Updated Characterization of Races of Plasmopara halstedii and Entomopathogenic Fungi as Endophytes of Sunflower Plants in Axenic Culture
Abstract
1. Introduction
2. Materials and Methods
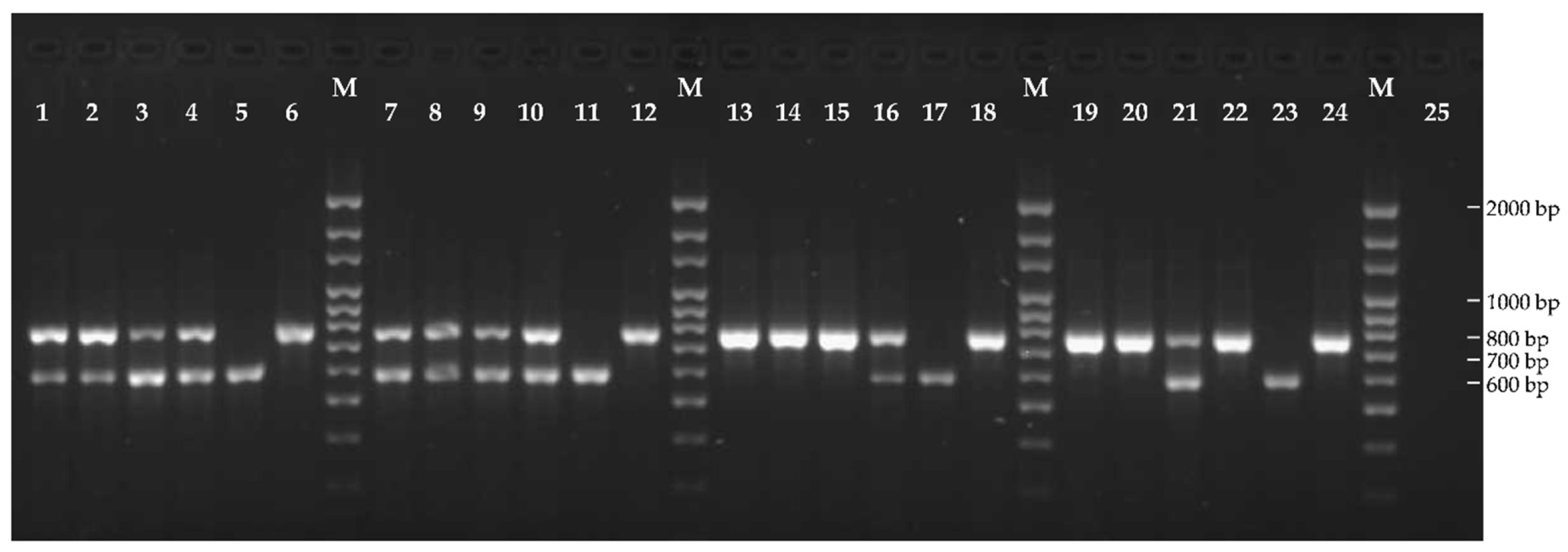
2.1. Racial Characterization of Plasmopara halstedii
2.2. Effect of Entomopathogenic Fungi on Sunflower Downy Mildew and on the Growth of Sunflower
2.2.1. Biological Materials: Plasmopara halstedii and Entomopathogenic Fungi
2.2.2. Effect of Entomopathogenic Fungi on Sunflower Downy Mildew
2.2.3. Effect of Entomopathogenic Fungi on Sunflower Growth
2.2.4. Microbiological and Molecular Detection
2.2.5. Data Analysis
3. Results
3.1. Racial Characterization of Plasmopara halstedii
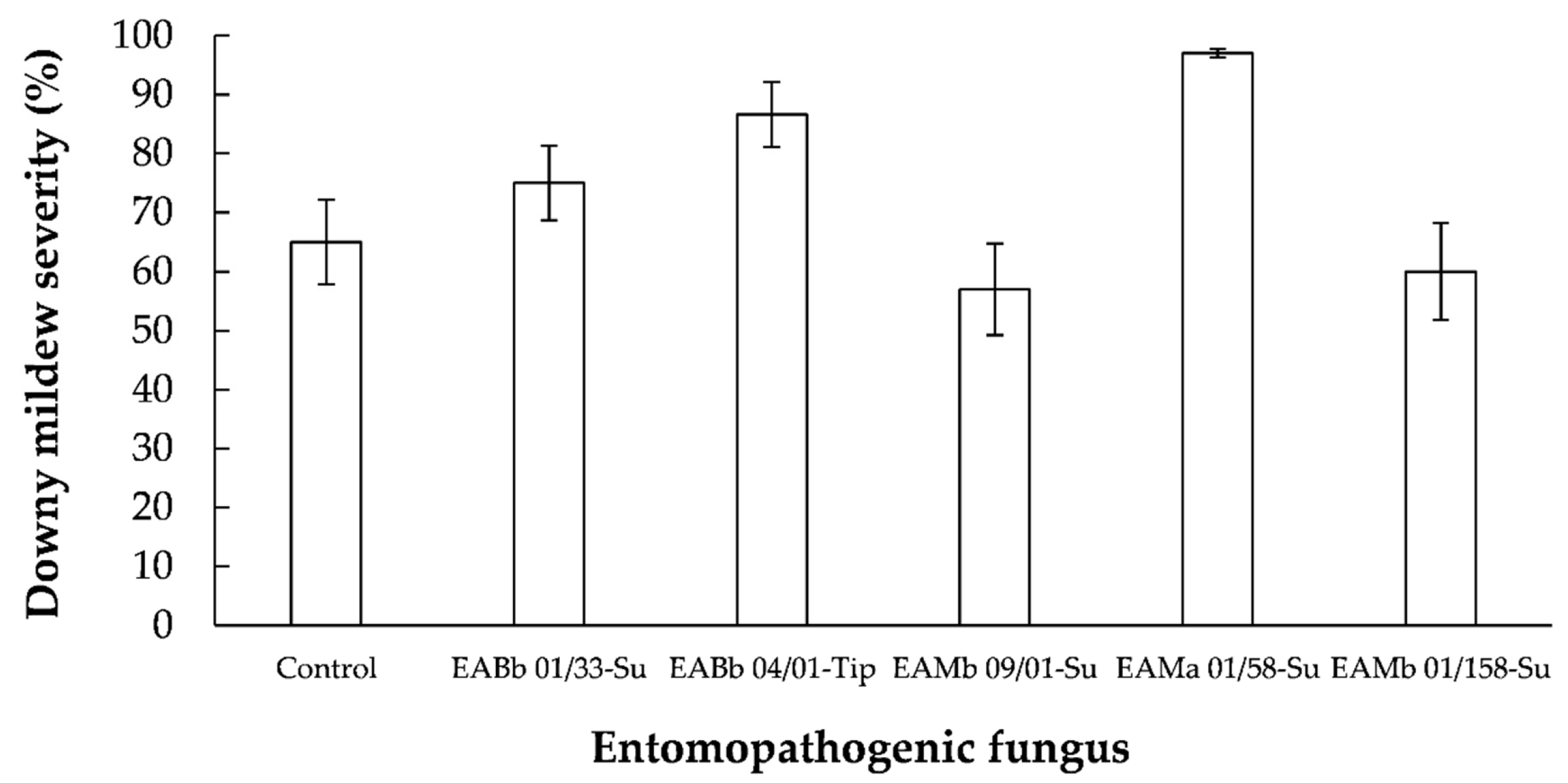
3.2. Effect of Entomopathogenic Fungi on Sunflower Downy Mildew
3.3. Effect of Entomopathogenic Fungi on Sunflower Growth
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EPPO. Plasmopara halstedii datasheet. Available online: https://gd.eppo.int/taxon/PLASHA (accessed on 4 June 2020).
- Molinero-Ruiz, L. Recent advances on the characterization and control of sunflower soilborne pathogens under climate change conditions. OCL 2019, 26, 2. [Google Scholar] [CrossRef]
- Gulya, T.; Harveson, R.; Mathew, F.; Block, C.; Thompson, S.; Kandel, H.; Berglund, D.; Sandbakken, J.; Kleingartner, L.; Markell, S. Comprehensive Disease Survey of U.S. Sunflower: Disease Trends, Research Priorities and Unanticipated Impacts. Plant Dis. 2019, 103, 601–618. [Google Scholar] [CrossRef]
- Gascuel, Q.; Martinez, Y.; Boniface, M.C.; Vear, F.; Pichon, M.; Godiard, L. The sunflower downy mildew pathogen Plasmopara halstedii. Mol. Plant Pathol. 2015, 16, 109–122. [Google Scholar] [CrossRef]
- Achbani, E.H.; Lamrhari, A.; Serrhini, M.N.; Douira, A.; de Labrouche, D.T. Evaluation of the efficacy of seed treatments against Plasmopara halstedii. Bull. OEPP 1999, 29, 443–449. [Google Scholar] [CrossRef]
- Molinero-Ruiz, M.L.; Cordon-Torres, M.M.; Martinez-Aguilar, J.; Melero-Vara, J.M.; Dominguez, J. Resistance to metalaxyl and to metalaxyl-M in populations of Plasmopara halstedii causing downy mildew in sunflower. Can. J. Plant Pathol. 2008, 30, 97–105. [Google Scholar] [CrossRef]
- Molinero-Ruiz, M.L.; Melero-Vara, J.M.; Dominguez, J. Inheritance of resistance to race 330 of Plasmopara halstedii in three sunflower lines. Plant Breed. 2002, 121, 61–65. [Google Scholar] [CrossRef]
- Molinero-Ruiz, M.L.; Melero-Vara, J.M.; Gulya, T.J.; Dominguez, J. First Report of Resistance to Metalaxyl in Downy Mildew of Sunflower Caused by Plasmopara halstedii in Spain. Plant Dis. 2003, 87, 749. [Google Scholar] [CrossRef]
- Molinero-Ruiz, M.L.; Dominguez, J.; Melero-Vara, J.M. Races of isolates of Plasmopara halstedii from Spain and studies on their virulence. Plant Dis. 2002, 86, 736–740. [Google Scholar] [CrossRef]
- Garcia-Carneros, A.B.; Molinero-Ruiz, L. First report of the highly virulent race 705 of Plasmopara halstedii (downy mildew of sunflower) in Portugal and in Spain. Plant Dis. 2017, 101, 1555. [Google Scholar] [CrossRef]
- Gilley, M.A.; Gulya, T.J.; Seiler, G.; Underwood, W.; Hulke, B.S.; Misar, C.G.; Markell, S.G. Determination of virulence phenotypes of Plasmopara halstedii in the United States. Plant Dis 2020, 104, 2823–2831. [Google Scholar] [CrossRef]
- Menzler-Hokkanen, I. Socioeconomic significance of biological control. In An Ecological and Societal Approach to Biological Control; Eilenberg, G., Menzler-Hokkanen, I., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 13–25. [Google Scholar]
- Quesada-Moraga, E.; Yousef-Naef, M.; Garrido-Jurado, I. Advances in the use of entomopathogenic fungi as biopesticides in suppressing crop pests. In Biopesticides for Sustainable Agriculture; Birch, N., Glare, T., Eds.; Burleigh Dodds Science Publishing Limited: Cambridge, UK, 2020; pp. 1–36. [Google Scholar]
- Jaber, L.R. Grapevine leaf tissue colonization by the fungal entomopathogen Beauveria bassiana s.l. and its effect against downy mildew. BioControl 2015, 60, 103–112. [Google Scholar] [CrossRef]
- Keyser, C.A.; Jensen, B.; Meyling, N.V. Dual effects of Metarhizium spp. and Clonostachys rosea against an insect and a seed-borne pathogen in wheat. Pest Manag. Sci. 2016, 72, 517–526. [Google Scholar] [CrossRef]
- Lozano-Tovar, M.D.; Garrido-Jurado, I.; Quesada-Moraga, E.; Raya-Ortega, M.C.; Trapero-Casas, A. Metarhizium brunneum and Beauveria bassiana release secondary metabolites with antagonistic activity against Verticillium dahliae and Phytophthora megasperma olive pathogens. Crop Prot. 2017, 100, 186–195. [Google Scholar] [CrossRef]
- Jaber, L.R.; Ownley, B.H. Can we use entomopathogenic fungi as endophytes for dual biological control of insect pests and plant pathogens? Biol. Control 2018, 116, 36–45. [Google Scholar] [CrossRef]
- Vega, F.E. The use of fungal entomopathogens as endophytes in biological control: A review. Mycologia 2018, 110, 4–30. [Google Scholar] [CrossRef]
- Miranda-Fuentes, P.; Garcia-Carneros, A.B.; Montilla-Carmona, A.M.; Molinero-Ruiz, L. Evidence of soil-located competition as the cause of the reduction of sunflower verticillium wilt by entomopathogenic fungi. Plant Pathol. 2020, 69, 1492–1503. [Google Scholar] [CrossRef]
- Barker, J.F. Laboratory evaluation of the pathogenicity of Beauveria bassiana and Metarhizium anisopliae to larvae of the banded sunflower moth, Cochylis hospes (Lepidoptera: Cochylidae). Great Lakes Entomol. 1999, 32, 101–106. [Google Scholar]
- Takov, D.I.; Draganova, S.A.; Toshova, T.B. Gregarine and Beauveria bassiana infections of the grey corn weevil, Tanymecus dilaticollis (Coleoptera: Curculionidae). Acta Phytopathol. Entomol. Hung. 2013, 48, 309–319. [Google Scholar] [CrossRef]
- Ortiz-Bustos, C.M.; Molinero-Ruiz, L.; Quesada-Moraga, E.; Garrido-Jurado, I. Natural occurrence of wireworms (Coleoptera: Elateridae) and entomopathogenic fungi in sunflower fields of Spain, and evaluation of their pathogenicity toward wireworms. In Abstract Book 49th Annual Meeting of the Society for Invertebrate Pathology, Proceedings of the 49th Annual Meeting of the Society for Invertebrate Pathology, Tours, France, 24–28 July 2016; Society for Invertebrate Pathology: Verona, WI, USA, 2016; p. 109. [Google Scholar]
- Raya-Diaz, S.; Quesada-Moraga, E.; Barron, V.; del Campillo, M.C.; Sanchez-Rodriguez, A.R. Redefining the dose of the entomopathogenic fungus Metarhizium brunneum (Ascomycota, Hypocreales) to increase Fe bioavailability and promote plant growth in calcareous and sandy soils. Plant Soil 2017, 418, 387–404. [Google Scholar] [CrossRef]
- Nagaraju, A.; Sudisha, J.; Murthy, S.M.; Ito, S. Seed priming with Trichoderma harzianum isolates enhances plant growth and induces resistance against Plasmopara halstedii, an incitant of sunflower downy mildew disease. Australas. Plant Pathol. 2012, 41, 609–620. [Google Scholar] [CrossRef]
- Rondot, Y.; Reineke, A. Endophytic Beauveria bassiana activates expression of defence genes in grapevine and prevents infections by grapevine downy mildew Plasmopara viticola. Plant Pathol. 2019, 68, 1719–1731. [Google Scholar] [CrossRef]
- Martin-Sanz, A.; Rueda, S.; Garcia-Carneros, A.B.; Molinero-Ruiz, L. First Report of a New Highly Virulent Pathotype of Sunflower Downy Mildew (Plasmopara halstedii) Overcoming the Pl8 Resistance Gene in Europe. Plant Dis. 2020, 104, 597–598. [Google Scholar] [CrossRef]
- Gulya, T.J.; de Labrouhe, D.T.; Masirevic, S.; Penaud, A.; Rashid, K.; Viranyi, F. Proposal for standardized nomenclature and identification of races of Plasmopara halstedii (sunflower downy mildew). In Proceedings of the ISA Symposium III Sunflower Downy Mildew, Fargo, ND, USA, 13–14 January 1998; Gulya, T., Vear, F., Eds.; ISA: Fargo, ND, USA, 1998; pp. 130–136. [Google Scholar]
- Miller, J.F.; Gulya, T.J. Registration of Six Downy Mildew Resistant Sunflower Germplasm Lines. Crop Sci. 1988, 28, 1040–1041. [Google Scholar] [CrossRef]
- Riethmüller, A.; Voglmayr, H.; Göker, M.; Weiß, M.; Oberwinkler, F. Phylogenetic Relationships of the Downy Mildews (Peronosporales) and Related Groups Based on Nuclear Large Subunit Ribosomal DNA Sequences. Mycologia 2002, 94, 834–849. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- McIntosh, M.S. Analysis of combined experiments. Agron. J. 1983, 75, 153–155. [Google Scholar] [CrossRef]
- Sedlarova, M.; Pospichalova, R.; Drabkova Trojanova, Z.; Bartusek, T.; Slobodianova, L.; Lebeda, A. First Report of Plasmopara halstedii New Races 705 and 715 on sunflower from the Czech Republic—Short Communication. Plant Prot. Sci. 2016, 52, 182–187. [Google Scholar]
- Viranyi, F.; Gulya, T.J.; Tourvieille, D.L. Recent Changes in the Pathogenic Variability of Plasmopara Halstedii (Sunflower Downy Mildew) Populations from Different Continents. Helia 2015, 38, 149–162. [Google Scholar] [CrossRef]
- Spring, O. Spreading and global pathogenic diversity of sunflower downy mildew—Review. Plant Prot. Sci. 2019, 55, 149–158. [Google Scholar] [CrossRef]
- Trojanova, Z.; Sedlarova, M.; Pospichalova, R.; Lebeda, A. Pathogenic variability of Plasmopara halstedii infecting sunflower in the Czech Republic. Plant Pathol. 2018, 67, 136–144. [Google Scholar] [CrossRef]
- Nisha, N.; Körösi, K.; Perczel, M.; Yousif, A.I.A.; Bán, R. First report on the occurrence of an aggressive pathotype, 734, of Plasmopara halstedii causing sunflower downy mildew in Hungary. Plant Dis. 2020. Available online: https://doi.org/10.1094/PDIS-05-20-1054-PDN (accessed on 22 December 2020). [CrossRef]
- Gonzalez-Guzman, A.; Sacristan, D.; Quesada-Moraga, E.; Torrent, J.; del Campillo, M.C.; Sanchez-Rodriguez, A.R. Effects of entomopathogenic fungi on growth and nutrition in wheat grown on two calcareous soils: Influence of the fungus application method. Ann. Appl. Biol. 2020, 177, 26–40. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, A.; Sacristan, D.; Sanchez-Rodriguez, A.R.; Barron, V.; Torrent, J.; del Campillo, M.C. Soil Nutrients Effects on the Performance of Durum Wheat Inoculated with Entomopathogenic Fungi. Agronomy 2020, 10, 589. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Heil, M. The microbe-free plant: Fact or artifact. Front. Plant Sci. 2011, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Behie, S.W.; Moreira, C.C.; Sementchoukova, I.; Barelli, L.; Zelisko, P.M.; Bidochka, M.J. Carbon translocation from a plant to an insect-pathogenic endophytic fungus. Nat. Commun. 2017, 8, 5. [Google Scholar] [CrossRef]
- Raya-Diaz, S.; Sanchez-Rodriguez, A.R.; Segura-Fernandez, J.M.; del Campillo, M.C.; Quesada-Moraga, E. Entomopathogenic fungi-based mechanisms for improved Fe nutrition in sorghum plants grown on calcareous substrates. PLoS ONE 2017, 12, e0185903. [Google Scholar] [CrossRef]
- Garrido-Jurado, I.; Resquin-Romero, G.; Amarilla, S.P.; Rios-Moreno, A.; Carrasco, L.; Quesada-Moraga, E. Transient endophytic colonization of melon plants by entomopathogenic fungi after foliar application for the control of Bemisia tabaci Gennadius (Hemiptera: Aleyrodidae). J. Pest Sci. 2017, 90, 319–330. [Google Scholar] [CrossRef]
- Miranda-Fuentes, P.; Yousef, M.; Valverde-Garcia, P.; Rodriguez-Gomez, I.M.; Garrido-Jurado, I.; Quesada-Moraga, E. Entomopathogenic fungal endophyte-mediated tritrophic interactions between Spodoptera littoralis and its parasitoid Hyposoter didymator. J. Pest Sci. 2020. [Google Scholar] [CrossRef]
- Resquin-Romero, G.; Garrido-Jurado, I.; Delso, C.; Rios-Moreno, A.; Quesada-Moraga, E. Transient endophytic colonizations of plants improve the outcome of foliar applications of mycoinsecticides against chewing insects. J. Invertebr. Pathol. 2016, 136, 23–31. [Google Scholar] [CrossRef]
| Sample Number | Sample ID | N. of Plants | Collection Year | Sunflower Genotype | Geographic Origin. Location (Province). Country a |
|---|---|---|---|---|---|
| 1 | DM01-11 | 2 | 2011 | Transol/2-4 | Alameda del Obispo (Cordoba). SP |
| 2 | DM02-11 | 1 | 2011 | Transol/18-2 | Alameda del Obispo (Cordoba). SP |
| 3 | DM03-11 | 2 | 2011 | 102/SIg | Marchena (Seville). SP |
| 4 | DM04-11 | 3 | 2011 | Bosfora | Fuentes de Andalucía (Seville). SP |
| 5 | DM05-11 | 3 | 2011 | Transol | Fuentes de Andalucía (Seville). SP |
| 6 | DM01-13 | 1 | 2013 | C1/DBa | Las Cabezas de San Juan (Seville). SP |
| 7 | DM02-13 | 2 | 2013 | B2/DBa | Las Cabezas de San Juan (Seville). SP |
| 8 | 1978-R1 | 2 | 2014 | - b | Tomejil (Seville). SP |
| 9 | DB0 | - | 2014 | - | Las Cabezas de San Juan (Seville). SP |
| 10 | LGB | 1 | 2014 | LG560440 | Bornos (Cadiz). SP |
| 11 | 1SM | 3 | 2014 | Experimental line | Sao Matias (Beja). PO |
| 12 | 2SEP | 2 | 2014 | Experimental line | Serpa (Beja), PO |
| 13 | DM01-15 | 2 | 2015 | - | Cerro Perea (Seville). SP |
| 14 | DM02-15 | 1 | 2015 | - | Fuentes de Andalucía (Seville). SP |
| 15 | DM03-15 | 1 | 2015 | - | Fuentes de Andalucía (Seville). SP |
| 16 | DM04-15 | 4 | 2015 | - | Jerez de la Frontera (Cadiz). SP |
| 17 | DM01-16 | 2 | 2016 | - | Fuentes de Andalucía (Seville). SP |
| 18 | DM02-16 | - | 2016 | - | Alcalá del Río (Seville). SP |
| 19 | DM01-17 | 2 | 2017 | LG5485 | Montellano (Seville). SP |
| 20 | B-B190418 | 4 | 2018 | Experimental line | Utrera (Seville). SP |
| 21 | B-S040518-1 | 4 | 2018 | Experimental line | Alcalá del Río (Seville). SP |
| 22 | B-S040518-2 | 4 | 2018 | Experimental line | Viso del Alcor (Seville). SP |
| 23 | B-S100518 | 4 | 2018 | Experimental line | El Arahal (Seville). SP |
| 24 | B-B180518 | 4 | 2018 | Experimental line | Cañada Rosal (Seville). SP |
| 25 | B-B210518-1 | - | 2018 | Solnet | Espera (Cadiz). SP |
| 26 | B-B210518-2 | - | 2018 | Solnet | Alcalá Guadaira (Seville). SP |
| 27 | B-B210518-3 | - | 2018 | Bonasol | Utrera (Seville). SP |
| 28 | B-B220518 | 1 | 2018 | Solnet | Huévar (Seville). SP |
| 29 | B-S230518 | 2 | 2018 | Syedison | Palma del Río (Cordoba). SP |
| 30 | B-B290518-1 | 1 | 2018 | Solnet | Trigueros (Seville). SP |
| 31 | B-B290518-2 | 1 | 2018 | Solnet | El Trobal (Seville). SP |
| 32 | B-B300518 | - | 2018 | Solnet | Paradas (Seville). SP |
| 33 | B-LG310518-1 | 3 | 2018 | Experimental line | Las Cabezas de San Juan (Seville). SP |
| 34 | B-LG310518-2 | 3 | 2018 | Experimental line | Lebrija (Seville). SP |
| 35 | B-LG310518-3 | 2 | 2018 | Experimental line | Maribáñez (Seville). SP |
| 36 | B-F050618 | 1 | 2018 | - | Escacena del Campo (Huelva). SP |
| 37 | B-E050618-1 | 2 | 2018 | Artic | Paterna del Campo (Huelva). SP |
| 38 | B-E050618-2 | 1 | 2018 | Artic-T | Paterna del Campo (Huelva). SP |
| 39 | B-P080618 | 2 | 2018 | - | Tulcea, RO |
| 40 | B-B120618 | - | 2018 | Bonasol | La Campana (Seville). SP |
| 41 | B-P130618-1 | 1 | 2018 | 64LE25 | Cortona, IT |
| 42 | B-P130618-2 | 1 | 2018 | 64LE25-T | Cortona, IT |
| 43 | B-P130618-3 | 3 | 2018 | XF16942 | Cortona, IT |
| 44 | B-P130618-4 | 2 | 2018 | P64LE99 | Mauroux, FR |
| 45 | B-P130618-5 | 3 | 2018 | P64LE25 | Taybosc, FR |
| 46 | B-P130618-6 | 1 | 2018 | P64LE25 | Lectoure, FR |
| 47 | B-LG140618 | 2 | 2018 | Experimental line | Écija (Seville). SP |
| 48 | B-S190618 | 2 | 2018 | Experimental line | Olivares (Seville). SP |
| 49 | B-S280618 | 1 | 2018 | - | Escacena del Campo (Huelva). SP |
| 50 | B-S230419 (SYT-1) | - | 2019 | M4 | Villamanrique de la Condesa (Seville). SP |
| 51 | B-S080519-1 (2019HU01) | 3 | 2019 | - | Palma del Condado (Seville). SP |
| 52 | B-S080519-2 (2019-SE01) | 3 | 2019 | - | Gerena (Seville). SP |
| 53 | B-S230519-1 (GIB-1) | - | 2019 | M4 | Villarrasa (Huelva). SP |
| 54 | B-S230519-2 (GIB-2) | - | 2019 | M4-T | Villarrasa (Huelva). SP |
| 55 | B-S190520-1 | 3 | 2020 | M4 | Fernán Núñez (Cordoba). SP |
| 56 | B-S190520-2 | 2 | 2020 | M9 | Pedro Abad (Cordoba). SP |
| 57 | B-S270520 | 1 | 2020 | - | Andújar (Jaen). SP |
| 58 | Ph04-20 | 2 | 2020 | M4 | Calzada de Bureba (Burgos). SP |
| Sample ID | Sample Number | Isolate Reference Number 1 | Race 2 | Reaction of RHA-340 Line 3 | Sample ID | Sample Number | Isolate Reference Number | Race | Reaction of RHA-340 Line |
|---|---|---|---|---|---|---|---|---|---|
| DM01-11 | 1 | Ph01-11 | 100 | R | B-B290518-1 | 30 | Ph11-18 | 110 | R |
| DM02-11 | 2 | Ph02-11 | 310 | R | B-B290518-2 | 31 | Ph12-18 | 310 | R |
| DM03-11 | 3 | Ph03-11 | 310 | R | B-B300518 | 32 | Ph13-18 | 114 | R |
| DM04-11 | 4 | Ph04-11 | 710 | R | B-LG310518-1 | 33 | Ph14-18 | 315 | R |
| DM05-11 | 5 | Ph05-11 | 710 | R | B-LG310518-2 | 34 | Ph15-18 | 311 | R |
| DM01-13 | 6 | Ph01-13 | 304 | R | B-LG310518-3 | 35 | Ph16-18 | 711 | R |
| DM02-13 | 7 | Ph02-13 | 304 | R | B-F050618 | 36 | Ph17-18 | 304 | R |
| 1978-R1 | 8 | 1978-R1 | 710 | R | B-E050618-1 | 37 | Ph18-18 | 312 | R |
| DB0 | 9 | DB0 | 700 | R | B-E050618-2 | 38 | Ph19-18 | 312 | R |
| LGB | 10 | LGB | 713 | R | B-P080618 | 39 | Ph20-18 * | 705 | R |
| 1SM | 11 | 1SM * 4 | 304 | R | B-B120618 | 40 | Ph21-18 | 711 | R |
| 2SEP | 12 | 2SEP * | 700 | R | B-P130618-1 | 41 | Ph22-18 * | 301 | R |
| DM01-15 | 13 | Ph01-15 | 310 | R | B-P130618-2 | 42 | Ph23-18 * | 715 | S |
| DM02-15 | 14 | Ph02-15 | 310 | R | B-P130618-3 | 43 | Ph24-18 * | 715 | S |
| DM03-15 | 15 | Ph03-15 | 310 | R | B-P130618-4 | 44 | Ph25-18 * | 715 | S |
| DM04-15 | 16 | Ph04-15 | 314 | R | B-P130618-5 | 45 | Ph26-18 * | 715 | S |
| DM01-16 | 17 | Ph01-16 | 705 | R | B-P130618-6 | 46 | Ph27-18 * | 715 | S |
| DM02-16 | 18 | Ph02-16 | 704 | R | B-LG140618 | 47 | Ph28-18 | 114 | R |
| DM01-17 | 19 | Ph01-17 | 317 | R | B-S190618 | 48 | Ph29-18 | 714 | R |
| B-B190418 | 20 | Ph01-18 | 305 | R | B-S280618 | 49 | Ph33-18 | 304 | R |
| B-S040518-1 | 21 | Ph02-18 | 715 | R | B-S230419 (SYT-1) | 50 | Ph01-19 | 705 | R |
| B-S040518-2 | 22 | Ph03-18 | 714 | R | B-S080519-1 (2019HU01) | 51 | Ph02-19 | 715 | R |
| B-S100518 | 23 | Ph04-18 | 315 | R | B-S080519-2 (2019-SE01) | 52 | Ph03-19 | 715 | R |
| B-B180518 | 24 | Ph05-18 | 311 | R | B-S230519-1 (GIB-1) | 53 | Ph06-19 | 505 | R |
| B-B210518-1 | 25 | Ph06-18 | 311 | R | B-S230519-2 (GIB-2) | 54 | Ph07-19 | 705 | R |
| B-B210518-2 | 26 | Ph07-18 | 712 | R | B-S190520-1 | 55 | Ph01-20 | 304 | R |
| B-B210518-3 | 27 | Ph08-18 | 313 | R | B-S190520-2 | 56 | Ph02-20 | 714 | R |
| B-B220518 | 28 | Ph09-18 | 313 | R | B-S270520 | 57 | Ph03-20 | 715 | R |
| B-S230518 | 29 | Ph10-18 | 315 | R | Ph04-20 | 58 | Ph04-20 | 705 | R |
| Entomopathogenic Fungus (EF) | Shoot Height 1 (cm) | Root Length (cm) | Shoot Dry Weight 2 (g) | Root Dry Weight (g) |
|---|---|---|---|---|
| Control | 9.06 ± 1.55 3 | 9.11 ± 0.86 | 0.62 ± 0.15 | 0.16 ± 0.05 |
| EABb 01/33-Su | 7.33 ± 1.26 | 8.78 ± 0.67 | 0.55 ± 0.12 | 0.15 ± 0.03 |
| EABb 04/01-Tip | 7.33 ± 1.44 | 9.20 ± 1.03 | 0.58 ± 0.16 | 0.18 ± 0.04 |
| EABb 09/01-Su | 9.83 ± 1.70 | 9.30 ± 0.67 | 0.84 ± 0.19 | 0.21 ± 0.04 |
| EAMa 01/58-Su | 10.23 ± 1.99 | 7.73 ± 0.86 | 0.58 ± 0.15 | 0.15 ± 0.03 |
| EAMb 01/158-Su | 8.69 ± 1.84 | 6.91 ± 0.86 | 0.58 ± 0.16 | 0.13 ± 0.04 |
| Entomopathogenic Fungus (EF) | Shoot Height 1 (cm) | Root Length (cm) | Shoot Dry Weight 2 (g) | Root Weight (g) |
|---|---|---|---|---|
| Control | 14.72 ± 0.83 3 a | 12.33 ± 0.93 a | 1.15 ± 0.06 a | 0.39 ± 0.03 a |
| EABb 01/33-Su | 9.83 ± 1.54 b | 9.17 ± 1.16 c | 0.67 ± 0.09 b | 0.24 ± 0.03 c |
| EABb 04/01-Tip | 13.03 ± 0.94 a | 11.10 ± 0.49 abc | 0.98 ± 0.09 a | 0.28 ± 0.02 bc |
| EABb 09/01-Su | 15.44 ± 0.73 a | 9.99 ± 0.62 bc | 1.20 ± 0.13 a | 0.32 ± 0.05 abc |
| EAMa 01/58-Su | 15.07 ± 0.70 a | 11.64 ± 0.29 ab | 1.09 ± 0.07 a | 0.32 ± 0.04 abc |
| EAMb 01/158-Su | 14.80 ± 0.74 a | 11.22 ± 0.49 abc | 1.13 ± 0.05 a | 0.33 ± 0.02 ab |
| Entomopathogenic Fungus | Roots | Stem | Leaves |
|---|---|---|---|
| Control | 0.0 1 | 0.0 | 0.0 |
| EABb 01/33-Su | 87.5 | 0.0 | 0.0 |
| EABb 04/01-Tip | 30.0 | 20.0 | 10.0 |
| EABb 09/01-Su | 20.0 | 22.22 | 0.0 |
| EAMa 01/58-Su | 20.0 | 0.0 | 0.0 |
| EAMb 01/158-Su | 20.0 | 30.0 | 20.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miranda-Fuentes, P.; García-Carneros, A.B.; Molinero-Ruiz, L. Updated Characterization of Races of Plasmopara halstedii and Entomopathogenic Fungi as Endophytes of Sunflower Plants in Axenic Culture. Agronomy 2021, 11, 268. https://doi.org/10.3390/agronomy11020268
Miranda-Fuentes P, García-Carneros AB, Molinero-Ruiz L. Updated Characterization of Races of Plasmopara halstedii and Entomopathogenic Fungi as Endophytes of Sunflower Plants in Axenic Culture. Agronomy. 2021; 11(2):268. https://doi.org/10.3390/agronomy11020268
Chicago/Turabian StyleMiranda-Fuentes, Pedro, Ana B. García-Carneros, and Leire Molinero-Ruiz. 2021. "Updated Characterization of Races of Plasmopara halstedii and Entomopathogenic Fungi as Endophytes of Sunflower Plants in Axenic Culture" Agronomy 11, no. 2: 268. https://doi.org/10.3390/agronomy11020268
APA StyleMiranda-Fuentes, P., García-Carneros, A. B., & Molinero-Ruiz, L. (2021). Updated Characterization of Races of Plasmopara halstedii and Entomopathogenic Fungi as Endophytes of Sunflower Plants in Axenic Culture. Agronomy, 11(2), 268. https://doi.org/10.3390/agronomy11020268







