -------- | GC-MS; LC-MS | PCA | --------------- | ------------------- | [142] |
| Triticeae | Drought stress | Methanol-water | GC-MS | PCA, PLS-DA; HCA | R | NIST, Wiley | [65] |
| Hordeum spp. | Salinity Stress | ------- | GC-MS, HPLC-MS | ANOVA, t Student, FDR | MetaboAnalyst 3.0 | Uninformed | [109] |
| Arabidopsis thaliana | Low phosphorus stress | Methanol-water | GC-TOF-MS, LC-q-TOF-MS | PCA, FDR | R and Cytoscape | Uninformed | [110] |
| T. durum Desf. Cv. Ofanto | Light stress and high salinity | Methanol-water Ethanol-water | LC-UV-ESI-MS | ANOVA y Pearson, heat map, PCA, Venn diagrams. | Multibase 2015 | In-house database | [143] |
| Lycopersicon esculentum Mill. Cv. Puhong 968 | High-Temperature stress |
Trichloroacetic acid acetone. Urea, dithiothreitol, CHAPS y Tris. Urea, thiourea, CHAPS. | BE y MS MALDI-TOF/TOF | Duncan | Uninformed | Bruker Online Client software suite | [144] |
| Arabidopsis thaliana | Drought stress | ------- | GC-MS | ANOVA, t de Student | TAGFINDER 4.0 | ArMet | [64] |
| Zea mays | Drought, heat and combined stress | -------- | GC-MS | ANOVA, PCA, Tuckey, t de Student, Bonferroni correction, heat map. | R v3.1.1 | Golm Metabolome Database | [6] |
| Allium cepa | Abiotic stress | Methanol | LC-MS/MS | PCA, heat map | Uninformed | In-house database | [4] |
| Lycopersicum esculentum var cerasiforme | Low oxygen stress | Methanol-chloroform-water | GC-MS | t
de Student, PLS-DA | Unscrambler v.10.2, AMDIS | In-house database | [145] |
| Arabidopsis thaliana | Drought stress | Methanol | LC-MS/MS; LC-PDA-MS | PCA, t de Student, heat map. | SIMCA-P 11.5 MassLynx ver. 4.1 | DROP Met in PRIMe | [49] |
| Vitis vinífera L. cv. Shiraz y Cabernet Sauvignon | Drought stress | Methanol-chloroform-water | LC-MS/GC-MS | t de Student, PCA, OPLS-DA | R 3.0.1; R. Cytoscape | NIST; RI libraries | [146] |
| Haberlea rhodopensis, Thellungiella halophyla y Arabidopsis thaliana | Low temperature stress | HCl, methanol, chloroform | GC-MS | PCA | Metaboanalyst | MPI Golm Metabolome | [147] |
| Oryza sativa L. | Salt stress | Methanol | GC-MS | t
de Student | Xcalibur AMDIS | NIST; Mass spectra library of the Max-Planck-Institute | [148] |
| Selaginella lepidophylla | Desiccation stress | Methanol | GC-MS, UHLC-MS-MS | PCA, PLS-DA, Welch’s t-test and FDR | KEGG Plant Metabolic Network (PMN) | In-house plant database. | [149] |
| Zea mays | Drought stress | Ribitol, chloroform, water | GC-TOF-MS | PCA, ANOVA, Bonferroni test | R-2.13.0 | Golm Metabolome; ArMet | [150] |
| Triticum cultivars | Drought stress | Methanol/ribitol-norleucine | GC-MS | t Student, Bonferroni test. | Xcalibur VANTED | NIST; Mass spectra library of the Max-Planck-Institute | [90] |
| Thellungiella salsuginea Arabidopsis thaliana | No stress | Ethanol, toluene | HPAEC, HPLC | Pearson in R statistic | imzML | Uninformed | [151] |
| Arabidopsis thaliana | Various abiotic stresses | --------- | GC-MS | PCA, heat map, ANOVA, FDR, t de Student | R | NIST; Wiley | [152] |
| Arabidopsis thaliana | No stress | --------- | GC-TOF-MS | PCA, FDR | R, TargetSearch package | Uninformed | [153] |
| Arabidopsis thaliana | Salt stress | Methanol, ribitol and chloroform, water | UPLC, GC-MS | PCA | AMDIS Statistica Software v7.1 | Uninformed | [154] |
| Arabidopsis thaliana | Freezing stress | ------------ | GC-MS | PCA, heat map, PLS, FDR | R | Golm Metabolome | [155] |
OPLS-DA = orthogonal projections to discriminant analysis of latent structure; HCA = hierarchical cluster analysis; BE = two-dimensional electrophoresis; LC-PDA-MS = Liquid Chromatography Photodiode Matrix Mass Spectrometry; HPAEC = high performance anion exchange chromatograph.
6. Metabolomic Assessment of Abiotic Stress in Plants
Abiotic stress causes innumerable transformations in plant metabolisms, such as disturbances in enzyme activities, high requirement for various metabolites, high levels of reactive oxygen species (ROS) or a composition of them [156,157]. Consequently, abiotic stresses alter cellular structures and impair key functions of plant physiology [158]. Among the predominant alterations, low photosynthetic capacity, attenuated development, decrease in fertility and interruption of reproduction are found, causing a decrease in crop yield [156,157]. Stress influences each species in different ways, and even under some circumstances can be positive to obtain a desired crop response [159,160]. There are an abundance of plant response mechanisms against environmental stresses [156].
Metabolomics is able to assist prominently in the analysis of stress biology in plants [15] and has the potential to elucidate the mechanisms for tolerance to abiotic stress in plants [161]. Qualitative and quantitative studies of the metabolites of plants subjected to biotic and abiotic stresses are not only descriptive, but may also reveal deep genetic and biochemical mechanisms as responses of stressed plants, as well as distinguish the ability of plants to resist and tolerate stress [15]. Plants under stressful abiotic factors can react with tolerance or adaptation. Most metabolomics studies have focused on comparing the response of stress-susceptible and stress-tolerant cultivars. In most studies, the role of amino acids as osmoprotectants was confirmed but the importance of other metabolites including organic acids, sugars and phenolic compounds in abiotic stress have been proposed for various plants [162,163]. In general, the stressful situation appears to activate proline production, while proline catabolism improves during stress recovery [156].
In the coming paragraphs, we briefly comment on topics about metabolomic studies on important abiotic stress factors (drought, temperature, salt and oxidative stresses) with emphasis on the most recent years.
7. Water Scarcity and Drought Stress
Drought stress is one of the most damaging stresses in plants, particularly in regions with rain-based plant irrigation, generating dramatic changes in metabolism. Under water scarcity, physiological adjustments tend to reduce water loss and increase water uptake, which leads to metabolic consequences. Thereby, among biochemical responses is found the accumulation of osmoregulators in order to maintain cell turgor [164], including sugars, poyalcohols, polyamines and amino acids, mainly proline [163]. Thus, several metabolomic studies have been carried out in leaf tissues regarding drought stress and proline accumulation in dehydrated leaves resulted frequent [150,165,166]. Proline accumulation has been also reported in a wide variety of plants under stresses which may lead to low water availability, such as high salinity, heavy metals and low temperatures [156,165,167,168].
Most of the metabolomics analyses in plants were performed on aerial parts, mainly leaves. Dehydration at the metabolomic level has been well studied in Arabidopsis thaliana L. Under dehydration, ABA is generated, and the aerial part of this species accumulated amino acids and polyamines under an ABA-dependent manner, as well as raffinose, which was produced independently of this hormone [165]. Together with those biomarkers of drought stress, under water scarcity, A. thaliana aerial parts also accumulated flavonols and anthocyanins, which could indicate that these molecules can alleviate such stress [49]. Leaves are more affected by drought stress than other parts of the plant. For example, drought led to stronger changes in composition of corn leaves when compared to other organs [32,150]. In corn, the metabolites that accumulated most in the leaves were the ringed amino acids (proline, tryptophan, phenylalanine and histidine), while pyruvic and quinic acids decreased.
Metabolome affectation under water stress is dependent on genotype. Thus, although some metabolite contents showed similar alterations in leaves of two wine cultivars (Shiraz and Cabernet Sauvignon) [168], more metabolites were affected in the cv. Shiraz [168], which showed less-adjusted stomatal regulation than the other cultivar. Nicotinate was the only organic acid that increased in both cultivars, whereas glycerate and galactonate decreased. Furthermore, both genotypes experienced a marked increase in certain amino acids (the drought stress-associated proline, and also threonine, tryptophan, valine, leucine and phenylalanine) and changes in the phenylpropanoid pathway. Nevertheless, glutamine increased in Shiraz and decreased in Cabernet Sauvignon.
Metabolomic tools have permitted the characterization of the response to stress of specifically drought-susceptible (DS) and drought-tolerant (DT) cultivars and have allowed the identification of potential biomarkers related with this type of stress. Thus, metabolomics was used to compare DT and DS in wheat varieties. After drought stress treatment, thymine, the aminoacids L-cysteinylglycine and fructoselysine, as well as a series of phenolic compounds, accumulated higher in leaves in the DT variety than in the DS one, whereas increased proline levels were observed in the DS variety only [34]. Another study on leaves of wheat cultivars under water scarcity [32] showed that tryptophan, valine, and the TCA-cycle acids citric, fumaric and malic appeared in higher levels in the DT cultivar when compared to the DS cultivar. Similarly, the DT cultivar significantly accumulated alpha-tocopherol and the acids 3-hydroxy propanoic, gluconic, glycolic, citric and isocitric, whereas the DS cultivar accumulated both alpha- and gamma- tocopherols as well as gluconic and malic acids. Regarding sugars, the concentrations of nigrose, seduheptose and galactose were enhanced in DS, whereas glucose, fructose and galactose levels increased in DT. Similarly, in both leaves and roots of peanuts submitted to drought stress, metabolomics analysis showed that pentitol, phytol, xylonic acid, d-xylopyranose, stearic acid and D-ribose were important drought-response metabolites [33]. Agmatine and cadaverine were present only in DT during drought. Additionally, polyphenols such as syringic acid and vanillic acid were more accumulated in DT than in DS peanuts, while catechin production was higher in DS than in DT during drought.
Regarding metabolic pathways, a metabolite profiling analysis in leaves of wild soybean suggested that the TCA-cycle was enhanced in DT whereas it was inhibited in the common DS under drought stress [169]. Ɣ-aminobutyric acid (GABA), asparagine and methionine increased significantly in DT but not in DS. Organic acids, such as galactonic, glucoheptonic, malonic and glycolic acids, increased significantly in DT. Unsaturated fatty acids, including linolenic and linoleic acids, accumulated significantly in DT. In addition, secondary antioxidant metabolites, including 5-methoxytryptamine and fluorine, accumulated significantly in DT. Furthermore, the aromatic aminoacid phenylalanine and phenolic compounds (ferulic acid, salicylic acid and 4-hydroxycinnamic acid) increased significantly in DT. Other metabolites, including glucose-1-phosphate, D-fructose 1, 6-bisphosphate, pyruvic acid, D-glyceric acid, oxalic acid and 2-methylfumarate, increased significantly in both DT and DS cultivars. Similarly, amino acids, including proline, glycine, serine, valine, beta-alanine, threonine and isoleucine, accumulated significantly in both DT and DS.
The temporal dynamics of metabolite reprograming of DT and DS cultivars under drought stress has also been studied by metabolomic tools. In Tibetan hulless barley, both types of cultivars responded to drought stress by accumulating flavonoids and glycerophospholipids compounds within one hour [31]. However, the number of differentially accumulated metabolites decreased in DS over time but increased in DT. In DT, the differentially accumulated metabolites after 8 h of drought stress were quite different from those identified at the previous hours suggesting a specific metabolite reprograming aimed to cope with drought stress.
Under osmotic stress (associated to drought stress), various plant species such as barley usually show an increase in L-proline together with other metabolites, including various osmoprotectants like mannitol [170]. Interestingly, a DS barley genotype showed the highest increase in the levels of most metabolite-except mannitol-and decreased levels of maltose, when compared to the stable contents in other genotypes. In both DS and DT wild genotypes tested, there was a decrease of the TCA cycle metabolite 2 α–-ketoglutaric – acid from which proline proceeds- and xylitol.
The nature of drought (cyclic vs. acute), as well as its frequency and severity, may also affect the degree of osmotic adjustment and the nature of the organic solutes that are accumulated. In Populus deltoides L. leaves, whereas cyclic drought induced the largest responses in primary metabolism (soluble sugars, organic acids and amino acids), acute onset of prolonged drought induced the greatest osmotic adjustment and largest responses in secondary metabolism, especially populosides (hydroxycinnamic acid conjugates of salicin). [30]. In rice, a combination of metabolomics with proteomics revealed that, under abrupt drought-flood alternation stress, energy metabolism pathways and reactive oxygen species (ROS) changed strongly, leading to grain yield reduction [171].
Metabolomics has been the basis of several mixed omics studies. The combination of metabolomics and proteomics allowed to assess the recovery of Eucaliptus globulus L. [125] or the response of DT and DS spring-wheat cultivars [172] to water stress. In this case, leaves of the DS cultivar showed increased levels of amino acids such as proline, methionine, arginine and lysine. However, only two pathways were affected in the DT cultivar, one of them being purine metabolism. Similarly, transcriptomics and metabolomics studies revealed that ABA, aminoacids and organic acids accumulated in leaves of both DS and DT sesame varieties but the DT variety showed higher levels of ABA, proline, arginine, lysine, aromatic and branched chain amino acids, GABA, saccharopine, 2-aminoadipate and allantoin than the DS under the stress condition [173].
From a practical point of view, drought favors the obtaining of several secondary metabolites like complex phenols, terpenes and alkaloids. For example, under drought stress, phenolics increased in Hypericum polyanthemum [174], Oryza sativa [175], Salvia officinalis [176] and Hordeum vulgare [177], and monoterpenes or terpenoids rose in these last two species, respectively [177,178]. However, the stress generally caused a reduction in the plant biomass.
Drought stress usually leads to heat stress due to the decrease of transpirational cooling. Most of the physiological, biochemical and metabolic changes observed under drought and heat stresses were reversed upon recovery in Eucalyptus globulus Labill [125] and rice (Oryza sativa L.) [179]. In rice flag leaves at the flowering statage, many compounds with after-stress altered concentration reached original levels after rehydration. Thus, 60 h after rewatering, some primary organic acids (such as the TCAs isocitric, and citric, and glyceric acid) increased, and glucose, raffinose, glycine, N-carboxiglycine and proline decreased. Proline decreased more in drought-tolerant cultivars. The behavior of several metabolites was generally shared in early grain-filling flag leaves after stress and rewatering (the TCAs isocitric and citric, and proline).
Drought and heat stress affected soybean (Glycine max L.) leaf metabolomics [180]. The levels of glycolisis intermediates (pyruvate, glucose, dihydroxyacetone phosphate) were dramatically reduced, whereas those from TCA-cycle metabolites (malate, succinate, alpha-ketoglutarate, citrate) were significantly decreased. Pyruvate decrease can be the cause of the affectation of the biosynthesis of phenylalanine, tryptophan, tyrosine, isoleucine and alanine. Nevertheless, the oxaloacetate (a metabolite from TCA cycle) and its amino-acidic derivatives methionine and lysine also decreased. Similarly, sugar alcohols such as mannitol and galactitol were reduced as a result of temperature stress. In maize, the combination of drought and cold stresses was assayed and led to a multi-faceted metabolic response that revealed an ABA-dependent acclimation mechanism [34].
8. Temperature Stress
Plants are committed to a wide variety of factors, and one of the primary agents that constitute the organization and fate of plants is temperature stress, which comprises both high and low temperature stresses and is judged to be the major abiotic stress for seedlings [181,182]. Temperature stress is of interest to plant researchers due to climate change, which adversely affects crop productivity worldwide [183,184]. The increase in temperature is capable of damage essential physiological issues like the balanced relationship between primary and secondary metabolites with hormones, or the correspondence between water and membrane consistency, respiration and photosynthesis [185]. The generated disruptions impair metabolic development reducing plant growth and altering its development, and ultimately results in little economic benefit.
Various studies have been carried out recently on metabolomic responses to cold stress (tomato, wheat, maize, miscanthus) [29,186,187,188,189], including several medicinal plants [190], but Arabidopsis thaliana is the species traditionally more studied on such respects. In this grass, cold stress generates increases in most of its metabolites, including proline, sugars and TCA-cycle intermediates [191]. A biomarker candidate of cold tolerance in Arabidopsis resulted to be raffinose [155,192], although the general response depends on ecotypes. In this species, most of its heat shock metabolite responses were shared with those caused by cold, such as increases of amino acids derivated from pyruvate and the TCA-cycle [193].
Heat is an important abiotic stress continuously affecting crops in many countries, as worldwide temperatures are increasing. Heat stress in plants causes the overproduction of phenolic metabolites, phenylpropanoids and flavonoids [28]. In tomato (Solanum lycopersicum L.) leaves, both the suppression and overexpression of the heat stress factor B1 (HsfB1) enhanced the plant thermotolerance by different means. The overexpression of HsfB1 caused the accumulation of products of the phenylpropanoid and flavonoid pathways, including several caffeoyl quinic acid isomers [29]. However, tomato leaves with suppressed HsfB1 showed an accumulation of the polyamine putrescine and the sugars sucrose and glucose. Therefore, heat-tolerance was not specific to one metabolite group. Thus, affectations in respiratory pathways at substrate level were detected by means of metabolomics and transcriptomics, which revealed the differential response of Arabidopsis thaliana leaves to heat stress caused by prolonged warming or by heat shocks. Prolonged warming enhanced the glycolysis pathway but inhibited the TCA-cycle, while a heat shock significantly limited the conversion of pyruvate into acetyl coenzyme A [194], then also negatively affecting TCA-cycle, but not, in such a measure, glycolysis. Moreover, in wheat (Triticum aestivum L.) subjected to high temperature stress, the flag leaves after 10 days of anthesis showed a general increase in pipecolate and L-tryptophan, whereas anthranilate and drummondol decreased. The metabolic pathways most impaired by high temperature stress were the biosynthesis of secondary metabolites and the aminoacyl-tRNA pathway [195].
Similarly, metabolomic tools have permitted the assessment of the metabolite dynamics of heat-tolerant and heat-sensitive plants in response to heat stress. A recent metabolomic study showed a two-step response of Pinus radiata to heat stress with the metabolite profile of leaves significantly changing after a 3-day heat-stress to activate a long-term plant response to stress. Cytokinins (CKs), fatty acid metabolism and flavonoid and terpenoid biosynthesis were the most important pathways involved in heat-stress response. Additionally, L-phenylalanine, hexadecanoic acid, and dihydromyricetin were identified as potential biomarkers for heat toletrant plants.
In spite of the described metabolic changes, it seems that pollen is more resistant to heat stress than tissues. In tomato pollen heat stress did not lead to the detection of differential metabolites [186].
Heat stress has been studied in combination with salt stress in lablab bean (Dolichos lablab) seedlings, which were pretreated at 35 °C and 100 mM NaCl and then exposed to 45 °C for 5 h and restored to 25 °C. Such treatment generated higher levels of proline, ascorbate peroxidase, glutathione reductase, peroxidase, ascorbate, glutathione, and sugars [196].
9. Salt Stress
High concentrations of salts in the soil trigger various damages to the plant, avoiding absorption of water (thus generating secondarily water deficiency stress) and nutrients (for example, potassium), thereby hindering anabolic pathways like photosynthesis and protein biosynthesis [197]. Salinity is one of the main factors causing abiotic stress in plants.
Targeted metabolomics has assessed the phytohormone response to salt stress in roots and shoots form Arabidopsis seedlings [198], while a non-targeted approach in maize (Zea mays L.), revealed that the metablomic changes in roots were negligible [199]. Similarly, metabolomics allowed to understand the response to salt stress of salinity-resistant transgenic tobacco plants [200]. The transgenic plants increased the contents of proline, glutathione and trehalose, whereas the pentose phosphate pathway was stimulated, which may be associated to the observed decrease in fructose.
Metabolomes in response to salt stress were compared between species. Thus, the metabolome of 1-month cucumber and tomato plantlets under stress was different as the content of saponins, proline and total antioxidant capacity was reduced more notably in tomato as compared to cucumber. However, the levels of phenolics and flavonoids increased in both plants with a higher increase in tomato compared to cucumber [201].
Comparative metabolomics was also performed between two related species, Thellungiella halophila L. (extremophyla) and Arabidopsis thaliana, under high salt concentrations. Both species showed increased levels of proline and sugars, but with higher concentrations in the halophyte, suggesting its priming for salt tolerance [202]. Comparisons were made in halophyte species of the same genus. Thus, a differential response to salinity was observed in roots of Salicornia brachiata, S. maritima and S. portulacastrum. The level of proline increased with NaCl concentration in S. portulacastrum and S. maritima, whereas glycine, betaine and polyols were accumulated in S. maritima and S. brachiata. Moreover, in S. brachiata, the amount of flavonoids and other phenolic compounds increased in presence of NaCl, whereas these metabolites were down regulated in S. portulacastrum, who accumulated carotenoids [36].
Genotypes belonging to the same species were also contrasted. Recently, [35] investigated the metabolomic changes in five genotypes of Triticum durum Desf. exposed to different concentrations of NaCl. All genotypes showed an accumulation in shoots of proline and the depletion of organic acids, including TCA-cycle intermediates, at the highest salt concentration (200 mM), a situation that resembles oxygen depletion as that caused by flood. Also, there were a number of metabolites that accumulated depending on the genotype, especially GABA, threonine, leucine, glutamic acid, glycine, mannose and fructose. This fact suggests an association of the accumulated metabolites with the different tolerance of genotypes to salinity.
Similarly, metabolomics have contributed to a characterization of salt-tolerant (ST) varieties. However, ST biomarkers vary for different cultivars as each cultivar can cope differently under stress conditions. Sánchez et al. [203] subjected aerial parts of Lotus japonicus L. seedlings to salinity and observed a decrease in asparagine and glutamine–-which are the primary products of nitrogen assimilation–and an increase in glucuronic and gulonic acids, as well as ononitol, threonine and serine. These last three analytes also increased in a later experiment [204], together with other analytes, in both ST and salt sensitive (SS) Lotus species (three tolerant, three sensitive) under salinity stress. However, the levels of organic acids, including threonic acid and the TCA-cycle acids malic, succinic and citric, decreased with low salinity in all genotypes. In spite of the generalities, SS genotypes had few specific alterations (for example, gulonic acid increased and aspartic acid decreased) and the tolerant genotypes increased in asparagine. In a similar experiment in barley [205] the more tolerant cultivar showed increased concentrations of hexose phosphates and TCA-cycle intermediates. In another study in barley, roots of the ST genotype showed increased proline and carbohydrates [98]. In rice leaves, salinity stress increased the levels of serotonin and gentisic acid of ST varieties [206].
Metabolomics has also been used to specifically characterize the plant response to stress caused by the presence of different levels of salt, showing a differential accumulation of metabolites according to the levels of salt. In a study in maize, the salt-resistance mechanisms in leaves were the most effective at low salt-stress levels in a a SS genotype when compared with the ST [199]. Additionally, the levels of quinic, 1,3-O-dicaffeoylquinic, 1,4-O-dicaffeoylquinic, chlorogenic and neochlorogenic acids were increased in flower samples of Lonicerae Japonicae Flos under low salt conditions (100 mM NaCl). Similarly, the levels of flavonoids such as luteoloside, quercetin, luteolin-5-O-β-D-glucopyranoside, lonicerin and flavoyadorinin-B increased under low salt conditions. In addition to the changes observed under low salt conditions, accumulation of caffeic acid, 1,3-O-dicaffeoylquinic acid, chlorogenic acid, 1,4-O-dicaffeoylquinic acid and the flavonoids, luteolin-5-O-β-D-glucopyranoside, lonicerin, and flavoyadorinin-B, were observed under medium salt conditions (200 Mm NaCl). High salt conditions (300 mM NaCl) caused a reduction in quinic acid, caffeic acid, ferulic acid, isochlorogenic acid A, isochlorogenic acid C, luteolin-5-O-β-D-glucopyranoside, lonicerin, flavoyadorinin-B, isoquercitrin and astragalin [207]. Further, in a recent study in bananas (Musa acuminata L. Barangan), in vitro cultured shoots were submitted to 100 mM NaCl, and alcohols disappeared [208]. With 150 mM NaCl, phenols also disappeared. It is possible that such absences were due processes of esterification produced under such conditions, since when 0.50, 100, 150 and 200 mM of NaCl were applied, the metabolites that most increased were the esters.
The effects of the salt-exposure length on plants have also been assessed by metabolomics. In general, metabolite reprograming after an initial exposure to salt stress has been the key difference between ST and SS plant varieties. A recent study [209] showed a total of 13 stress-related metabolites including piperidine, L-tryptophan, L-glutamic acid, L-saccharopine, L-phenylalanine, 6-methylcoumarin, cinnamic acid, inosine 5′-monophosphate, aminomalonic acid, 6-aminocaproic acid, putrescine, tyramine and abscisic acid were the main metabolites differentially accumulated in hulless barley leaves regardless of the salt-tolerance level. However, the high tolerance of the ST cultivar was due to a metabolic reprogramming at key stress times (48–72 h exposure to stress) and nine salt-tolerance biomarkers including pyridoxine O-glucoside, L-alanine, hesperetin O-malonylhexoside, kynurenic acid O-hexside, 2′-deoxyadenosine-5′-monophosphate, 4-hydroxy-7-methoxycoumarin-beta-rhamnoside, nicotinate ribonucleoside, chrysoeriol 6-C-hexoside 8-C-hexoside-O-hexoside and 7-hydroxycoumarin-beta-rhamnoside were proposed [210]. Similarly, the content of sucrose decreased, whereas the content of organic acids such as L-malic acid and 2-oxoglutaric acid increased in roots of sugar beet seedlings after 1 day of salt stress. However, the levels of betaine, melatonin, and (S)-2-aminobutyric acid increased significantly after 7 days of salt stress [211]. Results suggest a metabolite reprograming from short-term to long-term mechanisms for tolerance to salt stress, similar to what was reported in plants as Pinus radiata under heat stress [212,213]. In a 7-day study of the metabolomic profile rice cells cultured in 10 mM NaCl solution, the more prominent metabolite content changes were a rapid increase in glucose and a later-end decrease in 2-amino butyric acid [148]. The amino acids that had the highest growth were ornithine at the beginning and proline at the end of the studied period. With 100 mM salt, results were not the same, registering increases in proline, cysteine, threonine, methionine, isoleucine, mannose, gentibiose and fructose 6-P. In roots of tolerant rice cultivars, there was an increase in amino acids and a decrease in organic acids (including TCA intermediates) [206]. In in vitro Spinacia oleracea L. sprouts, as salt stress increased, the contents of sodium ions also increased, but not that of K+ and Ca++ ions significantly decreased. However, in the cultures with higher salt concentration, Na+ and K+ ions in tissues increased [210]. Other changes observed in these in vitro cultures were increased osmotic adjustment due to elevated levels of soluble sugars and antioxidant enzyme activity. The metabolite that accumulated significantly more was 20-hydroxyecdysone, when the salt concentration was high.
Metabolomic techniques have also been used to assess the effect of treatments to increase salt tolerance in plats. Metabolic profiling of tomato leaves allowed the identification of the reprogramming of a wide range of metabolites in response to Omeprazole treatment [214]. Omeprazole increased the plant’s tolerance to salt-stress and caused in leaves hormonal changes such as an increase in abscisic acid, decrease in auxins and cytokinin, and a reduction in gibberellic acid. Additionally, polyamine conjugates, alkaloids and sesquiterpene lactones accumulated in response to Omeprazole. Results suggested that Omeprazole acted as a hormone that elicited an improved tolerance to salinity stress.
10. Oxidative Stress
Oxidative stress is a critical latent factor which can be caused secondarily by abiotic and biotic stresses and is also an important conditioning agent of plant development in the field [215]. Most of the metabolites induced by abiotic stress show antioxidative activity in vitro [216] and their roles in vivo are being studied. Oxidative stress in plants is produced by metabolic changes due to the overproduction of reactive oxygen species (ROS), which cause the oxidative deterioration of cellular structures such as lipids, proteins and DNA [217]. Glutathione is an important antioxidant and protect cells against ROS [218]. As a consequence, glutathione is an important mediator in several plant stresses [219,220,221]. Glutathione lacking and overexpressing Arabidopsis thaliana genotypes exposed to direct oxidative stress (induced by methyl violet) [110] showed the same growth as control plants. Glutathione lacking mutants overexpressed, for example, flavonoids, and transcriptomic analyses demonstrated a glutathione-independent reconfiguration of their metabolic networks.
Among abiotic stresses generating a secondary oxidative stress is found UV-radiation. This generates ROS and the biochemical responses of the plant include protection against both radiation and oxidation, as occurs in Pinus radiata [222]. In oxidative stress studies, metabolomic tools have allowed the assessment of plants tolerant to mineral stresses. Minerals such as cadmium can produce oxidative stress in plants and cause accumulation of reactivate oxygen species that induce damage in macromolecules such as DNA [223]. Species such as Brassica napus L. are tolerant to high concentrations of cadmium and can be used for soil bioremediation. Recently, metabolomic studies showed that cadmium-tolerant plants usually accumulate higher concentrations of unsaturated fatty acids and inositol-derived signaling metabolites in leaves when compared to sensitive varieties [224]. A phytotoxicity analysis using metabolomics have also allowed identification of plants that can cause damage to human health. Thus, the exposure of wheat plants to typical chlorinated organophosphate esters caused the translocation of the compounds in different parts of the plants and an increase in respiration and production of antioxidative metabolites of different compounds [225]. In the same way, leaves of cucumber exposed to silver ions and nanoparticles showed oxidative stress and responded with antioxidant production and enhanced oxygen consumption [101].
11. Future Perspectives
The integration of libraries of different omics to construct systematic backgrounds has lead, for example, to the development of metabolomic quantitative trait loci maps [226]. Metabolomics-assisted biological engineering to obtain plant stress tolerances is a promising field (reviewed by [227]). More concretely, the studies of plant metabolomics in these two decades have developed considerably thanks to technologies such as mass spectrometry, which has become a powerful tool that contributes to the explanation of essential biological processes related to the plant world. Future perspectives of metabolomics are oriented to the construction of mega-libraries of metabolites to determine and interpret the configuration of compounds, establishing and adjusting the spectral singularities of the metabolites evaluated with cured spectra of observation patterns. Another activity, in the same line of action that researchers should carry out, is to provide databases that allow functional comparisons and integration of metabolites of higher plant species to improve productivity and adaptability of crops to the environment. One of the most pressing perspectives for metabolomics is the development of bioinformatic software to purify the metabolites in order to confirm the nature of the compounds in the existing databases. Last, but not least, researchers of this omic technology must strengthen the research groups dedicated to the study of plant metabolites by being more careful and stricter in the whole process of analysis and identification of plant metabolites.
Author Contributions
Conceptualization, J.M.C.-C.; writing—original draft preparation, F.P.C. by support of C.N. and M.G.M.-Z.; writing—review and editing and supervision, C.N. and J.M.C.-C.; project administration and funding acquisition, C.N. and J.M.C.-C. All authors have read and agreed to the published version of the manuscript.
Funding
The present work was partially financed by VLIR-UOS (grant VLIR Network Ecuador) and the REDU Project 2018-PREDU-2016-002 (Ecuador), “Indicadores metabolómicos de variación somaclonal en banano (Musa AAA) y expresión de genes implicados” (2017).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets analyzed during the current study are available from corresponding author upon reasonable request.
Acknowledgments
We acknowledge the Prometeo Program (SENESCYT, Ecuador) and to the BIOALI-CYTED network for facilitating the interaction between authors.
Conflicts of Interest
The authors declare no conflict of interests.
References
- Raza, A.; Zahra, N.; Hafeez, M.B.; Ahmad, M.; Iqbal, S.; Shaukat, K.; Ahmad, G. Nitrogen Fixation of Legumes: Biology and Physiology. In The Plant Family Fabaceae; Springer: Singapore, 2020. [Google Scholar]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef]
- Newton, A.C.; Johnson, S.N.; Gregory, P.J. Implications of climate change for diseases, crop yields and food security. Euphytica 2011, 179, 3–18. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Sawada, Y.; Nakabayashi, R.; Sato, S.; Hirakawa, H.; El-Sayed, M.; Hirai, M.Y.; Saito, K.; Yamauchi, N.; Shigyo, M. Integrating transcriptome and target metabolome variability in doubled haploids of Allium cepa for abiotic stress protection. Mol. Breed. 2015, 35, 1–11. [Google Scholar] [CrossRef]
- Costa, M.C.D.; Farrant, J.M. Plant Resistance to Abiotic Stresses. Plants 2019, 8, 553. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Witt, S.; Lisec, J.; Palacios-Rojas, N.; Florez-Sarasa, I.; Yousfi, S.; Fernie, A.R. Metabolite profiles of maize leaves in drought, heat and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol. 2015, 169, 2665–2683. [Google Scholar] [CrossRef] [PubMed]
- Rosato, A.; Tenori, L.; Cascante, M.; Carulla, P.R.D.A.; Dos Santos, V.A.P.M.; Saccenti, E. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 1–20. [Google Scholar] [CrossRef]
- Jorge, T.F.; Rodrigues, J.A.; Caldana, C.; Schmidt, R.; van Dongen, J.T.; Thomas-Oates, J.; António, C. Mass spectrometry-based plant metabolomics: Metabolite responses to abiotic stress. Mass Spectrom. Rev. 2016, 35, 620–649. [Google Scholar] [CrossRef]
- Jorge, T.F.; Mata, A.T.; António, C. Mass spectrometry as a quantitative tool in plant metabolomics. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150370. [Google Scholar] [CrossRef]
- Jorge, T.F.; António, C. Plant Metabolomics in a Changing World: Metabolite Responses to Abiotic Stress Combinations. In Plant, Abiotic Stress and Responses to Climate Change; IntechOpen: London, UK, 2018. [Google Scholar]
- Alseekh, S.; Bermudez, L.; De Haro, L.A.; Fernie, A.R.; Carrari, F. Crop metabolomics: From diagnostics to assisted breeding. Metabolomics 2018, 14, 148. [Google Scholar] [CrossRef]
- Sharma, K.; Sarma, S.; Bohra, A.; Mitra, A.; Sharma, N.K.; Kumar, A. Plant Metabolomics: An Emerging Technology for Crop Improvement. New Vis. Plant Sci. 2018. [Google Scholar] [CrossRef]
- Cambiaghi, A.; Ferrario, M.; Masseroli, M. Analysis of metabolomic data: Tools, current strategies and future challenges for omics data integration. Briefings Bioinform. 2017, 18, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, J.; Khatri, P.; Singla, P.; Kumawat, S.; Kumari, A.; Vikram, A.; Jindal, S.K.; Kardile, H.; Kumar, R.; Sonah, H.; et al. Advances in Omics Approaches for Abiotic Stress Tolerance in Tomato. Biology 2019, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Yang, L.; Zhang, D.; Shi, J. Plant Metabolomics: An Indispensable System Biology Tool for Plant Science. Int. J. Mol. Sci. 2016, 17, 767. [Google Scholar] [CrossRef]
- Scossa, F.; Brotman, Y.; Lima, F.D.A.; Willmitzer, L.; Nikoloski, Z.; Tohge, T.; Fernie, A.R. Genomics-based strategies for the use of natural variation in the improvement of crop metabolism. Plant Sci. 2016, 242, 47–64. [Google Scholar] [CrossRef]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef]
- Patil, G.; Jeong-Dong, L.; Vuong, T.D.; Valliyodan, B.; Lee, J.-D.; Chaudhary, J.; Shannon, J.G.; Nguyen, H.T. Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 2016, 6, 19199. [Google Scholar] [CrossRef]
- Yan, M.; Xu, G. Current and future perspectives of functional metabolomics in disease studies—A review. Anal. Chim. Acta 2018, 1037, 41–54. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Nielsen, J.; Fernie, A.R. Engineering central metabolism—A grand challenge for plant biologists. Plant J. 2017, 90, 749–763. [Google Scholar] [CrossRef]
- Peters, K.; Worrich, A.; Weinhold, A.; Alka, O.; Balcke, G.; Birkemeyer, C.; Bruelheide, H.; Calf, O.W.; Dietz, S.; Dührkop, K.; et al. Current Challenges in Plant Eco-Metabolomics. Int. J. Mol. Sci. 2018, 19, 1385. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.M.; Miguel, C.; Chaves, I.; António, C. Mass spectrometry-based forest tree metabolomics. Mass Spectrom. Rev. 2021, 40, 126–157. [Google Scholar] [CrossRef]
- Jacyna, J.; Kordalewska, M.; Markuszewski, M.J. Design of Experiments in metabolomics-related studies: An overview. J. Pharm. Biomed. Anal. 2019, 164, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Dudzik, D.; Barbas-Bernardos, C.; García, A.; Barbas, C. Quality assurance procedures for mass spectrometry untargeted metabolomics. a review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, J.; Want, E.J. From Samples to Insights into Metabolism: Uncovering Biologically Relevant Information in LC-HRMS Metabolomics Data. Metabolites 2019, 9, 308. [Google Scholar] [CrossRef]
- Gullberg, J.; Jonsson, P.; Nordström, A.; Sjöström, M.; Moritz, T. Design of experiments: An efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal. Biochem. 2004, 331, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Delaroza, F.; Rakocevic, M.; Malta, G.B.; Sanchez, P.M.; Bruns, R.E.; Scarminio, I.S. Factorial design effects of plant density, pattern and light availability on the caffeine, chlorogenic acids, lipids, reducing sugars and ash contents of Coffea arabica L. beans and leaves. Anal. Methods 2017, 9, 3612–3618. [Google Scholar] [CrossRef]
- Masouleh, S.S.S.; Sassine, Y.N. Molecular and biochemical responses of horticultural plants and crops to heat stress. Ornam. Hortic. 2020, 26, 148–158. [Google Scholar] [CrossRef]
- Paupière, M.J.; Tikunov, Y.; Schleiff, E.; Bovy, A.; Fragkostefanakis, S. Reprogramming of Tomato Leaf Metabolome by the Activity of Heat Stress Transcription Factor HsfB. Front Plant Sci. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Tschaplinski, T.J.; Abraham, P.; Jawdy, S.S.; Gunter, L.; Martin, M.Z.; Engle, N.L.; Yang, X.; Tuskan, G. The nature of the progression of drought stress drives differential metabolomic responses in Populus deltoides. Ann. Bot. 2019, 124, 617–626. [Google Scholar] [CrossRef]
- Yuan, H.; Zeng, X.; Shi, J.; Xu, Q.; Wang, Y.; Jabu, D.; Sang, Z.; Nyima, T. Time-Course Comparative Metabolite Profiling under Osmotic Stress in Tolerant and Sensitive Tibetan Hulless Barley. BioMed Res. Int. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
- Kang, Z.; Babar, A.; Khan, N.; Guo, J.; Khan, J.; Islam, S.; Shrestha, S.; Shahi, D. Comparative metabolomic profiling in the roots and leaves in contrasting genotypes reveals complex mechanisms involved in post-anthesis drought tolerance in wheat. PLoS ONE 2019, 14, e0213502. [Google Scholar] [CrossRef]
- Gundaraniya, S.A.; Ambalam, P.S.; Tomar, R.S. Metabolomic Profiling of Drought-Tolerant and Susceptible Peanut (Arachis hypogaea L.) Genotypes in Response to Drought Stress. ACS Omega 2020, 5, 31209–31219. [Google Scholar] [CrossRef]
- Guo, X.; Xin, Z.; Yang, T.; Ma, X.; Zhang, Y.; Wang, Z.; Ren, Y.; Lin, T. Metabolomics Response for Drought Stress Tolerance in Chinese Wheat Genotypes (Triticum aestivum). Plants 2020, 9, 520. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Fragasso, M.; Nigro, F.; Platani, C.; Papa, R.; Beleggia, R.; Trono, D. Analysis of metabolic and mineral changes in response to salt stress in durum wheat (Triticum turgidum ssp. durum) genotypes, which differ in salinity tolerance. Plant Physiol. Biochem. 2018, 133, 57–70. [Google Scholar] [CrossRef]
- Benjamin, J.J.; Lucini, L.; Jothiramshekar, S.; Parida, A. Metabolomic insights into the mechanisms underlying tolerance to salinity in different halophytes. Plant Physiol. Biochem. 2019, 135, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.C.M.; Caldana, C.; Wolf, L.D.; De Abreu, L.G.F. The Importance of Experimental Design, Quality Assurance, and Control in Plant Metabolomics Experiments. Methods Mol. Biol. 2018, 1778, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Blaise, B.J.; Correia, G.; Tin, A.; Young, J.H.; Vergnaud, A.-C.; Lewis, M.; Pearce, J.T.M.; Elliott, P.; Nicholson, J.K.; Holmes, E.; et al. Power Analysis and Sample Size Determination in Metabolic Phenotyping. Anal. Chem. 2016, 88, 5179–5188. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]
- Nyamundanda, G.; Gormley, I.C.; Fan, Y.; Gallagher, W.M.; Brennan, L. MetSizeR: Selecting the optimal sample size for metabolomic studies using an analysis based approach. BMC Bioinform. 2013, 14, 338. [Google Scholar] [CrossRef]
- Hendriks, M.M.; Eeuwijk, F.A.; Jellema, R.H.; Westerhuis, J.A.; Reijmers, T.H.; Hoefsloot, H.C.; Smilde, A.K. Data-processing strategies for metabolomics studies. TrAC Trends Anal. Chem. 2011, 30, 1685–1698. [Google Scholar] [CrossRef]
- Fiehn, O.; Wohlgemuth, G.; Scholz, M. Setup and Annotation of Metabolomic Experiments by Integrating Biological and Mass Spectrometric Metadata. Trans. Petri Nets Other Models Concurr. XV 2005, 3615, 224–239. [Google Scholar] [CrossRef]
- Bénard, C.; Bernillon, S.; Biais, B.; Osorio, S.; Maucourt, M.; Ballias, P.; Deborde, C.; Colombié, S.; Cabasson, C.; Jacob, D.; et al. Metabolomic profiling in tomato reveals diel compositional changes in fruit affected by source–sink relationships. J. Exp. Bot. 2015, 66, 3391–3404. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based metabolomic analysis of plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Ric, C.H.; De Vos, A.M.; Deborde, C.; Erban, A.; Kopka, J.; Goodacre, R.; Robert, D.H. Plant Metabolomics and Its Potential for Systems Biology Research. In Methods in Enzymology, 1st ed.; Jameson, D., Verma, M., Hans, V.W., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2011; Volume 500. [Google Scholar]
- Glauser, G.; Boccard, J.; Wolfender, J.-L.; Rudaz, S. Metabolomics: Application in Plant Sciences. Metab. Pract. 2013, 313–343. [Google Scholar] [CrossRef]
- T’Kindt, R.; Morreel, K.; Deforce, D.; Boerjan, W.; Van Bocxlaer, J. Joint GC–MS and LC–MS platforms for comprehensive plant metabolomics: Repeatability and sample pre-treatment. J. Chromatogr. B 2009, 877, 3572–3580. [Google Scholar] [CrossRef]
- Duvane, J.A.; Jorge, T.F.; Maquia, I.; Ribeiro, N.; Ribeiro-Barros, A.I.F.; António, C. Characterization of the Primary Metabolome of Brachystegia boehmii and Colophospermum mopane under Different Fire Regimes in Miombo and Mopane African Woodlands. Front. Plant Sci. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Cevallos, J.M.; Jines, C.; Maridueña-Zavala, M.G.; Molina-Miranda, M.J.; Ochoa, D.E.; Flores-Cedeno, J.A. GC-MS metabolite profiling for specific detection of dwarf somaclonal variation in banana plants. Appl. Plant Sci. 2018, 6, e01194. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Cevallos, J.M.; Reyes-De-Corcuera, J.I.; Etxeberria, E.; Danyluk, M.D.; Rodrick, G.E. Metabolomic analysis in food science: A review. Trends Food Sci. Technol. 2009, 20, 557–566. [Google Scholar] [CrossRef]
- De Koning, W.; van Dam, K.; de Koning, W. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal. Biochem. 1992, 204, 118–123. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. TrAC Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar] [CrossRef]
- Pongsuwan, W.; Bamba, T.; Harada, K.; Yonetani, T.; Kobayashi, A.; Fukusaki, E. High-Throughput Technique for Comprehensive Analysis of Japanese Green Tea Quality Assessment Using Ultra-performance Liquid Chromatography with Time-of-Flight Mass Spectrometry (UPLC/TOF MS). J. Agric. Food Chem. 2008, 56, 10705–10708. [Google Scholar] [CrossRef] [PubMed]
- Maridueña-Zavala, M.G.; Freire-Peñaherrera, A.; Cevallos-Cevallos, J.M.; Peralta, E.L. GC-MS metabolite profiling of Phytophthora infestans resistant to metalaxyl. Eur. J. Plant Pathol. 2017, 149, 563–574. [Google Scholar] [CrossRef]
- Verpoorte, R.; Choi, Y.H.; Mustafa, N.R.; Kim, H.K. Metabolomics: Back to basics. Phytochem. Rev. 2008, 7, 525–537. [Google Scholar] [CrossRef]
- Berini, J.L.; Brockman, S.A.; Hegeman, A.D.; Reich, P.B.; Muthukrishnan, R.; Montgomery, R.A.; Forester, J.D. Combinations of Abiotic Factors Differentially Alter Production of Plant Secondary Metabolites in Five Woody Plant Species in the Boreal-Temperate Transition Zone. Front. Plant Sci. 2018, 9, 1257. [Google Scholar] [CrossRef] [PubMed]
- Sedio, B.E.; Echeverri, J.C.R.; Boya, C.; Wright, S.J.S.J.; Boya, C.A.P. Sources of variation in foliar secondary chemistry in a tropical forest tree community. Ecology 2017, 98, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Kopka, J. Current challenges and developments in GC–MS based metabolite profiling technology. J. Biotechnol. 2006, 124, 312–322. [Google Scholar] [CrossRef]
- Shinbo, Y.; Nakamura, Y.; Altaf-Ul-Amin, M.; Asahi, H.; Kurokawa, K.; Arita, M.; Saito, K.; Ohta, D.; Shibata, D.; Kanaya, S. KNApSAcK: A Comprehensive Species-Metabolite Relationship Database. Biotechnol. Agric. For. 2006, 57, 165–181. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2006, 26, 51–78. [Google Scholar] [CrossRef]
- Pires, M.V.; Júnior, A.A.P.; Medeiros, D.B.; Daloso, D.M.; Pham, P.A.; Barros, K.A.; Engqvist, M.K.M.; Florian, A.; Krahnert, I.; Maurino, V.G.; et al. The influence of alternative pathways of respiration that utilize branched-chain amino acids following water shortage in Arabidopsis. Plant Cell Environ. 2016, 39, 1304–1319. [Google Scholar] [CrossRef]
- Ullah, N.; Yüce, M.; Gökçe, Z.N.Ö.; Budak, H. Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Genom. 2017, 18, 969. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Cevallos, J.M.; García-Torres, R.; Etxeberria, E.; Reyes-De-Corcuera, J.I. GC-MS Analysis of Headspace and Liquid Extracts for Metabolomic Differentiation of Citrus Huanglongbing and Zinc Deficiency in Leaves of ‘Valencia’ Sweet Orange from Commercial Groves. Phytochem. Anal. 2010, 22, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Shawky, E.; Selim, D.A. Evaluation of the effect of extraction solvent and organ selection on the chemical profile of Astragalus spinosus using HPTLC- multivariate image analysis. J. Chromatogr. B 2017, 1061–1062, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fan, G.; Zhang, J.; Zhang, Y.; Li, J.; Xiong, C.; Zhang, Q.; Li, X.; Lai, X. Metabolic discrimination of sea buckthorn from different Hippophaë species by 1H NMR based metabolomics. Sci. Rep. 2017, 7, 1585. [Google Scholar] [CrossRef]
- Deborde, C.; Moing, A.; Roch, L.; Jacob, D.; Rolin, D.; Giraudeau, P. Plant metabolism as studied by NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 102–103, 61–97. [Google Scholar] [CrossRef] [PubMed]
- Segers, K.; Declerck, S.; Mangelings, D.; Heyden, Y.V.; Van Eeckhaut, A. Analytical techniques for metabolomic studies: A review. Bioanalysis 2019, 11, 2297–2318. [Google Scholar] [CrossRef] [PubMed]
- De Raad, M.; Fischer, C.R.; Northen, T.R. High-throughput platforms for metabolomics. Curr. Opin. Chem. Biol. 2016, 30, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.-L.; Marti, G.; Thomas, A.; Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef]
- Rodríguez-Calcerrada, J.; Rodrigues, A.M.; Perdiguero, P.; António, C.; Atkin, O.K.; Li, M.; Collada, C.; Gil, L. A molecular approach to drought-induced reduction in leaf CO2 exchange in drought-resistant Quercus ilex. Physiol. Plant. 2017, 162, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Schiffmann, C.; Hansen, R.; Baumann, S.; Kublik, A.; Nielsen, P.H.; Adrian, L.; von Bergen, M.; Jehmlich, N.; Seifert, J. Comparison of targeted peptide quantification assays for reductive dehalogenases by selective reaction monitoring (SRM) and precursor reaction monitoring (PRM). Anal. Bioanal. Chem. 2014, 406, 283–291. [Google Scholar] [CrossRef]
- Antonio, C.; Mustafa, N.R.; Osorio, S.; Tohge, T.; Giavalisco, P.; Willmitzer, L.; Rischer, H.; Oksman-Caldentey, K.-M.; Verpoorte, R.; Fernie, A.R. Analysis of the Interface between Primary and Secondary Metabolism in Catharanthus roseus Cell Cultures Using 13C-Stable Isotope Feeding and Coupled Mass Spectrometry. Mol. Plant 2013, 6, 581–584. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.-L.; Morgan, J.A. Metabolic flux analysis of secondary metabolism in plants. Metab. Eng. Commun. 2020, 10, e00123. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Tohge, T. The Genetics of Plant Metabolism. Annu. Rev. Genet. 2017, 51, 287–310. [Google Scholar] [CrossRef]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Hanhineva, K.; Rogachev, I.; Kokko, H.; Mintz-Oron, S.; Venger, I.; Kärenlampi, S.; Aharoni, A. Non-targeted analysis of spatial metabolite composition in strawberry (Fragaria×ananassa) flowers. Phytochemistry 2008, 69, 2463–2481. [Google Scholar] [CrossRef]
- Moco, S.; Bino, R.J.; Vorst, O.; Verhoeven, H.A.; De Groot, J.; Van Beek, T.A.; Vervoort, J.; De Vos, C.R. A Liquid Chromatography-Mass Spectrometry-Based Metabolome Database for Tomato. Plant Physiol. 2006, 141, 1205–1218. [Google Scholar] [CrossRef]
- Schweiggert, U.; Kammerer, D.R.; Carle, R.; Schieber, A. Characterization of carotenoids and carotenoid esters in red pepper pods (Capsicum annuum L.) by high-performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 2617–2628. [Google Scholar] [CrossRef]
- Qin, Z.; Liao, D.; Chen, Y.; Zhang, C.; An, R.; Zeng, Q.; Li, X. A Widely Metabolomic Analysis Revealed Metabolic Alterations of Epimedium Pubescens Leaves at Different Growth Stages. Molecules 2019, 25, 137. [Google Scholar] [CrossRef]
- Olennikov, D.N.; Gadimli, A.I.; Isaev, J.I.; Kashchenko, N.I.; Prokopyev, A.S.; Kataeva, T.N.; Chirikova, N.K.; Vennos, C. Caucasian Gentiana Species: Untargeted LC-MS Metabolic Profiling, Antioxidant and Digestive Enzyme Inhibiting Activity of Six Plants. Metabolites 2019, 9, 271. [Google Scholar] [CrossRef]
- Berman, P.; Futoran, K.; Lewitus, G.M.; Mukha, D.; Benami, M.; Shlomi, T.; Meiri, D. A new ESI-LC/MS approach for comprehensive metabolic profiling of phytocannabinoids in Cannabis. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Ramautar, R.; Somsen, G.W.; De Jong, G.J. CE-MS in metabolomics. Electrophoresis 2009, 30, 276–291. [Google Scholar] [CrossRef] [PubMed]
- Ramautar, R.; Somsen, G.W.; De Jong, G.J. The Role of CE-MS in Metabolomics. Metab. Pract. 2013, 177–208. [Google Scholar] [CrossRef]
- Moreno, D.; Berli, F.; Bottini, R.; Piccoli, P.N.; Silva, M.F. Grapevine tissues and phenology differentially affect soluble carbohydrates determination by capillary electrophoresis. Plant Physiol. Biochem. 2017, 118, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Monton, M.R.N.; Soga, T. Metabolome analysis by capillary electrophoresis–mass spectrometry. J. Chromatogr. A 2007, 1168, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Hirayama, A.; Robert, M.; Abe, S.; Soga, T.; Tomita, M. Prediction of metabolite identity from accurate mass, migration time prediction and isotopic pattern information in CE-TOFMS data. Electrophoresis 2010, 31, 2311–2318. [Google Scholar] [CrossRef]
- Bowne, J.B.; Erwin, T.A.; Juttner, J.; Schnurbusch, T.; Langridge, P.; Bacic, A.; Roessner, U. Drought Responses of Leaf Tissues from Wheat Cultivars of Differing Drought Tolerance at the Metabolite Level. Mol. Plant 2012, 5, 418–429. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; AlAhmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Huhman, D.V.; Sumner, L.W. Mass Spectrometry Strategies in Metabolomics. J. Biol. Chem. 2011, 286, 25435–25442. [Google Scholar] [CrossRef]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. 2009, 21, 4–13. [Google Scholar] [CrossRef]
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. [Google Scholar] [CrossRef]
- Chèze, M.; Gaulier, J.-M. Drugs Involved in Drug-Facilitated Crimes (DFC). Toxicol. Aspects Drug Facil. Crimes 2014, 181–222. [Google Scholar] [CrossRef]
- Cajka, T. Gas Chromatography–Time-of-Flight Mass Spectrometry in Food and Environmental Analysis; Elsevier: Amsterdam, The Netherlands, 2013; Volume 61, pp. 271–302. [Google Scholar]
- Wu, H.; Guo, J.; Chen, S.; Liu, X.; Zhou, Y.; Zhang, X.; Xu, X. Recent developments in qualitative and quantitative analysis of phytochemical constituents and their metabolites using liquid chromatography–mass spectrometry. J. Pharm. Biomed. Anal. 2013, 72, 267–291. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.-J.; Schultz, A.W.; Wang, J.; Johnson, C.H.; Yannone, S.M.; Patti, G.J.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013, 8, 451–460. [Google Scholar] [CrossRef]
- Motilva, M.-J.; Serra, A.; Macià, A. Analysis of food polyphenols by ultra high-performance liquid chromatography coupled to mass spectrometry: An overview. J. Chromatogr. A 2013, 1292, 66–82. [Google Scholar] [CrossRef]
- Zhang, H.; Du, W.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L.; White, J.C.; Keller, A.A.; Guo, H.; Ji, R.; Zhao, L. Metabolomics Reveals How Cucumber (Cucumis sativus) Reprograms Metabolites To Cope with Silver Ions and Silver Nanoparticle-Induced Oxidative Stress. Environ. Sci. Technol. 2018, 52, 8016–8026. [Google Scholar] [CrossRef]
- Fang, L.-H.; Wang, R.-P.; Hu, S.-Y.; Teng, Y.-H.; Xie, W.-B. The Effect of Tou Nong San on Transplanted Tumor Growth in Nude Mice. Evid.-Based Complement. Altern. Med. 2015, 2015, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Zhu, Y.; Lu, L.; Gu, F.; Chen, H. The Applications and Features of Liquid Chromatography-Mass Spectrometry in the Analysis of Traditional Chinese Medicine. Evid.-Based Complement. Altern. Med. 2016, 2016, 1–7. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, M.; Li, Y.; Xu, R.; Li, X. Comprehensive analysis of 61 characteristic constituents from Siraitiae fructus using ultrahigh-pressure liquid chromatography with time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2016, 125, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Reeve, M. MALDI-TOF MS-Based Analysis of Seed Proteins from Catalogue Varieties of Solanum lycopersicum/Lycopersicon esculentum. Horticulturae 2019, 5, 48. [Google Scholar] [CrossRef]
- Okazaki, Y.; Saito, K. Recent advances of metabolomics in plant biotechnology. Plant Biotechnol. Rep. 2011, 6, 1–15. [Google Scholar] [CrossRef]
- Papetti, A.; Colombo, R. High-Performance Capillary Electrophoresis for Food Quality Evaluation; Elsevier: Amsterdam, The Netherlands, 2019; pp. 301–377. [Google Scholar]
- Cocuron, J.-C.; Anderson, B.; Boyd, A.; Alonso, A.P. Targeted Metabolomics of Physaria fendleri, an Industrial Crop Producing Hydroxy Fatty Acids. Plant Cell Physiol. 2014, 55, 620–633. [Google Scholar] [CrossRef]
- Cao, D.; Lutz, A.; Hill, C.B.; Callahan, D.L.; Roessner, U. A Quantitative Profiling Method of Phytohormones and Other Metabolites Applied to Barley Roots Subjected to Salinity Stress. Front. Plant Sci. 2017, 7, 2070. [Google Scholar] [CrossRef]
- Fukushima, A.; Iwasa, M.; Nakabayashi, R.; Kobayashi, M.; Nishizawa, T.; Okazaki, Y.; Saito, K.; Kusano, M. Effects of Combined Low Glutathione with Mild Oxidative and Low Phosphorus Stress on the Metabolism of Arabidopsis thaliana. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; Van Der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef]
- García-Pérez, I.; Vallejo, M.; García, A.; Legido-Quigley, C.; Barbas, C. Metabolic fingerprinting with capillary electrophoresis. J. Chromatogr. A 2008, 1204, 130–139. [Google Scholar] [CrossRef]
- Brodsky, L.; Moussaieff, A.; Shahaf, N.; Aharoni, A.; Rogachev, I. Evaluation of Peak Picking Quality in LC−MS Metabolomics Data. Anal. Chem. 2010, 82, 9177–9187. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Kawakami, M.; Robert, M.; Soga, T. Bioinformatics Tools for Mass Spectroscopy-Based Metabolomic Data Processing and Analysis. Curr. Bioinform. 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Pedrioli, P.G.; Eng, J.K.; Hubley, R.; Vogelzang, M.; Deutsch, E.W.; Raught, B.; Pratt, B.; Nilsson, E.; Angeletti, R.H.; Apweiler, R.; et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 2004, 22, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, L. Sample normalization methods in quantitative metabolomics. J. Chromatogr. A. 2016, 1430, 80–95. [Google Scholar] [CrossRef] [PubMed]
- Warrack, B.M.; Hnatyshyn, S.; Ott, K.-H.; Reily, M.D.; Sanders, M.; Zhang, H.; Drexler, D.M. Normalization strategies for metabonomic analysis of urine samples. J. Chromatogr. B 2009, 877, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Senn, H. Probabilistic Quotient Normalization as Robust method to aacount for dilution of complex biuological mixtures. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B. Data normalization strategies in metabolomics: Current challenges, approaches, and tools. Eur. J. Mass Spectrom. 2020, 26, 165–174. [Google Scholar] [CrossRef]
- Sas, K.M.; Karnovsky, A.; Michailidis, G.; Pennathur, S. Metabolomics and Diabetes: Analytical and Computational Approaches. Diabetes 2015, 64, 718–732. [Google Scholar] [CrossRef]
- Moncayo Martín, S. Desarrollo y aplicación de métodos quimiométricos para el estudio de muestras mediante Espectroscopia de Ablación Láser (LIBS). Ph.D. Thesis, Universidad Complutense de Madrid, Departamento de Química Analítica, Madrid, Spain, 2017; pp. 40–58. [Google Scholar]
- Wang, Y.; Xu, L.; Shen, H.; Wang, J.; Liu, W.; Zhu, X.; Wang, R.; Sun, X.; Liu, L. Metabolomic analysis with GC-MS to reveal potential metabolites and biological pathways involved in Pb & Cd stress response of radish roots. Sci. Rep. 2015, 5, 18296. [Google Scholar] [CrossRef]
- Correia, B.; Hancock, R.D.; Amaral, J.; Gomez-Cadenas, A.; Valledor, L.; Pinto, G. Combined Drought and Heat Activates Protective Responses in Eucalyptus globulus That Are Not Activated When Subjected to Drought or Heat Stress Alone. Front. Plant Sci. 2018, 9, 819. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis—Amarriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Duarte, L.M.; Filgueiras, P.R.; Silva, S.R.; Dias, J.C.; Oliveira, L.M.; Castro, E.V.; De Oliveira, M.A. Determination of some physicochemical properties in Brazilian crude oil by 1H NMR spectroscopy associated to chemometric approach. Fuel 2016, 181, 660–669. [Google Scholar] [CrossRef]
- Skubel, S.A.; Su, X.; Poulev, A.; Foxcroft, L.C.; Dushenkov, V.; Raskin, I. Metabolomic differences between invasive alien plants from native and invaded habitats. Sci. Rep. 2020, 10, 9749. [Google Scholar] [CrossRef]
- Peñaloza, E.; Holandino, C.; Scherr, C.; De Araujo, P.I.P.; Borges, R.M.; Urech, K.; Baumgartner, S.; Garrett, R. Comprehensive Metabolome Analysis of Fermented Aqueous Extracts of Viscum album L. by Liquid Chromatography−High Resolution Tandem Mass Spectrometry. Molecules 2020, 25, 4006. [Google Scholar] [CrossRef]
- Lee, S.; Oh, D.-G.; Singh, D.; Lee, J.S.; Lee, S.; Lee, C.H. Exploring the metabolomic diversity of plant species across spatial (leaf and stem) components and phylogenic groups. BMC Plant Biol. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Bartel, J.; Krumsiek, J.; Theis, F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013, 4, e201301009. [Google Scholar] [CrossRef]
- Wu, M.; Ge, Y.; Xu, C.; Wang, J. Metabolome and Transcriptome Analysis of Hexaploid Solidago canadensis Roots Reveals its Invasive Capacity Related to Polyploidy. Genes 2020, 11, 187. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-F.; Yao, M.-Z.; Ma, C.-L.; Ma, J.-Q.; Jin, J.-Q.; Chen, L. Differential Metabolic Profiles during the Albescent Stages of ‘Anji Baicha’ (Camellia sinensis). PLoS ONE 2015, 10, e0139996. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [PubMed]
- Cavill, R.; Jennen, D.G.J.; Kleinjans, J.C.S.; Briedé, J.J. Transcriptomic and metabolomic data integration. Briefings Bioinform. 2016, 17, 891–901. [Google Scholar] [CrossRef]
- Novak, J.M.; Busscher, W.J. Selection and Use of Designer Biochars to Improve Characteristics of Southeastern USA Coastal Plain Degraded Soils. In Advanced Biofuels and Bioproducts; Springer: New York, NY, USA, 2013; pp. 69–96. [Google Scholar]
- Du, X.; Smirnov, A.; Pluskal, T.; Jia, W.; Sumner, S. Metabolomics Data Preprocessing Using ADAP and MZmine. Methods Mol. Biol. 2020, 2104, 25–48. [Google Scholar]
- Yi, Y.; Fang, Y.; Wu, K.; Liu, Y.; Zhang, W. Comprehensive gene and pathway analysis of cervical cancer progression. Oncol. Lett. 2020, 19, 3316–3332. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Zhang, Z.-Y.; Quan, H.; Li, M.-J.; Zhao, F.-Y.; Xu, Y.-J.; Liu, J.; Sai, M.; Zheng, W.-L.; Lan, X.-Z. Integrated analysis of transcriptomic and metabolomic data reveals critical metabolic pathways involved in rotenoid biosynthesis in the medicinal plant Mirabilis himalaica. Mol. Genet. Genom. 2017, 293, 635–647. [Google Scholar] [CrossRef]
- Guo, R.; Shi, L.; Jiao, Y.; Li, M.; Zhong, X.; Gu, F.; Liu, Q.; Xia, X.; Li, H. Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB Plants 2018, 10, ply016. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S.; et al. Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 2017, 3, 17097. [Google Scholar] [CrossRef]
- Todaka, D.; Zhao, Y.; Yoshida, T.; Kudo, M.; Kidokoro, S.; Mizoi, J.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Fernie, A.R.; Sato, M.; et al. Temporal and spatial changes in gene expression, metabolite accumulation and phytohormone content in rice seedlings grown under drought stress conditions. Plant J. 2016, 90, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Woodrow, P.; Ciarmiello, L.F.; Annunziata, M.G.; Pacifico, S.; Iannuzzi, F.; Mirto, A.; D’Amelia, L.; Dell’Aversana, E.; Piccolella, S.; Fuggi, A.; et al. Durum wheat seedling responses to simultaneous high light and salinity involve a fine reconfiguration of amino acids and carbohydrate metabolism. Physiol. Plant. 2016, 159, 290–312. [Google Scholar] [CrossRef] [PubMed]
- Sang, Q.; Shan, X.; An, Y.; Shu, S.; Sun, J.; Guo, S. Proteomic Analysis Reveals the Positive Effect of Exogenous Spermidine in Tomato Seedlings’ Response to High-Temperature Stress. Front. Plant Sci. 2017, 8, 120. [Google Scholar] [CrossRef]
- Ampofo-Asiama, J.; Baiye, V.M.M.; Hertog, M.L.A.T.M.; Waelkens, E.; Geeraerd, A.; Nicolai, B.M. The metabolic response of cultured tomato cells to low oxygen stress. Plant Biol. 2013, 16, 594–606. [Google Scholar] [CrossRef]
- Hochberg, U.; Degu, A.; Toubiana, D.; Gendler, T.; Nikoloski, Z.; Rachmilevitch, S.; Fait, A. Near isohydric grapevine cultivar displays higher photosynthetic efficiency and photorespiration rates under drought stress as compared with near anisohydric grapevine cultivar. Physiol. Plant. 2012, 147, 443–452. [Google Scholar] [CrossRef]
- Benina, M.; Obata, T.; Mehterov, N.; Ivanov, I.; Petrov, V.; Toneva, V.; Fernie, A.R.; Gechev, T.S. Comparative metabolic profiling of Haberlea rhodopensis, Thellungiella halophyla, and Arabidopsis thaliana exposed to low temperature. Front. Plant Sci. 2013, 4, 499. [Google Scholar] [CrossRef]
- Liu, D.; Ford, K.L.; Roessner, U.; Natera, S.; Cassin, A.M.; Patterson, J.H.; Bacic, A. Rice suspension cultured cells are evaluated as a model system to study salt responsive networks in plants using a combined proteomic and metabolomic profiling approach. Proteomics 2013, 13, 2046–2062. [Google Scholar] [CrossRef] [PubMed]
- Yobi, A.; Wone, B.W.; Xu, W.; Alexander, D.C.; Guo, L.; Ryals, J.A.; Oliver, M.J.; Cushman, J.C. Metabolomic Profiling in Selaginella lepidophylla at Various Hydration States Provides New Insights into the Mechanistic Basis of Desiccation Tolerance. Mol. Plant 2013, 6, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Witt, S.; Galicia, L.; Lisec, J.; Cairns, J.; Tiessen, A.; Araus, J.L.; Palacios-Rojas, N.; Fernie, A.R. Metabolic and Phenotypic Responses of Greenhouse-Grown Maize Hybrids to Experimentally Controlled Drought Stress. Mol. Plant 2012, 5, 401–417. [Google Scholar] [CrossRef]
- Lee, Y.J.; Perdian, D.C.; Song, Z.; Yeung, E.S.; Nikolau, B.J. Use of mass spectrometry for imaging metabolites in plants. Plant J. 2012, 70, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Caldana, C.; Degenkolbe, T.; Cuadros-Inostroza, A.; Klie, S.; Sulpice, R.; Leisse, A.; Steinhauser, D.; Fernie, A.R.; Willmitzer, L.; Hannah, M.A. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011, 67, 869–884. [Google Scholar] [CrossRef]
- Espinoza, C.; Degenkolbe, T.; Caldana, C.; Zuther, E.; Leisse, A.; Willmitzer, L.; Hincha, D.K.; Hannah, M.A. Interaction with Diurnal and Circadian Regulation Results in Dynamic Metabolic and Transcriptional Changes during Cold Acclimation in Arabidopsis. PLoS ONE 2010, 5, e14101. [Google Scholar] [CrossRef] [PubMed]
- Renault, H.; Roussel, V.; El Amrani, A.; Arzel, M.; Renault, D.; Bouchereau, A.; Deleu, C. The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 2010, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Korn, M.; Gärtner, T.; Erban, A.; Kopka, J.; Selbig, J.; Hincha, D.K. Predicting Arabidopsis Freezing Tolerance and Heterosis in Freezing Tolerance from Metabolite Composition. Mol. Plant 2010, 3, 224–235. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of Photosynthesis during Abiotic Stress-Induced Photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [PubMed]
- Larcher, W. Physiological Plant Ecology, 4th ed.; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Mohanapriya, G.; Bharadwaj, R.; Noceda, C.; Costa, J.H.; Kumar, S.R.; Sathishkumar, R.; Thiers, K.L.L.; Macedo, E.S.; Silva, S.; Annicchiarico, P.; et al. Alternative Oxidase (AOX) Senses Stress Levels to Coordinate Auxin-Induced Reprogramming From Seed Germination to Somatic Embryogenesis—A Role Relevant for Seed Vigor Prediction and Plant Robustness. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Panda, A.; Rangani, J. Metabolomics-Guided Elucidation of Abiotic Stress Tolerance Mechanisms in Plants. In Plant Metabolites and Regulation under Environmental Stress; Elsevier BV: Amsterdam, The Netherlands, 2018; pp. 89–131. [Google Scholar]
- Dawid, C.; Hille, K. Functional Metabolomics—A Useful Tool to Characterize Stress-Induced Metabolome Alterations Opening New Avenues towards Tailoring Food Crop Quality. Agronomy 2018, 8, 138. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Metabolomics in Plant Stress Physiology. Plant Genet. Mol. Biol. 2018, 164, 187–236. [Google Scholar] [CrossRef]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—From genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Urano, K.; Maruyama, K.; Ogata, Y.; Morishita, Y.; Takeda, M.; Sakurai, N.; Suzuki, H.; Saito, K.; Shibata, D.; Kobayashi, M.; et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009, 57, 1065–1078. [Google Scholar] [CrossRef]
- Skirycz, A.; De Bodt, S.; Obata, T.; De Clercq, I.; Claeys, H.; De Rycke, R.; Andriankaja, M.; Van Aken, O.; Van Breusegem, F.; Fernie, A.R.; et al. Developmental Stage Specificity and the Role of Mitochondrial Metabolism in the Response of Arabidopsis Leaves to Prolonged Mild Osmotic Stress. Plant Physiol. 2010, 152, 226–244. [Google Scholar] [CrossRef]
- Lugan, R.; Niogret, M.-F.; Leport, L.; Guégan, J.-P.; Larher, F.R.; Savoure, A.; Kopka, J.; Bouchereau, A. Metabolome and water homeostasis analysis of Thellungiella salsuginea suggests that dehydration tolerance is a key response to osmotic stress in this halophyte. Plant J. 2010, 64, 215–229. [Google Scholar] [CrossRef]
- Hochberg, U.; Degu, A.; Toubiana, D.; Gendler, T.; Nikoloski, Z.; Rachmilevitch, S.; Fait, A. Metabolite profiling and network analysis reveal coordinated changes in grapevine water stress response. BMC Plant Biol. 2013, 13, 184. [Google Scholar] [CrossRef]
- Wang, X.; Guo, R.; Li, M.; Liu, Y.; Zhao, M.; Fu, H.; Liu, X.; Wang, S.; Shi, L. Metabolomics reveals the drought-tolerance mechanism in wild soybean (Glycine soja). Acta Physiol. Plant. 2019, 41, 1–11. [Google Scholar] [CrossRef]
- Wu, X.; Cai, K.; Zhang, G.; Zeng, F. Metabolite Profiling of Barley Grains Subjected to Water Stress: To Explain the Genotypic Difference in Drought-Induced Impacts on Malting Quality. Front. Plant Sci. 2017, 8, 1547. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.-Q.; Shen, T.-H.; Zhong, L.; Zhu, C.-L.; Peng, X.-S.; He, X.-P.; Fu, J.-R.; Ouyang, L.-J.; Bian, J.-M.; Hu, L.-F.; et al. Comprehensive metabolomic, proteomic and physiological analyses of grain yield reduction in rice under abrupt drought–flood alternation stress. Physiol. Plant. 2018, 167, 564–584. [Google Scholar] [CrossRef] [PubMed]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat. Sci. Rep. 2018, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Wang, X.; Dossa, K.; Zhou, R.; Yu, J.; Zhang, Y.; Wang, L.; et al. Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef]
- Nunes, J.D.M.; Bertodo, L.; Da Rosa, L.; Von Poser, G.; Rech, S.B. Stress induction of valuable secondary metabolites in Hypericum polyanthemum acclimatized plants. South Afr. J. Bot. 2014, 94, 182–189. [Google Scholar] [CrossRef]
- Quan, N.T.; Anh, L.H.; Khang, D.T.; Tuyen, P.T.; Toan, N.P.; Minh, T.N.; Minh, L.T.; Bach, D.T.; Ha, P.T.T.; Elzaawely, A.A.; et al. Involvement of Secondary Metabolites in Response to Drought Stress of Rice (Oryza sativa L.). Agriculture 2016, 6, 23. [Google Scholar] [CrossRef]
- Bettaieb, I.; Hamrouni-Sellami, I.; Bourgou, S.; Limam, F.; Marzouk, B. Drought effects on polyphenol composition and antioxidant activities in aerial parts of Salvia officinalis L. Acta Physiol. Plant. 2011, 33, 1103–1111. [Google Scholar] [CrossRef]
- Piasecka, A.; Sawikowska, A.; Kuczyńska, A.; Ogrodowicz, P.; Mikołajczak, K.; Krystkowiak, K.; Gudyś, K.; Guzy-Wróbelska, J.; Krajewski, P.; Kachlicki, P. Drought-related secondary metabolites of barley ( Hordeum vulgare L.) leaves and their metabolomic quantitative trait loci. Plant J. 2017, 89, 898–913. [Google Scholar] [CrossRef]
- Radwan, A.; Kleinwächter, M.; Selmar, D. Impact of drought stress on specialised metabolism: Biosynthesis and the expression of monoterpene synthases in sage (Salvia officinalis). Phytochemistry 2017, 141, 20–26. [Google Scholar] [CrossRef]
- Lawas, L.M.F.; Erban, A.; Kopka, J.; Jagadish, S.V.K.; Zuther, E.; Hincha, D.K. Metabolic responses of rice source and sink organs during recovery from combined drought and heat stress in the field. GigaScience 2019, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Rushton, P.J.; Rohila, J.S. Metabolomic Profiling of Soybeans (Glycine max L.) Reveals the Importance of Sugar and Nitrogen Metabolism under Drought and Heat Stress. Plants 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Furumoto, T. Cold Signaling and Cold Response in Plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, R.; Bhandari, K.; Nayyar, H. Temperature stress and redox homeostasis in agricultural crops. Front. Environ. Sci. 2015, 3, 1–24. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather. Clim. Extremes 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Hemantaranjan, A. Heat Stress Responses and Thermotolerance. Adv. Plants Agric. Res. 2014, 1, 1–10. [Google Scholar] [CrossRef]
- Paupière, M.J.; Müller, F.; Li, H.; Rieu, I.; Tikunov, Y.M.; Visser, R.G.F.; Bovy, A.G. Untargeted metabolomic analysis of tomato pollen development and heat stress response. Plant Reprod. 2017, 30, 81–94. [Google Scholar] [CrossRef]
- Qi, X.; Xu, W.; Zhang, J.; Guo, R.; Zhao, M.; Hu, L.; Wang, H.; Dong, H.; Li, Y. Physiological characteristics and metabolomics of transgenic wheat containing the maize C4 phosphoenolpyruvate carboxylase (PEPC) gene under high temperature stress. Protoplasma 2017, 254, 1017–1030. [Google Scholar] [CrossRef]
- Sun, C.X.; Gao, X.X.; Li, M.Q.; Fu, J.Q.; Zhang, Y.L. Plastic responses in the metabolome and functional traits of maize plants to temperature variations. Plant Biol. 2016, 18, 249–261. [Google Scholar] [CrossRef]
- Le Gall, H.; Fontaine, J.-X.; Molinié, R.; Pelloux, J.; Mesnard, F.; Gillet, F.; Fliniaux, O. NMR-based Metabolomics to Study the Cold-acclimation Strategy of TwoMiscanthusGenotypes. Phytochem. Anal. 2017, 28, 58–67. [Google Scholar] [CrossRef]
- Ghassemi, S.; Delangiz, N.; Lajayer, B.A.; Saghafi, D.; Maggi, F. Review and future prospects on the mechanisms related to cold stress resistance and tolerance in medicinal plants. Acta Ecol. Sin. 2020. [Google Scholar] [CrossRef]
- Cook, D.; Fowler, S.; Fiehn, O.; Thomashow, M.F. From the Cover: A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 15243–15248. [Google Scholar] [CrossRef]
- Wienkoop, S.; Morgenthal, K.; Wolschin, F.; Scholz, M.; Selbig, J.; Weckwerth, W. Integration of Metabolomic and Proteomic Phenotypes. Mol. Cell. Proteom. 2008, 7, 1725–1736. [Google Scholar] [CrossRef]
- Kaplan, F.; Kopka, J.; Haskell, D.W.; Zhao, W.; Schiller, K.C.; Gatzke, N.; Guy, C.L. Exploring the Temperature-Stress Metabolome of Arabidopsis. Plant Physiol. 2004, 136, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ma, K.B.; Lu, Z.G.; Ren, S.X.; Jiang, H.R.; Cui, J.W.; Jin, B. Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 2020, 20, 1–86. [Google Scholar] [CrossRef] [PubMed]
- Thomason, K.; Babar, A.; Erickson, J.E.; Mulvaney, M.; Beecher, C.; Macdonald, G. Comparative physiological and metabolomics analysis of wheat (Triticum aestivum L.) following post-anthesis heat stress. PLoS ONE 2018, 13, e0197919. [Google Scholar] [CrossRef] [PubMed]
- D’souza, M.R.; Devaraj, V.R. Induction of thermotolerance through heat acclimation in lablab bean (Dolichos lablab). Afr. J. Biotechnol. 2013, 12, 5695–5704. [Google Scholar] [CrossRef]
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ratio in leaves during salinity stress. Plant, Cell Environ. 2010, 33, 552–565. [Google Scholar] [CrossRef]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant Hormonomics: Multiple Phytohormone Profiling by Targeted Metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef]
- Richter, J.A.; Erban, A.; Kopka, J.; Zörb, C. Metabolic contribution to salt stress in two maize hybrids with contrasting resistance. Plant Sci. 2015, 233, 107–115. [Google Scholar] [CrossRef]
- Kissoudis, C.; Kalloniati, C.; Flemetakis, E.; Madesis, P.; Labrou, N.E.; Tsaftaris, A.; Nianiou-Obeidat, I. Stress-inducible GmGSTU4 shapes transgenic tobacco plants metabolome towards increased salinity tolerance. Acta Physiol. Plant. 2015, 37, 1–11. [Google Scholar] [CrossRef]
- Abdel-Farid, I.B.; Marghany, M.R.; Rowezek, M.M.; Sheded, M.G. Effect of Salinity Stress on Growth and MetabolomicProfiling of Cucumis sativus and Solanum lycopersicum. Plants 2020, 9, 1626. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Li, P.; Ma, S.; Rupassara, S.I.; Bohnert, H.J. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005, 44, 826–839. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Lippold, F.; Redestig, H.; Hannah, M.A.; Erban, A.; Krämer, U.; Udvardi, M.K. Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J. 2007, 53, 973–987. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, D.H.; Lippold, F.; Redestig, H.; Hannah, M.A.; Erban, A.; Krämer, U.; Kopka, J.; Udvardi, M.K. Comparative Functional Genomics of Salt Stress in Related Model and Cultivated Plants Identifies and Overcomes Limitations to Translational Genomics. PLoS ONE 2011, 6, e17094. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.H.; Newbigin, E.; Tester, M.; Bacic, A.; Roessner, U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 2009, 60, 4089–4103. [Google Scholar] [CrossRef]
- Gupta, P.; De, B. Metabolomics analysis of rice responses to salinity stress revealed elevation of serotonin, and gentisic acid levels in leaves of tolerant varieties. Plant Signal. Behav. 2017, 12, e1335845. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Chen, H.; Chen, J.; Yang, R.; Zou, L.; Wang, C.; Chen, J.; Tan, M.; Mei, Y.; Wei, L.; et al. Metabolomics characterizes the metabolic changes of Lonicerae Japonicae Flos under different salt stresses. PLoS ONE 2020, 15, e0243111. [Google Scholar] [CrossRef]
- Zubair, A.; Nuraini, A.; Qosim, W.A. Effect of salinity stress on shoot musa acuminata L. Barangan cultivar in vitro culture. Pakistan J. Biol. Sci. 2019, 22, 201–205. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, X.; Xu, Q.; Mei, X.; Yuan, H.; Jiabu, D.; Nyima, T. Metabolite profiling in two contrasting Tibetan hulless barley cultivars revealed the core salt-responsive metabolome and key salt-tolerance biomarkers. AoB Plants 2019, 11, plz021. [Google Scholar] [CrossRef]
- Muchate, N.S.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. NaCl induced salt adaptive changes and enhanced accumulation of 20-hydroxyecdysone in the in vitro shoot cultures of Spinacia oleracea (L.). Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, B.; Liu, D.; Zou, C.; Wu, P.; Wang, Z.; Li, C. Transcriptomic and metabolomic analyses reveal mechanisms of adaptation to salinity in which carbon and nitrogen metabolism is altered in sugar beet roots. BMC Plant Biol. 2020, 20, 1–21. [Google Scholar] [CrossRef]
- Escandón, M.; Valledor, L.; Pascual, J.; Pinto, G.; Cañal, M.J.; Meijón, M. System-wide analysis of short-term response to high temperature in Pinus radiata. J. Exp. Bot. 2017, 68, 3629–3641. [Google Scholar] [CrossRef] [PubMed]
- Escandón, M.; Meijon, M.; Valledor, L.; Pascual, J.; Pinto, G.; Cañal, M.J. Metabolome Integrated Analysis of High-Temperature Response in Pinus radiata. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Rouphael, Y.; Raimondi, G.; Lucini, L.; Carillo, P.; Kyriacou, M.C.; Colla, G.; Cirillo, V.; Pannico, A.; El-Nakhel, C.; De Pascale, S. Physiological and Metabolic Responses Triggered by Omeprazole Improve Tomato Plant Tolerance to NaCl Stress. Front. Plant Sci. 2018, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef]
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative Modifications to Cellular Components in Plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2011, 35, 454–484. [Google Scholar] [CrossRef]
- Gullner, G.; Kömives, T.; Rennenberg, H. Enhanced tolerance of transgenic poplar plants overexpressing γ-glutamylcysteine synthetase towards chloroacetanilide herbicides. J. Exp. Bot. 2001, 52, 971–979. [Google Scholar] [CrossRef]
- Gomez, L.D. Regulation of calcium signalling and gene expression by glutathione. J. Exp. Bot. 2004, 55, 404. [Google Scholar] [CrossRef]
- Liedschulte, V.; Wachter, A.; Zhigang, A.; Rausch, T. Exploiting plants for glutathione (GSH) production: Uncoupling GSH synthesis from cellular controls results in unprecedented GSH accumulation. Plant Biotechnol. J. 2010, 8, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Cañal, M.J.; Escandón, M.; Meijón, M.; Weckwerth, W.; Valledor, L. Integrated Physiological, Proteomic, and Metabolomic Analysis of Ultra Violet (UV) Stress Responses and Adaptation Mechanisms in Pinus radiata. Mol. Cell. Proteom. 2017, 16, 485–501. [Google Scholar] [CrossRef]
- El Rasafi, T.; Oukarroum, A.; Haddioui, A.; Song, H.; Kwon, E.E.; Bolan, N.; Tack, F.M.G.; Sebastian, A.; Prasad, M.N.V.; Rinklebe, J. Cadmium stress in plants: A critical review of the effects, mechanisms, and tolerance strategies. Crit. Rev. Environ. Sci. Technol. 2020, 1–52. [Google Scholar] [CrossRef]
- Mwamba, T.; Islam, F.; Ali, B.; Lwalaba, J.; Gill, R.; Zhang, F.; Farooq, M.; Ali, S.; Ulhassan, Z.; Huang, Q.; et al. Comparative metabolomic responses of low- and high-cadmium accumulating genotypes reveal the cadmium adaptive mechanism in Brassica napus. Chemosphere 2020, 250, 126308. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, M.; Wu, S.; Xiao, B.; Wang, X.; Sun, B.; Zhu, L. Metabolomics Reveals Antioxidant Stress Responses of Wheat (Triticum aestivum L.) Exposed to Chlorinated Organophosphate Esters. J. Agric. Food Chem. 2020, 68, 6520–6529. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Burritt, D.J.; Tran, L.-S.P.; Adbelrahman, M. The use of metabolomic quantitative trait locus mapping and osmotic adjustment traits for the improvement of crop yields under environmental stresses. Semin. Cell Dev. Biol. 2018, 83, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Gupta, P.; Priscilla, K.; Kumar, S.; Hangargi, B.; Veershetty, A.; Ramrao, D.; Suresh, S.; Narasanna, R.; Naik, G.; et al. Metabolomics Intervention Towards Better Understanding of Plant Traits. Cells 2021, 10, 346. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).