KRAS Exon 2 Mutations in Patients with Sporadic Colorectal Cancer: Prevalence Variations in Mexican and Latin American Populations
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Methods
2.3. Statistical Analysis
3. Results
3.1. Allele Frequency for KRAS Exon 2 Mutations in 150 CRC Patients from Western Mexico
3.2. Comparison of Patients’ Clinical–Pathological Features
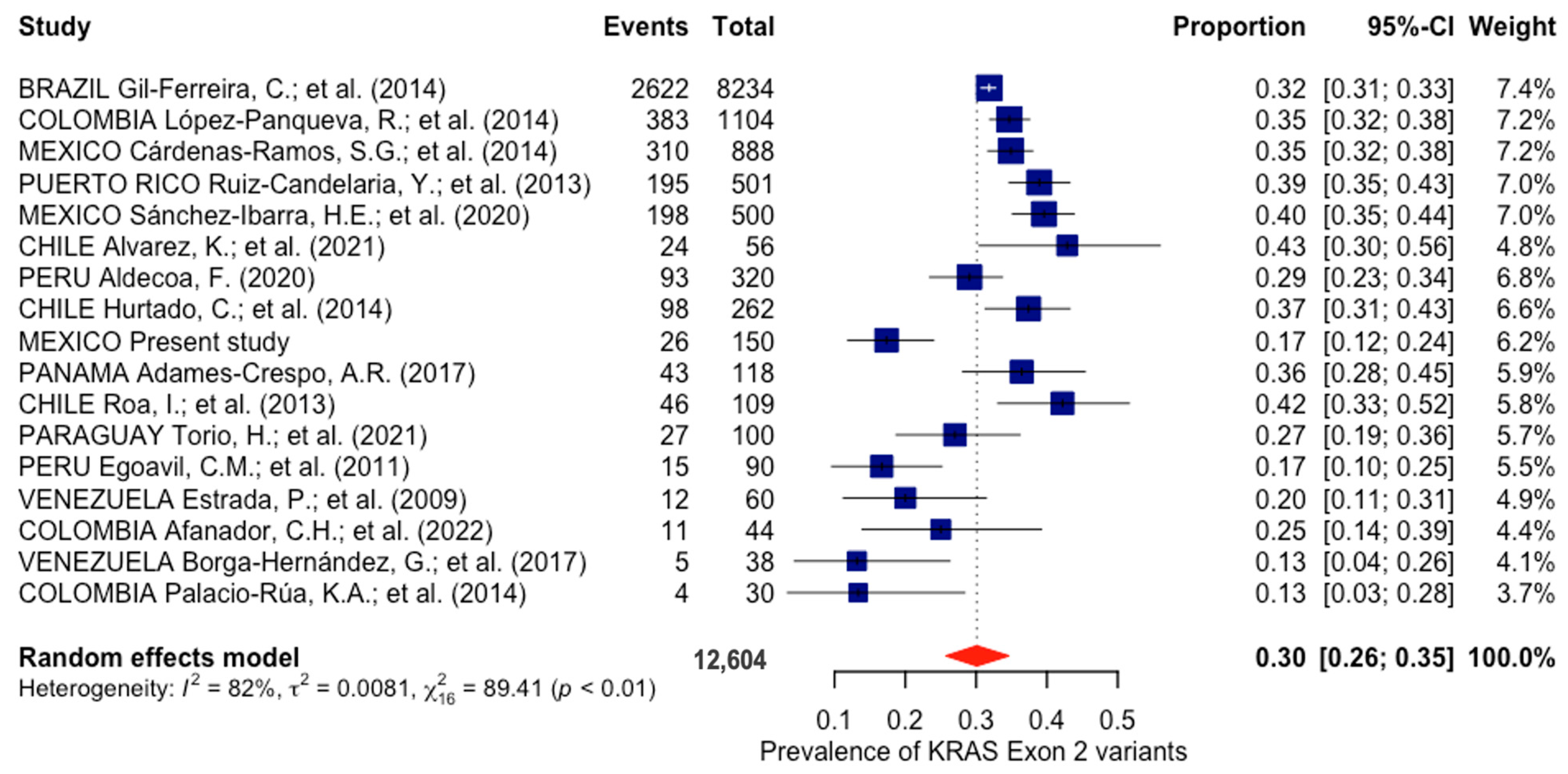
3.3. Prevalence of KRAS Exon 2 Mutations in Latin America
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today (Version 1.1); International Agency for Research on Cancer: Lyon, France. Available online: https://gco.iarc.who.int/today (accessed on 23 February 2024).
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Nguyen, L.H.; Goel, A.; Chung, D.C. Pathways of colorectal carcinogenesis. Gastroenterology 2020, 158, 291–302. [Google Scholar] [CrossRef]
- Pantsar, T. The current understanding of KRAS protein structure and dynamics. Comput. Struct. Biotechnol. J. 2020, 18, 189–198. [Google Scholar] [CrossRef]
- Ternet, C.; Kiel, C. Signaling pathways in intestinal homeostasis and colorectal cancer: KRAS at centre stage. Cell Commun. Signal. 2021, 19, 31. [Google Scholar] [CrossRef]
- Li, Z.-N.; Zhao, L.; Yu, L.-F.; Wei, M.-J. BRAF and KRAS mutations in metastatic colorectal cancer: Future perspectives for personalized therapy. Gastroenterol. Rep. 2020, 8, 192–205. [Google Scholar] [CrossRef] [PubMed]
- De Roock, W.; Jonker, D.J.; Di Nicolantonio, F.; Sartore-Bianchi, A.; Tu, D.; Siena, S.; Lamba, S.; Arena, S.; Frattini, M.; Piessevaux, H.; et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 2010, 304, 1812–1820. [Google Scholar] [CrossRef]
- Cefalì, M.; Epistolio, S.; Palmarocchi, M.C.; Frattini, M.; De Dosso, S. Research progress on KRAS mutations in colorectal cancer. J. Cancer Metastasis Treat. 2021, 7, 26. [Google Scholar] [CrossRef]
- Coppedè, F.; Lopomo, A.; Spisni, R.; Migliore, L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World. J. Gastroenterol. 2014, 20, 943–956. [Google Scholar] [CrossRef]
- Ensembl Genome Browser 111. Available online: https://www.ensembl.org/index.html (accessed on 16 February 2024).
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The frequency of Ras mutations in cancer. Cancer Res. 2020, 80, 2969–2974. [Google Scholar] [CrossRef]
- Saeed, O.; Lopez-Beltran, A.; Fisher, K.W.; Scarpelli, M.; Montironi, R.; Cimadamore, A.; Massari, F.; Santoni, M.; Cheng, L. RAS genes in colorectal carcinoma: Pathogenesis, testing guidelines and treatment implications. J. Clin. Pathol. 2019, 72, 135–139. [Google Scholar] [CrossRef]
- Sadough, A.; Afshari, M.; Rostami, F.; Barzegari, S.; Janbabaee, G.; Tabrizi, R.A.; Akbari, M.; Alizadeh-Navaei, R.; Hedayatizadeh-Omran, A.; Moosazadeh, M. Systematic review and meta-analysis on the prevalence of KRAS gene mutation in samples of colorectal cancer. WCRJ 2020, 7, e1522. [Google Scholar] [CrossRef]
- Li, J.; Gan, S.; Blair, A.; Min, K.; Rehage, T.; Hoeppner, C.; Halait, H.; Brophy, V.H. A highly verified assay for KRAS mutation detection in tissue and plasma of lung, colorectal, and pancreatic cancer. Arch. Pathol. Lab. Med. 2019, 143, 183–189. [Google Scholar] [CrossRef]
- Bohorquez, M.; Sahasrabudhe, R.; Criollo, A.; Sanabria-Salas, M.C.; Vélez, A.; Castro, J.M.; Marquez, J.R.; Mateus, G.; Bolaños, F.; Panqueva, C.; et al. Clinical manifestations of colorectal cancer patients from a large multicenter study in Colombia. Medicine 2016, 95, e4883. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef]
- Pita-Fernández, S.; González-Sáez, L.; López-Calviño, B.; Seoane-Pillado, T.; Rodríguez-Camacho, E.; Pazos-Sierra, A.; González-Santamaría, P.; Pértega-Díaz, S. Effect of diagnostic delay on survival in patients with colorectal cancer: A retrospective cohort study. BMC Cancer 2016, 16, 664. [Google Scholar] [CrossRef]
- Hultcrantz, R. Aspects of colorectal cancer screening, methods, age and gender. J. Intern. Med. 2021, 289, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Aldecoa, F. Supervivencia global del cáncer colorrectal metastásico en Lima Metropolitana: Relación con el estado mutacional del gen KRAS. Acta Méd. Peru. 2020, 37, 122–129. [Google Scholar] [CrossRef]
- Gil-Ferreira, C.; Aran, V.; Zalcberg-Renault, I.; Victorino, A.P.; Salem, J.H.; Bonamino, M.H.; Vieira, F.M.; Zalis, M. KRAS mutations: Variable incidences in a Brazilian cohort of 8,234 metastatic colorectal cancer patients. BMC Gastroenterol. 2014, 14, 73. [Google Scholar] [CrossRef]
- López-Panqueva, R.; Torres-Carvajal, M.M.; Barrera, L.; Alvarez, J.; Canon, D.; Ospina, N. Kras mutation status in Colombian patients with colorectal cancer. Analysis of 1104 consecutive cases. Ann. Oncol. 2014, 25, ii67. [Google Scholar] [CrossRef]
- Cárdenas-Ramos, S.G.; Alcázar-González, G.; Reyes-Cortés, L.M.; Torres-Grimaldo, A.A.; Calderón-Garcidueñas, A.L.; Morales-Casas, J.; Flores-Sánchez, P.; De León-Escobedo, R.; Gómez-Díaz, A.; Moreno-Bringas, C.; et al. The frequency and type of K-RAS mutations in Mexican patients with colorectal cancer: A national study. Am. J. Clin. Oncol. 2017, 40, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Candelaria, Y.; Miranda-Diaz, C.; Hunter-Mellado, R.F. K-RAS mutation profile in Puerto Rican patients with colorectal cancer: Trends from april 2009 to january 2011. Int. J. Biol. Markers 2013, 28, 393–397. [Google Scholar] [CrossRef]
- Sanchez-Ibarra, H.E.; Jiang, X.; Gallegos-Gonzalez, E.Y.; Cavazos-González, A.C.; Chen, Y.; Morcos, F.; Barrera-Saldaña, H.A. KRAS, NRAS, and BRAF mutation prevalence, clinicopathological association, and their application in a predictive model in Mexican patients with metastatic colorectal cancer: A retrospective cohort study. PLoS ONE 2020, 15, e0235490. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, K.; Cassana, A.; De La Fuente, M.; Canales, T.; Abedrapo, M.; López-Köstner, F. Clinical, pathological and molecular characteristics of Chilean patients with early-, intermediate- and late-onset colorectal cancer. Cells 2021, 10, 631. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, C.; Encina, G.; Wielandt, A.M.; Zárate, A.J.; Castro, M.; Carrillo, K.; Kronberg, U.; López-Köstner, F. KRAS gene somatic mutations in Chilean patients with colorectal cancer. Rev. Med. Chil. 2014, 142, 1407–1414. [Google Scholar] [CrossRef][Green Version]
- Adames-Crespo, A.R. Identificación de Variantes del Gen KRAS Asociadas a Cáncer Colorrectal en la Población Panameña. Master’s Thesis, Facultad de Ciencias Naturales Exactas y Tecnología, Universidad de Panamá, Panama City, Panama, December 2017. [Google Scholar]
- Roa, I.; Sánchez, T.; Majlis, A.; Schalper, K. Mutación del gen KRAS en el cáncer de colon y recto. Rev. Med. Chil. 2013, 141, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Torio, H.; Rodríguez, I.; Molina, H.; Romero, B.A.; Rojas, A.; Zárate, R.; Martínez, M.T. Detection of KRAS gene mutations in paraguayan patients with colorectal cancer. An. Fac. Cienc. Méd. 2021, 54, 33–40. [Google Scholar] [CrossRef]
- Egoavil, C.M.; Montenegro, P.; Soto, J.L.; Casanova, L.; Sanchez-Lihon, J.; Castillejo, M.I.; Martinez-Canto, A.; Perez-Carbonell, L.; Castillejo, A.; Guarinos, C.; et al. Clinically important molecular features of Peruvian colorectal tumours: High prevalence of DNA mismatch repair deficiency and low incidence of KRAS mutations. Pathology 2011, 43, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Estrada, P.; Rojas-Atencio, A.; Zabala, W.; Borjas, L.; Soca, L.; Urdaneta, K.; Alvarez-Nava, F.; Cañizales, J.; Rojas, J.; Soto, M. Frecuencia y asociación clínicopatológico de las mutaciones del oncogen K-Ras en pacientes venezolanos con cáncer colorectal. Investig. Clin. 2009, 50, 55–63. [Google Scholar]
- Afanador, C.H.; Palacio, K.A.; Isaza, L.F.; Ahumada, E.; Ocampo, C.M.; Muñetón, C.M. Caracterización molecular de pacientes con cáncer colorrectal. Biomédica 2022, 42, 154–171. [Google Scholar] [CrossRef]
- Borga-Hernández, G.; Martínez-Alfonzo, B.; Maldonado, R.J. Frecuencia mutacional KRAS-NRAS en cáncer de colon metastásico/recaída implicaciones en la supervivencia. Rev. Venez. Oncol. 2017, 29, 15–21. [Google Scholar]
- Palacio-Rúa, K.A.; Isaza-Jiménez, L.F.; Ahumada-Rodríguez, E.; Ceballos-García, H.; Muñetón-Peña, C.M. Análisis genético en APC, KRAS y TP53 en pacientes con cáncer de estómago y colon. Rev. Gastroenterol. Mex. 2014, 79, 79–89. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Timar, J.; Kashofer, K. Molecular epidemiology and diagnostics of KRAS mutations in human cancer. Cancer Metastasis Rev. 2020, 39, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.-M.; Kim, D.-W.; Oh, H.J.; Kim, H.K.; Lee, H.S.; Lee, T.G.; Shin, H.-R.; Yang, I.J.; Lee, J.; Suh, J.W.; et al. Different oncological features of colorectal cancer codon-specific KRAS mutations: Not codon 13 but codon 12 have prognostic value. World. J. Gastroenterol. 2023, 29, 4883–4899. [Google Scholar] [CrossRef]
- Dolatkhah, R.; Dastgiri, S.; Eftekhar Sadat, A.T.; Farassati, F.; Nezamdoust, M.; Somi, M.H. Impact of RAS/RAF mutations on clinical and prognostic outcomes in metastatic colorectal cancer. Bioimpacts 2021, 11, 5–14. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, J.; Yang, Y.; Lu, J.; Gao, J.; Lu, T.; Sun, J.; Jiang, H.; Zhu, Y.; Zheng, Y.; et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: Analysis of 1,110 cases. Sci. Rep. 2015, 5, 18678. [Google Scholar] [CrossRef]
- Staudacher, J.J.; Yazici, C.; Bul, V.; Zeidan, J.; Khalid, A.; Xia, Y.; Krett, N.; Jung, B. Increased frequency of KRAS mutations in African Americans compared with Caucasians in sporadic colorectal cancer. Clin. Transl. Gastroenterol. 2017, 8, e124. [Google Scholar] [CrossRef]
- Gurjao, C.; Zhong, R.; Haruki, K.; Li, Y.Y.; Spurr, L.F.; Lee-Six, H.; Reardon, B.; Ugai, T.; Zhang, X.; Cherniack, A.D.; et al. Discovery and features of an alkylating signature in colorectal cancer. Cancer Discov. 2021, 11, 2446–2455. [Google Scholar] [CrossRef]
- Judd, J.; Abdel Karim, N.; Khan, H.; Naqash, A.R.; Baca, Y.; Xiu, J.; VanderWalde, A.M.; Mamdani, H.; Raez, L.E.; Nagasaka, M.; et al. Characterization of KRAS mutation subtypes in non-small cell lung cancer. Mol. Cancer Ther. 2021, 20, 2577–2584. [Google Scholar] [CrossRef]
- Moldvay, J.; Tímár, J. KRASG12C mutant lung adenocarcinoma: Unique biology, novel therapies and new challenges. Pathol. Oncol. Res. 2024, 29, 1611580. [Google Scholar] [CrossRef]
- Frost, M.G.; Jensen, K.J.; Gotfredsen, D.R.; Sørensen, A.M.S.; Ankarfeldt, M.Z.; Louie, K.S.; Sroczynski, N.; Jakobsen, E.; Andersen, J.L.; Jimenez-Solem, E.; et al. KRAS G12C mutated advanced non-small cell lung cancer (NSCLC): Characteristics, treatment patterns and overall survival from a Danish nationwide observational register study. Lung Cancer 2023, 178, 172–182. [Google Scholar] [CrossRef]
- Woodward, A.A.; Urbanowicz, R.J.; Naj, A.C.; Moore, J.H. Genetic heterogeneity: Challenges, impacts, and methods through an associative lens. Genet. Epidemiol. 2022, 46, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.H.; Melloni, G.E.M.; Gulhan, D.C.; Park, P.J.; Haigis, K.M. The origins and genetic interactions of KRAS mutations are allele- and tissue-specific. Nat. Commun. 2021, 12, 1808. [Google Scholar] [CrossRef]
- Lee, J.K.; Sivakumar, S.; Schrock, A.B.; Madison, R.; Fabrizio, D.; Gjoerup, O.; Ross, J.S.; Frampton, G.M.; Napalkov, P.; Montesion, M.; et al. Comprehensive pan-cancer genomic landscape of KRAS altered cancers and real-world outcomes in solid tumors. NPJ Precis. Oncol. 2022, 6, 91. [Google Scholar] [CrossRef]
- Yaeger, R.; Weiss, J.; Pelster, M.S.; Spira, A.I.; Barve, M.; Ou, S.-H.I.; Leal, T.A.; Bekaii-Saab, T.S.; Paweletz, C.P.; Heavey, G.A.; et al. Adagrasib with or without Cetuximab in Colorectal Cancer with Mutated KRAS G12C. N. Engl. J. Med. 2023, 388, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Kuboki, Y.; Fakih, M.; Strickler, J.; Yaeger, R.; Masuishi, T.; Kim, E.J.; Bestvina, C.M.; Kopetz, S.; Falchook, G.S.; Langer, C.; et al. Sotorasib with panitumumab in chemotherapy-refractory KRASG12C-mutated colorectal cancer: A phase 1b trial. Nat. Med. 2024, 30, 265–270. [Google Scholar] [CrossRef]
- Neumann, J.; Zeindl-Eberhart, E.; Kirchner, T.; Jung, A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol. Res. Pract. 2009, 205, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.H.; Tougeron, D.; Shi, Q.; Alberts, S.R.; Mahoney, M.R.; Nelson, G.D.; Nair, S.G.; Thibodeau, S.N.; Goldberg, R.M.; Sargent, D.J.; et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance). Clin. Cancer Res. 2014, 20, 3033–3043. [Google Scholar] [CrossRef]
- Henry, J.T.; Coker, O.; Chowdhury, S.; Shen, J.P.; Morris, V.K.; Dasari, A.; Raghav, K.; Nusrat, M.; Kee, B.; Parseghian, C.; et al. Comprehensive clinical and molecular characterization of KRASG12C-mutant colorectal cancer. JCO Precis. Oncol. 2021, 5, 613–621. [Google Scholar] [CrossRef]
- Wang, X.; Allen, S.; Blake, J.F.; Bowcut, V.; Briere, D.M.; Calinisan, A.; Dahlke, J.R.; Fell, J.B.; Fischer, J.P.; Gunn, R.J.; et al. Identification of MRTX1133, a noncovalent, potent, and selective KRASG12D inhibitor. J. Med. Chem. 2021, 65, 3123–3133. [Google Scholar] [CrossRef]
- Study of MRTX1133 in Patients with Advanced Solid Tumors Harboring a KRAS G12D Mutation. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT05737706 (accessed on 28 February 2024).
| Reference SNP | Mutation | Allele Number/Frequency |
|---|---|---|
| rs121913529 | c.35G>A(p.Gly12Asp) | 12/0.04 |
| c.35G>T(p.Gly12Val) | 4/0.013 | |
| c.35G>C(p.Gly12Ala) | 1/0.0033 | |
| rs112445441 | c.38G>A(p.Gly13Asp) | 9/0.03 |
| Features | KRAS wt | KRAS mut | p-Value * | OR (CI) |
|---|---|---|---|---|
| n = 124 | n = 26 | |||
| Sex | ||||
| Female | 55 (44%) | 9 (35%) | 0.202 | 0.558 |
| Male | 69 (56%) | 17 (65%) | (0.226–1.378) | |
| Age | ||||
| <50 | 26 (21%) | 9 (35%) | 0.13 | 0.495 |
| ≥50 | 98 (79%) | 17 (65%) | (0.197–1.243) | |
| Tumor Localization | ||||
| Colon | 76 (61%) | 12 (46%) | 0.218 | 1.71 |
| Rectum | 48 (39%) | 13 (50%) | (0.723–4.06) | |
| ND | 1 (4%) | |||
| Histological grade | ||||
| Poorly differentiated | 28 (23%) | 4 (15%) | 0.482 | 1.5 |
| Well + moderately differentiated | 92 (74%) | 22 (85%) | (0.477–4.77) | |
| ND | 4 (3%) | |||
| Tumor stage | ||||
| I–II | 41 (33%) | 9 (35%) | 0.819 | 0.899 |
| III–IV | 76 (61%) | 15 (58%) | (0.362–2.23) | |
| ND | 7 (6%) | 2 (7%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venegas-Rodríguez, J.L.; Hernández-Sandoval, J.A.; Gutiérrez-Angulo, M.; Moreno-Ortiz, J.M.; González-Mercado, A.; Peregrina-Sandoval, J.; Ramírez-Plascencia, H.H.F.; Flores-López, B.A.; Alvizo-Rodríguez, C.R.; Valenzuela-Pérez, J.A.; et al. KRAS Exon 2 Mutations in Patients with Sporadic Colorectal Cancer: Prevalence Variations in Mexican and Latin American Populations. Cancers 2024, 16, 2323. https://doi.org/10.3390/cancers16132323
Venegas-Rodríguez JL, Hernández-Sandoval JA, Gutiérrez-Angulo M, Moreno-Ortiz JM, González-Mercado A, Peregrina-Sandoval J, Ramírez-Plascencia HHF, Flores-López BA, Alvizo-Rodríguez CR, Valenzuela-Pérez JA, et al. KRAS Exon 2 Mutations in Patients with Sporadic Colorectal Cancer: Prevalence Variations in Mexican and Latin American Populations. Cancers. 2024; 16(13):2323. https://doi.org/10.3390/cancers16132323
Chicago/Turabian StyleVenegas-Rodríguez, José Luis, Jesús Arturo Hernández-Sandoval, Melva Gutiérrez-Angulo, José Miguel Moreno-Ortiz, Anahí González-Mercado, Jorge Peregrina-Sandoval, Helen Haydee Fernanda Ramírez-Plascencia, Beatriz Armida Flores-López, Carlos Rogelio Alvizo-Rodríguez, Jesús Alonso Valenzuela-Pérez, and et al. 2024. "KRAS Exon 2 Mutations in Patients with Sporadic Colorectal Cancer: Prevalence Variations in Mexican and Latin American Populations" Cancers 16, no. 13: 2323. https://doi.org/10.3390/cancers16132323
APA StyleVenegas-Rodríguez, J. L., Hernández-Sandoval, J. A., Gutiérrez-Angulo, M., Moreno-Ortiz, J. M., González-Mercado, A., Peregrina-Sandoval, J., Ramírez-Plascencia, H. H. F., Flores-López, B. A., Alvizo-Rodríguez, C. R., Valenzuela-Pérez, J. A., Cervantes-Ortiz, S., & Ayala-Madrigal, M. d. l. L. (2024). KRAS Exon 2 Mutations in Patients with Sporadic Colorectal Cancer: Prevalence Variations in Mexican and Latin American Populations. Cancers, 16(13), 2323. https://doi.org/10.3390/cancers16132323









