Kernel-Based Regularized EEGNet Using Centered Alignment and Gaussian Connectivity for Motor Imagery Discrimination
Abstract
1. Introduction
2. Materials and Methods
2.1. Centered Kernel Alignment Fundamentals
2.2. Gaussian Functional Connectivity from EEG Records
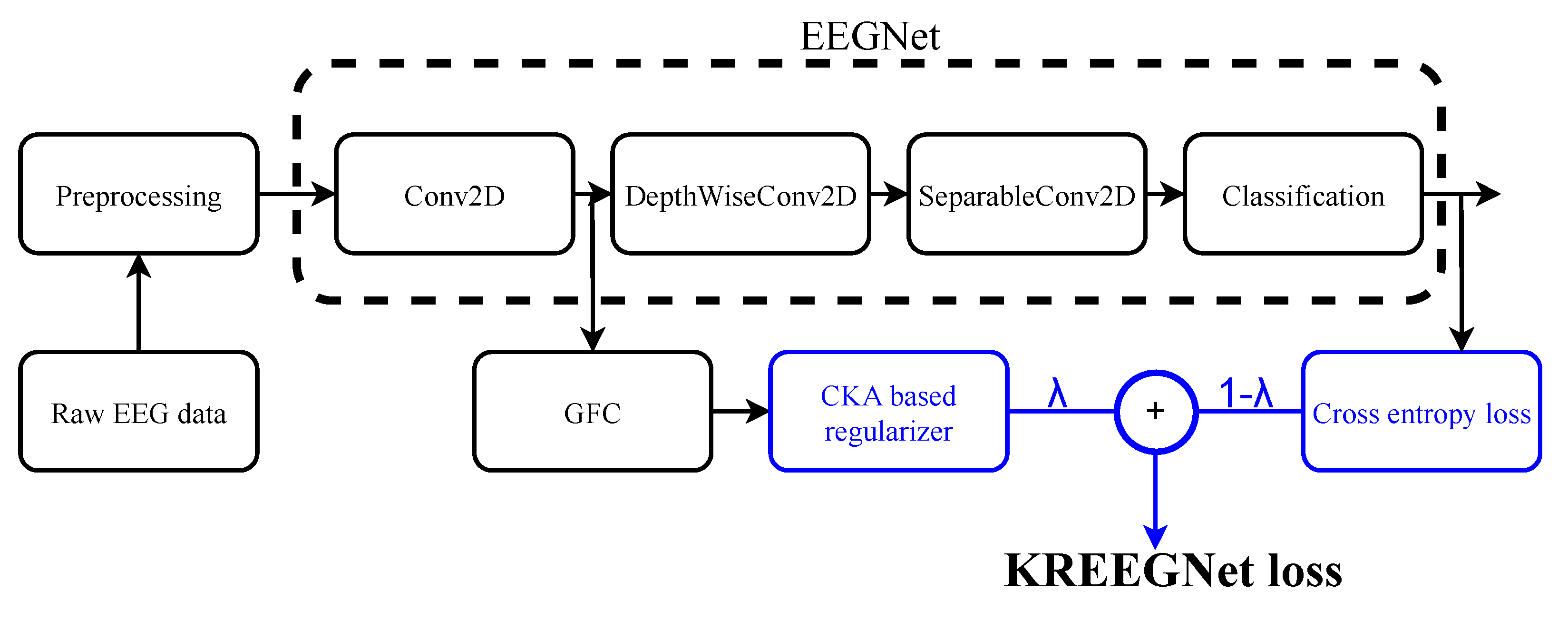
2.3. KREEGNet: Kernel-Based Regularized EEG Network
- -
- is a convolutional layer holding filters, a batch normalization, and a linear activation.
- -
- is a depthwise convolutional layer holding ELU activation ( gathers the number of spatial filters), followed by an average pooling and a dropout operation.
- -
- is a separable convolutional layer with ELU activation ( is the number of pointwise filters), setting a batch normalization, an average pooling, and a dropout.
- -
- is a fully connected classification layer fixing a flatten operation and a softmax activation.
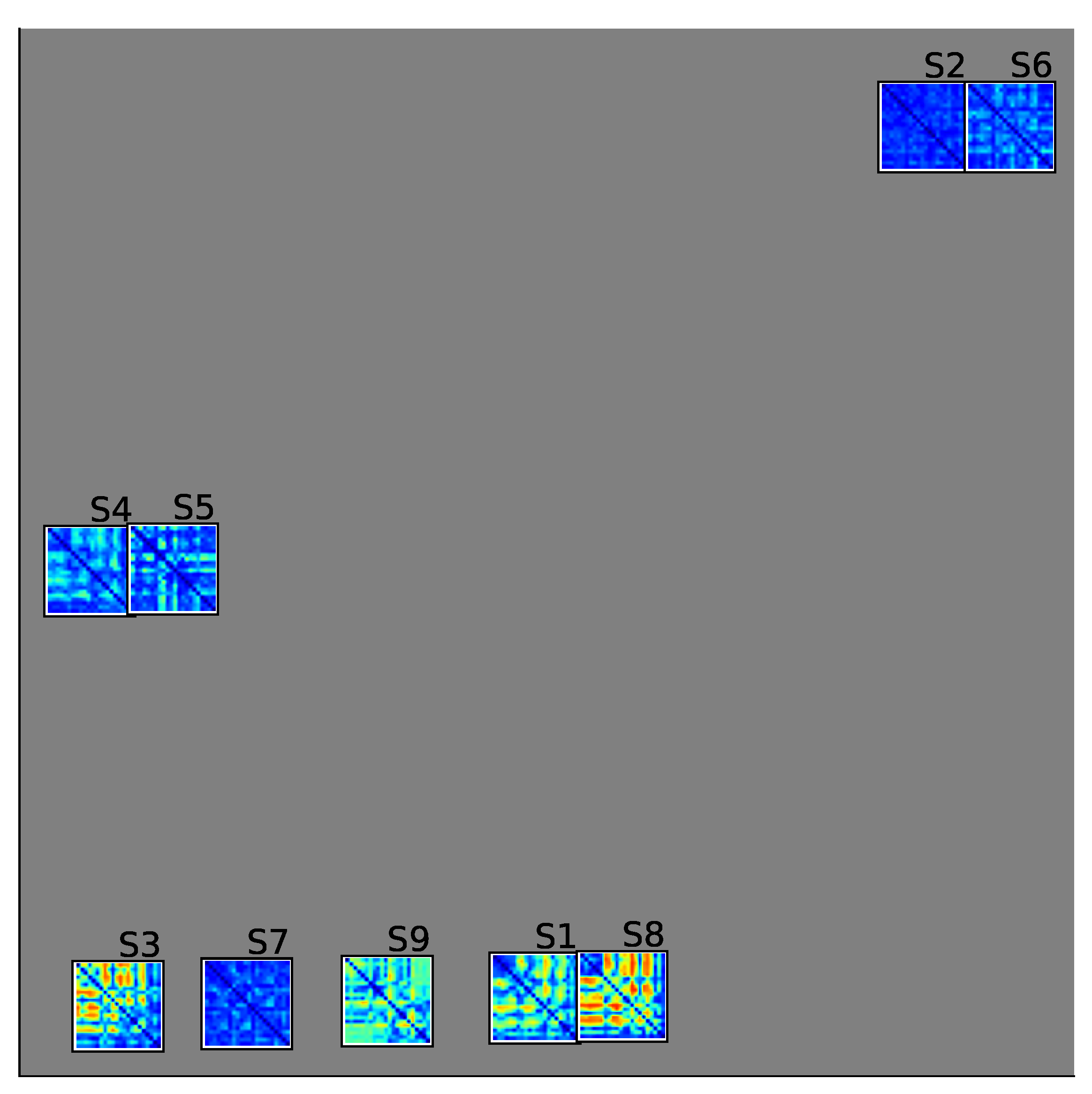
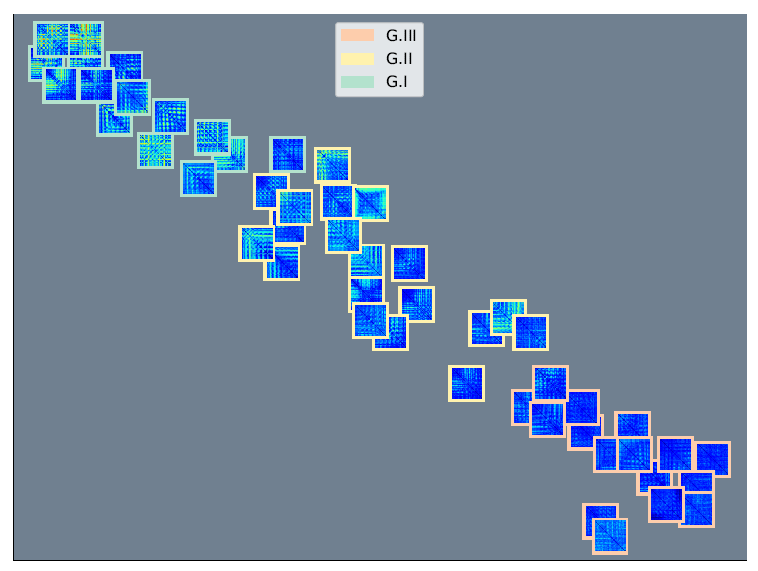
2.4. Group Analysis from EEGNet and KREEGNet Performance
3. Experimental Setup
3.1. Dataset Description
3.2. KREEGNet Training Details and Assessment
3.3. Method Comparison
4. Results and Discussion
4.1. Baseline EEGNet vs. KREEGNet: Subject and Group-Level Results
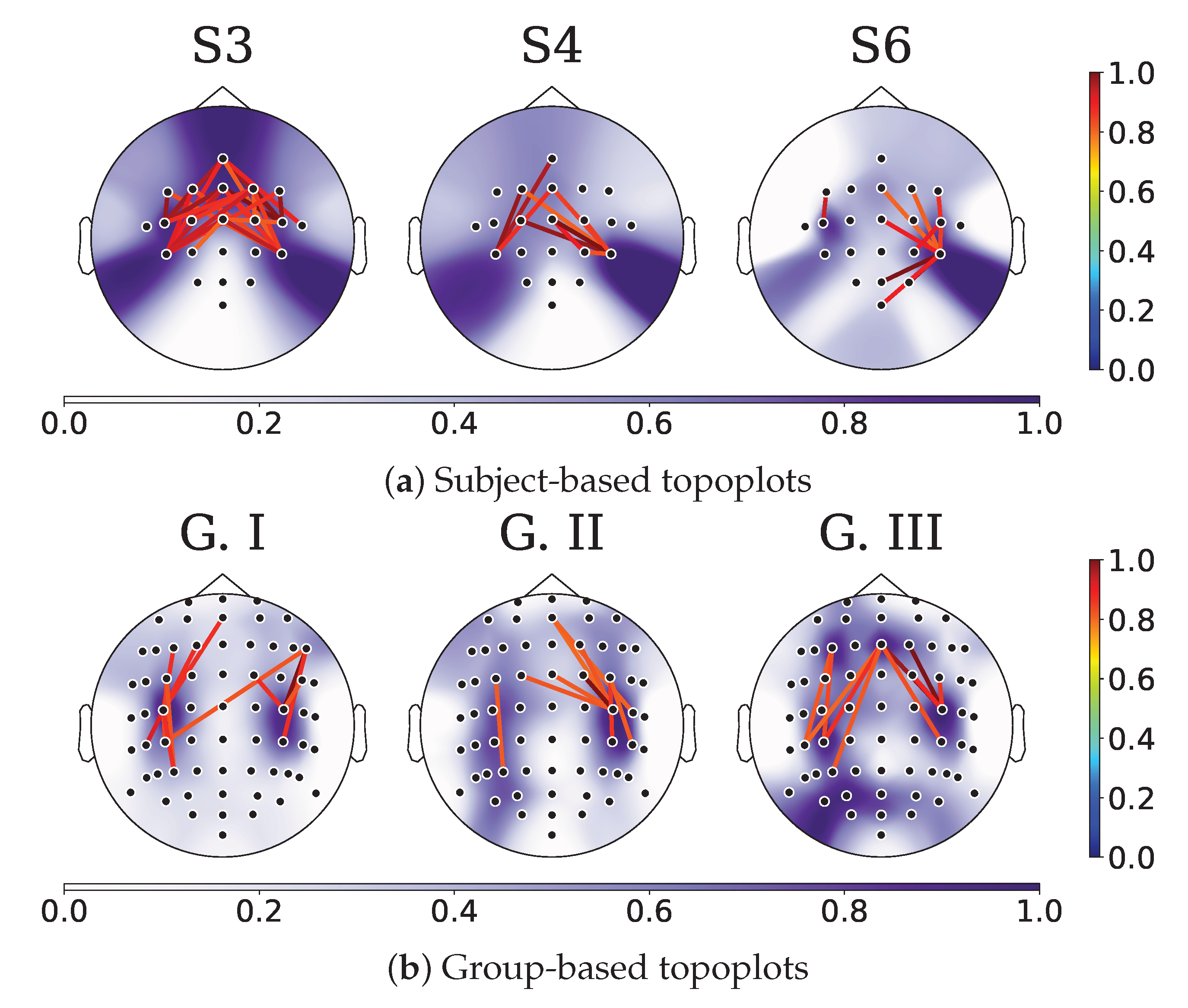
4.2. Relevance Analysis Results
- -
- We categorized each connection’s trials for an individual based on the label, forming the right and left sample sets.
- -
- Following this, we calculated the KS statistic for the connectivity between each pair of EEG channels along the training set trials. A KS value nearing 1 signifies a high level of distinguishability for the connectivity between two channels, whereas a value approaching 0 suggests a low level of separability. Here, the two-sample KS test compares the underlying distributions of two independent samples regarding the MI classes.
- -
- Moreover, we utilized the maximum operator across the estimated feature maps to establish a KS statistic matrix. This matrix denotes the class separability of each connectivity.
- -
- In order to illustrate the variations in each KS statistic matrix across subjects and groups, we depicted each matrix of KS statistic values on a two-dimensional scatter representation. Both dimensions were calculated employing the widely accepted t-SNE algorithm [66].
- -
- Lastly, to fully comprehend the key connectivities and channels involved in the MI classification, we used topoplots from the KS statistic matrix.
4.3. Method Comparison Results: Binary and Multiclass MI Classification
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Venkatachalam, K.; Devipriya, A.; Maniraj, J.; Sivaram, M.; Ambikapathy, A.; Iraj, S.A. A Novel Method of motor imagery classification using eeg signal. Artif. Intell. Med. 2020, 103, 101787. [Google Scholar]
- Dai, G.; Zhou, J.; Huang, J.; Wang, N. HS-CNN: A CNN with hybrid convolution scale for EEG motor imagery classification. J. Neural Eng. 2020, 17, 016025. [Google Scholar] [CrossRef] [PubMed]
- Gaur, P.; McCreadie, K.; Pachori, R.B.; Wang, H.; Prasad, G. An automatic subject specific channel selection method for enhancing motor imagery classification in EEG-BCI using correlation. Biomed. Signal Process. Control 2021, 68, 102574. [Google Scholar] [CrossRef]
- Khan, M.A.; Das, R.; Iversen, H.K.; Puthusserypady, S. Review on motor imagery based BCI systems for upper limb post-stroke neurorehabilitation: From designing to application. Comput. Biol. Med. 2020, 123, 103843. [Google Scholar] [CrossRef] [PubMed]
- Kanna, R.K.; Vasuki, R. Classification of Brain Signals Using Classifiers for Automated Wheelchair Application. Int. J. Mod. Agric. 2021, 10, 2426–2431. [Google Scholar]
- Miao, M.; Hu, W.; Yin, H.; Zhang, K. Spatial-frequency feature learning and classification of motor imagery EEG based on deep convolution neural network. Comput. Math. Methods Med. 2020, 2020, 1981728. [Google Scholar] [CrossRef]
- Padfield, N.; Zabalza, J.; Zhao, H.; Masero, V.; Ren, J. EEG-based brain-computer interfaces using motor-imagery: Techniques and challenges. Sensors 2019, 19, 1423. [Google Scholar] [CrossRef]
- Kaur, J.; Kaur, A. A review on analysis of EEG signals. In Proceedings of the 2015 International Conference on Advances in Computer Engineering and Applications, Ghaziabad, India, 19–20 March 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 957–960. [Google Scholar]
- Ghosh, P.; Mazumder, A.; Bhattacharyya, S.; Tibarewala, D.N.; Hayashibe, M. Functional connectivity analysis of motor imagery EEG signal for brain-computer interfacing application. In Proceedings of the 2015 7th International IEEE/EMBS Conference on Neural Engineering (NER), Montpellier, France, 22–24 April 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 210–213. [Google Scholar]
- Han, C.H.; Kim, Y.W.; Kim, D.Y.; Kim, S.H.; Nenadic, Z.; Im, C.H. Electroencephalography-based endogenous brain–computer interface for online communication with a completely locked-in patient. J. Neuroeng. Rehabil. 2019, 16, 18. [Google Scholar] [CrossRef]
- Collazos-Huertas, D.F.; Álvarez-Meza, A.M.; Castellanos-Dominguez, G. Spatial interpretability of time-frequency relevance optimized in motor imagery discrimination using Deep&Wide networks. Biomed. Signal Process. Control 2021, 68, 102626. [Google Scholar]
- Saha, S.; Baumert, M. Intra-and inter-subject variability in EEG-based sensorimotor brain computer interface: A review. Front. Comput. Neurosci. 2020, 13, 87. [Google Scholar] [CrossRef]
- Pérez-Velasco, S.; Santamaria-Vazquez, E.; Martinez-Cagigal, V.; Marcos-Martinez, D.; Hornero, R. EEGSym: Overcoming Inter-Subject Variability in Motor Imagery Based BCIs With Deep Learning. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 30, 1766–1775. [Google Scholar] [CrossRef]
- Seghier, M.L.; Price, C.J. Interpreting and utilising intersubject variability in brain function. Trends Cogn. Sci. 2018, 22, 517–530. [Google Scholar] [CrossRef]
- Sannelli, C.; Vidaurre, C.; Müller, K.R.; Blankertz, B. A large scale screening study with a SMR-based BCI: Categorization of BCI users and differences in their SMR activity. PLoS ONE 2019, 14, e0207351. [Google Scholar] [CrossRef]
- Vidaurre, C.; Blankertz, B. Towards a cure for BCI illiteracy. Brain Topogr. 2010, 23, 194–198. [Google Scholar] [CrossRef]
- Caicedo-Acosta, J.; Castaño, G.A.; Acosta-Medina, C.; Alvarez-Meza, A.; Castellanos-Dominguez, G. Deep Neural Regression Prediction of Motor Imagery Skills Using EEG Functional Connectivity Indicators. Sensors 2021, 21, 1932. [Google Scholar] [CrossRef]
- LK Jaya Shree, B. Automatic Detection of EEG as Biomarker using Deep Learning: A review. Ann. Rom. Soc. Cell Biol. 2021, 25, 6502–6511. [Google Scholar]
- Shoka, A.; Dessouky, M.; El-Sherbeny, A.; El-Sayed, A. Literature review on EEG preprocessing, feature extraction, and classifications techniques. Menoufia J. Electron. Eng. Res 2019, 28, 292–299. [Google Scholar] [CrossRef]
- Salami, A.; Andreu-Perez, J.; Gillmeister, H. EEG-ITNet: An explainable inception temporal convolutional network for motor imagery classification. IEEE Access 2022, 10, 36672–36685. [Google Scholar] [CrossRef]
- Somers, B.; Francart, T.; Bertrand, A. A generic EEG artifact removal algorithm based on the multi-channel Wiener filter. J. Neural Eng. 2018, 15, 036007. [Google Scholar] [CrossRef]
- Kwon, M.; Han, S.; Kim, K.; Jun, S.C. Super-resolution for improving EEG spatial resolution using deep convolutional neural network—Feasibility study. Sensors 2019, 19, 5317. [Google Scholar] [CrossRef]
- Kotte, S.; Dabbakuti, J.K. Methods for removal of artifacts from EEG signal: A review. Proc. J. Phys. Conf. Ser. 2020, 1706, 012093. [Google Scholar] [CrossRef]
- Singh, A.; Hussain, A.A.; Lal, S.; Guesgen, H.W. A comprehensive review on critical issues and possible solutions of motor imagery based electroencephalography brain-computer interface. Sensors 2021, 21, 2173. [Google Scholar] [CrossRef] [PubMed]
- Stergiadis, C.; Kostaridou, V.D.; Klados, M.A. Which BSS method separates better the EEG Signals? A comparison of five different algorithms. Biomed. Signal Process. Control 2022, 72, 103292. [Google Scholar] [CrossRef]
- Rashid, M.; Sulaiman, N.; Majeed, A.P.P.A.; Musa, R.M.; Nasir, A.F.A.; Bari, B.S.; Khatun, S. Current status, challenges, and possible solutions of EEG-based brain-computer interface: A comprehensive review. Front. Neurorobot. 2020, 14, 25. [Google Scholar] [CrossRef]
- Uribe, L.F.S.; Stefano Filho, C.A.; de Oliveira, V.A.; da Silva Costa, T.B.; Rodrigues, P.G.; Soriano, D.C.; Boccato, L.; Castellano, G.; Attux, R. A correntropy-based classifier for motor imagery brain-computer interfaces. Biomed. Phys. Eng. Express 2019, 5, 065026. [Google Scholar] [CrossRef]
- Mridha, M.F.; Das, S.C.; Kabir, M.M.; Lima, A.A.; Islam, M.R.; Watanobe, Y. Brain-Computer Interface: Advancement and Challenges. Sensors 2021, 21, 5746. [Google Scholar] [CrossRef]
- Xu, B.; Zhang, L.; Song, A.; Wu, C.; Li, W.; Zhang, D.; Xu, G.; Li, H.; Zeng, H. Wavelet transform time-frequency image and convolutional network-based motor imagery EEG classification. IEEE Access 2018, 7, 6084–6093. [Google Scholar] [CrossRef]
- Tobón-Henao, M.; Álvarez-Meza, A.; Castellanos-Domínguez, G. Subject-dependent artifact removal for enhancing motor imagery classifier performance under poor skills. Sensors 2022, 22, 5771. [Google Scholar] [CrossRef]
- dos Santos, E.M.; Cassani, R.; Falk, T.H.; Fraga, F.J. Improved motor imagery brain-computer interface performance via adaptive modulation filtering and two-stage classification. Biomed. Signal Process. Control 2020, 57, 101812. [Google Scholar] [CrossRef]
- Rajabioun, M. Motor imagery classification by active source dynamics. Biomed. Signal Process. Control 2020, 61, 102028. [Google Scholar] [CrossRef]
- Taran, S.; Bajaj, V. Motor imagery tasks-based EEG signals classification using tunable-Q wavelet transform. Neural Comput. Appl. 2019, 31, 6925–6932. [Google Scholar] [CrossRef]
- Sadiq, M.T.; Yu, X.; Yuan, Z.; Fan, Z.; Rehman, A.U.; Li, G.; Xiao, G. Motor imagery EEG signals classification based on mode amplitude and frequency components using empirical wavelet transform. IEEE Access 2019, 7, 127678–127692. [Google Scholar] [CrossRef]
- Zhang, R.; Xiao, X.; Liu, Z.; Jiang, W.; Li, J.; Cao, Y.; Ren, J.; Jiang, D.; Cui, L. A new motor imagery EEG classification method FB-TRCSP+ RF based on CSP and random forest. IEEE Access 2018, 6, 44944–44950. [Google Scholar] [CrossRef]
- Hsu, W.Y. Improving classification accuracy of motor imagery EEG using genetic feature selection. Clin. EEG Neurosci. 2014, 45, 163–168. [Google Scholar] [CrossRef]
- Bashashati, A.; Fatourechi, M.; Ward, R.K.; Birch, G.E. A survey of signal processing algorithms in brain–computer interfaces based on electrical brain signals. J. Neural Eng. 2007, 4, R32. [Google Scholar] [CrossRef]
- Van Erp, J.; Lotte, F.; Tangermann, M. Brain-computer interfaces: Beyond medical applications. Computer 2012, 45, 26–34. [Google Scholar] [CrossRef]
- Altaheri, H.; Muhammad, G.; Alsulaiman, M.; Amin, S.U.; Altuwaijri, G.A.; Abdul, W.; Bencherif, M.A.; Faisal, M. Deep learning techniques for classification of electroencephalogram (EEG) motor imagery (MI) signals: A review. Neural Comput. Appl. 2021, 35, 14681–14722. [Google Scholar] [CrossRef]
- Sun, B.; Liu, Z.; Wu, Z.; Mu, C.; Li, T. Graph Convolution Neural Network based End-to-end Channel Selection and Classification for Motor Imagery Brain-computer Interfaces. IEEE Trans. Ind. Inform. 2022, 1–10. [Google Scholar] [CrossRef]
- Ma, Y.; Bian, D.; Xu, D.; Zou, W.; Wang, J.; Hu, N. A Spatio-Temporal Interactive Attention Network for Motor Imagery EEG Decoding. In Proceedings of the 2022 IEEE International Conference on Signal Processing, Communications and Computing (ICSPCC), Xi’an, China, 25–27 October 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–6. [Google Scholar]
- Song, Y.; Jia, X.; Yang, L.; Xie, L. Transformer-based spatial-temporal feature learning for EEG decoding. arXiv 2021, arXiv:2106.11170. [Google Scholar]
- He, Y.; Lu, Z.; Wang, J.; Shi, J. A channel attention based MLP-Mixer network for motor imagery decoding with EEG. In Proceedings of the ICASSP 2022-2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 23–27 May 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1291–1295. [Google Scholar]
- Song, Y.; Wang, D.; Yue, K.; Zheng, N.; Shen, Z.J.M. EEG-based motor imagery classification with deep multi-task learning. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–8. [Google Scholar]
- Berton, L.; Valverde-Rebaza, J.; de Andrade Lopes, A. Link prediction in graph construction for supervised and semi-supervised learning. In Proceedings of the 2015 International Joint Conference on Neural Networks (IJCNN), Killarney, Ireland, 12–17 July 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 1–8. [Google Scholar]
- Kong, Q.; Wu, Y.; Yuan, C.; Wang, Y. Ct-cad: Context-aware transformers for end-to-end chest abnormality detection on x-rays. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1385–1388. [Google Scholar]
- Yang, L.; Song, Y.; Ma, K.; Xie, L. Motor imagery EEG decoding method based on a discriminative feature learning strategy. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 368–379. [Google Scholar] [CrossRef]
- Phunruangsakao, C.; Achanccaray, D.; Hayashibe, M. Deep adversarial domain adaptation with few-shot learning for motor-imagery brain-computer interface. IEEE Access 2022, 10, 57255–57265. [Google Scholar] [CrossRef]
- Ganin, Y.; Ustinova, E.; Ajakan, H.; Germain, P.; Larochelle, H.; Laviolette, F.; Marchand, M.; Lempitsky, V. Domain-adversarial training of neural networks. J. Mach. Learn. Res. 2016, 17, 1–35. [Google Scholar]
- Halme, H.L.; Parkkonen, L. Across-subject offline decoding of motor imagery from MEG and EEG. Sci. Rep. 2018, 8, 10087. [Google Scholar] [CrossRef] [PubMed]
- Marquand, A.F.; Brammer, M.; Williams, S.C.; Doyle, O.M. Bayesian multi-task learning for decoding multi-subject neuroimaging data. NeuroImage 2014, 92, 298–311. [Google Scholar] [CrossRef]
- Roy, S.; Chowdhury, A.; McCreadie, K.; Prasad, G. Deep learning based inter-subject continuous decoding of motor imagery for practical brain-computer interfaces. Front. Neurosci. 2020, 14, 918. [Google Scholar] [CrossRef]
- Huang, Y.C.; Chang, J.R.; Chen, L.F.; Chen, Y.S. Deep neural network with attention mechanism for classification of motor imagery EEG. In Proceedings of the 2019 9th International IEEE/EMBS Conference on Neural Engineering (NER), San Francisco, CA, USA, 20–23 March 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1130–1133. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Cai, D.; He, X.; Han, J.; Huang, T.S. Graph regularized nonnegative matrix factorization for data representation. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 33, 1548–1560. [Google Scholar]
- Cortes, C.; Mohri, M.; Rostamizadeh, A. Algorithms for learning kernels based on centered alignment. J. Mach. Learn. Res. 2012, 13, 795–828. [Google Scholar]
- Alvarez-Meza, A.M.; Orozco-Gutierrez, A.; Castellanos-Dominguez, G. Kernel-based relevance analysis with enhanced interpretability for detection of brain activity patterns. Front. Neurosci. 2017, 11, 550. [Google Scholar] [CrossRef]
- Géron, A. Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2022. [Google Scholar]
- García-Murillo, D.G.; Álvarez-Meza, A.M.; Castellanos-Dominguez, C.G. KCS-FCnet: Kernel Cross-Spectral Functional Connectivity Network for EEG-Based Motor Imagery Classification. Diagnostics 2023, 13, 1122. [Google Scholar] [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain–computer interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Zhang, A.; Lipton, Z.C.; Li, M.; Smola, A.J. Dive into deep learning. arXiv 2021, arXiv:2106.11342. [Google Scholar]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef]
- Musallam, Y.K.; AlFassam, N.I.; Muhammad, G.; Amin, S.U.; Alsulaiman, M.; Abdul, W.; Altaheri, H.; Bencherif, M.A.; Algabri, M. Electroencephalography-based motor imagery classification using temporal convolutional network fusion. Biomed. Signal Process. Control 2021, 69, 102826. [Google Scholar] [CrossRef]
- Berger, V.W.; Zhou, Y. Kolmogorov–smirnov test: Overview. In Wiley Statsref: Statistics Reference Online; Wiley Online Library: Hoboken, NJ, USA, 2014. [Google Scholar]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Giraldo, L.G.S.; Rao, M.; Principe, J.C. Measures of entropy from data using infinitely divisible kernels. IEEE Trans. Inf. Theory 2014, 61, 535–548. [Google Scholar] [CrossRef]
- Hou, Y.; Jia, S.; Lun, X.; Hao, Z.; Shi, Y.; Li, Y.; Zeng, R.; Lv, J. GCNs-net: A graph convolutional neural network approach for decoding time-resolved eeg motor imagery signals. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–12. [Google Scholar] [CrossRef]
- De La Pava Panche, I.; Gómez-Orozco, V.; Álvarez-Meza, A.; Cárdenas-Peña, D.; Orozco-Gutiérrez, Á. Estimating Directed Phase-Amplitude Interactions from EEG Data through Kernel-Based Phase Transfer Entropy. Appl. Sci. 2021, 11, 9803. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, Q.; Wang, Q.; Gao, X.; Heng, P.A. Global Adaptive Transformer for Cross-Subject Enhanced EEG Classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2023, 31, 2767–2777. [Google Scholar] [CrossRef]



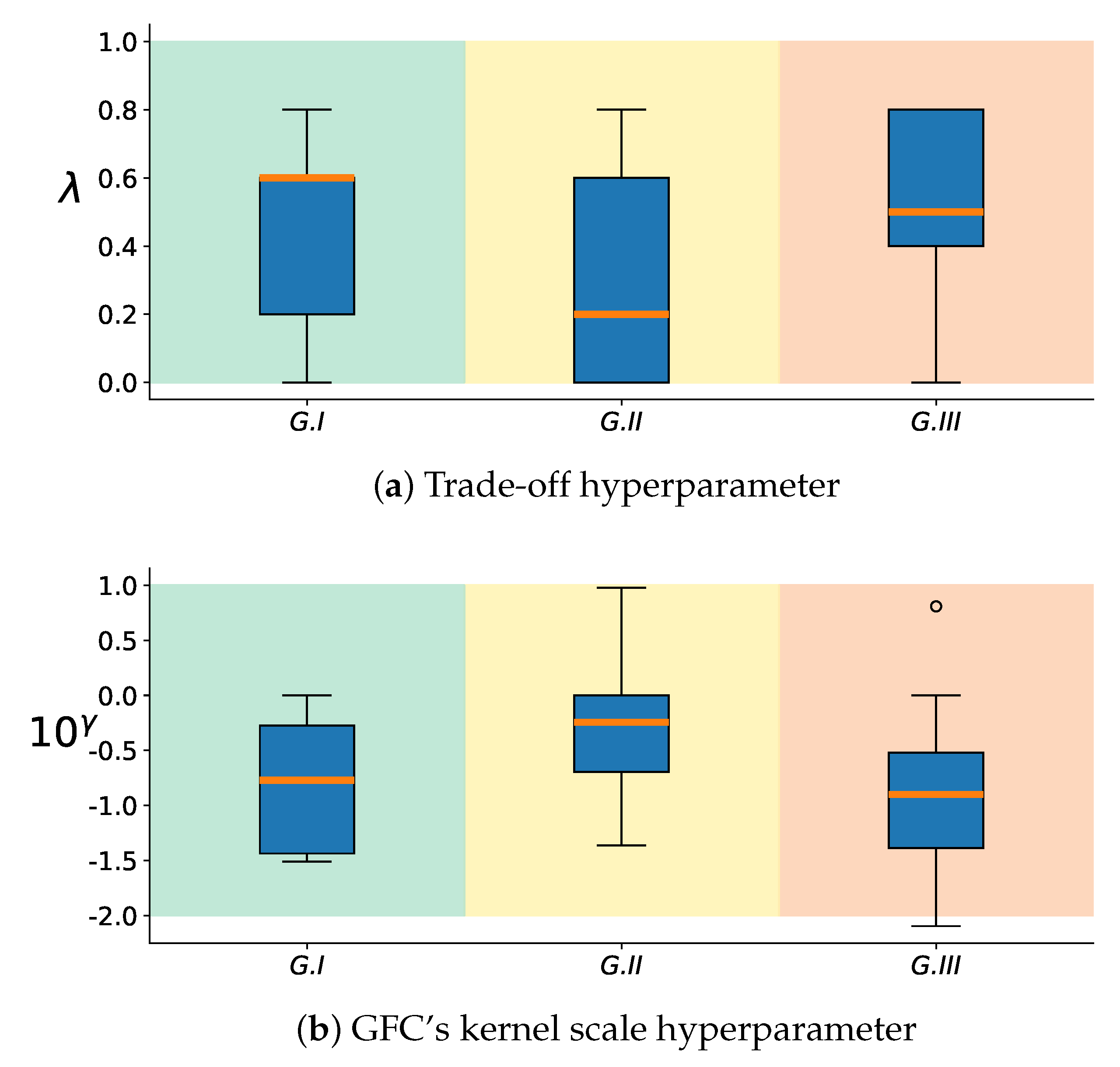

| Approach | Accuracy | Kappa | AUC |
|---|---|---|---|
| DeepConvNet [63] | |||
| ShallowConvNet [63] | |||
| EEGNet [61] | |||
| TCFussionnet [64] | |||
| KREEGNet (ours) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tobón-Henao, M.; Álvarez-Meza, A.M.; Castellanos-Dominguez, C.G. Kernel-Based Regularized EEGNet Using Centered Alignment and Gaussian Connectivity for Motor Imagery Discrimination. Computers 2023, 12, 145. https://doi.org/10.3390/computers12070145
Tobón-Henao M, Álvarez-Meza AM, Castellanos-Dominguez CG. Kernel-Based Regularized EEGNet Using Centered Alignment and Gaussian Connectivity for Motor Imagery Discrimination. Computers. 2023; 12(7):145. https://doi.org/10.3390/computers12070145
Chicago/Turabian StyleTobón-Henao, Mateo, Andrés Marino Álvarez-Meza, and Cesar German Castellanos-Dominguez. 2023. "Kernel-Based Regularized EEGNet Using Centered Alignment and Gaussian Connectivity for Motor Imagery Discrimination" Computers 12, no. 7: 145. https://doi.org/10.3390/computers12070145
APA StyleTobón-Henao, M., Álvarez-Meza, A. M., & Castellanos-Dominguez, C. G. (2023). Kernel-Based Regularized EEGNet Using Centered Alignment and Gaussian Connectivity for Motor Imagery Discrimination. Computers, 12(7), 145. https://doi.org/10.3390/computers12070145








