Integrative Bioinformatic Analysis of Transcriptomic Data Identifies Conserved Molecular Pathways Underlying Ionizing Radiation-Induced Bystander Effects (RIBE)
Abstract
:1. Introduction
2. Results
2.1. Statistical Inference and Differential Expression
2.2. Functional Enrichment Analysis
2.3. Rank Aggregation of Linker Genes
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
- Regarding the datasets GSE12435, GSE18760, GSE55869, GSE3201, and GSE21059, a method of the inner-outer dish was used, with the outer dish having a 6-micron Mylar strip base for the formation of the irradiated cells and the inner dish having one of 38-micron Mylar strips (which shields the cell from the IR) for the formation of the bystander cells [35,39].
- Regarding the dataset GSE25772, another experimental design was used, with the transference of conditioned medium from the irradiated cells to the “bystander” cells [37].
- Lastly, in the dataset GSE8993, micro beam and broad beam irradiation was used so as to form bystander and irradiated cells, respectively [40].
4.2. Computational Pipeline and Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Desouky, O.; Ding, N.; Zhou, G. Targeted and non-targeted effects of ionizing radiation. J. Radiat. Res. Appl. Sci. 2015, 8, 247–254. [Google Scholar] [CrossRef]
- Bray, F.N.; Simmons, B.J.; Wolfson, A.H.; Nouri, K. Acute and Chronic Cutaneous Reactions to Ionizing Radiation Therapy. Dermatol. Ther. (Heidelb). 2016, 6, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Yamamori, T.; Yasui, H.; Yamazumi, M.; Wada, Y.; Nakamura, Y.; Nakamura, H.; Inanami, O. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic. Biol. Med. 2012, 53, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Azzam, E.I.; Jay-Gerin, J.P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-J.; Randers-Pehrson, G.; Xu, A.; Waldren, C.A.; Geard, C.R.; Yu, Z.; Hei, T.K. Targeted cytoplasmic irradiation with alpha particles induces mutations in mammalian cells. Proc. Natl. Acad. Sci. USA 1999, 96, 4959–4964. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, H.; Little, J.B. Induction of sister chromatid exchanges by extremely low doses of α-particles. Cancer Res. 1992, 52, 6394–6396. [Google Scholar] [PubMed]
- Kaminaga, K.; Noguchi, M.; Narita, A.; Hattori, Y.; Usami, N.; Yokoya, A. Cell cycle tracking for irradiated and unirradiated bystander cells in a single colony with exposure to a soft X-ray microbeam. Int. J. Radiat. Biol. 2016, 92, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Nagasawa, H.; Little, J.B. HPRT mutants induced in bystander cells by very low fluences of alpha particles result primarily from point mutations. Radiat. Res. 2001, 156, 521–525. [Google Scholar] [CrossRef]
- Fournier, C.; Becker, D.; Winter, M.; Barberet, P.; Heiss, M.; Fischer, B.; Topsch, J.; Taucher-Scholz, G. Cell cycle-related bystander responses are not increased with LET after heavy-ion irradiation. Radiat. Res. 2007, 167, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Buonanno, M.; de Toledo, S.M.; Azzam, E.I. Increased frequency of spontaneous neoplastic transformation in progeny of bystander cells from cultures exposed to densely ionizing radiation. PLoS ONE 2011, 6, e21540. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Michael, D.; Alex, E.L. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Pilalis, E.; Valavanis, I.; Chatziioannou, A. Weblet Importer. Available online: https://bioinfominer.com/login (accessed on 10 October 2017).
- Blake, J.A.; Christie, K.R.; Dolan, M.E.; Drabkin, H.J.; Hill, D.P.; Ni, L.; Sitnikov, D.; Burgess, S.; Buza, T.; Gresham, C.; et al. Gene ontology consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.; Mundo, A.; Haw, R.; Milacic, M. The Reactome pathway knowledgebase. Nucleic Acids 2014, 42, D472–D477. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef] [PubMed]
- Blake, J.A.; Eppig, J.T.; Kadin, J.A.; Richardson, J.E.; Smith, C.L.; Bult, C.J.; Anagnostopoulos, A.; Baldarelli, R.M.; Beal, J.S.; Bello, S.M.; et al. Mouse Genome Database (MGD)-2017: Community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017, 45, D723–D729. [Google Scholar] [CrossRef] [PubMed]
- Bult, C.J.; Krupke, D.M.; Begley, D.A.; Richardson, J.E.; Neuhauser, S.B.; Sundberg, J.P.; Eppig, J.T. Mouse Tumor Biology (MTB): A database of mouse models for human cancer. Nucleic Acids Res. 2015, 43, D818–D824. [Google Scholar] [CrossRef] [PubMed]
- Finger, J.H.; Smith, C.M.; Hayamizu, T.F.; McCright, I.J.; Eppig, J.T.; Kadin, J.A.; Richardson, J.E.; Ringwald, M. The mouse Gene Expression Database (GXD): 2011 update. Nucleic Acids Res. 2011, 39, D835–D841. [Google Scholar] [CrossRef] [PubMed]
- Piccioli, P.; Rubartelli, A. The secretion of IL-1β and options for release. Semin. Immunol. 2013, 25, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, J.; De Ley, M.; Opdenakker, G.; Billiau, A.; De Sommer, P. Homogeneous interferon-inducing 22K factor is related to endogenous pyrogen and interleukin-1. Nature 1985, 314, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Andrei, C.; Margiocco, P.; Poggi, A.; Lotti, L.V.; Torrisi, M.R.; Rubartelli, A. Phospholipases C and A2 control lysosome-mediated IL-1 β secretion: Implications for inflammatory processes. Proc. Natl. Acad. Sci. USA 2004, 101, 9745–9750. [Google Scholar] [CrossRef] [PubMed]
- Rouault, C.; Pellegrinelli, V.; Schilch, R.; Cotillard, A.; Poitou, C.; Tordjman, J.; Sell, H.; Clément, K.; Lacasa, D. Roles of chemokine ligand-2 (CXCL2) and neutrophils in influencing endothelial cell function and inflammation of human adipose tissue. Endocrinology 2013, 154, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, S.; Reyes-Aldasoro, C.C.; Candel, S.; Renshaw, S.A.; Mulero, V.; Calado, A. Cxcl8 (IL-8) Mediates Neutrophil Recruitment and Behavior in the Zebrafish Inflammatory Response. J. Immunol. 2013, 190, 4349–4359. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Tran, V.; Nishikawa, K.; Kaneda, T.; Hamada, Y.; Kawaguchi, N.; Fujita, M.; Takada, Y.K.; Matsuura, N.; Zhao, M.; Takada, Y. A Dominant-Negative FGF1 Mutant (the R50E Mutant) Suppresses Tumorigenesis and Angiogenesis. PLoS ONE 2013, 8, e57927. [Google Scholar] [CrossRef] [PubMed]
- Decker, C.G.; Wang, Y.; Paluck, S.J.; Shen, L.; Loo, J.A.; Levine, A.J.; Miller, L.S.; Maynard, H.D. Fibroblast growth factor 2 dimer with superagonist in vitro activity improves granulation tissue formation during wound healing. Biomaterials 2016, 81, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.F. Inducible Nitric Oxide Synthase Binds, S-Nitrosylates, and Activates Cyclooxygenase-2. Science 2005, 310, 1966–1970. [Google Scholar] [CrossRef] [PubMed]
- Goodman, J.E.; Bowman, E.D.; Chanock, S.J.; Alberg, A.J.; Harris, C.C. Arachidonate lipoxygenase (ALOX) and cyclooxygenase (COX) polymorphisms and colon cancer risk. Carcinogenesis 2004, 25, 2467–2472. [Google Scholar] [CrossRef] [PubMed]
- Düwel, M.; Welteke, V.; Oeckinghaus, A.; Baens, M.; Kloo, B.; Ferch, U.; Darnay, B.G.; Ruland, J.; Marynen, P.; Krappmann, D. A20 Negatively Regulates T Cell Receptor Signaling to NF-κB by Cleaving Malt1 Ubiquitin Chains. J. Immunol. 2009, 182, 7718–7728. [Google Scholar] [CrossRef] [PubMed]
- Opipari, A.W.; Boguski, M.S.; Dixit, V.M. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J. Biol. Chem. 1990, 265, 14705–14708. [Google Scholar] [PubMed]
- Eto, A.; Muta, T.; Yamazaki, S.; Takeshige, K. Essential roles for NF-κB and a Toll/IL-1 receptor domain-specific signal(s) in the induction of IκB-ζ. Biochem. Biophys. Res. Commun. 2003, 301, 495–501. [Google Scholar] [CrossRef]
- Totzke, G.; Essmann, F.; Pohlmann, S.; Lindenblatt, C.; Jänicke, R.U.; Schulze-Osthoff, K. A novel member of the IκB family, human IκB-ζ, inhibits transactivation of p65 and its DNA binding. J. Biol. Chem. 2006, 281, 12645–12654. [Google Scholar] [CrossRef] [PubMed]
- Ghandhi, S.A.; Ponnaiya, B.; Panigrahi, S.K.; Hopkins, K.M.; Cui, Q.; Hei, T.K.; Amundson, S.A.; Lieberman, H.B. RAD9 deficiency enhances radiation induced bystander DNA damage and transcriptomal response. Radiat. Oncol. 2014, 9, 206. [Google Scholar] [CrossRef] [PubMed]
- Kalanxhi, E.; Dahle, J. Transcriptional responses in irradiated and bystander fibroblasts after low dose α-particle radiation. Int. J. Radiat. Biol. 2012, 88, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Ghandhi, S.A.; Sinha, A.; Markatou, M.; Amundson, S.A. Time-series clustering of gene expression in irradiated and bystander fibroblasts: An application of FBPA clustering. BMC Genomics 2011, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Kalanxhi, E.; Dahle, J. Genome-Wide Microarray Analysis of Human Fibroblasts in Response to γ Radiation and the Radiation-Induced Bystander Effect. Radiat. Res. 2012, 177, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Ghandhi, S.A.; Ming, L.; Ivanov, V.N.; Hei, T.K.; Amundson, S.A. Regulation of early signaling and gene expression in the alpha-particle and bystander response of IMR-90 human fibroblasts. BMC Med. Genomics 2010, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Ghandhi, S.A.; Yaghoubian, B.; Amundson, S.A. Global gene expression analyses of bystander and alpha particle irradiated normal human lung fibroblasts: Synchronous and differential responses. BMC Med. Genomics 2008, 1, 63. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, M.; Hamada, N.; Imadome, K.; Funayama, T.; Sakashita, T.; Kobayashi, Y.; Imai, T. Expression profiles are different in carbon ion-irradiated normal human fibroblasts and their bystander cells. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2008, 642, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Goehe, R.W.; Di, X.; Hicks, M.A.; Torti, S.V.; Torti, F.M.; Harada, H.; Gewirtz, D.A. A novel cytostatic form of autophagy in sensitization of non-small cell lung cancer cells to radiation by vitamin D and the vitamin D analog, EB 1089. Autophagy 2014, 10, 2346–2361. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Wang, S.-J.; Li, D.; Chu, B.; Gu, W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. USA 2016, 113, E6806–E6812. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mandal, A.; Park, M.H. Depletion of the polyamines spermidine and spermine by overexpression of spermidine/spermine N1-acetyltransferase 1 (SAT1) leads to mitochondria-mediated apoptosis in mammalian cells. Biochem. J. 2015, 468, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Spermidine/spermine-N 1-acetyltransferase: A key metabolic regulator. Am. J. Physiol. Endocrinol. Metab. 2008, 294, E995–E1010. [Google Scholar] [CrossRef] [PubMed]
- Beenken, A.; Mohammadi, M. The FGF family: Biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 2009, 8, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Nikitaki, Z.; Mavragani, I.V.; Laskaratou, D.A.; Gika, V.; Moskvin, V.P.; Theofilatos, K.; Vougas, K.; Stewart, R.D.; Georgakilas, A.G. Systemic mechanisms and effects of ionizing radiation: A new “old” paradigm of how the bystanders and distant can become the players. Semin. Cancer Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kavalali, E.T.; Nelson, E.D.; Monteggia, L.M. Role of MeCP2, DNA methylation, and HDACs in regulating synapse function. J. Neurodev. Disord. 2011, 3, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Bektas, M.; Allende, M.L.; Lee, B.G.; Chen, W.; Amar, M.J.; Remaley, A.T.; Saba, J.D.; Proia, R.L. Sphingosine 1-Phosphate Lyase Deficiency Disrupts Lipid Homeostasis in Liver. J. Biol. Chem. 2010, 285, 10880–10889. [Google Scholar] [CrossRef] [PubMed]
- Heckmann, B.L.; Zhang, X.; Xie, X.; Liu, J. The G0/G1 switch gene 2 (G0S2): Regulating metabolism and beyond. Biochim. Biophys. Acta 2013, 1831, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Pajonk, F.; McBride, W.H. Ionizing radiation affects 26s proteasome function and associated molecular responses, even at low doses. Radiother. Oncol. 2001, 59, 203–212. [Google Scholar] [CrossRef]
- Rolfe, M.; Chiu, M.I.; Pagano, M. The ubiquitin-mediated proteolytic pathway as a therapeutic area. J. Mol. Med. 1997, 75, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Grune, T. Oxidative stress, aging and the proteasomal system. Biogerontology 2000, 1, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Grune, T.; Reinheckel, T.; Joshi, M.; Davies, K.J.A. Proteolysis in cultured liver epithelial cells during oxidative stress: Role of the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1995, 270, 2344–2351. [Google Scholar] [CrossRef] [PubMed]
- Dallaporta, B.; Pablo, M.; Maisse, C.; Daugas, E.; Loeffler, M.; Zamzami, N.; Kroemer, G. Proteasome activation as a critical event of thymocyte apoptosis. Cell Death Differ. 2000, 7, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Grimm, L.M.; Goldberg, A.L.; Poirier, G.G.; Schwartz, L.M.; Osborne, B.A. Proteasomes play an essential role in thymocyte apoptosis. EMBO J. 1996, 15, 3835–3844. [Google Scholar] [PubMed]
- Sean, D.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- The Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 6 February 2017).
- Bioconductor. Available online: https://www.bioconductor.org/ (accessed on 6 February 2017).
- Ritchie, M.E.; Silver, J.; Oshlack, A.; Holmes, M.; Diyagama, D.; Holloway, A.; Smyth, G.K. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007, 23, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Thorne, N. Normalization for Two-Color cDNA Microarray Data. Lect. Notes-Monograph Ser. 2003, 40, 403–418. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.K. limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V., Dudoit, S., Irizarry, R., Huber, W., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef] [PubMed]
- Illumina HumanHT12v3 Annotation Data (Chip IlluminaHumanv3), 2015. Available online: http://bioconductor.org/packages/illuminaHumanv3.db/ (accessed on 1 December 2016).
- Ritchie, M.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.; Shi, W.; Smyth, G. Bioconductor—Limma. Available online: https://bioconductor.org/packages/release/bioc/html/limma.html (accessed on 9 October 2017).
- Hochberg, Y.; Benjaminit, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Koutsandreas, T.; Pilalis, E.; Vlachavas, E.I.; Koczan, D.; Klippel, S.; Dimitrakopoulou-Strauss, A.; Valavanis, I.; Chatziioannou, A. Making sense of the biological complexity through the platform-driven unification of the analytical and visualization tasks. In Proceedings of the 2015 IEEE 15th International Conference on Bioinformatics and Bioengineering, BIBE, Belgrade, Serbia, 2–4 November 2015; pp. 1–6. [Google Scholar]
- Pihur, V.; Datta, S.; Datta, S. RankAggreg, an R package for weighted rank aggregation. BMC Bioinform. 2009, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn Diagrams. 2007–2015. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 1 December 2016).
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard, S.G.; Brouwer, C. Pathview Web: User friendly pathway visualization and data integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef] [PubMed]
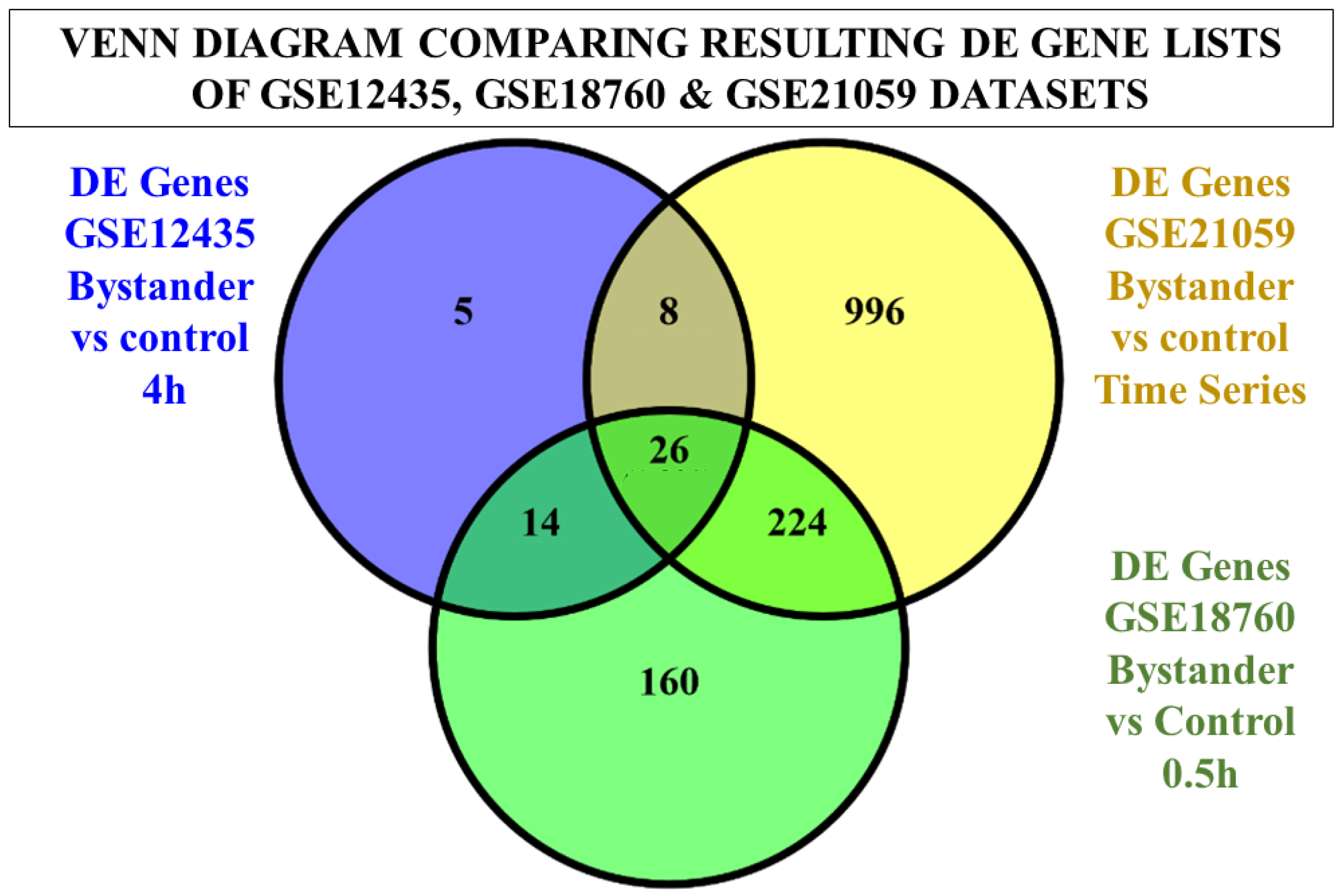
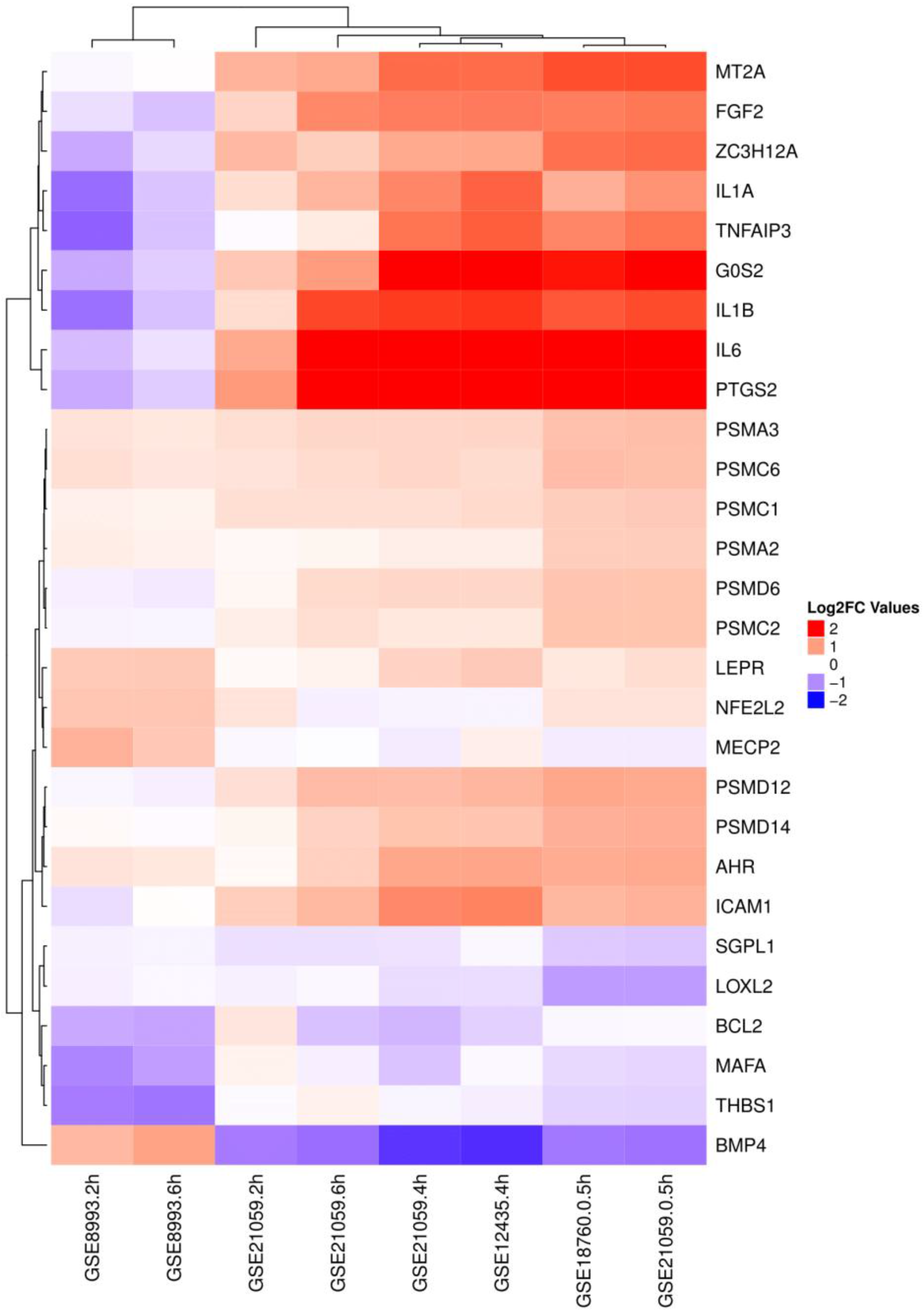
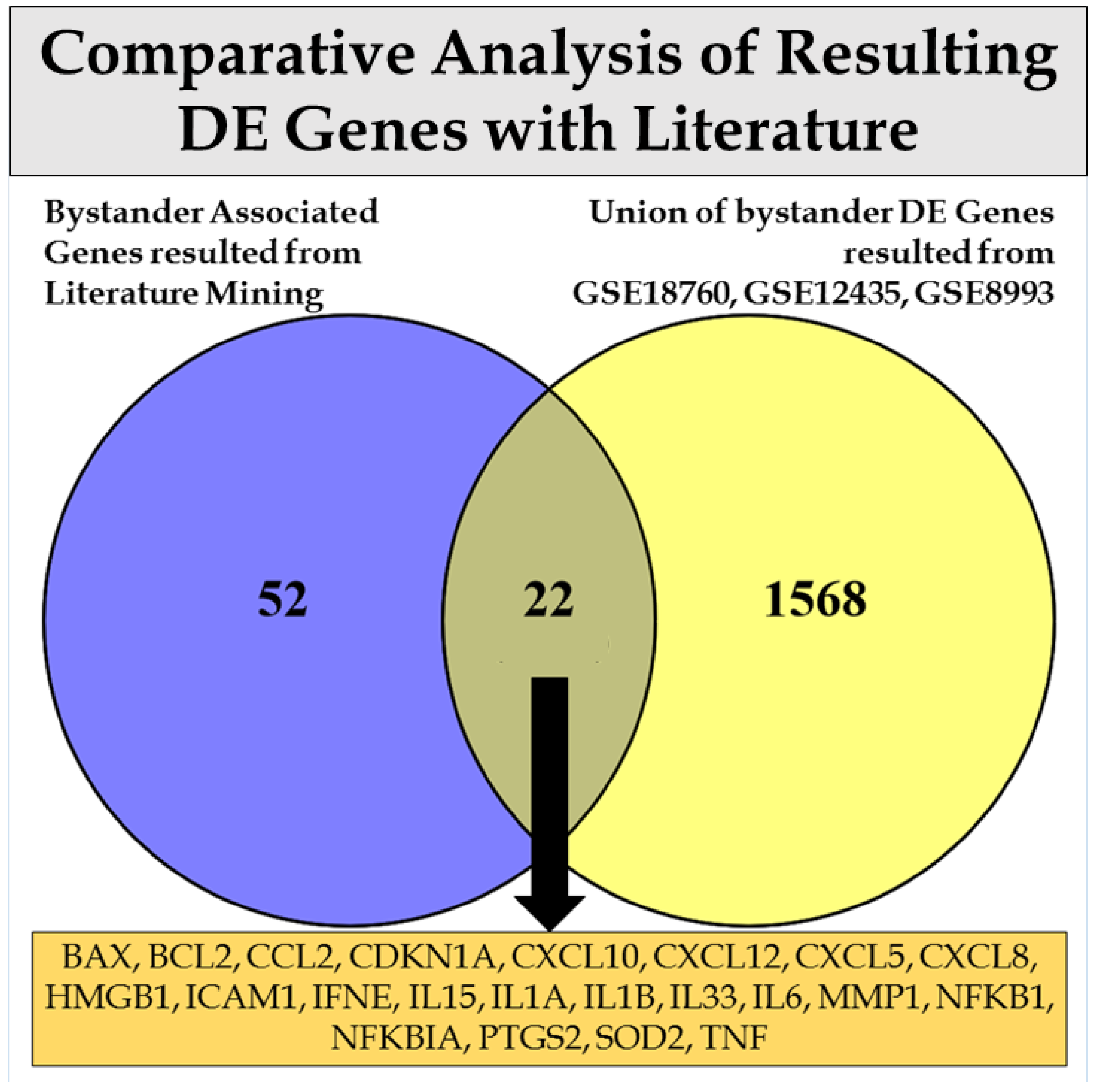
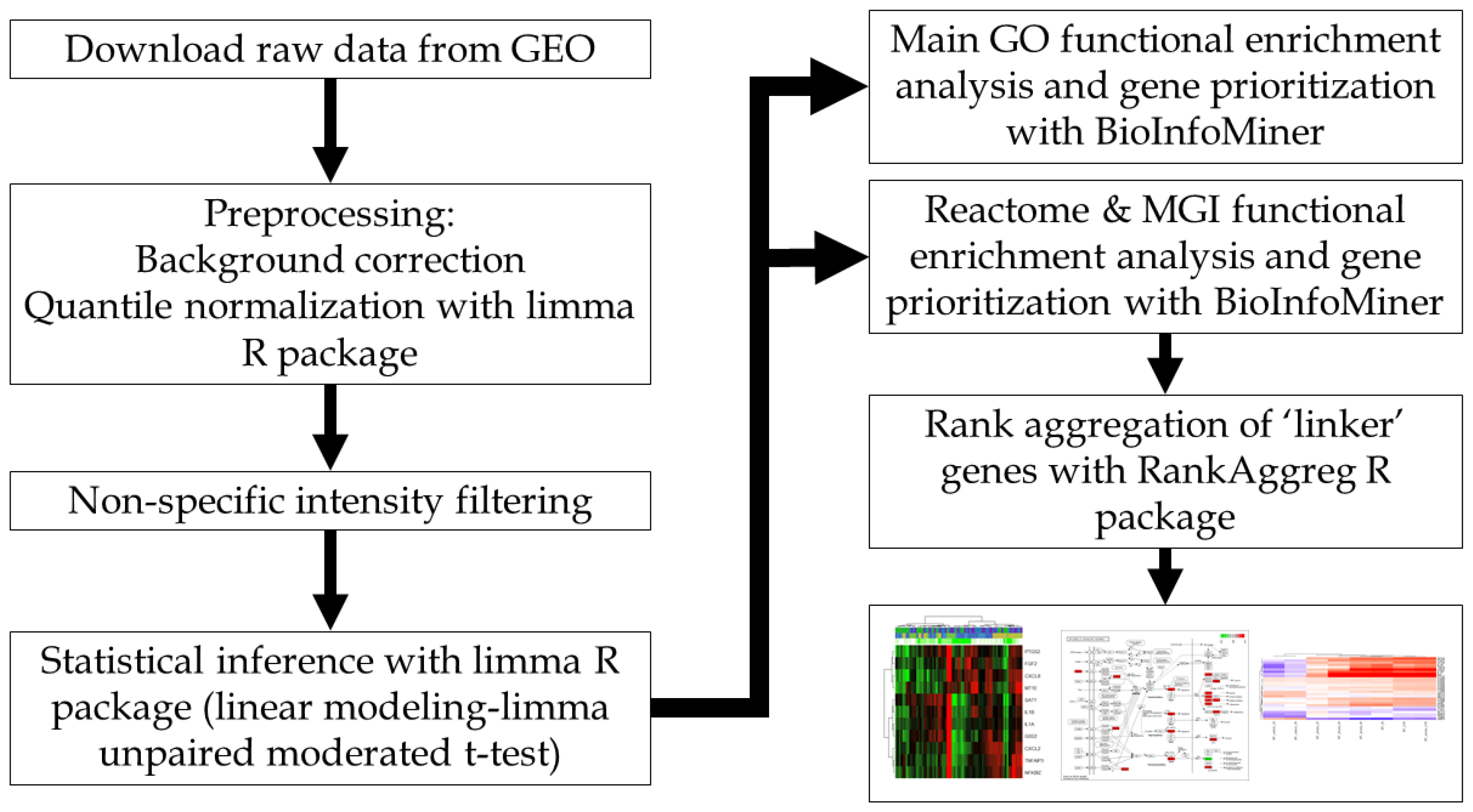
| Dataset | GSE12435 | GSE18760 | GSE21059 | GSE55869 | GSE32091 | GSE25772 | GSE8993 |
|---|---|---|---|---|---|---|---|
| Type of Radiation | α-particles | γ-rays | carbon-ions | ||||
| DE Bystander vs. Control | 53 (4 h) | 424 (0.5 h) | 1254 (ANOVA-time-series) | 0 | 0 | 0 | 1003 (2 h) 796 (6 h) |
| DE Irradiated vs. Control | 76 (4 h) | 481 (0.5 h) | 2399 (ANOVA-time-series) | 47 (4 h) | 3 (4 h) 0 (8 h) 0 (26 h) | 271 (4 h) 223 (8 h) 1977 (26 h) | 1502 (2 h) 1897 (6 h) |
| DE Common | 39 | 339 | 1169 | - | - | - | 264 (2 h) 324 (6 h) |
| Common DE Genes | Fold Change in Expression | |||||||
|---|---|---|---|---|---|---|---|---|
| Datasets | GSE18760 | GSE12435 | GSE21059 | |||||
| Time Points | 0.5 h | 4 h | 0.5 h | 1 h | 2 h | 4 h | 6 h | 24 h |
| MT1B | 2.421 | 1.905 | 2.456 | 0.898 | 1.122 | 1.927 | 1.244 | 1.185 |
| MT1E | 2.574 | 2.165 | 2.620 | 0.964 | 1.143 | 2.178 | 1.209 | 1.114 |
| MT1H | 2.380 | 2.001 | 2.424 | 0.982 | 1.076 | 2.028 | 1.186 | 1.205 |
| MT1X | 2.528 | 2.002 | 2.480 | 1.013 | 1.048 | 2.033 | 1.173 | 1.196 |
| MT2A | 1.690 | 1.450 | 1.704 | 0.678 | 0.790 | 1.455 | 0.885 | 0.975 |
| PTGS2 | 2.615 | 2.401 | 2.769 | 0.842 | 1.036 | 2.259 | 2.616 | 0.323 |
| CXCL5 | 1.589 | 2.063 | 1.975 | 0.383 | 0.133 | 1.772 | 2.335 | 1.154 |
| MMP3 | 2.582 | 1.932 | 2.690 | 1.143 | 0.963 | 1.901 | 3.335 | 2.023 |
| MT1L | 2.364 | 1.931 | 2.404 | 0.898 | 1.014 | 1.958 | 1.192 | 1.280 |
| ARC | 2.102 | 1.904 | 2.778 | 0.603 | −0.374 | 1.289 | 1.244 | 0.163 |
| TSLP | 0.618 | 1.407 | 0.703 | 0.628 | 0.466 | 1.354 | 0.829 | 1.043 |
| CXCL1 | 1.518 | 1.420 | 1.508 | 0.673 | 0.761 | 1.453 | 1.160 | 0.836 |
| GPR68 | 0.824 | 1.709 | 0.893 | 0.690 | 0.810 | 1.707 | 2.082 | 1.441 |
| MMP1 | 2.154 | 1.648 | 2.187 | 1.078 | 0.941 | 1.662 | 2.827 | 1.366 |
| MMP10 | 1.098 | 1.666 | 1.262 | 0.726 | 0.699 | 1.549 | 1.663 | 0.892 |
| KYNU | 1.963 | 1.806 | 2.121 | 1.220 | 0.876 | 1.622 | 1.385 | 1.332 |
| SLC16A6 | 1.723 | 1.709 | 1.888 | 0.796 | 0.839 | 1.579 | 2.431 | 1.493 |
| SLC7A11 | 1.445 | 1.259 | 1.522 | 1.076 | 0.946 | 1.224 | 0.887 | 1.033 |
| NAMPT | 1.393 | 1.486 | 1.426 | 0.659 | 0.524 | 1.571 | 0.736 | 0.639 |
| HSD11B1 | 1.509 | 1.500 | 1.620 | 0.718 | 0.607 | 1.442 | 1.491 | 1.074 |
| LAMB3 | 1.548 | 1.443 | 1.702 | 0.644 | 0.564 | 1.383 | 1.580 | 1.153 |
| PLA2G4A | 1.115 | 1.199 | 1.229 | 0.665 | 0.468 | 1.138 | 0.881 | 0.724 |
| C8orf4 | 1.277 | 1.486 | 1.353 | 0.734 | 0.586 | 1.432 | 0.780 | 1.036 |
| EPHA4 | −0.881 | −1.109 | −0.893 | −0.937 | −0.727 | −0.704 | −0.628 | −0.947 |
| ADGRG1 | 1.022 | 0.873 | 1.086 | 0.540 | 0.131 | 0.841 | 0.548 | 1.123 |
| CCK | 1.048 | 1.065 | 1.208 | 0.570 | 0.273 | 0.995 | 0.869 | 0.867 |
| Gene Ontology | Datasets/Enrichments | |||||
|---|---|---|---|---|---|---|
| GSE12435 | GSE18760 | GSE21059 | ||||
| Bystander 4 h | Irradiated 4 h | Bystander 0.5 h | Irradiated 0.5 h | Bystander Time-Series | Irradiated Time-Series | |
| Cellular Response to zinc ion | 5/18 | 6/18 | 9/18 | 9/18 | 10/18 | 11/18 |
| Response to Zinc Ion | 5/53 | 6/53 | 11/53 | 12/53 | 14/53 | 16/53 |
| Cellular Response to Cadmium Ion | 3/15 | 4/15 | 6/15 | 6/15 | 7/15 | 8/15 |
| Cellular Response to Metal Ion | 5/126 | 8/126 | 15/126 | 16/126 | 23/126 | 29/126 |
| Response to Inorganic Substance | 10/428 | 12/428 | 33/428 | 34/428 | 54/428 | - |
| Cellular Response to Inorganic Substance | 6/146 | 9/146 | 16/146 | 17/146 | 25/146 | - |
| Response to Metal Ion | 8/298 | 11/298 | 26/298 | 27/298 | 41/298 | - |
| Protein Folding | - | - | - | 17/211 | 34/211 | 54/211 |
| Cytokine-Mediated Signalling Pathway | 8/440 | - | 31/440 | 32/440 | - | - |
| Regulation of NF-kappaB Import into Nucleus | 3/44 | - | 7/44 | 7/44 | - | - |
| Positive Regulation of Reactive Oxygen Species Biosynthetic Process | 3/46 | - | 7/46 | 7/46 | - | - |
| Cytokine-mediated Signalling Pathway | 8/440 | - | 31/440 | 32/440 | - | - |
| Regulation of Anatomical Structure Morphogenesis | - | - | 57/934 | 56/934 | 105/934 | 163/934 |
| Extracellular Matrix Disassembly | 4/73 | - | - | - | 15/73 | 21/73 |
| Embryonic Skeletal System Development | - | - | - | - | 10/43 | 14/43 |
| Regulation of Protein Modification Process | - | - | 79/1616 | - | 155/1616 | 279/1616 |
| Response to Unfolded Protein | - | - | 7/45 | 8/45 | 10/45 | 15/45 |
| Wnt Signalling Pathway, Planar Cell Polarity Pathway | - | - | 11/99 | 11/99 | 17/99 | 26/99 |
| Gene Ontology | Dataset/Enrichments | |||
|---|---|---|---|---|
| GSE8993 | ||||
| Bystander 2 h | Irradiated 2 h | Bystander 6 h | Irradiated 6 h | |
| Negative Regulation of Nucleobase-containing Compound Metabolic Process | 112/1310 | - | 84/1310 | 188/1310 |
| Negative Regulation of Cellular Biosynthetic Process | 117/1394 | - | 88/1394 | 196/1394 |
| Negative Regulation of Nitrogen Compound Metabolic Process | 119/1425 | - | 90/1425 | 202/1425 |
| Negative Regulation of RNA Metabolic Process | 99/1178 | - | 79/1178 | 170/1178 |
| Regulation of Cell Migration | 62/662 | 91/662 | - | 113/662 |
| Regulation of Epithelial Cell Migration | 20/165 | 27/165 | - | 34/165 |
| Negative Regulation of Cell Migration | - | 34/206 | 19/206 | 39/206 |
| Negative Regulation of Cellular Component Movement | - | 39/247 | 22/247 | 44/247 |
| Negative Regulation of Cell Motility | - | - | 20/218 | 39/218 |
| Unique Gene Ontology Terms α-Particles IR (GSE12435, GSE18760) | |
|---|---|
| Bystander | Irradiated |
| positive regulation of vasoconstriction | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest |
| polyamine catabolic process | activation of cysteine-type endopeptidase activity involved in apoptotic signalling pathway |
| cell chemotaxis | extrinsic apoptotic signalling pathway via death domain receptors |
| regulation of response to external stimulus | negative regulation of G1/S transition of mitotic cell cycle |
| cell migration | regulation of apoptotic process |
| inflammatory response | nucleic acid phosphodiester bond hydrolysis |
| regulation of defence response to virus by host | activation of MAPKKK activity |
| regulation of response to wounding | atrioventricular valve morphogenesis |
| positive regulation of leukocyte migration | atrial septum development |
| positive regulation of cell-matrix adhesion | embryo development |
| Unique Gene Ontology Terms Carbon-Ion IR (GSE8993) | |||
|---|---|---|---|
| Bystander | Enrichment | Irradiated | Enrichment |
| positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signalling pathway | 9/35 | positive regulation of protein binding | 24/75 |
| positive regulation of protein homooligomerization | 4/8 | cell cycle arrest | 34/148 |
| negative regulation of intracellular protein transport | 13/84 | cellular component disassembly involved in execution phase of apoptosis | 10/25 |
| positive regulation of release of cytochrome c from mitochondria | 7/28 | cellular response to transforming growth factor β stimulus | 16/53 |
| regulation of oxidative phosphorylation | 5/15 | regulation of cell migration | 123/662 |
| regulation of steroid hormone secretion | 5/19 | response to transforming growth factor β | 17/59 |
| mitochondrial membrane organization | 12/90 | regulation of p38MAPK cascade | 10/26 |
| cellular response to oxygen levels | 14/111 | regulation of TOR signalling | 19/70 |
| regulation of excretion | 6/25 | positive regulation of extrinsic apoptotic signalling pathway | 15/52 |
| multicellular organismal response to stress | 9/59 | regulation of cell-matrix adhesion | 22/91 |
| Common Genes | Bystander | |||||
|---|---|---|---|---|---|---|
| α-Particles | Carbon Ion | |||||
| GSE18760 | GSE12435 | GSE21059 | GSE8993 | |||
| 0.5 h | 4 h | 2 h | 6 h | 2 h | 6 h | |
| IL1A | 0.81 * | 1.53 * | 0.34 | 0.76 | −1.27 | −0.5 * |
| IL1B | 1.62 * | 1.85 * | 0.36 | 1.74 | −1.23 * | −0.54 * |
| NFKBIZ | 1.32 | 1.44 | 0.51 | 0.85 | −1.41 | −0.53 |
| SAT1 | 1.16 | 0.91 * | - | 0.4 | 0.52 | 0.54 |
| TNFAIP3 | 1.22 * | 1.58 * | - | 0.22 | −1.35 | −0.52 |
| CXCL2 | 2.42 * | 2.64 | 0.64 | 1.14 | −0.92 | - |
| G0S2 | 1.96 | 2.15 | 0.57 | 1.02 | −0.73 | - |
| MT1E | 2.57 | 2.16 | 1.1 | 1.2 | −0.5 | - |
| PTGS2 | 2.61 * | 2.4 * | 1.03 * | 2.61 * | −0.73* | - |
| CXCL8 | 3.53 * | - | 1.3 | 3.6 | −1.36 | −0.69 |
| FGF2 | 1.29 | 1.31 | - | - | - | −0.53 * |
| Ranked Linker DE Genes | ||
|---|---|---|
| GO | MGI | Reactome Pathways |
| IL6 | PTGS2 | PSMD6 |
| ZC3H12A | BMP4 | PSMA2 |
| PTGS2 | IL6 | PSMA3 |
| BCL2 | LEPR | PSMD14 |
| BMP4 | IL1B | PSMC1 |
| THBS1 | NFE2L2 | PSMC2 |
| IL1A | AHR | PSMC6 |
| IL1B | MECP2 | IL1B |
| TNFAIP3 | SGPL1 | FGF2 |
| ICAM1 | G0S2 | PSMD12 |
| MT2A | LOXL2 | |
| MAFA | ||
| Top 5 Ranked Linker Genes GO | Enriched Clusters | Top 5 Ranked Linker Genes MGI | Enriched Clusters | Top 5 Ranked Linker Genes Reactome | Enriched Clusters |
|---|---|---|---|---|---|
| IL6 | inflammatory response, cytokine-mediated signaling pathway, cellular response to oxidative stress | PTGS2 | abnormal wound healing, increased IgA level, abnormal IgG3 level | PSMD6 | Hedgehog “on” state, Degradation of beta-catenin by the destruction complex, Beta-catenin independent WNT signaling, PCP/CE pathway, Regulation of activated PAK-2p34 by proteasome mediated degradation, CLEC7A (Dectin-1) signaling, Metabolism of polyamines |
| ZC3H12A | negative regulation of cell death, cellular response to oxidative stress, inflammatory response, regulation of apoptotic process | BMP4 | increased apoptosis | PSMA2 | |
| PTGS2 | cellular response to oxidative stress, cellular response to metal ion, cellular response to fluid shear stress, regulation of apoptotic process | IL6 | increased IgA level, abnormal interferon-gamma secretion, abnormal circulating interleukin level | PSMA3 | |
| BCL2 | negative regulation of extrinsic apoptotic signaling pathway, response to hypoxia | LEPR | increased apoptosis, abnormal interferon-gamma secretion, abnormal circulating interleukin level | PSMD14 | |
| BMP4 | system development, positive regulation of cell migration, positive regulation of protein modification process | IL1B | abnormal wound healing, abnormal macrophage physiology, decreased interleukin-6 secretion | PSMC1 |
| GEO Accession Number | GSE18760 | GSE12435 | GSE21059 | GSE55869 | GSE32091 | GSE25772 | GSE8993 |
|---|---|---|---|---|---|---|---|
| Type of Radiation | α-particles | γ-rays | carbon-ion | ||||
| Time of Extraction of Total RNA after Irradiation (h) | 0.5 | 4 | 0.5, 1, 2, 4, 6, 24 | 4 | 4, 8, 26 | 2, 6 | |
| Irradiation Dose (Gy) | 0.5 | 1 | 0.1 | 2 | 1.3, 0.13, 0.013 | ||
| Cell Line | IMR-90 primary lung fibroblasts | H1299 non-small cell lung carcinoma | F11-hTERT immortalized foreskin fibroblasts | AG01522D primary normal human diploid skin fibroblasts | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeles, C.; Vlachavas, E.-I.; Papadodima, O.; Pilalis, E.; Vorgias, C.E.; Georgakilas, A.G.; Chatziioannou, A. Integrative Bioinformatic Analysis of Transcriptomic Data Identifies Conserved Molecular Pathways Underlying Ionizing Radiation-Induced Bystander Effects (RIBE). Cancers 2017, 9, 160. https://doi.org/10.3390/cancers9120160
Yeles C, Vlachavas E-I, Papadodima O, Pilalis E, Vorgias CE, Georgakilas AG, Chatziioannou A. Integrative Bioinformatic Analysis of Transcriptomic Data Identifies Conserved Molecular Pathways Underlying Ionizing Radiation-Induced Bystander Effects (RIBE). Cancers. 2017; 9(12):160. https://doi.org/10.3390/cancers9120160
Chicago/Turabian StyleYeles, Constantinos, Efstathios-Iason Vlachavas, Olga Papadodima, Eleftherios Pilalis, Constantinos E. Vorgias, Alexandros G. Georgakilas, and Aristotelis Chatziioannou. 2017. "Integrative Bioinformatic Analysis of Transcriptomic Data Identifies Conserved Molecular Pathways Underlying Ionizing Radiation-Induced Bystander Effects (RIBE)" Cancers 9, no. 12: 160. https://doi.org/10.3390/cancers9120160
APA StyleYeles, C., Vlachavas, E.-I., Papadodima, O., Pilalis, E., Vorgias, C. E., Georgakilas, A. G., & Chatziioannou, A. (2017). Integrative Bioinformatic Analysis of Transcriptomic Data Identifies Conserved Molecular Pathways Underlying Ionizing Radiation-Induced Bystander Effects (RIBE). Cancers, 9(12), 160. https://doi.org/10.3390/cancers9120160







