Fusarium Wilt Management in Legume Crops
Abstract
1. Introduction
2. Fusarium oxysporum Diversity
3. Fusarium Wilt Integrated Disease Management
3.1. Chemical Control
3.2. Biological Control
3.3. Cultural Control
4. Breeding for Fusarium Wilt Resistance in Legumes
4.1. Resistance Screening Methods
4.1.1. Mass Screening Methods
4.1.2. Detailed Screening Methods
4.2. Resistance Mechanisms Against Fo
4.3. Genetic Basis of Resistance Against Fo
4.3.1. Monogenic Resistance
4.3.2. Oligogenic/Polygenic Resistance
4.4. Gene Regulation Upon Legume Fo Infection
4.4.1. Fusarium oxysporum Effector Genes
4.5. Conventional Breeding
4.6. Precision Breeding Aprroaches
5. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Moss, M.O.; Smith, J.E. The Applied Mycology of Fusarium; Cambridge University Press: Cambridge, UK, 1984. [Google Scholar]
- Ma, L.-J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef]
- Di Pietro, A.; Madrid, M.P.; Caracuel, Z.; Delgado-Jarana, J.; Roncero, M.I.G. Fusarium oxysporum: Exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 2003, 4, 315–325. [Google Scholar] [CrossRef]
- Agrios, G.N. Plant Pathology; Elsevier Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- European Commission, Commission Implementing Regulation (EU) 2019/2072 of 28 November 2019 establishing uniform conditions for the implementation of Regulation (EU) 2016/2031 of the European Parliament and the Council, as regards protective measures against pests of plants, a. Off. J. Eur. Union 2019, 62, 1–319.
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef]
- Roncero, M.I.G.; Hera, C.; Ruiz-Rubio, M.; Maceira, F.I.G.; Madrid, M.P.; Caracuel, Z.; Calero, F.; Delgado-Jarana, J.; Roldán-Rodríguez, R.; Martínez-Rocha, A.L.; et al. Fusarium as a model for studying virulence in soilborne plant pathogens. Physiol. Mol. Plant Pathol. 2003, 62, 87–98. [Google Scholar] [CrossRef]
- Nelson, E.B. Exudate molecules initiating fungal responses to seeds and roots. In The Rhizosphere and Plant Growth; Springer: Dordrecht, The Netherlands, 1991; pp. 197–209. [Google Scholar]
- Turrà, D.; El Ghalid, M.; Rossi, F.; Di Pietro, A. Fungal pathogen uses sex pheromone receptor for chemotropic sensing of host plant signals. Nature 2015, 527, 521–524. [Google Scholar] [CrossRef]
- Perez-Nadales, E.; Di Pietro, A. The membrane mucin Msb2 regulates invasive growth and plant infection in Fusarium oxysporum. Plant Cell 2011, 23, 1171–1185. [Google Scholar] [CrossRef]
- Perez-Nadales, E.; Almeida Nogueira, M.F.; Baldin, C.; Castanheira, S.; El Ghalid, M.; Grund, E.; Lengeler, K.; Marchegiani, E.; Mehrotra, P.V.; Moretti, M.; et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 2014, 70, 42–67. [Google Scholar] [CrossRef]
- Bishop, C.D.; Cooper, R.M. An ultrastructural study of vascular colonization in three vascular wilt diseases I. Colonization of susceptible cultivars. Physiol. Plant Pathol. 1983, 23, 323–343. [Google Scholar] [CrossRef]
- Beckman, C.H. The nature of wilt diseases of plants; APS Press: St. Paul, MN, USA, 1987. [Google Scholar]
- Jiménez-Díaz, R.M.; Castillo, P.; Jiménez-Gasco, M.d.M.; Landa, B.B.; Navas-Cortés, J.A. Fusarium wilt of chickpeas: Biology, ecology and management. Crop Prot. 2015, 73, 16–27. [Google Scholar] [CrossRef]
- Vance, C.P.; Graham, P.H.; Allan, D.L. Biological Nitrogen Fixation: Phosphorus—A Critical Future Need? In Nitrogen Fixation: From Molecules to Crop Productivity; Pedrosa, F.O., Hungria, M., Yates, G., Newton, W.E., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 509–514. [Google Scholar]
- Azooz, M.M.; Ahmad, P. Legumes under Environmental Stress: Yield, Improvement and Adaptations; Wiley-Blackwell: Hoboken, NJ, USA, 2015. [Google Scholar]
- Rubiales, D.; Fondevilla, S.; Chen, W.; Gentzbittel, L.; Higgins, T.J.V.; Castillejo, M.A.; Singh, K.B.; Rispail, N. Achievements and challenges in legume breeding for pest and disease resistance. CRC Crit. Rev. Plant Sci. 2015, 34, 195–236. [Google Scholar] [CrossRef]
- Williams, A.H.; Sharma, M.; Thatcher, L.F.; Azam, S.; Hane, J.K.; Sperschneider, J.; Kidd, B.N.; Anderson, J.P.; Ghosh, R.; Garg, G.; et al. Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genom. 2016, 17, 191. [Google Scholar] [CrossRef]
- Haglund, W.A.; Kraft, J.M. Fusarium wilt. In Compendium of Pea Diseases and Pests; Kraft, J.M., Pfleger, F.L., Eds.; APS Press: St. Paul, MN, USA, 2001; pp. 14–16. [Google Scholar]
- FAOSTAT. Food and Agriculture Organization of the United Nations, FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 21 May 2020).
- Alves-Santos, F.M.; Cordeiro-Rodrigues, L.; Sayagués, J.M.; Martín-Domínguez, R.; Garcia-Benavides, P.; Crespo, M.C.; Díaz-Mínguez, J.M.; Eslava, A.P. Pathogenicity and race characterization of Fusarium oxysporum f. sp. phaseoli isolates from Spain and Greece. Plant Pathol. 2002, 51, 605–611. [Google Scholar] [CrossRef]
- Navas-Cortés, J.A.; Hau, B.; Jiménez-Díaz, R.M. Yield loss in chickpeas in relation to development of Fusarium wilt epidemics. Phytopathology 2000, 90, 1269–1278. [Google Scholar] [CrossRef]
- Taylor, P.; Lindbeck, K.; Chen, W.; Ford, R. Lentil Diseases. In Lentil; Springer: Dordrecht, The Netherlands, 2007; pp. 291–313. [Google Scholar]
- Ramírez-Suero, M.; Khanshour, A.; Martinez, Y.; Rickauer, M. A study on the susceptibility of the model legume plant Medicago truncatula to the soil-borne pathogen Fusarium oxysporum. Eur. J. Plant Pathol. 2010, 126, 517–530. [Google Scholar] [CrossRef]
- Panth, M.; Hassler, S.C.; Baysal-Gurel, F. Methods for management of soilborne diseases in crop production. Agriculture 2020, 10, 16. [Google Scholar] [CrossRef]
- Armstrong, G.M.; Armstrong, J.K. Fusarium: Diseases, biology, and taxonomy. In Formae Speciales and Races of Fusarium Oxysporum Causing Wilt Diseases; Nelson, P.E., Toussoun, T.A., Cook, R., Eds.; The Pennsylvania State University Press: University Park, PA, USA, 1981; pp. 391–399. [Google Scholar]
- Edel-Hermann, V.; Lecomte, C. Current status of Fusarium oxysporum formae speciales and races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef]
- Infantino, A.; Kharrat, M.; Riccioni, L.; Coyne, C.J.; Mcphee, K.E.; Grünwald, N.J. Screening techniques and sources of resistance to root diseases in cool season food legumes. Euphytica 2006, 147, 201–221. [Google Scholar] [CrossRef]
- Demers, J.E.; Garzón, C.D.; Jiménez-Gasco, M.d.M. Striking genetic similarity between races of Fusarium oxysporum f. sp. ciceris confirms a monophyletic origin and clonal evolution of the chickpea vascular wilt pathogen. Eur. J. Plant Pathol. 2014, 139, 309–324. [Google Scholar] [CrossRef]
- Pottorff, M.; Wanamaker, S.; Ma, Y.Q.; Ehlers, J.D.; Roberts, P.A.; Close, T.J. Genetic and physical mapping of candidate genes for resistance to Fusarium oxysporum f.sp. tracheiphilum race 3 in cowpea [Vigna unguiculata (L.) Walp]. PLoS ONE 2012, 7, e41600. [Google Scholar] [CrossRef]
- Jendoubi, W.; Bouhadida, M.; Boukteb, A.; Béji, M.; Kharrat, M. Fusarium Wilt Affecting Chickpea Crop. Agriculture 2017, 7, 23. [Google Scholar] [CrossRef]
- Armstrong, G.M.; Armstrong, J.K. Lupinus species—Common hosts for wilt Fusaria from alfalfa, bean cassia, cowpea, lupine and U.S. cotton. Phytopathology 1964, 54, 1232–1234. [Google Scholar] [CrossRef]
- Lamberts, H. Broadening the basis for the breeding of yellow sweet lupine. Euphytica 1955, 4, 97–196. [Google Scholar] [CrossRef]
- Venuto, B.C.; Smith, R.R.; Grau, C.R. Virulence, legume hostspecificity, and genetic relatedness of isolates of Fusarium oxysporum from red clover. Plant Dis. 1995, 79, 406–410. [Google Scholar] [CrossRef]
- Grajal-Martín, M.J.; Simon, C.J.; Muehlbauer, F.J. Use of Random Ampfified Polymorphic DNA (RAPD) to characterize race 2 of Fusarium oxysporum f. sp. pisi. Mol. Plant Pathol. 1993, 83, 612–614. [Google Scholar] [CrossRef]
- Haglund, W.A.; Kraft, J.M. Fusarium oxysporum f. sp. pisi, race 6: Occurrence and distribution. Phytopathology 1979, 69, 818. [Google Scholar] [CrossRef]
- Haware, M.P.; Nene, Y.L. Races of Fusarium oxysporum f. sp. ciceri. Plant Dis. 1982, 66, 809–810. [Google Scholar] [CrossRef]
- Jiménez-Díaz, R.M.; Basallote-Ureba, M.J.; Rapoport, H. Colonization and pathogenesis in chickpeas infected by races of Fusarium oxysporum f. sp. ciceri. In Vascular Wilt Diseases of Plants; Springer: Berlin/Heidelberg, Germany, 1989; pp. 113–121. [Google Scholar]
- Jiménez-Díaz, R.M.; Alcala, A.; Hervas, A.; Trapero Casas, J.L. Pathogenic variability and host resistance in the Fusarium oxysporum f.sp. ciceris/Cicer arietinum pathosystem. Hod. Rosl. Aklim. i Nasienn. 1993, 37, 87–94. [Google Scholar]
- Gordon, W.L. Pathogenic strains of Fusarium oxysporum. Can. J. Bot. 1965, 43, 1309–1318. [Google Scholar] [CrossRef]
- Pouralibaba, H.R.; Rubiales, D.; Fondevilla, S. Identification of pathotypes in Fusarium oxysporum f. sp. lentis. Eur. J. Plant Pathol. 2016, 144, 539–549. [Google Scholar] [CrossRef]
- Hiremani, N.S.; Dubey, S.C. Race profiling of Fusarium oxysporum f. sp. lentis causing wilt in lentil. Crop Prot. 2018, 108, 23–30. [Google Scholar] [CrossRef]
- Hare, W.W. A new race of Fusarium causing wilt of cowpea. Phytopathology 1953, 43, 291. [Google Scholar]
- Smith, S.N.; Helms, D.M.; Temple, S.R.; Frate, C. The distribution of Fusarium wilt of blackeyed cowpeas within California caused by Fusarium oxysporum f. sp. tracheiphilum race 4. Plant Dis. 1999, 83, 694. [Google Scholar] [CrossRef]
- Armstrong, G.M.; Armstrong, J.K. A wilt of soybean caused by a new form of Fusarium oxysporum. Phytopathology 1965, 55, 237–239. [Google Scholar]
- Brayford, D. Fusarium oxysporum f. sp. medicaginis. IMI Descr. Fungi Bact. 1996, 1268, 1–2. [Google Scholar]
- Synder, W.C.; Hansen, H.N. The species concept in Fusarium. Am. J. Bot. 1940, 27, 64–67. [Google Scholar]
- Yu, T.F.; Fang, C.T. Fusarium diseases of broad beans. III. Root-rot and wilt of broad beans caused by two new forms of Fusarium. Phytopathology 1948, 38, 587–594. [Google Scholar]
- Wunsch, M.J.; Baker, A.H.; Kalb, D.W.; Bergstrom, G.C. Characterization of Fusarium oxysporum f. sp. loti Forma Specialis nov., a Monophyletic Pathogen Causing Vascular Wilt of Birdsfoot Trefoil. Plant Dis. 2009, 93, 58–66. [Google Scholar] [CrossRef]
- De Curtis, F.; Palmieri, D.; Vitullo, D.; Lima, G. First report of Fusarium oxysporum f.sp. pisi as causal agent of root and crown rot on chickpea (Cicer arietinum) in Southern Italy. Plant Dis. 2014, 98, 995. [Google Scholar] [CrossRef]
- Sumner, D.R.; Minton, N.A. Interaction of Fusarium wilt and nematodes in Cobb soybean. Plant Dis. 1987, 71, 20–23. [Google Scholar]
- Dhingra, O.D.; Netto, R.A.C. Reservoir and non-reservoir hosts of bean-wilt pathogen, Fusarium oxysporum f. sp. phaseoli. J. Phytopathol. 2001, 149, 463–467. [Google Scholar] [CrossRef]
- Yadeta, K.A.; Thomma, B.P.H.J. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef]
- Gullino, M.L.; Camponogara, A.; Gasparrini, G.; Rizzo, V.; Clini, C.; Garibaldi, A. Replacing methyl bromide for soil disinfection: The Italian experience and implications for other countries. Plant Dis. 2003, 87, 1012–1021. [Google Scholar] [CrossRef]
- Ebbels, D.L. Effect of soil fumigants on Fusarium wilt and nodulation of peas (Pisum sativum L.). Ann. Appl. Biol. 1967, 60, 391–398. [Google Scholar] [CrossRef]
- Zhao, J.; Mei, Z.; Zhang, X.; Xue, C.; Zhang, C.; Ma, T.; Zhang, S. Suppression of Fusarium wilt of cucumber by ammonia gas fumigation via reduction of Fusarium population in the field. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Barzman, M.; Bàrberi, P.; Birch, A.N.E.; Boonekamp, P.; Dachbrodt-Saaydeh, S.; Graf, B.; Hommel, B.; Jensen, J.E.; Kiss, J.; Kudsk, P.; et al. Eight principles of integrated pest management. Agron. Sustain. Dev. 2015, 35, 1199–1215. [Google Scholar] [CrossRef]
- El-Hassan, S.A.; Gowen, S.R.; Pembroke, B. Use of Trichoderma hamatum for biocontrol of lentil vascular wilt disease: Efficacy, mechanisms of interaction and future prospects. J. Plant Prot. Res. 2013, 53, 12–26. [Google Scholar] [CrossRef]
- Carvalho, D.D.C.; de Mello, S.C.M.; Martins, I.; Lobo, M. Biological control of Fusarium wilt on common beans by in-furrow application of Trichoderma harzianum. Trop. Plant Pathol. 2015, 40, 375–381. [Google Scholar] [CrossRef]
- Hervás, A.; Landa, B.; Datnoff, L.E.; Jiménez-Díaz, R.M. Effects of commercial and indigenous microorganisms on Fusarium wilt development in chickpea. Biol. Control 1998, 13, 166–176. [Google Scholar] [CrossRef]
- Landa, B.B.; Navas-Cortés, J.A.; Hervás, A.; Jiménez-Díaz, R.M. Influence of temperature and inoculum density of Fusarium oxysporum f. sp. ciceris on suppression of Fusarium wilt of chickpea by rhizosphere bacteria. Phytopathology 2001, 91, 807. [Google Scholar] [CrossRef]
- Bubici, G. Streptomyces spp. as biocontrol agents against Fusarium species. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2018, 13, 1–15. [Google Scholar] [CrossRef]
- Bubici, G.; Kaushal, M.; Prigigallo, M.I.; Cabanás, C.G.L.; Mercado-Blanco, J. Biological control agents against Fusarium wilt of banana. Front. Microbiol. 2019, 10, 616. [Google Scholar] [CrossRef]
- Anusha, B.G.; Gopalakrishnan, S.; Naik, M.K.; Sharma, M. Evaluation of Streptomyces spp. and Bacillus spp. for biocontrol of Fusarium wilt in chickpea (Cicer arietinum L.). Arch. Phytopathol. Plant Prot. 2019, 52, 417–442. [Google Scholar] [CrossRef]
- Palmieri, D.; Vitullo, D.; De Curtis, F.; Lima, G. A microbial consortium in the rhizosphere as a new biocontrol approach against fusarium decline of chickpea. Plant Soil 2017, 412, 425–439. [Google Scholar] [CrossRef]
- Abdul Wahid, O.A. Improving control of Fusarium wilt of leguminous plants by combined application of biocontrol agents. Phytopathol. Mediterr. 2006, 45, 231–237. [Google Scholar] [CrossRef]
- Bapat, S.; Shah, A.K. Biological control of fusarial wilt of pigeon pea by Bacillus brevis. Can. J. Microbiol. 2000, 46, 125–132. [Google Scholar] [CrossRef]
- Das, K.; Prasanna, R.; Saxena, A.K. Rhizobia: A potential biocontrol agent for soilborne fungal pathogens. Folia Microbiol. 2017, 62, 425–435. [Google Scholar] [CrossRef]
- Demir, S.; Akkopru, A. Using of arbuscular mycorrhizal fungi (AMF) for biocontrol of soil-borne fungal plant pathogens. In Biological Control of Plant Diseases; Chincholkar, S.B., Mukerji, K.G., Eds.; Haworth Press: Philadelphia, PA, USA, 2007; pp. 17–37. [Google Scholar]
- Singh, P.K.; Singh, M.; Vyas, D. Biocontrol of fusarium wilt of chickpea using arbuscular mycorrhizal fungi and rhizobium leguminosorum biovar. Caryologia 2010, 63, 349–353. [Google Scholar] [CrossRef]
- Arfaoui, A.; Sifi, B.; Boudabous, A.; El Hadrami, I.; Chérif, M. Identification of Rhizobium isolates possessing antagonistic activity against Fusarium oxysporum f. sp. ciceris, the causal agent of Fusarium wilt of chickpea. J. Plant Pathol. 2006, 88, 67–75. [Google Scholar] [CrossRef]
- Sajeena, A.; Nair, D.S.; Sreepavan, K. Non-pathogenic Fusarium oxysporm as a biocontrol agent. Indian Phytopathol. 2020, 73, 177–183. [Google Scholar] [CrossRef]
- Benhamou, N.; Garand, C. Cytological analysis of defense-related mechanisms induced in pea root tissues in response to colonization by nonpathogenic Fusarium oxysporum Fo47. Phytopathology 2001, 91, 730–740. [Google Scholar] [CrossRef]
- Landa, B.B.; Navas-Cortés, J.A.; Jiménez-Díaz, R.M. Integrated management of Fusarium wilt of chickpea with sowing date, host resistance, and biological control. Phytopathology 2004, 94, 946–960. [Google Scholar] [CrossRef]
- Gordon, T.R.; Martyn, R.D. The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 1997, 35, 111–128. [Google Scholar] [CrossRef]
- Pande, S.; Rao, J.N.; Sharma, M. The plant pathology journal establishment of the chickpea wilt pathogen Fusarium oxysporum f. sp. ciceris in the soil through seed transmission. Plant Pathol. J. 2007, 23, 3–6. [Google Scholar]
- de Sousa, M.V.; Machado, J.d.C.; Simmons, H.E.; Munkvold, G.P. Real-time quantitative PCR assays for the rapid detection and quantification of Fusarium oxysporum f. sp. phaseoli in Phaseolus vulgaris (common bean) seeds. Plant Pathol. 2015, 64, 478–488. [Google Scholar] [CrossRef]
- Haware, M.P. Diseases of chickpea. In The Pathology of Food and Pasture Legumes; Allen, D.J., Lenné, J.M., Eds.; CAB International: Wallingford, UK, 1998; pp. 473–561. [Google Scholar]
- Chauhan, Y.S.; Nene, Y.L.; Johansen, C.; Haware, M.P.; Saxena, N.P.; Sardar, S.; Sharma, S.B.; Sahrawat, K.L.; Burford, J.R.; Rupela, O.P.; et al. Effects of Soil Solarization on Pigeonpea and Chickpea; International Crops Research Institute for the Semi-Arid Tropics: Patancheru, India, 1988. [Google Scholar]
- Larkin, R.P. Green manures and plant disease management. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2013, 8. [Google Scholar] [CrossRef]
- Prasad, P.; Kumar, J. Management of fusarium wilt of chickpea using brassicas as biofumigants. Legum. Res. 2017, 40, 178–182. [Google Scholar] [CrossRef]
- Gordon, T.R. Colonization of muskmelon and nonsusceptible crops by Fusarium oxysporum f. sp. melonis and other species of Fusarium. Phytopathology 1989, 79, 1095. [Google Scholar] [CrossRef]
- Haware, M.P.; Nene, Y.L.; Natarajan, M. The survival of Fusarium oxysporum f. sp. ciceri in the soil in the absence of chickpea. Phytopathol. Mediterr. 1996, 35, 9–12. [Google Scholar]
- Helbig, J.B.; Carroll, R.B. Dicotyledonous weeds as a source of Fusarium oxysporum pathogenic on soybean. Plant Dis. 1984, 68, 694–696. [Google Scholar] [CrossRef]
- Jiménez-Fernández, D.; Montes-Borrego, M.; Jiménez-Díaz, R.M.; Navas-Cortés, J.A.; Landa, B.B. In planta and soil quantification of Fusarium oxysporum f. sp. ciceris and evaluation of fusarium wilt resistance in chickpea with a newly developed quantitative polymerase chain reaction assay. Phytopathology 2011, 101, 250–262. [Google Scholar] [CrossRef]
- Sharma, K.D.; Rathour, R.; Kapila, R.K.; Paul, Y.S. Detection of pea wilt pathogen Fusarium oxysporum f. sp. pisi using DNA-based markers. J. Plant Biochem. Biotechnol. 2018, 27, 342–350. [Google Scholar] [CrossRef]
- Pouralibaba, H.R.; Rubiales, D.; Fondevilla, S. Identification of resistance to Fusarium oxysporum f. sp. lentis in Spanish lentil germplasm. Eur. J. Plant Pathol. 2015, 143, 399–405. [Google Scholar] [CrossRef]
- Bayaa, B.; Erskine, W.; Singh, M. Screening lentil for resistance to fusarium wilt: Methodology and sources of resistance. Euphytica 1997, 98, 69–74. [Google Scholar] [CrossRef]
- Castillo, P.; Navas-Cortés, J.A.; Gomar-Tinoco, D.; Di Vito, M.; Jiménez-Díaz, R.M. Interactions between Meloidogyne artiellia, the cereal and legume root-knot nematode, and Fusarium oxysporum f. sp. ciceris race 5 in chickpea. Phytopathology 2003, 93, 1513–1523. [Google Scholar] [CrossRef]
- Castillo, P.; Mora-Rodríguez, M.P.; Navas-Cortés, J.A.; Jiménez-Díaz, R.M. Interactions of Pratylenchus thornei and Fusarium oxysporum f. sp. ciceris on chickpea. Phytopathology 1998, 88, 828–836. [Google Scholar] [CrossRef]
- Krishna Rao, V.; Krishnappa, K. Interaction of Fusarium oxysporum f.sp. ciceri with Meloidogyne incognita on chickpea in two soil types. Indian Phytopathol. 1996, 49, 142–147. [Google Scholar]
- Navas-Cortés, J.A.; Landa, B.B.; Rodríguez-López, J.; Jiménez-Díaz, R.M.; Castillo, P. Infection by Meloidogyne artiellia does not break down resistance to races 0, 1A, and 2 of Fusarium oxysporum f. sp. ciceris in chickpea genotypes. Phytopathology 2008, 98, 709–718. [Google Scholar] [CrossRef]
- Haglund, W.A. A rapid method for inoculating pea seedlings with Fusarium oxysporum f. sp. pisi. Plant Dis. 1989, 73, 457. [Google Scholar] [CrossRef]
- Eynck, C.; Koopmann, B.; von Tiedemann, A. Identification of Brassica accessions with enhanced resistance to Verticillium longisporum under controlled and field conditions. J. Plant Dis. Prot. 2009, 116, 63–72. [Google Scholar] [CrossRef]
- Landa, B.B.; Navas-Cortés, J.A.; del Mar Jiménez-Gasco, M.; Katan, J.; Retig, B.; Jiménez-Díaz, R.M. Temperature response of chickpea cultivars to races of Fusarium oxysporum f. sp. ciceris, causal agent of Fusarium wilt. Plant Dis. 2006, 90, 365–374. [Google Scholar] [CrossRef]
- Haware, M.P.; Nene, Y.L.; Pundir, R.P.S.; Narayana Rao, J. Screening of world chickpea germplasm for resistance to fusarium wilt. Field Crop. Res. 1992, 30, 147–154. [Google Scholar] [CrossRef]
- Jiménez-Díaz, R.M. Resistance in Kabuli chickpeas to Fusarium wilt. Plant Dis. 1991, 75, 914. [Google Scholar] [CrossRef]
- Sharma, M.; Kiran Babu, T.; Gaur, P.M.; Ghosh, R.; Rameshwar, T.; Chaudhary, R.G.; Upadhyay, J.P.; Gupta, O.; Saxena, D.R.; Kaur, L.; et al. Identification and multi-environment validation of resistance to Fusarium oxysporum f. sp. ciceris in chickpea. Field Crop. Res. 2012, 135, 82–88. [Google Scholar] [CrossRef]
- Sharma, M.; Ghosh, R.; Tarafdar, A.; Rathore, A.; Chobe, D.R.; Kumar, A.V.; Gaur, P.M.; Samineni, S.; Gupta, O.; Singh, N.P.; et al. Exploring the genetic cipher of chickpea (Cicer arietinum L.) through identification and multi-environment validation of resistant sources against Fusarium wilt (Fusarium oxysporum f. sp. ciceris). Front. Sustain. Food Syst. 2019, 3, 78. [Google Scholar] [CrossRef]
- Mohammadi, N.; Puralibaba, H.; Goltapeh, E.M.; Ahari, A.B.; Sardrood, B.P. Advanced lentil lines screened for resistance to Fusarium oxysporum f. sp. lentis under greenhouse and field conditions. Phytoparasitica 2012, 40, 69–76. [Google Scholar] [CrossRef]
- Navas-Cortés, J.A.; Hau, B.; Jiménez-Díaz, R.M. Effect of sowing date, host cultivar, and race of Fusarium oxysporum f. sp. ciceris on development of Fusarium wilt of chickpea. Phytopathology 1998, 44, 1338–1346. [Google Scholar] [CrossRef]
- Singh, M.; Rana, J.C.; Singh, B.; Kumar, S.; Saxena, D.R.; Saxena, A.; Rizvi, A.H.; Sarker, A. Comparative agronomic performance and reaction to Fusarium wilt of Lens culinaris × L. orientalis and L. culinaris × L. ervoides derivatives. Front. Plant Sci. 2017, 8, 1162. [Google Scholar] [CrossRef]
- Emberger, G.; Welty, R.E. Evaluation of virulence of Fusarium oxysporum f. sp. medicaginis and Fusarium wilt resistance in alfalfa. Plant Dis. 1983, 67, 94. [Google Scholar] [CrossRef]
- McPhee, K.E.; Tullu, A.; Kraft, J.M.; Muehlbauer, F.J. Resistance to Fusarium wilt race 2 in the Pisum core collection. J. Am. Soc. Hortic. Sci. 1999, 124, 28–31. [Google Scholar] [CrossRef]
- McPhee, K.E.; Inglis, D.A.; Gundersen, B.; Coyne, C.J. Mapping QTL for Fusarium wilt race 2 partial resistance in pea (Pisum sativum). Plant Breed. 2012, 131, 300–306. [Google Scholar] [CrossRef]
- Stoilova, T.; Chavdarov, P. Evaluation of lentil germplasm for disease resistance to Fusarium wilt (Fusarium oxysporum f. sp. lentis). J. Cent. Eur. Agric. 2006, 7, 121–126. [Google Scholar]
- Bani, M.; Rubiales, D.; Rispail, N. A detailed evaluation method to identify sources of quantitative resistance to Fusarium oxysporum f. sp. pisi race 2 within a Pisum spp. germplasm collection. Plant Pathol. 2012, 61, 532–542. [Google Scholar] [CrossRef]
- Rispail, N.; Bani, M.; Rubiales, D. Resistance reaction of Medicago truncatula genotypes to Fusarium oxysporum: Effect of plant age, substrate and inoculation method. Crop Pasture Sci. 2015, 66, 506. [Google Scholar] [CrossRef]
- Bayaa, B.; Erskine, W.; Hamdi, A. Evaluation of a wild lentil collection for resistance to vascular wilt. Genet. Resour. Crop Evol. 1995, 42, 231–235. [Google Scholar] [CrossRef]
- Buruchara, R.A.; Camacho, L. Common bean reaction to Fusarium oxysporum f. sp. phaseoli, the cause of severe vascular wilt in Central Africa. J. Phytopathol. 2000, 148, 39–45. [Google Scholar] [CrossRef]
- Fall, A.L.; Byrne, P.F.; Jung, G.; Coyne, D.P.; Brick, M.A.; Schwartz, H.F. Detection and mapping of a major locus for Fusarium wilt resistance in common bean. Crop Sci. 2001, 41, 1494. [Google Scholar] [CrossRef]
- Rispail, N.; Rubiales, D. Identification of sources of quantitative resistance to Fusarium oxysporum f. sp. medicaginis in Medicago truncatula. Plant Dis. 2014, 98, 667–673. [Google Scholar] [CrossRef]
- Charchart, M.; Kraft, J.M. Response of near-isogenic pea cultivars to infection by Fusarium oxysporum f. sp. pisi races 1 and 5. Can. J. Plant Sci. 1989, 69, 1335–1346. [Google Scholar]
- Merzoug, A.; Belabid, L.; Youcef-Benkada, M.; Benfreha, F.; Bayaa, B. Pea Fusarium wilt races in Western Algeria. Plant Prot. Sci. 2014, 50, 70–77. [Google Scholar] [CrossRef]
- Neumann, S.; Xue, A.G. Reactions of field pea cultivars to four races of Fusarium oxysporum f. sp. pisi. Can. J. Plant Sci. 2003, 83, 377–379. [Google Scholar] [CrossRef]
- Arvayo-Ortiz, R.M.; Martin, E.; Acedo-Felix, E.; Gonzalez-Rios, H.; Vargas-Rosales, G. New lines of chickpea against Fusarium oxysporum f. sp. ciceris wilt. Am. J. Appl. Sci. 2012, 9, 686–693. [Google Scholar] [CrossRef]
- Leitão, S.T.; Malosetti, M.; Song, Q.; van Eeuwijk, F.; Rubiales, D.; Vaz Patto, M.C. Natural variation in Portuguese common bean germplasm reveals new sources of resistance against Fusarium oxysporum f. sp. phaseoli and resistance-associated candidate genes. Phytopathology 2020, 110, 633–647. [Google Scholar] [CrossRef]
- Altier, N.A.; Groth, J.V. Characterization of aggressiveness and vegetative compatibility diversity of Fusarium oxysporum associated with crown and root rot of birdsfoot trefoil. Lotus Newsl. 2005, 35, 59–74. [Google Scholar]
- Lebeda, A.; Svabova, L.; Dostalova, R. Screening of peas for resistance to fusarium wilt and root rot (Fusarium oxysporum, Fusarium solani). In Mass Screening Techniques for Selecting Crops Resistant to Disease; Spencer, M., Lebeda, A., Eds.; FAO/IAEA: Vienna, Austria, 2010; pp. 189–196. [Google Scholar]
- Zian, A.H.; El-Demardash, I.S.; El-Mouhamady, A.A.; El-Barougy, E. Studies the resistance of lupine for Fusarium oxysporum f. sp lupini) through molecular genetic technique. World Appl. Sci. J. 2013, 26, 1064–1069. [Google Scholar] [CrossRef]
- Rispail, N.; Rubiales, D. Rapid and efficient estimation of pea resistance to the soil-borne pathogen Fusarium oxysporum by infrared imaging. Sensors 2015, 15, 3988–4000. [Google Scholar] [CrossRef]
- Bishop, C.D.; Cooper, R.M. An ultrastructural study of root invasion in three vascular wilt diseases. Physiol. Plant Pathol. 1983, 22, 15–27. [Google Scholar] [CrossRef]
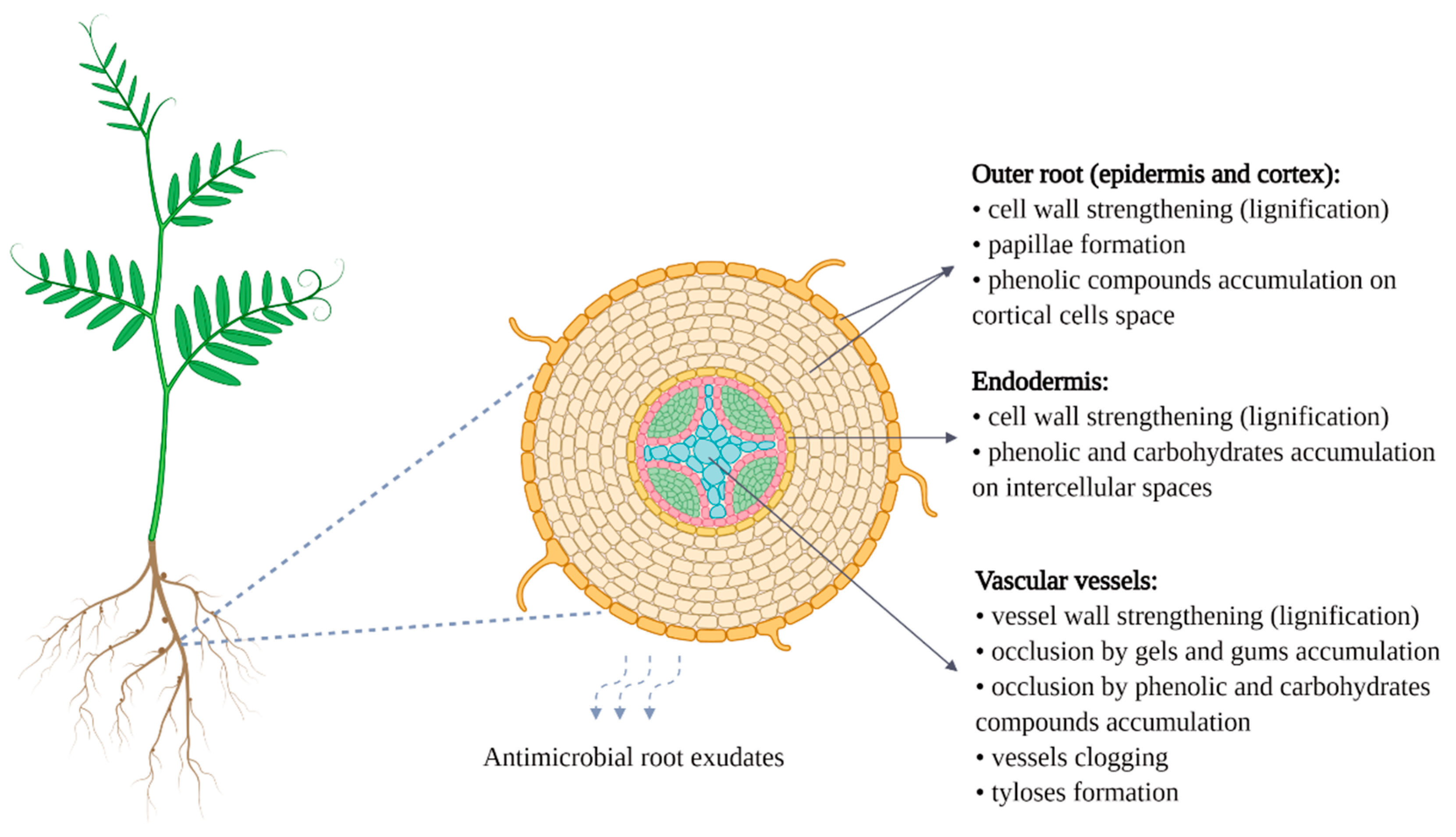
- Bani, M.; Pérez-De-Luque, A.; Rubiales, D.; Rispail, N. Physical and chemical barriers in root tissues contribute to quantitative resistance to Fusarium oxysporum f. sp. pisi in pea. Front. Plant Sci. 2018, 9, 199. [Google Scholar] [CrossRef]
- Tessier, B.J.; Mueller, W.C.; Morgham, A.T. Histopathology and ultrastructure of vascular-responses in peas resistant or susceptible to Fusarium oxysporum f. sp. pisi. Phytopathology 1990, 80, 756–764. [Google Scholar] [CrossRef]
- Pouralibaba, H.R.; Pérez-de-Luque, A.; Rubiales, D. Histopathology of the infection on resistant and susceptible lentil accessions by two contrasting pathotypes of Fusarium oxysporum f. sp. lentis. Eur. J. Plant Pathol. 2017, 148, 53–63. [Google Scholar] [CrossRef]
- Gupta, S.; Chakraborti, D.; Sengupta, A.; Basu, D.; Das, S. Primary metabolism of chickpea is the initial target of wound inducing early sensed Fusarium oxysporum f. sp. ciceri race I. PLoS ONE 2010, 5, e9030. [Google Scholar] [CrossRef]
- Joshi, N.S.; Rao, K.S.; Subramanian, R.B. Anatomical and biochemical aspects of interaction between roots of chickpea and Fusarium oxysporum f. sp. ciceris race 2. Arch. Phytopathol. Plant Prot. 2012, 45, 1773–1789. [Google Scholar] [CrossRef]
- Miranda, K.; Girard-Dias, W.; Attias, M.; De Souza, W.; Ramos, I.; Celular, U.; Meyer, H. Three dimensional reconstruction by electron microscopy in the life sciences: An introduction for cell and tissue biologists. Mol. Reprod. Dev. Mol. Reprod. Dev. 2015, 82, 530–547. [Google Scholar] [CrossRef]
- Pereira, A.C.; Cruz, M.F.A.; Paula Júnior, T.J.; Rodrigues, F.A.; Carneiro, J.E.S.; Vieira, R.F.; Carneiro, P.C.S. Infection process of Fusarium oxysporum f. sp. phaseoli on resistant, intermediate and susceptible bean cultivars. Trop. Plant Pathol. 2013, 38, 323–328. [Google Scholar] [CrossRef]
- Xue, R.; Wu, J.; Zhu, Z.; Wang, L.; Wang, X.; Wang, S.; Blair, M.W. Differentially expressed genes in resistant and susceptible common bean (Phaseolus vulgaris L.) genotypes in response to Fusarium oxysporum f. sp. phaseoli. PLoS ONE 2015, 10, e0127698. [Google Scholar] [CrossRef]
- Jiménez-Fernández, D.; Landa, B.B.; Kang, S.; Jiménez-Díaz, R.M.; Navas-Cortés, J.A. Quantitative and microscopic assessment of compatible and incompatible interactions between chickpea cultivars and Fusarium oxysporum f. sp. ciceris races. PLoS ONE 2013, 8, e61360. [Google Scholar] [CrossRef]
- Islam, M.N.; Nizam, S.; Verma, P.K. A highly efficient Agrobacterium mediated transformation system for chickpea wilt pathogen Fusarium oxysporum f. sp. ciceri using DsRed-Express to follow root colonisation. Microbiol. Res. 2012, 167, 332–338. [Google Scholar] [CrossRef]
- Upasani, M.L.; Gurjar, G.S.; Kadoo, N.Y.; Gupta, V.S. Dynamics of colonization and expression of pathogenicity related genes in Fusarium oxysporum f. sp. ciceri during chickpea vascular wilt disease progression. PLoS ONE 2016, 11, e0156490. [Google Scholar] [CrossRef]
- Niño-Sánchez, J.; Tello, V.; Casado-del Castillo, V.; Thon, M.R.; Benito, E.P.; Díaz-Mínguez, J.M. Gene expression patterns and dynamics of the colonization of common bean (Phaseolus vulgaris L.) by highly virulent and weakly virulent strains of Fusarium oxysporum. Front. Microbiol. 2015, 6, 234. [Google Scholar] [CrossRef][Green Version]
- Whalley, W.M.; Taylor, G.S. Influence of pea-root exudates on germination of conidia and chlamydospores of physiologic races of Fusarium oxysporum f. pisi. Ann. Appl. Biol. 1973, 73, 269–276. [Google Scholar]
- Stevenson, P.C.; Turner, H.C.; Haware, M.P. Phytoalexin accumulation in the roots of chickpea (Cicer arietinum L.) seedlings associated with resistance to fusarium wilt (Fusarium oxysporum f. sp. ciceri). Physiol. Mol. Plant Pathol. 1997, 50, 167–178. [Google Scholar] [CrossRef]
- Kumar, Y.; Zhang, L.; Panigrahi, P.; Dholakia, B.B.; Dewangan, V.; Chavan, S.G.; Kunjir, S.M.; Wu, X.; Li, N.; Rajmohanan, P.R.; et al. Fusarium oxysporum mediates systems metabolic reprogramming of chickpea roots as revealed by a combination of proteomics and metabolomics. Plant Biotechnol. J. 2016, 14, 1589–1603. [Google Scholar] [CrossRef]
- Bani, M.; Cimmino, A.; Evidente, A.; Rubiales, D.; Rispail, N. Pisatin involvement in the variation of inhibition of Fusarium oxysporum f. sp. pisi spore germination by root exudates of Pisum spp. germplasm. Plant Pathol. 2018, 67, 1046–1054. [Google Scholar] [CrossRef]
- Nóbrega, F.M.; Santos, I.S.; Da Cunha, M.; Carvalho, A.O.; Gomes, V.M. Antimicrobial proteins from cowpea root exudates: Inhibitory activity against Fusarium oxysporum and purification of a chitinase-like protein. Plant Soil 2005, 272, 223–232. [Google Scholar] [CrossRef]
- Stevenson, P.C.; Padgham, D.E.; Haware, M.P. Root exudates associated with the resistance of four chickpea cultivars (Cicer arietinum) to two races of Fusarium oxysporum f.sp. ciceri. Plant Pathol. 1995, 44, 686–694. [Google Scholar] [CrossRef]
- Castillejo, M.Á.; Bani, M.; Rubiales, D. Understanding pea resistance mechanisms in response to Fusarium oxysporum through proteomic analysis. Phytochemistry 2015, 115, 44–58. [Google Scholar] [CrossRef]
- Upasani, M.L.; Limaye, B.M.; Gurjar, G.S.; Kasibhatla, S.M.; Joshi, R.R.; Kadoo, N.Y.; Gupta, V.S. Chickpea-Fusarium oxysporum interaction transcriptome reveals differential modulation of plant defense strategies. Sci. Rep. 2017, 7, 7746. [Google Scholar] [CrossRef]
- Bishop, C.D.; Cooper, R.M. Ultrastructure of vascular colonization by fungal wilt pathogens. II. Invasion of resistant cultivars. Physiol. Plant Pathol. 1984, 24, 277–289. [Google Scholar] [CrossRef]
- Sharma, K.D.; Chen, W.; Muehlbauer, F.J. Genetics of chickpea resistance to five races of Fusarium wilt and a concise set of race differentials for Fusarium oxysporum f. sp. ciceris. Plant Dis. 2005, 89, 385–390. [Google Scholar] [CrossRef]
- Sharma, K.D.; Winter, P.; Kahl, G.; Muehlbauer, F.J. Molecular mapping of Fusarium oxysporum f. sp. ciceris race 3 resistance gene in chickpea. Theor. Appl. Genet. 2004, 108, 1243–1248. [Google Scholar] [CrossRef]
- Tullu, A.; Muehlbauer, F.J.; Simon, C.J.; Mayer, M.S.; Kumar, J.; Kaiser, W.J.; Kraft, J.M. Inheritance and linkage of a gene for resistance to race 4 of fusarium wilt and RAPD markers in chickpea. Euphytica 1998, 102, 227–232. [Google Scholar] [CrossRef]
- Tekeoglu, M.; Tullu, A.; Kaiser, W.J.; Muehlbauer, F.J. Inheritance and linkage of two genes that confer resistance to Fusarium wilt in chickpea. Crop Sci. 2000, 40, 1247. [Google Scholar] [CrossRef]
- Grajal-Martìn, M.J.; Muehlbauer, F.J. Genomic location of the Fw gene for resistance to Fusarium wilt race 1 in peas. J. Hered. 2002, 93, 291–293. [Google Scholar] [CrossRef]
- McClendon, M.T.; Inglis, D.A.; McPhee, K.E.; Coyne, C.J. DNA markers linked to Fusarium wilt race 1 resistance in pea. J. Am. Soc. Hortic. Sci. 2002, 127, 602–607. [Google Scholar] [CrossRef]
- Coyne, C.J.; Inglis, D.A.; Whitehead, S.J.; McClendon, M.T.; Muehlbauer, F.J. Chromosomal location of Fwf, the Fusarium wilt race 5 resistance gene in Pisum sativum. Pisum Genet. 2000, 32, 20–22. [Google Scholar]
- Okubara, P.; Ingis, D.; Muehlbauer, F.; Coyne, C. A novel RAPD marker linked to the Fusarium wilt race 5 resistance gene (Fwf) in Pisum sativum. Pisum Genet. 2002, 34, 6–8. [Google Scholar]
- Ribeiro, R.L.D.; Hagedorn, D.J. Inheritance and nature of resistance in beans to Fusarium oxysporum f. sp. phaseoli. Phytopathology 1979, 69, 859. [Google Scholar] [CrossRef]
- Eujayl, I.; Erskine, W.; Bayaa, B.; Baum, M.; Pehu, E. Fusarium vascular wilt in lentil: Inheritance and identification of DNA markers for resistance. Plant Breed. 1998, 117, 497–499. [Google Scholar] [CrossRef]
- Hamwieh, A.; Udupa, S.M.; Choumane, W.; Sarker, A.; Dreyer, F.; Jung, C.; Baum, M. A genetic linkage map of Lens sp. based on microsatellite and AFLP markers and the localization of fusarium vascular wilt resistance. Theor. Appl. Genet. 2005, 110, 669–677. [Google Scholar] [CrossRef]
- Rubio, J.; Hajj-Moussa, E.; Kharrat, M.; Moreno, M.T.; Millan, T.; Gil, J. Two genes and linked RAPD markers involved in resistance to Fusarium oxysporum f. sp. ciceris race 0 in chickpea. Plant Breed. 2003, 122, 188–191. [Google Scholar] [CrossRef]
- Halila, I.; Cobos, M.J.; Rubio, J.; Millán, T.; Kharrat, M.; Marrakchi, M.; Gil, J. Tagging and mapping a second resistance gene for Fusarium wilt race 0 in chickpea. Eur. J. Plant Pathol. 2009, 124, 87–92. [Google Scholar] [CrossRef]
- Singh, H.; Kumar, J.; Smithson, J.B.; Haware, M.P. Complementation between genes for resistance to race 1 of Fusarium oxysporum f. sp. ciceri in chickpea. Plant Pathol. 1987, 36, 539–543. [Google Scholar] [CrossRef]
- Singh, H.; Kumar, J.; Haware, M.P.; Smithson, J.B. Genetics of resistance to fusarium wilt in chickpeas. In Genetics and Plant Pathogenesis; Day, P.R., Jellis, G.J., Eds.; Blackwell Scientific Publications: Oxford, UK, 1987; pp. 339–342. [Google Scholar]
- Upadhyaya, H.D.; Haware, M.P.; Kumar, J.; Smithson, J.B. Resistance to wilt in chickpea. I. Inheritance of late-wilting in response to race 1. Euphytica 1983, 32, 447–452. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Smithson, J.B.; Haware, M.P.; Kumar, J. Resistance to wilt in chickpea. II. Further evidence for two genes for resistance to race 1. Euphytica 1983, 32, 749–755. [Google Scholar] [CrossRef]
- Tullu, A.; Kaiser, W.J.; Kraft, J.M.; Muehlbauer, F.J. A second gene for resistance to race 4 of Fusarium wilt in chickpea and linkage with a RAPD marker. Euphytica 1999, 109, 43–50. [Google Scholar] [CrossRef]
- Hijano, E.H.; Barnes, D.K.; Frosheiser, F.I. Inheritance of resistance to Fusarium wilt in alfalfa1. Crop Sci. 1983, 23, 31. [Google Scholar] [CrossRef]
- Kamboj, R.K.; Pandey, M.P.; Chaube, H.S. Inheritance of resistance to Fusarium wilt in Indian lentil germplasm (Lens culinaris Medik.). Euphytica 1990, 50, 113–117. [Google Scholar] [CrossRef]
- Cross, H.; Brick, M.A.; Schwartz, H.F.; Panella, L.W.; Byrne, P.F. Inheritance of resistance to Fusarium wilt in two common bean races. Crop Sci. 2000, 40, 954. [Google Scholar] [CrossRef]
- Salgado, M.O.; Schwartz, H.F.; Brick, M.A. Inheritance of resistance to a Colorado race of Fusarium oxysporum f. sp. phaseoli in common beans. Plant Dis. 1995, 79, 279. [Google Scholar] [CrossRef]
- Kwon, S.J.; Smýkal, P.; Hu, J.; Wang, M.; Kim, S.-J.; McGee, R.J.; McPhee, K.; Coyne, C.J. User-friendly markers linked to Fusarium wilt race 1 resistance Fw gene for marker-assisted selection in pea. Plant Breed. 2013, 132, 642–648. [Google Scholar] [CrossRef]
- Cobos, M.J.; Fernández, M.J.; Rubio, J.; Kharrat, M.; Moreno, M.T.; Gil, J.; Millán, T. A linkage map of chickpea (Cicer arietinum L.) based on populations from Kabuli × Desi crosses: Location of genes for resistance to fusarium wilt race 0. Theor. Appl. Genet. 2005, 110, 1347–1353. [Google Scholar] [CrossRef]
- Jendoubi, W.; Bouhadida, M.; Millan, T.; Kharrat, M.; Gil, J.; Rubio, J.; Madrid, E. Identification of the target region including the Foc01/foc01 gene and development of near isogenic lines for resistance to Fusarium wilt race 0 in chickpea. Euphytica 2016, 210, 119–133. [Google Scholar] [CrossRef]
- Gowda, S.J.M.; Radhika, P.; Kadoo, N.Y.; Mhase, L.B.; Gupta, V.S. Molecular mapping of wilt resistance genes in chickpea. Mol. Breed. 2009, 24, 177–183. [Google Scholar] [CrossRef]
- Sharma, D.K.; Muehlbauer, F.J. Fusarium wilt of chickpea: Physiological specialization, genetics of resistance and resistance gene tagging. Euphytica 2007, 157, 1–14. [Google Scholar] [CrossRef]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Chamarthi, S.K.; Singh, V.K.; Srinivasan, S.; Swapna, N.; Sharma, M.; Pande, S.; Singh, S.; et al. Marker-assisted backcrossing to introgress resistance to Fusarium wilt race 1 and Ascochyta blight in C 214, an elite cultivar of chickpea. Plant Genome 2014, 7, 1–11. [Google Scholar] [CrossRef]
- Pratap, A.; Chaturvedi, S.K.; Tomar, R.; Rajan, N.; Malviya, N.; Thudi, M.; Saabale, P.R.; Prajapati, U.; Varshney, R.K.; Singh, N.P. Marker-assisted introgression of resistance to fusarium wilt race 2 in Pusa 256, an elite cultivar of desi chickpea. Mol. Genet. Genom. 2017, 292, 1237–1245. [Google Scholar] [CrossRef]
- Winter, P.; Benko-Iseppon, A.M.; Hüttel, B.; Ratnaparkhe, M.; Tullu, A.; Sonnante, G.; Pfaff, T.; Tekeoglu, M.; Santra, D.; Sant, V.J.; et al. A linkage map of the chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum x C. reticulatum cross: Localization of resistance genes for fusarium wilt races 4 and 5. Theor. Appl. Genet. 2000, 101, 1155–1163. [Google Scholar] [CrossRef]
- Benko-Iseppon, A.M.; Winter, P.; Huettel, B.; Staginnus, C.; Muehlbauer, F.J.; Kahl, G. Molecular markers closely linked to fusarium resistance genes in chickpea show significant alignments to pathogenesis-related genes located on Arabidopsis chromosomes 1 and 5. Theor. Appl. Genet. 2003, 107, 379–386. [Google Scholar] [CrossRef]
- Caballo, C.; Madrid, E.; Gil, J.; Chen, W.; Rubio, J.; Millan, T. Saturation of genomic region implicated in resistance to Fusarium oxysporum f. sp. ciceris race 5 in chickpea. Mol. Breed. 2019, 39, 1–11. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Gupta, S.; Chakraborti, D.; Rangi, R.K.; Basu, D.; Das, S. A molecular insight into the early events of chickpea (Cicer arietinum) and Fusarium oxysporum f. sp. ciceri (race 1) interaction through cDNA-AFLP analysis. Phytopathology 2009, 99, 1245–1257. [Google Scholar] [CrossRef][Green Version]
- Gupta, S.; Bhar, A.; Chatterjee, M.; Ghosh, A.; Das, S.; Gupta, V. Transcriptomic dissection reveals wide spread differential expression in chickpea during early time points of Fusarium oxysporum f. sp. ciceri. PLoS ONE 2017, 12, e0178164. [Google Scholar] [CrossRef]
- Lanubile, A.; Muppirala, U.K.; Severin, A.J.; Marocco, A.; Munkvold, G.P. Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genom. 2015, 16, 1089. [Google Scholar] [CrossRef]
- Gurjar, G.S.; Giri, A.P.; Gupta, V.S. Gene expression profiling during wilting in chickpea caused by Fusarium oxysporum f. sp. ciceri. Am. J. Plant Sci. 2012, 3, 190–201. [Google Scholar] [CrossRef]
- Saabale, P.R.; Dubey, S.C.; Priyanka, K.; Sharma, T.R. Analysis of differential transcript expression in chickpea during compatible and incompatible interactions with Fusarium oxysporum f. sp. ciceris Race 4. 3 Biotech 2018, 8. [Google Scholar] [CrossRef]
- Chen, L.; Wu, Q.; He, W.; He, T.; Wu, Q.; Miao, Y. Combined de novo transcriptome and metabolome analysis of common bean response to Fusarium oxysporum f. sp. phaseoli infection. Int. J. Mol. Sci. 2019, 20, 6278. [Google Scholar] [CrossRef]
- Chang, C.; Tian, L.; Ma, L.; Li, W.; Nasir, F.; Li, X.; Tran, L.P.; Tian, C. Differential responses of molecular mechanisms and physiochemical characters in wild and cultivated soybeans against invasion by the pathogenic Fusarium oxysporum Schltdl. Physiol. Plant. 2019, 166, 1008–1025. [Google Scholar] [CrossRef]
- Caballo, C.; Castro, P.; Gil, J.; Millan, T.; Rubio, J.; Die, J.V. Candidate genes expression profiling during wilting in chickpea caused by Fusarium oxysporum f. sp. ciceris race 5. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Taylor, A.; Vágány, V.; Jackson, A.C.; Harrison, R.J.; Rainoni, A.; Clarkson, J.P. Identification of pathogenicity-related genes in Fusarium oxysporum f. sp. cepae. Mol. Plant Pathol. 2016, 17, 1032–1047. [Google Scholar] [CrossRef]
- Ramos, B.; Alves-Santos, F.M.; García-Sánchez, M.A.; Martín-Rodrigues, N.; Eslava, A.P.; Díaz-Mínguez, J.M. The gene coding for a new transcription factor (ftf1) of Fusarium oxysporum is only expressed during infection of common bean. Fungal Genet. Biol. 2007, 44, 864–876. [Google Scholar] [CrossRef]
- Niño-Sánchez, J.; Casado-Del Castillo, V.; Tello, V.; De Vega-Bartol, J.J.; Ramos, B.; Sukno, S.A.; Díaz Mínguez, J.M. The FTF gene family regulates virulence and expression of SIX effectors in Fusarium oxysporum. Mol. Plant Pathol. 2016, 17, 1124–1139. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Williams, A.H.; Garg, G.; Buck, S.A.G.; Singh, K.B. Transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. medicaginis during colonisation of resistant and susceptible Medicago truncatula hosts identifies differential pathogenicity profiles and novel candidate effector. BMC Genom. 2016, 17. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Kidd, B.N.; Singh, K.B. Tools and strategies for genetic and molecular dissection of Medicago truncatula resistance against Fusarium wilt disease. In The Model Legume Medicago Truncatula; Bruijn, F.J., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2020; pp. 331–339. [Google Scholar]
- Gaur, P.M.; Gowda, C.L.L.; Knights, E.J.; Warkentin, T.; Acikoz, N.; Yadav, S.S.; Kumar, J. Breeding achievements. In Chickpea Breeding and Management; Yadav, S.S., Reden, R.J., Chen, W., Sharma, B., Eds.; Centre for Agriculture and Bioscience International (CABI): Wallingford, UK, 2007. [Google Scholar]
- Salimath, P.M.; Toker, C.; Sandhu, J.S.; Kumar, J.; Suma, B.; Yadav, S.S.; Bahl, P.N. Conventional breeding methods. In Chickpea Breeding and Management; Yadav, S.S., Reden, R.J., Chen, W., Sharma, B., Eds.; Centre for Agriculture and Bioscience International (CABI): Wallingford, UK, 2007. [Google Scholar]
- Gaur, P.M.; Jukanti, A.K.; Varshney, R.K. Impact of genomic technologies on chickpea breeding strategies. Agronomy 2012, 2, 199–221. [Google Scholar] [CrossRef]
- Millán, T.; Madrid, E.; Cubero, J.I.; Amri, M.; Castro, P.; Rubio, J. Chickpea. In Grain Legumes; De Ron, A.M., Ed.; Springer: New York, NY, USA, 2015; pp. 85–109. [Google Scholar]
- Castro, P.; Pistón, F.; Madrid, E.; Millán, T.; Gil, J.; Rubio, J. Development of chickpea near-isogenic lines for fusarium wilt. Theor. Appl. Genet. 2010, 121, 1519–1526. [Google Scholar] [CrossRef]
- Jiang, G.-L. Molecular markers and marker-assisted breeding in plants. In Plant Breeding from Laboratories to Fields; InTech: London, UK, 2013. [Google Scholar]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Vleeshouwers, V.G.A.A.; Oliver, R.P. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 2014, 27, 196–206. [Google Scholar]
- Houterman, P.M.; Ma, L.; van Ooijen, G.; de Vroomen, M.J.; Cornelissen, B.J.C.; Takken, F.L.W.; Rep, M. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009, 58, 970–978. [Google Scholar] [CrossRef]
- Kankanala, P.; Nandety, R.S.; Mysore, K.S. Genomics of plant disease resistance in legumes. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Curtin, S.J. Editing the Medicago truncatula genome: Targeted mutagenesis using the CRISPR-Cas9 reagent. In Functional Genomics in Medicago Truncatula. Methods in Molecular Biology; Cañas, L., Beltrán, J., Eds.; Humana Press Inc.: Totowa, NJ, USA, 2018; Volume 1822, pp. 161–174. [Google Scholar]
- Curtin, S.J.; Xiong, Y.; Michno, J.M.; Campbell, B.W.; Stec, A.O.; Čermák, T.; Starker, C.; Voytas, D.F.; Eamens, A.L.; Stupar, R.M. CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 2018, 16, 1125–1137. [Google Scholar] [CrossRef]
- Ji, J.; Zhang, C.; Sun, Z.; Wang, L.; Duanmu, D.; Fan, Q. Genome editing in cowpea Vigna unguiculata using CRISPR-Cas9. Int. J. Mol. Sci. 2019, 20, 2471. [Google Scholar] [CrossRef]
- Almeida, N.F.; Leitão, S.T.; Krezdorn, N.; Rotter, B.; Winter, P.; Rubiales, D.; Vaz Patto, M.C. Allelic diversity in the transcriptomes of contrasting rust-infected genotypes of Lathyrus sativus, a lasting resource for smart breeding. BMC Plant Biol. 2014, 14, 376. [Google Scholar] [CrossRef]
- Vaz Patto, M.C.; Fernandez-Aparicio, M.; Moral, A.; Rubiales, D. Characterization of resistance to powdery mildew (Erysiphe pisi) in a germplasm collection of Lathyrus sativus. Plant Breed. 2006, 125, 308–310. [Google Scholar] [CrossRef]
- Lambein, F.; Travella, S.; Kuo, Y.-H.; Van Montagu, M.; Heijde, M. Grass pea (Lathyrus sativus L.): Orphan crop, nutraceutical or just plain food? Planta 2019, 250, 821–838. [Google Scholar] [CrossRef]
| Legume Species | Fo f. sp. | Race | Reference |
|---|---|---|---|
| Pea | pisi | 1,2,5,6 | [37,38] |
| Chickpea | ciceris | 0,1A,1B/C,2,3,4,5,6 | [39,40,41] |
| Common bean | phaseoli | 1,2,3,4,5,6,7 | [23] |
| Lentil | lentis | 1,2,3,4,5,6,7,8 | [42,43,44] |
| Cowpea | tracheiphilum | 1,2,3,4 | [45,46] |
| Lupin | lupini | 1,2,3 | [34,35] |
| Soybean | glycines | - | [47] |
| Barrel medic Alfalfa | medicaginis | - | [48] |
| Pigeon pea | udum | - | [49] |
| Red clover | - | - | [36] |
| Faba bean | fabae | [50] | |
| Birdsfoot trefoil | loti | - | [51] |
| Environment | Disease Assessment | Species and Reference |
|---|---|---|
| Field | Disease incidence (percentage of dead plants) | Chickpea [98,99,100,101]; Lentil [89,90,102] |
| Scale based on percentage of leaves showing symptoms | Chickpea [24,97,103] Lentil [104] | |
| Scale based on % of xylem discoloration | Alfalfa [105] | |
| Controlled conditions | Disease incidence (percentage of dead plants) | Pea [106,107]; Lentil [44,108] |
| Percentage infected leaves | Pea [109]; Barrel medic [110] | |
| Scale based on percentage of leaves showing symptoms | Chickpea [91,97]; Lentil [102,111]; Common bean [112,113]; Barrel medic [114] | |
| Visual scale at leaf level | Pea [109,115,116,117]; Chickpea [118]; Lentil [43,89]; Common bean [119]; Barrel medic [26]; Birdsfoot trefoil [120] | |
| Scale for root symptoms | Pea [121] | |
| Scale based on percentage xylem discoloration | Common bean [113]; Cowpea [32]; Alfalfa [105]; Lupin [122]; Red clover [36] | |
| Infra-red imaging (plant temperature) | Pea [123] |
| Screening Method | Species and Reference |
|---|---|
| Light microscopy | Pea [75,109,125,126]; Lentil [127]; Chickpea [128,129] |
| Scanning electron microscopy (SEM) | Common bean [131,132]; Chickpea [128,129] |
| Transmission electron microscopy (TEM) | Pea [75,124]; Common bean [132] |
| Fluorescence microscopy | Barrel medic [26] |
| Laser confocal microscopy | Chickpea [129,133,134,135]; Common bean [136]; Barrel medic [26] |
| Species | Fo f. sp. | Linkage Group/Chromosome | Resistance Genes/QTLs | Markers Linked to the Resistance Genes/QTLs (3) |
|---|---|---|---|---|
| Pea | pisi race 1 | LG3 (chr5 (1)) [150,151] | Fw [150,151] | ACG:CAT_222 (1.4) [151]; Fw_Trap_480 (1.2), Fw_Trap_340 (1.2) and Fw_Trap_220 (1.2) [168] |
| pisi race 2 | LG4 (chr4 (1)) [107] | Fnw4.1; | AC22_185 and AD171_197 [107]; | |
| LG3 (chr5 (1)) [107] | Fnw3.1; | AB70_203 and AD180_60 [107] | ||
| LG3 (chr5 (1)) [107] | Fnw3.2 [107] | - | ||
| pisi race 5 | LG2 (chr6 (1)) [152,153] | Fwf [152,153] | U693a (5.6) [153] | |
| Chickpea | ciceris race 0 | LG5 [169] | foc-01/Foc-01(2) [157]; | OPJ20600 (3.0) and TR59 (2.0) [157,169]; H2I20, CaGM20820, CaGM20889 and TS43 [170]; |
| LG2 [158] | foc-02/Foc 02(2) [158] | TA59 and TS47 [158] | ||
| ciceris race 1A | LG2 [158] | foc-1 (syn. H1) [159,160,161,162]; | H3A12 (3.9) and TA110 (2.1) [171]; TA59 (4.4), TA96 (4.9), TA27 (4.9) and CS27A (4.9) [172]; TR19, TA194 and TA660 [173] | |
| - | h2, H3 [159,160,161,162] | - | ||
| ciceris race 2 | LG2 [158] | foc-2 [146] | TA96 (1.5), TA27 (1.5), TR19 (4.9) and CS27A (1.5) [172]; H3A12 (2.7) [171]; TA110 and TA37 [174] | |
| ciceris race 3 | LG2 [158] | foc-3/Foc-3(2) [146,147] | TA59 (0.5), TA96 (0.5), TA27 (0.5) and CS27A (0.5) [147,172]; H1B06y (0.2) and TA194 (0.7) [171] | |
| ciceris race 4 | LG2 [158] | foc-4 [146]; | TA59 (3.8), TA96 (3.3), TA27 (3.3) and TR19 (3.1) [172]; CS27 (3.7) [175]; R-2609-1 (2.0) and OP-U17-1 (4.1) [176] | |
| - | Two recessive genes [163] | - | ||
| ciceris race 5 | LG2 [158] | foc-5/Foc-5(2) [146,149] | TA59 (2.4), TA96 (2.9), and CS27A (2.9) [172]; TA27 (2.9) [172,175]; SPA and PRP-RGA1 [177] | |
| Lentil | lentis | LG6 [156] | Fw [155]; | OP-KI5900 (10.8) [155]; SSR59-2B (8) and p17m30710 (3.5) [156] |
| - | Fuw1, Fuw2, Fuw3, Fuw4, Fuw5 [165] | - | ||
| Common bean | phaseoli race 1 | - | Fop1 [154] | - |
| phaseoli race 2 | - | Fop2 [154] | - | |
| phaseoli race 4 | LG10 [113] | - | U20.750 (1.0) [113] | |
| LG3 [113] | - | - | ||
| LG11 [113] | - | - | ||
| phaseoli race 6 | chr4 [119] | Phvul.004G000800, .004G006800, ng; | DART03480 (0.0), SNP01469 (0.0) and SNP01487 (0.0) [119]; | |
| chr5 [119] | .005G043100, ng; | DART04561 (0.0), SNP02051 (0.0) [119]; | ||
| chr7 [119] | ng, .007G270000 and .007G269900, 007G270500; | SNP03304 (0.0), SNP03305 (0.0) and SNP03306 (0.0) [119]; | ||
| chr8 [119] | .008G196600 [119] | DART07926 (0.0) [119] | ||
| Alfalfa | medicaginis | - | FW1, FW2 [164] | - |
| Fo f. sp. | Effector Genes |
|---|---|
| pisi race 1 | SIX7, SIX10, SIX11, SIX12 and SIX14 [187] |
| pisi race 2 | SIX13, SIX14 and CRX2 [187] |
| pisi race 5 | SIX13 [187] |
| ciceris race 1, 2 and 4 | Fgb1 (G protein subunit), Gas1 (glucanosyltransferase), chs7 (chitin synthase chaperonin) and Fow1 (mitochondrial carrier protein) [182] |
| phaseoli race 4 | SIX6, SIX8 and SIX11 [187] |
| phaseoli race 6 | SIX1 and SIX6 [136]; FTF1 [188,189] and FTF2 [189] |
| medicaginis | SIX1 [20], SIX8, SIX9 and SIX13 [20,190]; secreted proteins encoding a lysin motif implicated in chitin binding and proteases and peptidases involved in oxidative stress [190] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampaio, A.M.; Araújo, S.d.S.; Rubiales, D.; Vaz Patto, M.C. Fusarium Wilt Management in Legume Crops. Agronomy 2020, 10, 1073. https://doi.org/10.3390/agronomy10081073
Sampaio AM, Araújo SdS, Rubiales D, Vaz Patto MC. Fusarium Wilt Management in Legume Crops. Agronomy. 2020; 10(8):1073. https://doi.org/10.3390/agronomy10081073
Chicago/Turabian StyleSampaio, Ana Margarida, Susana de Sousa Araújo, Diego Rubiales, and Maria Carlota Vaz Patto. 2020. "Fusarium Wilt Management in Legume Crops" Agronomy 10, no. 8: 1073. https://doi.org/10.3390/agronomy10081073
APA StyleSampaio, A. M., Araújo, S. d. S., Rubiales, D., & Vaz Patto, M. C. (2020). Fusarium Wilt Management in Legume Crops. Agronomy, 10(8), 1073. https://doi.org/10.3390/agronomy10081073







