The Diagnostic Yield and Implications of Targeted Founder Pathogenic Variant Testing in an Israeli Cohort
Abstract
:Simple Summary
Abstract
1. Introduction
2. Patients and Methods
2.1. Study Population
2.2. Genotyping
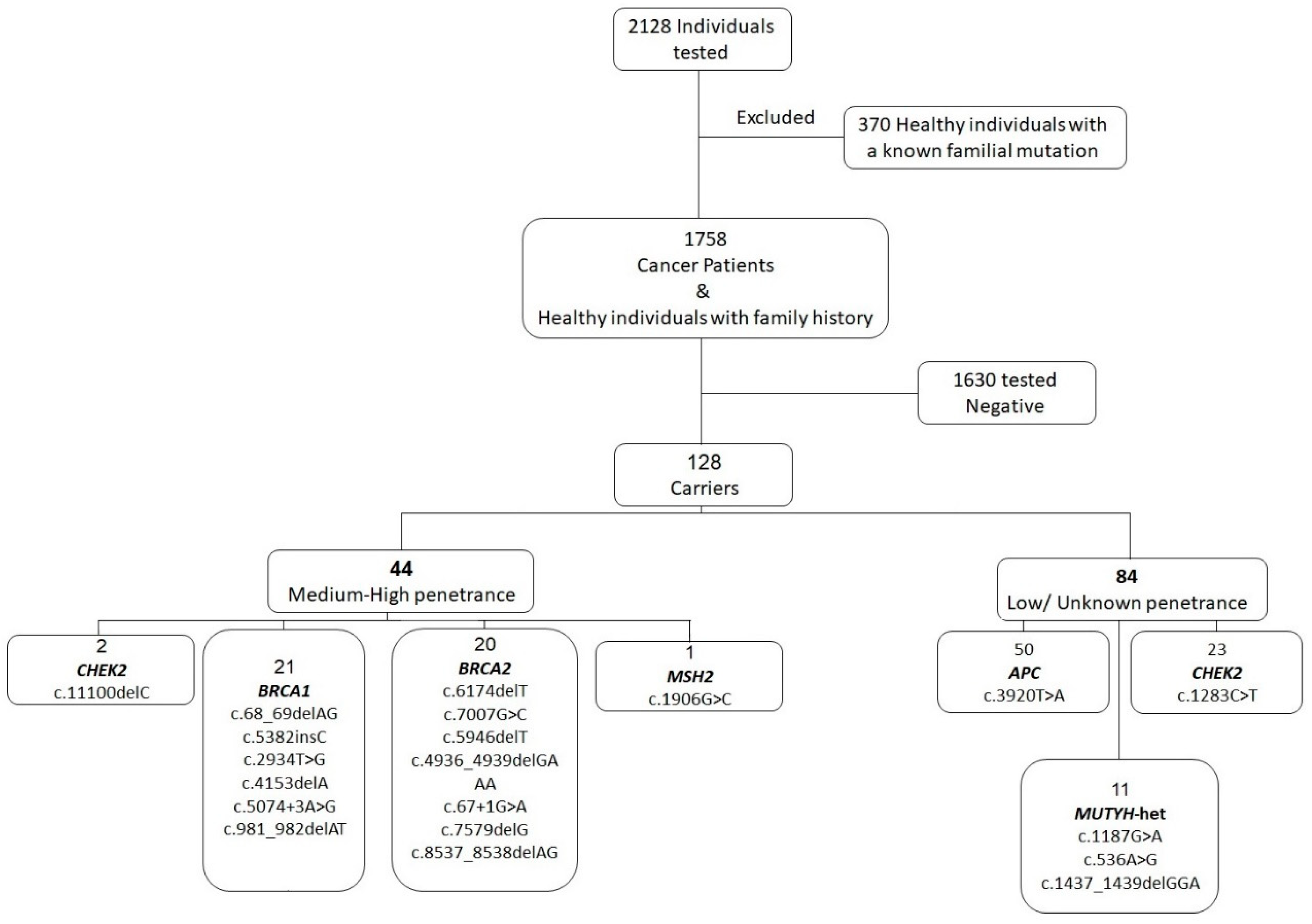
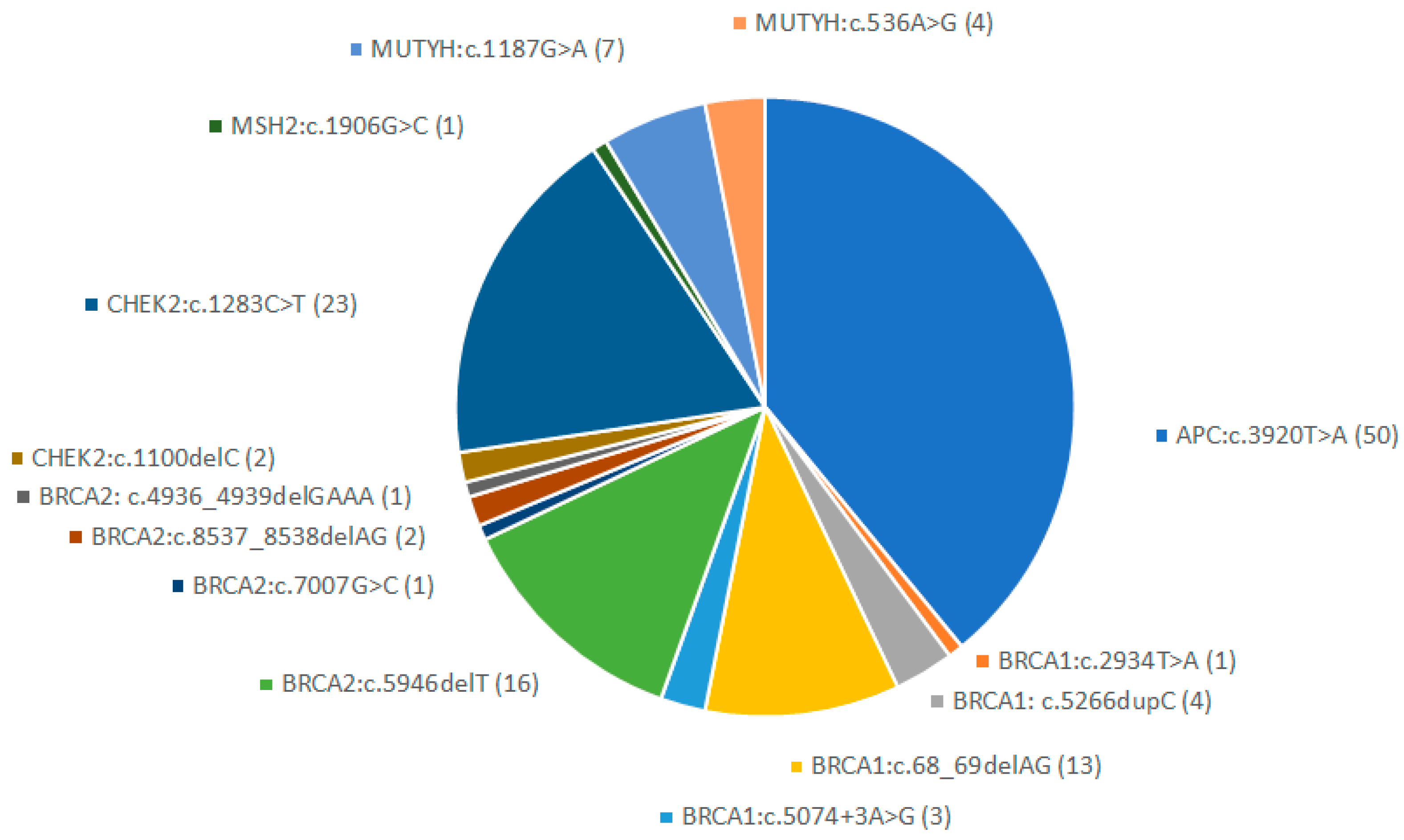
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| eGT | expanded genetic testing |
| FVT | founder variant testing |
| LPV | low-penetrance variant |
| MHPV | medium-high penetrance |
| MINAS | multilocus inherited neoplasia allele syndrome |
| NGS | next-generation sequencing |
| PV | pathogenic variant |
References
- Mao, R.; Krautscheid, P.; Graham, R.P.; Ganguly, A.; Shankar, S.; Ferber, M.; Hegde, M. Genetic testing for inherited colorectal cancer and polyposis, 2021 revision: A technical standard of the American College of Medical Genetics and Genomics (ACMG). Anesth. Analg. 2021, 23, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Marzuillo, C.; De Vito, C.; D’Andrea, E.; Rosso, A.; Villari, P. Predictive genetic testing for complex diseases: A public health perspective. QJM 2013, 107, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; van de Beld, M.C.J.; Jones, N.; Vogt, S.; Tops, C.M.; Vasen, H.F.; Sampson, J.R.; Aretz, S.; Hes, F.J. Analysis of MUTYH Genotypes and Colorectal Phenotypes in Patients With MUTYH-Associated Polyposis. Gastroenterology 2009, 136, 471–476. [Google Scholar] [CrossRef] [PubMed]
- De Braekeleer, M.; Hechtman, P.; Andermann, E.; Kaplan, F. The French Canadian Tay-Sachs disease deletion mutation: Identification of probable founders. Hum. Genet. 1992, 89, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Frisch, A.; Colombo, R.; Michaelovsky, E.; Karpati, M.; Goldman, B.; Peleg, L. Origin and spread of the 1278insTATC mutation causing Tay-Sachs disease in Ashkenazi Jews: Genetic drift as a robust and parsimonious hypothesis. Hum. Genet. 2004, 114, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Peleg, L.; Karpati, M.; Gazit, E.; Raasrothschild, A.; Goldman, B. Mutations of the Hexosaminidase A Gene in Ashkenazi and Non-Ashkenazi Jews. Biochem. Med. Metab. Biol. 1994, 52, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Olynyk, J.K.; Ramm, G.A. Hemochromatosis. New Engl. J. Med. 2022, 387, 2159–2170. [Google Scholar] [CrossRef]
- Foulkes, W.; Thiffault, I.; Gruber, S.; Horwitz, M.; Hamel, N.; Lee, C.; Shia, J.; Markowitz, A.; Figer, A.; Friedman, E.; et al. The Founder Mutation MSH2*1906G→C Is an Important Cause of Hereditary Nonpolyposis Colorectal Cancer in the Ashkenazi Jewish Population. Am. J. Hum. Genet. 2002, 71, 1395–1412. [Google Scholar] [CrossRef]
- Lieberman, S.; Tomer, A.; Ben-Chetrit, A.; Olsha, O.; Strano, S.; Beeri, R.; Koka, S.; Fridman, H.; Djemal, K.; Glick, I.; et al. Population screening for BRCA1/BRCA2 founder mutations in Ashkenazi Jews: Proactive recruitment compared with self-referral. Anesth. Analg. 2017, 19, 754–762. [Google Scholar] [CrossRef]
- Yablonski-Peretz, T.; Paluch-Shimon, S.; Gutman, L.S.; Kaplan, Y.; Dvir, A.; Barnes-Kedar, I.; Kadouri, L.; Semenisty, V.; Efrat, N.; Neiman, V.; et al. Screening for germline mutations in breast/ovarian cancer susceptibility genes in high-risk families in Israel. Breast Cancer Res. Treat. 2015, 155, 133–138. [Google Scholar] [CrossRef]
- Zick, A.; Kadouri, L.; Cohen, S.; Frohlinger, M.; Hamburger, T.; Zvi, N.; Plaser, M.; Avital, E.; Breuier, S.; Elian, F.; et al. Recurrent TP53 missense mutation in cancer patients of Arab descent. Fam. Cancer 2016, 16, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Locker, G.Y.; Lynch, H.T. Genetic factors and colorectal cancer in Ashkenazi Jews. Fam. Cancer 2004, 3, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Bernstein-Molho, R.; Friedman, E.; Kedar, I.; Laitman, Y.; Allweis, T.M.; Gal-Yam, E.N.; Feldman, H.B.; Grinshpun, A.; Halpern, N.; Hartmajer, S.; et al. Diagnostic yield of multigene panel testing in an Israeli cohort: Enrichment of low-penetrance variants. Breast Cancer Res. Treat. 2020, 181, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Lerner-Ellis, J.; Mighton, C.; Lazaro, C.; Watkins, N.; Di Gioacchino, V.; Wong, A.; Chang, M.C.; Charames, G.S. Multigene panel testing for hereditary breast and ovarian cancer in the province of Ontario. J. Cancer Res. Clin. Oncol. 2020, 147, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, E.; Blackford, A.; Parmigiani, G.; Biswas, S. Recent Enhancements to the Genetic Risk Prediction Model BRCAPRO. Cancer Inform. 2015, 14s2, CIN.S17292-157. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Hartley, S.; Lee, A.; Cunningham, A.P.; Archer, S.; de Villiers, C.B.; Roberts, J.; Ruston, R.; Walter, F.M.; Tischkowitz, M.; et al. CanRisk Tool—A Web Interface for the Prediction of Breast and Ovarian Cancer Risk and the Likelihood of Carrying Genetic Pathogenic Variants. Cancer Epidemiol. Biomark. Prev. 2021, 30, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Lindor, N.M.; Johnson, K.J.; Harvey, H.; Pankratz, V.S.; Domchek, S.M.; Hunt, K.; Wilson, M.; Smith, M.C.; Couch, F. Predicting BRCA1 and BRCA2 gene mutation carriers: Comparison of PENN II model to previous study. Fam. Cancer 2010, 9, 495–502. [Google Scholar] [CrossRef]
- Panchal, S.M.; Ennis, M.; Canon, S.; Bordeleau, L.J. Selecting a BRCA risk assessment model for use in a familial cancer clinic. BMC Med Genet. 2008, 9, 116. [Google Scholar] [CrossRef]
- Goverde, A.; Spaander, M.C.W.; Nieboer, D.; Ouweland, A.M.W.v.D.; Dinjens, W.N.M.; Dubbink, H.J.; Tops, C.J.; Broeke, S.W.T.; Bruno, M.J.; Hofstra, R.M.W.; et al. Evaluation of current prediction models for Lynch syndrome: Updating the PREMM5 model to identify PMS2 mutation carriers. Fam. Cancer 2017, 17, 361–370. [Google Scholar] [CrossRef]
- Bernstein-Molho, R.; Barnes-Kedar, I.; Ludman, M.D.; Reznik, G.; Feldman, H.B.; Samra, N.N.; Eilat, A.; Peretz, T.; Peretz, L.P.; Shapira, T.; et al. The yield of full BRCA1/2 genotyping in Israeli Arab high-risk breast/ovarian cancer patients. Breast Cancer Res. Treat. 2019, 178, 231–237. [Google Scholar] [CrossRef]
- Central Bureau of Statistics. Population of Israel on Eve of 2022. Available online: https://www.cbs.gov.il/en/mediarelease/Pages/2021/Population-of-Israel-on-the-Eve-of-2022.aspx (accessed on 1 November 2023).
- Lewin-Epstein, N.; Cohen, Y. Ethnic origin and identity in the Jewish population of Israel. J. Ethn. Migr. Stud. 2019, 45, 2118–2137. [Google Scholar] [CrossRef]
- Central Bureau of Statistics. Population. Available online: https://www.cbs.gov.il/en/subjects/Pages/Population.aspx (accessed on 1 November 2023).
- Zlotogora, J. The Israeli national population program of genetic carrier screening for reproductive purposes. How should it be continued? Isr. J. Heal. Policy Res. 2019, 8, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Goldfrank, D.J.; Kauff, N.D.; Robson, M.; Offit, K.; Robles-Díaz, L. Hereditary ovarian cancer in Ashkenazi Jews. Fam. Cancer 2004, 3, 259–264. [Google Scholar] [CrossRef]
- Moslehi, R.; Chu, W.; Karlan, B.; Fishman, D.; Risch, H.; Fields, A.; Smotkin, D.; Ben-David, Y.; Rosenblatt, J.; Russo, D.; et al. BRCA1 and BRCA2 Mutation Analysis of 208 Ashkenazi Jewish Women with Ovarian Cancer. Am. J. Hum. Genet. 2000, 66, 1259–1272. [Google Scholar] [CrossRef] [PubMed]
- Valle, L.; Katz, L.H.; Latchford, A.; Mur, P.; Moreno, V.; Frayling, I.M.; Heald, B.; Capellá, G. Position statement of the International Society for Gastrointestinal Hereditary Tumours (InSiGHT) onAPCI1307K and cancer risk. J. Med Genet. 2023, 60, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Laken, S.J.; Petersen, G.M.; Gruber, S.B.; Oddoux, C.; Ostrer, H.; Giardiello, F.M.; Hamilton, S.R.; Hampel, H.; Markowitz, A.; Klimstra, D.; et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat. Genet. 1997, 17, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Rozen, P.; Shomrat, R.; Strul, H.; Naiman, T.; Karminsky∥, N.; Legum, C.; Orr-Urtreger, A. Prevalence of the I1307K APC gene variant in Israeli Jews of differing ethnic origin and risk for colorectal cancer. Gastroenterology 1999, 116, 54–57. [Google Scholar] [CrossRef]
- Ceyhan-Birsoy, O.; Jayakumaran, G.; Kemel, Y.; Misyura, M.; Aypar, U.; Jairam, S.; Yang, C.; Li, Y.; Mehta, N.; Maio, A.; et al. Diagnostic yield and clinical relevance of expanded genetic testing for cancer patients. Genome Med. 2022, 14, 1–13. [Google Scholar] [CrossRef]
- Daly, M.B.; Rosenthal, E.; Cummings, S.; Bernhisel, R.; Kidd, J.; Hughes, E.; Gutin, A.; Meek, S.; Slavin, T.P.; Kurian, A.W. The association between age at breast cancer diagnosis and prevalence of pathogenic variants. Breast Cancer Res. Treat. 2023, 199, 617–626. [Google Scholar] [CrossRef]
- El Saghir, N.S.; Zgheib, N.K.; Assi, H.A.; Khoury, K.E.; Bidet, Y.; Jaber, S.M.; Charara, R.N.; Farhat, R.A.; Kreidieh, F.Y.; Decousus, S.; et al. BRCA1 and BRCA2 Mutations in Ethnic Lebanese Arab Women With High Hereditary Risk Breast Cancer. Oncol. 2015, 20, 357–364. [Google Scholar] [CrossRef]
- Abu-Helalah, M.; Azab, B.; Mubaidin, R.; Ali, D.; Jafar, H.; Alshraideh, H.; Drou, N.; Awidi, A. BRCA1 and BRCA2 genes mutations among high risk breast cancer patients in Jordan. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Laish, I.; Friedman, E.; Levi-Reznick, G.; Kedar, I.; Katz, L.; Levi, Z.; Halpern, N.; Parnasa, S.; Abu-Shatya, A.; Half, E.; et al. Double heterozygotes of BRCA1/BRCA2 and mismatch repair gene pathogenic variants: Case series and clinical implications. Breast Cancer Res. Treat. 2021, 188, 685–694. [Google Scholar] [CrossRef] [PubMed]
| Gene | Variant (DNA) | Variant (Protein) | Ethnicity (Frequency) | Gene | Variant (DNA) | Variant (Protein) | Ethnicity (Frequency) |
|---|---|---|---|---|---|---|---|
| APC | c.3920T>A | p.(Ile1307Lys) | AJ (6.5%) | BRCA2 | c.1813_1814insA | p.(Ile605?fs) | AJ |
| BRCA1 | c.2934T>G | p.(Tyr978Ter) | Iranian Jews (1%) | BRCA2 | c.771_775delTCAAA | p.(Asn257fs) | Muslim Arabs |
| BRCA1 | c.181T>G | p.(Cys61Gly) | Russian | BRCA2 | c.4284dupT | p.(Gln1429Serfs*9) | Muslim Arabs |
| BRCA1 | c.224_227delAAAG | p.(Glu75Valfs) | Jewish | BRCA2 | c.6685G>T | p.(Glu2229*) | Muslim Arabs |
| BRCA1 | c.5123C>A | p.(Ala1708Glu) | Jewish | BRCA2 | c.2482delGACT | Stop770 (exon 11) | Muslim Arabs |
| BRCA1 | c.5382insC/c.5266dupC | p.(Gln1756Profs) | AJ (0.45%) | CHEK2 | c.1100delC | p.(Thr367Metfs) | AJ |
| BRCA1 | c.68_69delAG (185delG) | p.(Glu23Valfs) | AJ (0.75%) | CHEK2 | c.1283C>T | p.(Ser428Phe) | AJ (2.2%) |
| BRCA1 | c.981_982delAT | p.(Cys328Terfs) | Jewish | CHEK2 | c.499G>A | p.(Gly167Arg) | Christian Arabs |
| BRCA1 | c.4065_4068delTCAA | p.(Asn1355_Gln1356fs) | AJ | MLH1 | c.1411_1414delAAGA | p.(Lys471AspfsX19) | AJ |
| BRCA1 | c.4153delA | p.(Glu1346fs) | Russian | MLH1 | c.1771-1772delGA | p.(Asp591Ter) | Jewish |
| BRCA1 | c.2311_2317delTTGGTAC | p.(Pro733fs) | Jewish | MSH2 | c.1906G>C | p.(Ala636Pro) | AJ (0.2%) |
| BRCA1 | c.5434 C>G | p.(Pro1812Ala) | Jewish | MSH2 | c.970_971delCA | p.(Gln324ValfsX8) | Jewish |
| BRCA1 | c.3607C>T | p.(Arg1203*) | AJ | MSH2 | c.1277-1 G>C | IVS7-1G->A | Jewish |
| BRCA1 | c.5074+3A>G | IVS17+3A > G | Muslim Arabs | MSH2 | c.1165C>T | p.(Arg389X) | Jewish |
| BRCA1 | c.5444G > A | p.(Trp1815*) | Jewish | MSH2 | c.705delA | p.(Asp236Thrfs) | Druze |
| BRCA1 | c.1224delA | p.(Val409*) | Muslim Arabs | MSH6 | c.3959_3962delCAAG | p.(Ala1320Glufs) | AJ (0.06%) |
| BRCA2 | c.7579delG | p.(Val2527*) | Ethiopian Jews (1%) | MSH6 | c.3984_3987dupGTCA | p.(Leu330ValfsX12) | AJ |
| BRCA2 | c.5946delT/c.6174delT | p.(Ser1982Argfs) | AJ | MSH6 | c.3603_3606delAGTC | p.(Arg870Serfs*) | Bedouin Arabs |
| BRCA2 | c.67+1G>A | IVS2 +1G>A | Jewish | MUTYH | c.1187G>A | p.(Gly396Asp) | AJ/Jewish |
| BRCA2 | c.7007G>C | p.(Arg2336His) | AJ/Jewish | MUTYH | c.536A>G | p.(Tyr179Cys) | AJ/Jewish |
| BRCA2 | c.8537_8538delAG/c.8765delAG | p.(Glu2846Glyfs) | Jewish | MUTYH | c.1437_1439delGGA | p.(Glu480del) | AJ/Jewish |
| BRCA2 | c.3751insA | p.(Thr1251Asnfs) | AJ/Jewish | PMS2 | c.1970dupA | p.(Asn657LysfsX6) | AJ/Jewish |
| BRCA2 | c.3847_3848del/4075delGT | p.(Val1283Lysfs) | AJ | PMS2 | c.2192T>G | p.(Leu731Ter) | AJ/Jewish |
| BRCA2 | c.4936_4939delGAAA | p.(Glu1646Glnfs) | AJ | PMS2 | c.686_687delCT | p.(Ser229fs) | Muslim Arabs |
| BRCA2 | c.4829_4830delTG/5057delTG | p.(Val1610Glyfs) | AJ/Jewish | TP53 | c.541C>T | p.(Arg181Cys) | Muslim Arabs |
| BRCA2 | c.2808_2811delACAA/3036delACAA | p.(Ala938fs) | Jewish |
| All individuals | Ashkenazi Jewish | Partly Ashkenazi Jewish | Non-Ashkenazi Jewish | Israeli Arabs | ||||||
| Total | % | Total | % | Total | % | Total | % | Total | % | |
| 1758 | 100 | 735 | 42% | 173 | 10% | 726 | 41% | 124 | 7% | |
| Gender | ||||||||||
| Male | 303 | 17% | 140 | 19% | 33 | 19% | 119 | 16% | 11 | 9% |
| Female | 1455 | 83% | 595 | 81% | 140 | 81% | 607 | 84% | 113 | 91% |
| Age | ||||||||||
| <40 | 281 | 16% | 125/735 | 17% | 52/173 | 30% | 80/726 | 11% | 24/124 | 19% |
| 40–50 | 317 | 18% | 126/735 | 17% | 48/173 | 28% | 101/726 | 14% | 42/124 | 34% |
| >50 | 1160 | 66% | 484/735 | 66% | 73/173 | 42% | 545/726 | 75% | 58/124 | 47% |
| Diagnosis | ||||||||||
| Any cancer | 1519 | 86% | 612/1519 | 40% | 127/1519 | 8% | 684/1519 | 45% | 96/1519 | 6% |
| Healthy with Family history | 239 | 14% | 123/239 | 51% | 46/239 | 19% | 42/239 | 18% | 28/239 | 12% |
| Type of cancer | ||||||||||
| Breast | 904 | 60% | 316/904 | 35% | 72/904 | 8% | 443/904 | 49% | 73/904 | 8% |
| Prostatic | 100 | 7% | 49/100 | 49% | 7/100 | 7% | 41/100 | 41% | 3/100 | 3% |
| Colorectal | 88 | 6% | 41/88 | 47% | 7/88 | 8% | 35/88 | 40% | 5/88 | 6% |
| Ovarian | 72 | 5% | 27/72 | 38% | 7/72 | 10% | 34/72 | 47% | 4/72 | 6% |
| Pancreatic | 64 | 4% | 21/64 | 33% | 7/64 | 11% | 31/64 | 48% | 5/64 | 8% |
| Other | 291 | 19% | 158/291 | 54% | 27/291 | 9% | 100/291 | 34% | 6/291 | 2% |
| 3a. Variant detection in patients with cancer—All carriers | ||||||||||
| TOTAL | AJ | PARTLY AJ | NON-AJ | ARABS | ||||||
| No | % | No | % | No | % | No | % | No | % | |
| Total | 128/1758 | 7% | 73/735 | 10% | 12/173 | 7% | 40/726 | 5% | 3/124 | 3% |
| Any Cancer | 111/1519 | 7% | 63/612 | 10% | 10/127 | 8% | 35/684 | 5% | 3/96 | 4% |
| Healthy & Family History | 17/239 | 7% | 10/123 | 1% | 2/46 | 4% | 5/42 | 12% | 0/28 | 0% |
| Breast | 67/904 | 7% | 36/316 | 11% | 6/72 | 8% | 22/443 | 5% | 4/73 | 5% |
| Prostatic | 9/100 | 9% | 6/49 | 12% | 1/7 | 14% | 2/41 | 5% | 0/3 | 0% |
| Colorectal | 6/88 | 7% | 2/41 | 5% | 0/7 | 0% | 4/35 | 11% | 0/5 | 0% |
| Ovarian | 7/72 | 10% | 3/27 | 11% | 0/7 | 0% | 4/34 | 12% | 0/4 | 0% |
| Pancreatic | 6/64 | 9% | 2/21 | 10% | 1/7 | 14% | 3/31 | 10% | 0/5 | 0% |
| Any Two Cancers | 10/76 | 13% | 7/38 | 18% | 1/10 | 10% | 2/19 | 11% | 0/9 | 0% |
| Breast and Ovarian | 1/6 | 17% | 1/4 | 25% | 0 | 0% | 0/2 | 0% | 0 | 0% |
| Breast and Pancreatic | 2/5 | 40% | 2/5 | 40% | 0 | 0% | 0 | 0% | 0 | 0% |
| Breast and Endometrial | 1/11 | 9% | 1/5 | 20% | 0/3 | 0% | 0/3 | 0% | 0 | 0% |
| Breast and CRC | 1/11 | 9% | 0/7 | 0% | 0 | 0% | 1/4 | 25% | 0 | 0% |
| CRC and Endometrial | 1/3 | 33% | 1/2 | 50% | 0/1 | 0% | 0 | 0% | 0 | 0% |
| CRC and Prostatic | 0/7 | 0% | 0/1 | 0% | 0/1 | 0% | 0/2 | 0% | 0 | 0% |
| Age < 40 | 64/281 | 23% | 42/125 | 34% | 8/52 | 25% | 13/80 | 10% | 1/24 | 4% |
| Age 40–50 | 43/317 | 14% | 25/126 | 20% | 2/48 | 4% | 15/101 | 15% | 1/42 | 2% |
| Age > 50 | 21/1160 | 2% | 6/484 | 1% | 2/73 | 3% | 12/545 | 2% | 1/58 | 2% |
| 3b. Variant detection in patients with cancer—Carriers excluding low/unknown penetrance variants (APC c.3920T>A, CHEK2 c.1283C>T, and MUTYH heterozygotes). | ||||||||||
| TOTAL | AJ | PARTLY AJ | NON-AJ | ARABS | ||||||
| No | % | No | % | No | % | No | % | No | % | |
| Total | 44/1758 | 3% | 29/735 | 4% | 6/173 | 3% | 7/726 | 1% | 2/124 | 2% |
| Any Cancer | 34/1519 | 2% | 21/612 | 3% | 4/127 | 3% | 7/684 | 1% | 2/96 | 2% |
| Healthy & Family history | 10/239 | 4% | 8/123 | 7% | 2/46 | 3% | 0/42 | 0% | 0/28 | 0% |
| Breast | 22/904 | 2% | 13/316 | 4% | 2/72 | 3% | 5/443 | 1% | 2/73 | 3% |
| Prostatic | 4/100 | 4% | 2/49 | 4% | 1/7 | 2% | 1/41 | 2% | 0/3 | 0% |
| Colorectal | 0/88 | 0% | 0/41 | 0% | 0/7 | 0% | 0/35 | 0% | 0/5 | 0% |
| Ovarian | 3/72 | 4% | 2/27 | 7% | 0/7 | 0% | 1/34 | 3% | 0/4 | 0% |
| Pancreatic | 3/64 | 5% | 2/21 | 10% | 0/7 | 0% | 1/31 | 3% | 0/5 | 0% |
| Any two cancers | 1/76 | 1% | 1/38 | 3% | 0/10 | 0% | 0/19 | 0% | 0/9 | 0% |
| Breast and ovarian | 0/6 | 0% | 0/4 | 0% | 0 | 0% | 0/2 | 0% | 0 | 0% |
| Breast and pancreatic | 0/5 | 0% | 0/5 | 0% | 0 | 0% | 0 | 0% | 0 | 0% |
| Breast and endometrial | 0/11 | 0% | 0/5 | 0% | 0/3 | 0% | 0/3 | 0% | 0 | 0% |
| Breast and CRC | 0/11 | 0% | 0/7 | 0% | 0 | 0% | 0/4 | 0% | 0 | 0% |
| CRC and endometrial | 0/3 | 0% | 0/2 | 0% | 0/1 | 0% | 0 | 0% | 0 | 0% |
| CRC and prostatic | 0/7 | 0% | 0/1 | 0% | 0/1 | 0% | 0/2 | 0% | 0 | 0% |
| Age < 40 | 20/281 | 7% | 12/125 | 10% | 4/52 | 8% | 3/80 | 4% | 1/24 | 4% |
| Age 40–50 | 17/317 | 5% | 13/126 | 10% | 1/48 | 2% | 3/101 | 3% | 0/42 | 0% |
| Age > 50 | 7/1160 | 1% | 4/484 | 1% | 1/73 | 1% | 1/545 | 0% | 1/58 | 2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abu Shtaya, A.; Kedar, I.; Mattar, S.; Mahamid, A.; Basel-Salmon, L.; Farage Barhom, S.; Naftaly Nathan, S.; Magal, N.; Azulay, N.; Levy Zalcberg, M.; et al. The Diagnostic Yield and Implications of Targeted Founder Pathogenic Variant Testing in an Israeli Cohort. Cancers 2024, 16, 94. https://doi.org/10.3390/cancers16010094
Abu Shtaya A, Kedar I, Mattar S, Mahamid A, Basel-Salmon L, Farage Barhom S, Naftaly Nathan S, Magal N, Azulay N, Levy Zalcberg M, et al. The Diagnostic Yield and Implications of Targeted Founder Pathogenic Variant Testing in an Israeli Cohort. Cancers. 2024; 16(1):94. https://doi.org/10.3390/cancers16010094
Chicago/Turabian StyleAbu Shtaya, Aasem, Inbal Kedar, Samar Mattar, Ahmad Mahamid, Lina Basel-Salmon, Sarit Farage Barhom, Sofia Naftaly Nathan, Nurit Magal, Noy Azulay, Michal Levy Zalcberg, and et al. 2024. "The Diagnostic Yield and Implications of Targeted Founder Pathogenic Variant Testing in an Israeli Cohort" Cancers 16, no. 1: 94. https://doi.org/10.3390/cancers16010094
APA StyleAbu Shtaya, A., Kedar, I., Mattar, S., Mahamid, A., Basel-Salmon, L., Farage Barhom, S., Naftaly Nathan, S., Magal, N., Azulay, N., Levy Zalcberg, M., Chen-Shtoyerman, R., Segol, O., Seri, M., Reznick Levi, G., Shkedi-Rafid, S., Vinkler, C., Netzer, I., Hagari Bechar, O., Chamma, L., ... Goldberg, Y. (2024). The Diagnostic Yield and Implications of Targeted Founder Pathogenic Variant Testing in an Israeli Cohort. Cancers, 16(1), 94. https://doi.org/10.3390/cancers16010094








