Practical Application of Deep Learning in Diagnostic Neuropathology—Reimagining a Histological Asset in the Era of Precision Medicine
Abstract
Simple Summary
Abstract
1. Introduction
2. History of Glass-Based and Molecular Features in Brain Cancer
3. Current Diagnostic Challenges and Shortcomings
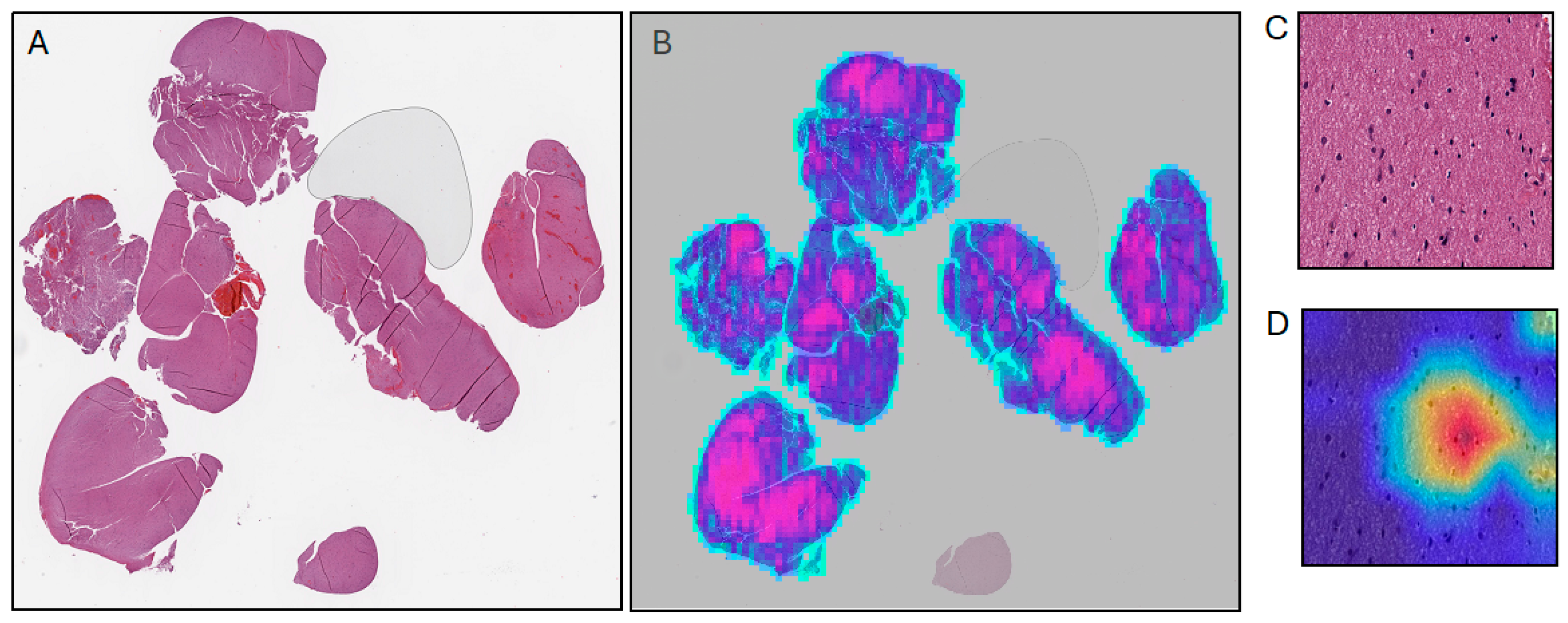
4. Supervised Machine Learning
5. Generative Machine Learning
6. Semi-Supervised and Self-Supervised Machine Learning
7. Challenges and Risks
8. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Zülch, K.J. International histological classification of tumours. In Histological Typing of Tumours of the Central Nervous System; WHO: Geneva, Switzerland, 1979; ISBN 978-92-4-176021-8. [Google Scholar]
- van den Bent, M.J. Interobserver Variation of the Histopathological Diagnosis in Clinical Trials on Glioma: A Clinician’s Perspective. Acta Neuropathol. 2010, 120, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Aldape, K.; Simmons, M.L.; Davis, R.L.; Miike, R.; Wiencke, J.; Barger, G.; Lee, M.; Chen, P.; Wrensch, M. Discrepancies in Diagnoses of Neuroepithelial Neoplasms: The San Francisco Bay Area Adult Glioma Study. Cancer 2000, 88, 2342–2349. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, B.W.; Priesterbach-Ackley, L.P.; Petersen, J.K.; Wesseling, P. Molecular Pathology of Tumors of the Central Nervous System. Ann. Oncol. 2019, 30, 1265–1278. [Google Scholar] [CrossRef]
- Weller, M.; Weber, R.G.; Willscher, E.; Riehmer, V.; Hentschel, B.; Kreuz, M.; Felsberg, J.; Beyer, U.; Löffler-Wirth, H.; Kaulich, K.; et al. Molecular Classification of Diffuse Cerebral WHO Grade II/III Gliomas Using Genome- and Transcriptome-Wide Profiling Improves Stratification of Prognostically Distinct Patient Groups. Acta Neuropathol. 2015, 129, 679–693. [Google Scholar] [CrossRef]
- Hartmann, C.; Hentschel, B.; Wick, W.; Capper, D.; Felsberg, J.; Simon, M.; Westphal, M.; Schackert, G.; Meyermann, R.; Pietsch, T.; et al. Patients with IDH1 Wild Type Anaplastic Astrocytomas Exhibit Worse Prognosis than IDH1-Mutated Glioblastomas, and IDH1 Mutation Status Accounts for the Unfavorable Prognostic Effect of Higher Age: Implications for Classification of Gliomas. Acta Neuropathol. 2010, 120, 707–718. [Google Scholar] [CrossRef]
- Aibaidula, A.; Chan, A.K.-Y.; Shi, Z.; Li, Y.; Zhang, R.; Yang, R.; Li, K.K.-W.; Chung, N.Y.-F.; Yao, Y.; Zhou, L.; et al. Adult IDH Wild-Type Lower-Grade Gliomas Should Be Further Stratified. Neuro-Oncology 2017, 19, 1327–1337. [Google Scholar] [CrossRef]
- Satgunaseelan, L.; Sy, J.; Shivalingam, B.; Sim, H.-W.; Alexander, K.L.; Buckland, M.E. Prognostic and Predictive Biomarkers in Central Nervous System Tumours: The Molecular State of Play. Pathology 2024, 56, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Jaunmuktane, Z.; Capper, D.; Jones, D.T.W.; Schrimpf, D.; Sill, M.; Dutt, M.; Suraweera, N.; Pfister, S.M.; Von Deimling, A.; Brandner, S. Methylation Array Profiling of Adult Brain Tumours: Diagnostic Outcomes in a Large, Single Centre. Acta Neuropathol. Commun. 2019, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Horbinski, C.; Ligon, K.L.; Brastianos, P.; Huse, J.T.; Venere, M.; Chang, S.; Buckner, J.; Cloughesy, T.; Jenkins, R.B.; Giannini, C.; et al. The Medical Necessity of Advanced Molecular Testing in the Diagnosis and Treatment of Brain Tumor Patients. Neuro-Oncology 2019, 21, 1498–1508. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, Y.; Teng, L.; Yan, J.; Guo, Y.; Qiu, Y.; Ji, Y.; Yu, B.; Pei, D.; Duan, W.; et al. Neuropathologist-Level Integrated Classification of Adult-Type Diffuse Gliomas Using Deep Learning from Whole-Slide Pathological Images. Nat. Commun. 2023, 14, 6359. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, K.J.; Löffler, C.M.L.; Muti, H.S.; Berghoff, A.S.; Eisenlöffel, C.; Van Treeck, M.; Carrero, Z.I.; El Nahhas, O.S.M.; Veldhuizen, G.P.; Weil, S.; et al. Direct Image to Subtype Prediction for Brain Tumors Using Deep Learning. Neuro-Oncology Adv. 2023, 5, vdad139. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.B.; Schlosser, C.; Grewal, J.; Coope, R.; Jones, S.J.M.; Yip, S. Rise of the Machines: Advances in Deep Learning for Cancer Diagnosis. Trends Cancer 2019, 5, 157–169. [Google Scholar] [CrossRef] [PubMed]
- El Nahhas, O.S.M.; Loeffler, C.M.L.; Carrero, Z.I.; Van Treeck, M.; Kolbinger, F.R.; Hewitt, K.J.; Muti, H.S.; Graziani, M.; Zeng, Q.; Calderaro, J.; et al. Regression-Based Deep-Learning Predicts Molecular Biomarkers from Pathology Slides. Nat. Commun. 2024, 15, 1253. [Google Scholar] [CrossRef]
- Fu, Y.; Karanian, M.; Perret, R.; Camara, A.; Le Loarer, F.; Jean-Denis, M.; Hostein, I.; Michot, A.; Ducimetiere, F.; Giraud, A.; et al. Deep Learning Predicts Patients Outcome and Mutations from Digitized Histology Slides in Gastrointestinal Stromal Tumor. NPJ Precis. Oncol. 2023, 7, 71. [Google Scholar] [CrossRef]
- Sahm, F.; Brandner, S.; Bertero, L.; Capper, D.; French, P.J.; Figarella-Branger, D.; Giangaspero, F.; Haberler, C.; Hegi, M.E.; Kristensen, B.W.; et al. Molecular Diagnostic Tools for the World Health Organization (WHO) 2021 Classification of Gliomas, Glioneuronal and Neuronal Tumors; an EANO Guideline. Neuro-Oncology 2023, 25, 1731–1749. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Ertosun, M.G.; Rubin, D.L. Automated Grading of Gliomas Using Deep Learning in Digital Pathology Images: A Modular Approach with Ensemble of Convolutional Neural Networks. AMIA Annu. Symp. Proc. 2015, 2015, 1899–1908. [Google Scholar]
- Jiang, S.; Zanazzi, G.J.; Hassanpour, S. Predicting Prognosis and IDH Mutation Status for Patients with Lower-Grade Gliomas Using Whole Slide Images. Sci. Rep. 2021, 11, 16849. [Google Scholar] [CrossRef]
- Truong, A.H.; Sharmanska, V.; Limbäck-Stanic, C.; Grech-Sollars, M. Optimization of Deep Learning Methods for Visualization of Tumor Heterogeneity and Brain Tumor Grading through Digital Pathology. Neuro-Oncology Adv. 2020, 2, vdaa110. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.; Liu, S.; Russo, C.; Cong, C.; Song, Y.; Rodriguez, M.; Di Ieva, A. Artificial Intelligence–Assisted Classification of Gliomas Using Whole Slide Images. Arch. Pathol. Lab. Med. 2023, 147, 916–924. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar] [CrossRef]
- Chunduru, P.; Phillips, J.J.; Molinaro, A.M. Prognostic Risk Stratification of Gliomas Using Deep Learning in Digital Pathology Images. Neuro-Oncology Adv. 2022, 4, vdac111. [Google Scholar] [CrossRef]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Velázquez Vega, J.E.; Brat, D.J.; Cooper, L.A.D. Predicting Cancer Outcomes from Histology and Genomics Using Convolutional Networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef]
- Rathore, S.; Iftikhar, M.A.; Gurcan, M.N.; Mourelatos, Z. Radiopathomics: Integration of Radiographic and Histologic Characteristics for Prognostication in Glioblastoma. arXiv 2019, arXiv:1909.07581. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Lipkova, J.; Noor, Z.; Shaban, M.; Shady, M.; Williams, M.; Joo, B.; et al. Pan-Cancer Integrative Histology-Genomic Analysis via Multimodal Deep Learning. Cancer Cell 2022, 40, 865–878.e6. [Google Scholar] [CrossRef]
- Liechty, B.; Xu, Z.; Zhang, Z.; Slocum, C.; Bahadir, C.D.; Sabuncu, M.R.; Pisapia, D.J. Machine Learning Can Aid in Prediction of IDH Mutation from H&E-Stained Histology Slides in Infiltrating Gliomas. Sci. Rep. 2022, 12, 22623. [Google Scholar] [CrossRef]
- Nicoll, J.A.R.; Bloom, T.; Clarke, A.; Boche, D.; Hilton, D. BRAIN UK: Accessing NHS Tissue Archives for Neuroscience Research. Neuropathol. Appl. Neurobiol. 2022, 48, e12766. [Google Scholar] [CrossRef]
- Roetzer-Pejrimovsky, T.; Moser, A.-C.; Atli, B.; Vogel, C.C.; Mercea, P.A.; Prihoda, R.; Gelpi, E.; Haberler, C.; Höftberger, R.; Hainfellner, J.A.; et al. The Digital Brain Tumour Atlas, an Open Histopathology Resource. Sci. Data 2022, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Gennatas, E.D.; Wu, A.; Braunstein, S.E.; Morin, O.; Chen, W.C.; Magill, S.T.; Gopinath, C.; Villaneueva-Meyer, J.E.; Perry, A.; McDermott, M.W.; et al. Preoperative and Postoperative Prediction of Long-Term Meningioma Outcomes. PLoS ONE 2018, 13, e0204161. [Google Scholar] [CrossRef]
- Sehring, J.; Dohmen, H.; Selignow, C.; Schmid, K.; Grau, S.; Stein, M.; Uhl, E.; Mukhopadhyay, A.; Németh, A.; Amsel, D.; et al. Leveraging Attention-Based Convolutional Neural Networks for Meningioma Classification in Computational Histopathology. Cancers 2023, 15, 5190. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Abas, F.S.; Gokozan, H.N.; Goksel, B.; Otero, J.J.; Gurcan, M.N. Intraoperative Neuropathology of Glioma Recurrence: Cell Detection and Classification; Gurcan, M.N., Madabhushi, A., Eds.; SPIE: San Diego, CA, USA, 2016; p. 979109. [Google Scholar]
- Orringer, D.A.; Pandian, B.; Niknafs, Y.S.; Hollon, T.C.; Boyle, J.; Lewis, S.; Garrard, M.; Hervey-Jumper, S.L.; Garton, H.J.L.; Maher, C.O.; et al. Rapid Intraoperative Histology of Unprocessed Surgical Specimens via Fibre-Laser-Based Stimulated Raman Scattering Microscopy. Nat. Biomed. Eng. 2017, 1, 0027. [Google Scholar] [CrossRef]
- Hollon, T.C.; Lewis, S.; Pandian, B.; Niknafs, Y.S.; Garrard, M.R.; Garton, H.; Maher, C.O.; McFadden, K.; Snuderl, M.; Lieberman, A.P.; et al. Rapid Intraoperative Diagnosis of Pediatric Brain Tumors Using Stimulated Raman Histology. Cancer Res. 2018, 78, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Hollon, T.C.; Pandian, B.; Adapa, A.R.; Urias, E.; Save, A.V.; Khalsa, S.S.S.; Eichberg, D.G.; D’Amico, R.S.; Farooq, Z.U.; Lewis, S.; et al. Near Real-Time Intraoperative Brain Tumor Diagnosis Using Stimulated Raman Histology and Deep Neural Networks. Nat. Med. 2020, 26, 52–58. [Google Scholar] [CrossRef]
- Reinecke, D.; Von Spreckelsen, N.; Mawrin, C.; Ion-Margineanu, A.; Fürtjes, G.; Jünger, S.T.; Khalid, F.; Freudiger, C.W.; Timmer, M.; Ruge, M.I.; et al. Novel Rapid Intraoperative Qualitative Tumor Detection by a Residual Convolutional Neural Network Using Label-Free Stimulated Raman Scattering Microscopy. Acta Neuropathol. Commun. 2022, 10, 109. [Google Scholar] [CrossRef]
- Kodali, N.; Abernethy, J.; Hays, J.; Kira, Z. On Convergence and Stability of GANs. arXiv 2017, arXiv:1705.07215. [Google Scholar] [CrossRef]
- Rombach, R.; Blattmann, A.; Lorenz, D.; Esser, P.; Ommer, B. High-Resolution Image Synthesis with Latent Diffusion Models. arXiv 2021, arXiv:2112.10752. [Google Scholar] [CrossRef]
- Levine, A.B.; Peng, J.; Farnell, D.; Nursey, M.; Wang, Y.; Naso, J.R.; Ren, H.; Farahani, H.; Chen, C.; Chiu, D.; et al. Synthesis of Diagnostic Quality Cancer Pathology Images by Generative Adversarial Networks. J. Pathol. 2020, 252, 178–188. [Google Scholar] [CrossRef]
- Moghadam, P.A.; Van Dalen, S.; Martin, K.C.; Lennerz, J.; Yip, S.; Farahani, H.; Bashashati, A. A Morphology Focused Diffusion Probabilistic Model for Synthesis of Histopathology Images. arXiv 2022, arXiv:2209.13167. [Google Scholar] [CrossRef]
- Truhn, D.; Loeffler, C.M.; Müller-Franzes, G.; Nebelung, S.; Hewitt, K.J.; Brandner, S.; Bressem, K.K.; Foersch, S.; Kather, J.N. Extracting Structured Information from Unstructured Histopathology Reports Using Generative Pre-trained Transformer 4 (GPT-4). J. Pathol. 2024, 262, 310–319. [Google Scholar] [CrossRef]
- Huang, Z.; Bianchi, F.; Yuksekgonul, M.; Montine, T.; Zou, J. Leveraging Medical Twitter to Build a Visual–Language Foundation Model for Pathology AI. Pathology 2023. [Google Scholar] [CrossRef]
- Wang, X.; Yang, S.; Zhang, J.; Wang, M.; Zhang, J.; Yang, W.; Huang, J.; Han, X. Transformer-Based Unsupervised Contrastive Learning for Histopathological Image Classification. Med. Image Anal. 2022, 81, 102559. [Google Scholar] [CrossRef] [PubMed]
- Filiot, A.; Ghermi, R.; Olivier, A.; Jacob, P.; Fidon, L.; Mac Kain, A.; Saillard, C.; Schiratti, J.-B. Scaling Self-Supervised Learning for Histopathology with Masked Image Modeling. Pathology 2023. [Google Scholar] [CrossRef]
- Vorontsov, E.; Bozkurt, A.; Casson, A.; Shaikovski, G.; Zelechowski, M.; Liu, S.; Severson, K.; Zimmermann, E.; Hall, J.; Tenenholtz, N.; et al. Virchow: A Million-Slide Digital Pathology Foundation Model. arXiv 2023, arXiv:2309.07778. [Google Scholar] [CrossRef]
- Chen, R.J.; Ding, T.; Lu, M.Y.; Williamson, D.F.K.; Jaume, G.; Chen, B.; Zhang, A.; Shao, D.; Song, A.H.; Shaban, M.; et al. A General-Purpose Self-Supervised Model for Computational Pathology. arXiv 2023, arXiv:2308.15474. [Google Scholar] [CrossRef]
- Baidoshvili, A.; Bucur, A.; Van Leeuwen, J.; Van Der Laak, J.; Kluin, P.; Van Diest, P.J. Evaluating the Benefits of Digital Pathology Implementation: Time Savings in Laboratory Logistics. Histopathology 2018, 73, 784–794. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Zhang, D.Y. Artificial Intelligence and Computational Pathology. Lab. Investig. 2021, 101, 412–422. [Google Scholar] [CrossRef]
- Hanna, M.G.; Reuter, V.E.; Samboy, J.; England, C.; Corsale, L.; Fine, S.W.; Agaram, N.P.; Stamelos, E.; Yagi, Y.; Hameed, M.; et al. Implementation of Digital Pathology Offers Clinical and Operational Increase in Efficiency and Cost Savings. Arch. Pathol. Lab. Med. 2019, 143, 1545–1555. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Pan, M.; Mo, K.; Mao, Y.; Zou, D. Emerging Role of Artificial Intelligence in Diagnosis, Classification and Clinical Management of Glioma. Semin. Cancer Biol. 2023, 91, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Khera, R.; Simon, M.A.; Ross, J.S. Automation Bias and Assistive AI: Risk of Harm From AI-Driven Clinical Decision Support. JAMA 2023, 330, 2255. [Google Scholar] [CrossRef] [PubMed]
- Nakhate, V.; Gonzalez Castro, L.N. Artificial Intelligence in Neuro-Oncology. Front. Neurosci. 2023, 17, 1217629. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. arXiv 2016, arXiv:1610.02391. [Google Scholar] [CrossRef]
- Roberts, K.F.; Dahiya, S.M. Mitotic Index Is (Still) Important for Grading Isocitrate Dehydrogenase (IDH)-Mutant Astrocytoma. Neuro-Oncology 2023, 25, 1450–1451. [Google Scholar] [CrossRef]
- Gu, H.; Yang, C.; Al-kharouf, I.; Magaki, S.; Lakis, N.; Williams, C.K.; Alrosan, S.M.; Onstott, E.K.; Yan, W.; Khanlou, N.; et al. Enhancing Mitosis Quantification and Detection in Meningiomas with Computational Digital Pathology. Acta Neuropathol. Commun. 2024, 12, 7. [Google Scholar] [CrossRef]
- Larrazabal, A.J.; Nieto, N.; Peterson, V.; Milone, D.H.; Ferrante, E. Gender Imbalance in Medical Imaging Datasets Produces Biased Classifiers for Computer-Aided Diagnosis. Proc. Natl. Acad. Sci. USA 2020, 117, 12592–12594. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, A.; Chen, R.J.; Williamson, D.F.K.; Song, A.H.; Jaume, G.; Yang, Y.; Hartvigsen, T.; Dyer, E.C.; Lu, M.Y.; Lipkova, J.; et al. Demographic Bias in Misdiagnosis by Computational Pathology Models. Nat. Med. 2024, 30, 1174–1190. [Google Scholar] [CrossRef] [PubMed]

| Adult-Type Diffusely Infiltrating Gliomas | ||||
|---|---|---|---|---|
| Astrocytoma | Oligodendroglioma | Glioblastoma | ||
| Molecular classification for diagnosis | IDH1/2 | Mutant | Mutant | Wildtype |
| 1p19q | Intact | Co-deleted | Intact | |
| H3 | Wildtype | Wildtype | Wildtype | |
| Other | ATRX, TP53 mutations | TERTp, CIC, FUBP1, NOTCH1 mutations | EGFR amplification, TERTp mutation, +7/−10 | |
| Morphologic and molecular features for grading | WHO grade 2 | Increased cellularity Nuclear atypia | Increased cellularity Nuclear atypia | N/A |
| WHO grade 3 | Elevated mitotic index | Elevated mitotic index MVP Necrosis | N/A | |
| WHO grade 4 | MVP Necrosis CDKN2A/B HD a | N/A | MVP b Necrosis b | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rich, K.; Tosefsky, K.; Martin, K.C.; Bashashati, A.; Yip, S. Practical Application of Deep Learning in Diagnostic Neuropathology—Reimagining a Histological Asset in the Era of Precision Medicine. Cancers 2024, 16, 1976. https://doi.org/10.3390/cancers16111976
Rich K, Tosefsky K, Martin KC, Bashashati A, Yip S. Practical Application of Deep Learning in Diagnostic Neuropathology—Reimagining a Histological Asset in the Era of Precision Medicine. Cancers. 2024; 16(11):1976. https://doi.org/10.3390/cancers16111976
Chicago/Turabian StyleRich, Katherine, Kira Tosefsky, Karina C. Martin, Ali Bashashati, and Stephen Yip. 2024. "Practical Application of Deep Learning in Diagnostic Neuropathology—Reimagining a Histological Asset in the Era of Precision Medicine" Cancers 16, no. 11: 1976. https://doi.org/10.3390/cancers16111976
APA StyleRich, K., Tosefsky, K., Martin, K. C., Bashashati, A., & Yip, S. (2024). Practical Application of Deep Learning in Diagnostic Neuropathology—Reimagining a Histological Asset in the Era of Precision Medicine. Cancers, 16(11), 1976. https://doi.org/10.3390/cancers16111976






