1. Introduction
Establishing treatment response is a crucial aspect of precision oncology [
1]. This determination involves categorizing the patient’s status into predefined discrete classes [
2,
3]. These classes are established by pooled assessment of the margins of variation of descriptive features extracted from medical data or images. Given the wide existing variety of medical data, imaging systems, and clinical protocols in oncology, there are standardized recommendations for defining treatment response. The World Health Organization (WHO) in 1979 determined four categories of response or non-response to treatment based on tumor volume [
4]: a complete, partial, stable, and progressive disease. In 2000, the Response Evaluation Criteria for Solid Tumors (RECIST) proposed to sum one-dimensional measurements of the greatest length of all lesions extracted from X-ray tomography (CT) or magnetic resonance imaging (MRI) images [
5]. The RECIST criteria have periodically been revised, and new versions have emerged to accommodate new targeted therapies. In 2009, the Positron Emission Tomography (PET) Response Criteria for Solid Tumours (PERCIST) was introduced to provide a continuous variable for categorizing patient response to treatment. This involves calculating the percentage change between pre-and post-treatment PET scans of the peak standard uptake value (SUL) corrected for body mass or the sum of all SULs of all lesions. The RECIST and PERCIST criteria provide classification labels that respond to the macroscopic characteristics of tumors and are robust and convenient for clinical practice. However, they provide little, if any, information about the effect of a precision treatment on its pharmacological target, e.g., immune checkpoint inhibition, anti-angiogenesis, and targeted immunotherapy. Therefore, RECIST and PERCIST are of limited interest for the evaluation of new treatments, which contrasts with the increasing availability of in vivo molecular and functional imaging approaches targeting tumor hallmarks [
6], and even the interaction of these hallmarks through hybrid imaging [
7,
8,
9]. Thus, new tumor response criteria specific to the pharmacological target being addressed are needed.
Artificial intelligence (AI), a term derived from the informatics field, has shown promising potential to accelerate the evolution of healthcare toward precision oncology [
3,
10]. In particular, machine learning (ML), a branch of AI that applies statistical methods to detect patterns within datasets, enables the assembly and analysis of large volumes of data and facilitates diagnosis, prognosis, and treatment response assessment [
3,
10,
11,
12,
13,
14,
15,
16,
17,
18]. Traditionally, unsupervised ML clustering methods have been used to cluster the molecular and/or genomics patient profiles and to analyze in response to treatment [
18] with posterior supervised learning generalization [
19]. These early “omics” studies have laid the groundwork for more recent analyses using profiles created with radiology imaging features, known as radiomics. Radiomics provide a large number of quantitative features that can be used by ML methods to detect high-dimensional patterns that correlate with relevant clinical endpoints. Because they can be applied to routinely acquired images at no cost, radiomics have expanded to almost all branches of molecular imaging [
20,
21,
22], anatomical imaging [
23,
24,
25] and hybrid imaging [
12]. However, radiomics techniques possess several limitations. Firstly, the biological significance of the imaging features extracted through radiomics is often unclear. To overcome this limitation, certain studies have attempted to establish correlations between radiomics features and manually crafted biological descriptors derived from the images [
17]. However, numerous radiomics features remain inadequately understood and their clinical applicability is hampered by a lack of interpretability [
26,
27]. Secondly, radiomics involves a vast number of features computed using predefined mathematical expressions [
12]. Given that translational research datasets are often limited in size, it is probable that employing numerous features may result in overfitting during machine learning (ML) training [
28]. Therefore, most radiomics studies concentrate on large clinical databases. On the other hand, preclinical studies, which, due to animal experimentation regulations, rely on small databases, often favor the use of a few handcrafted clinical image descriptors with direct biological interpretation [
29].
In this study, we investigate the response to an antitumoral treatment of paragangliomas (PGLs), rare neuroendocrine tumors arising from extra-adrenal chromaffin cells that originate from the neural crest cells and are characterized by high metabolism and extensive vascularization [
30]. Sunitinib is an anti-angiogenic drug used to treat patients with PGLs [
31]. In previous work by our team, we showed that the response to sunitinib treatment in experimental PGLs-bearing mice was highly variable [
32]. In some animals, the tumors responded well to sunitinib, while in other animals, the tumors resumed growth in just a couple of weeks [
32]. During treatment, we documented the vascular (using ultrafast Doppler imaging (UDI)), metabolic (using PET), and anatomical (using CT) responses of mice to sunitinib using a new hybrid imaging system that combines PET-registered ultrafast sonography (PETRUS) [
33]. Imaging with PETRUS sunitinib-treated or sham-treated mice documented the effect of sunitinib on tumor growth, vessels development, and 2
-[
F]fluoro-2
deoxy-D-glucose (FDG) uptake [
32].
Here, we combine hierarchical clustering analysis (HCA) and supervised ML classifiers to identify different stages of tumor progression and the response of PGLs undergoing sunitinib or sham treatments using a few longitudinal-handcrafted vascular–molecular–anatomical features with direct biological interpretation. Multiple classical ML classifiers exist with simplified models suitable for small preclinical databases such as ours, and to date, it has not been explored which classifier is best suited to the task of identifying response to the sunitinib treatment of PGL using multimodal descriptors. Therefore, in this work, we evaluated several ML classifiers and used the one with the best performance for the generalized classification of tumor progression stages. The concatenation of the resulting stages along the duration of anti-angiogenic or sham treatments resulted in the identification of trajectories of tumor evolution.
2. Materials and Methods
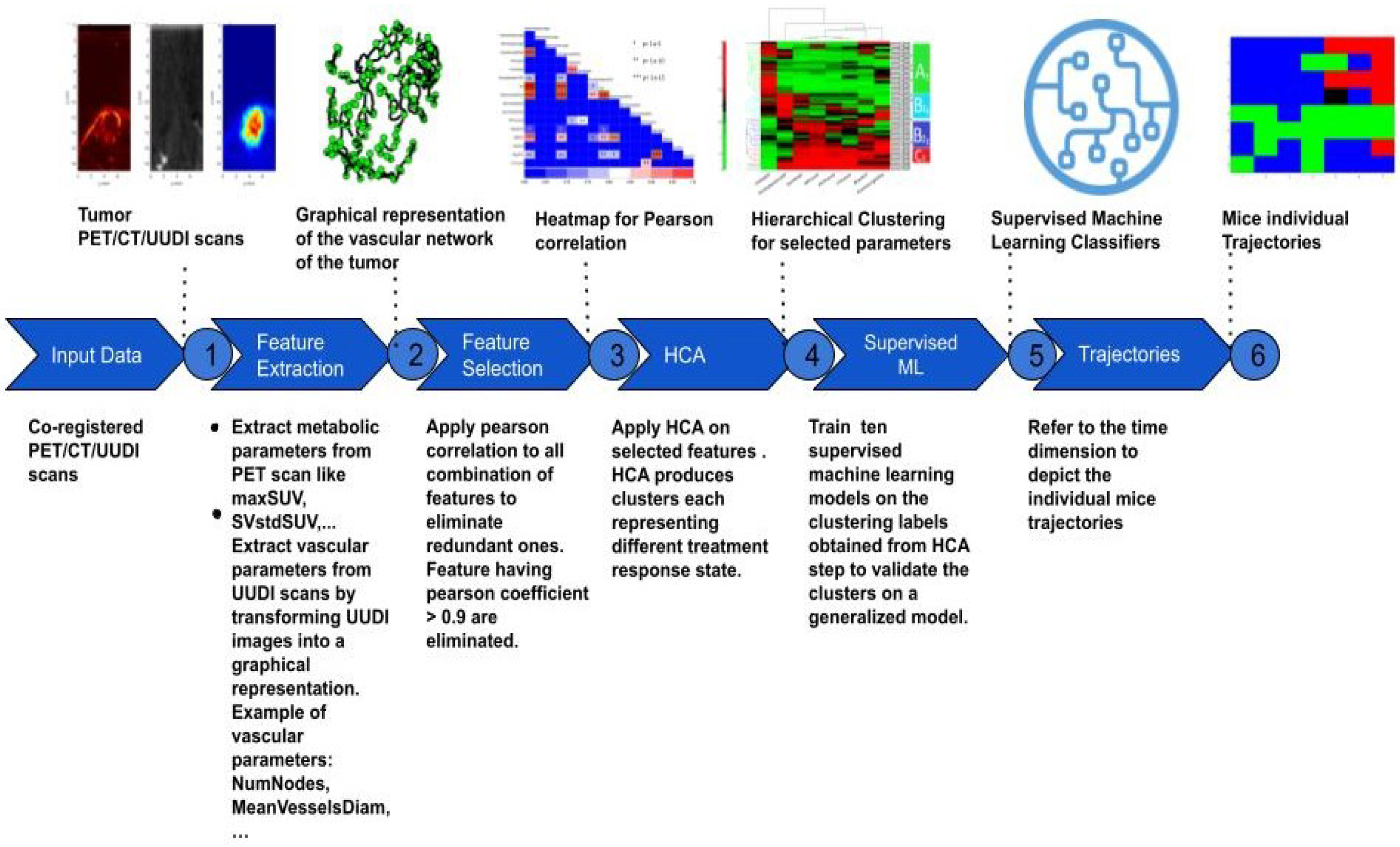
Figure 1 shows the pipeline of the framework implemented in this study that progresses from the acquisition of multi-modal image volumes to the definition of individual trajectories of response to treatment. Each element of this diagram will be described in the following sections.
2.1. Acquisition of Live Animal IMAGING Data
Two groups of mice followed the protocol of animal housing, tumor implantation, follow-up, and anti-angiogenic drug delivery described in [
32] and schematized in
Figure 2. The first group (training group) included 16 mice from [
32], while the second group (validation group) included another 11 mice that underwent the same experimental protocol. Imaging of the training group was performed at baseline, and then every week until week 3 for vehicle-treated animals (8 mice), and every week until week 6 for sunitinib-treated animals (8 mice). The validation group concerned only sunitinib-treated animals, and imaging was performed at the baseline, week 1, week 3 and week 6.
Animal experiments were approved by the French Ethical committee under reference No. 16-098 and performed by certified personnel following the French law on animal experimentation n°2013-118. In brief, adult female nude 6-week-old mice weighing 30 g (Janvier Labs, France) were implanted in the dorsal fat pad with tumors obtained from immortalized mouse chromaffin cells (imCC) carrying a homozygous knockout of the Sdhb gene (Sdhb
) as previously described [
32]. Mice were housed under controlled temperature (24 °C), relative humidity (50%), a 12/12 light/dark cycle, and free access to water and food. When the tumor volume reached 140 mm
, mice were randomly divided into a vehicle group (CON,
n = 8) and a sunitinib group (SUNI,
n = 8). The sunitinib group received sunitinib malate (Clinisciences, A10880-500) daily at a dose of 50 mg/kg body weight for 6 consecutive weeks, administered by oral gavage of 200 µL in a 10 mg/mL DMSO/PBS (1:4) solution. The control group received daily 200 μL doses of the DMSO-PBS solution (1:4) for 3 weeks. Mice were euthanized if the tumor volume exceeded UKCCCR recommendations [
34] or if they showed signs of advanced cancer disease.
The effect of sunitinib was monitored non-invasively using the hybrid In vivo imaging technology PETRUS (positron emission tomography registered ultrafast sonography) [
33], which allows for the simultaneous acquisition of tissue metabolism using [
F]Fluorodeoxyglucose (FDG) PET, computed tomography (CT) and ultrafast ultrasound Doppler imaging (UUDI) [
33]. PETRUS simultaneously reads the cellular metabolism activity alongside the micro-vascular architecture within the tumor, ensuring unimpaired physiological conditions for both sets of spatially co-registered features [
32].
2.2. Description of Database Formation
Each PETRUS acquisition comprised three image volumes registered in a common time and space reference frame that defined a multiparametric cube surrounding the animal tumor. The features describing the metabolic, vascular, and anatomical characteristics of the tumor were extracted from the PET, UUDI, and CT images, respectively (
Table 1). A volume of interest (VOI) covering the whole tumor was defined on the PET images by segmenting voxels with an FDG standard uptake value (SUV) greater than 30% of the tumor’s peak SUV at 50–60 min post-injection [
35]. This VOI was used to create a binary mask that was applied to the three spatiotemporal registered volumes. From the masked PET image, the following metabolic features were extracted: mean, coefficient of variance, minimum and maximum of standard uptake values (MeanSUV, CVstdSUV, MinSUV, MaxSuv), and PET volume (PETVolume). The masked UUDI volume was filtered using a Hessian-based vessel enhancement filter, and vessels were segmented using predefined thresholds [
36] and skeletonized using an iterative ordered thinning-based skeletonization method [
37,
38]. The skeletonized mask of vessels was transformed into a graph of nodes and edges representing the vascular network of the tumor. Using this graph, the following features describing the topology of the tumor vascularization were calculated: mean, minimum and maximum vessel length (MeanVesselsLength, MinVesselsLength, MaxVesselsLength), mean vessels tortuosity (Tort), which is the shortest distance between nodes divided by the vessel length), vessels length dispersion (VesselsLength-Disp), which is the standard deviation of the vessels length divided by the mean of the vessels length, number of nodes (NumNodes), density of nodes (DensityNodesinUSV), mean vessels diameter (MeanVesselsDiam) and ultrasound volume (USVolume), which is the number of voxels of the vascular skeleton multiplied by the voxel volume. The quantification of PETRUS images was performed using MATLAB version R2021b. The CT volume (CTVolume) was delineated from the fat pad surrounding the tumor.
The working database assembled all 15 features extracted from the imaging modalities, as well as a unique record number that defined the mouse, the week of the imaging session (where week zero (W0) is the pre-treatment imaging session and W1-6 is the rest of the treatment weeks), and the treatment group assignment (CON for sham-treated mice; SUNI for sunitinib-treated mice). Data were divided into 3 subgroups, (i) containing the SUNI mice in the training group, aggregating a total of 54 records (ii) containing the CON mice from the training group, forming a total of 27 records, and (iii) containing the SUNI mice from the validation group forming a total of 28 records.
2.3. Feature Selection
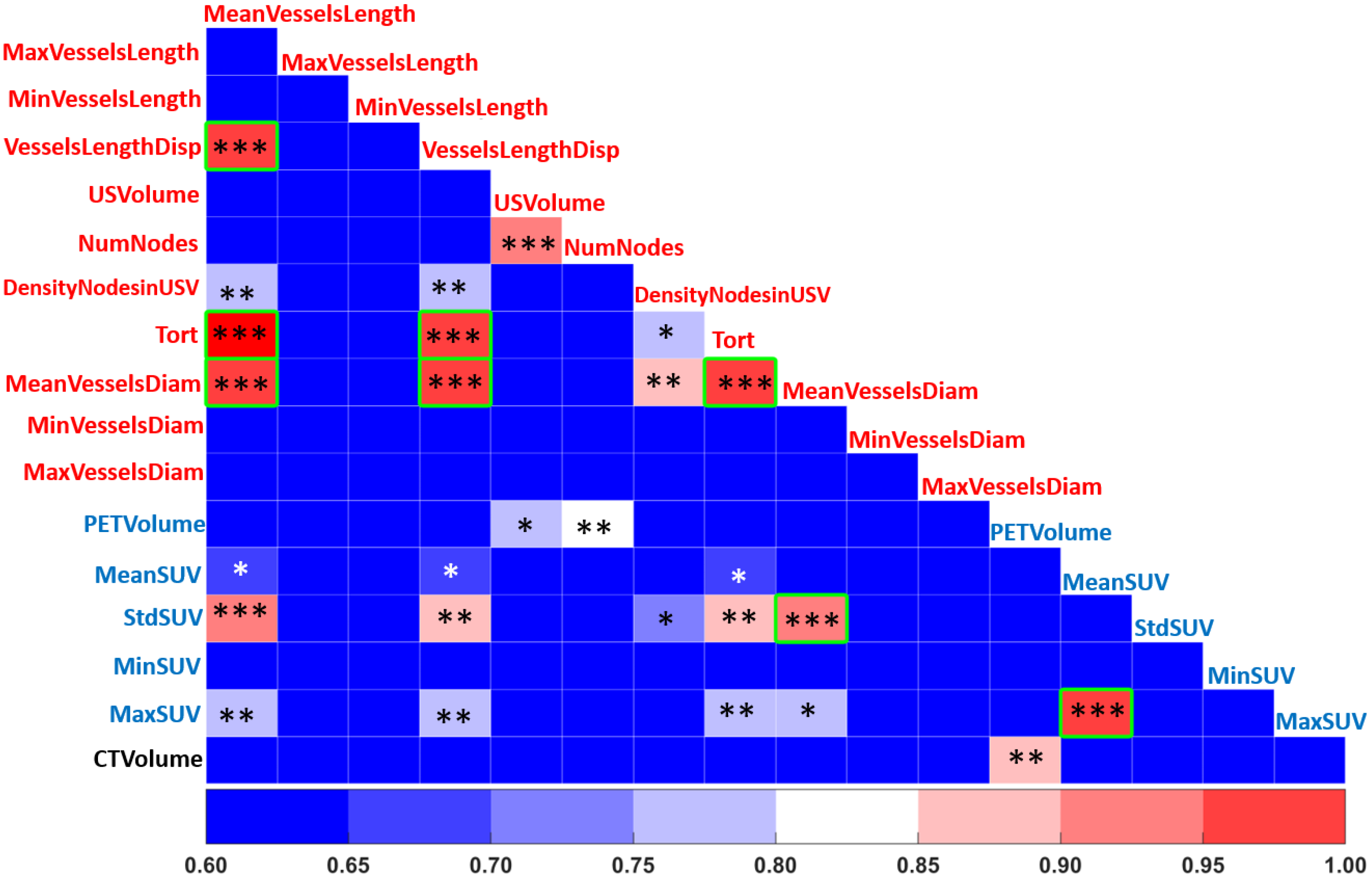
Feature selection is an important pre-processing step that affects the accuracy and decreases the training time of any classifier. By removing non-useful or redundant features, the dimensionality of the feature space can be reduced, an essential step to improve the performance of a classifier [
39]. In order to identify linear correlations between the different features, we applied a Pearson correlation using a Pearson coefficient
> 0.9 (
p-value < 0.05) to detect redundant features [
40]. In addition, non-informative features with a low coefficient of variation (CV < 0.1) were removed.
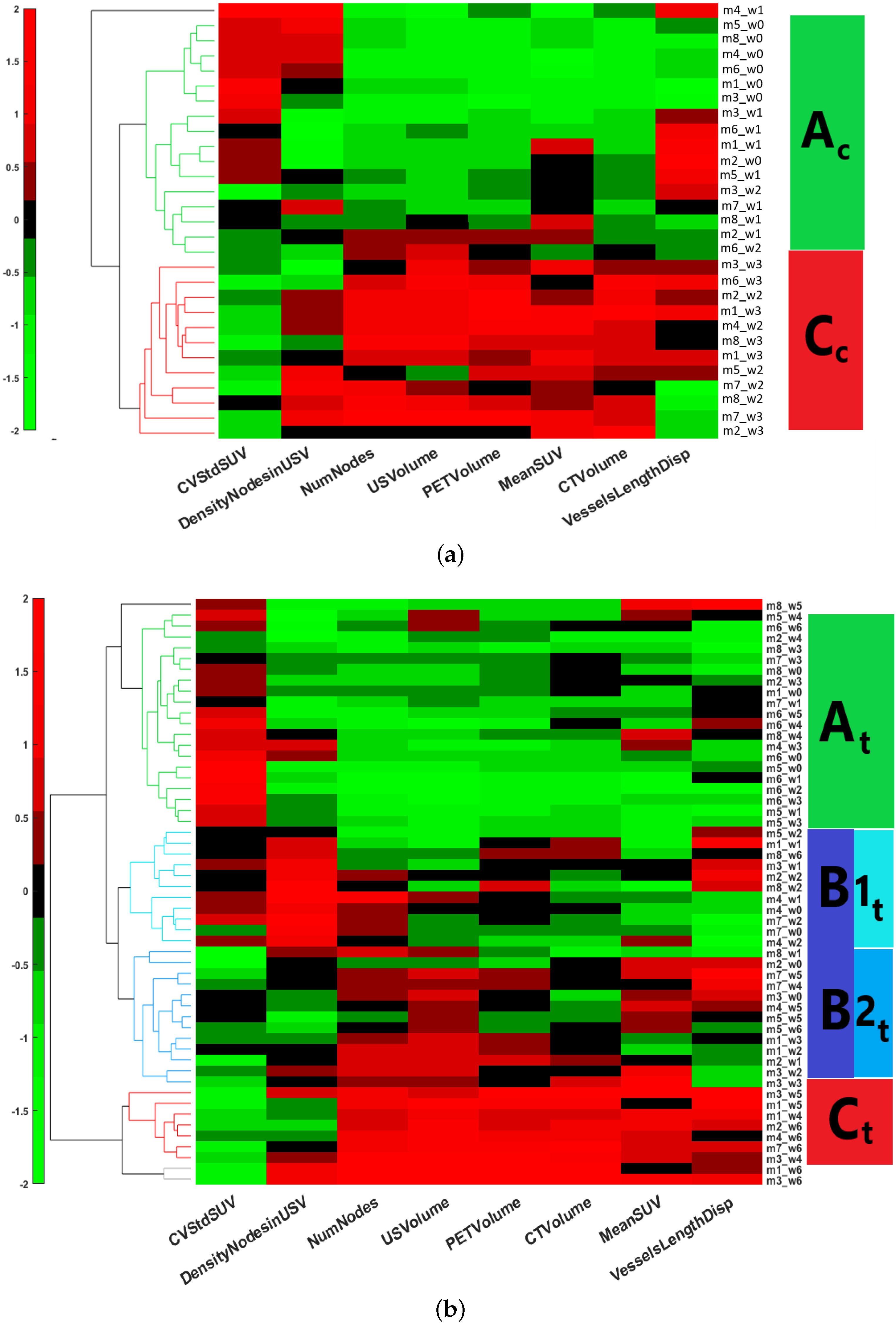
2.4. Unsupervised Classification: Hierarchical Clustering
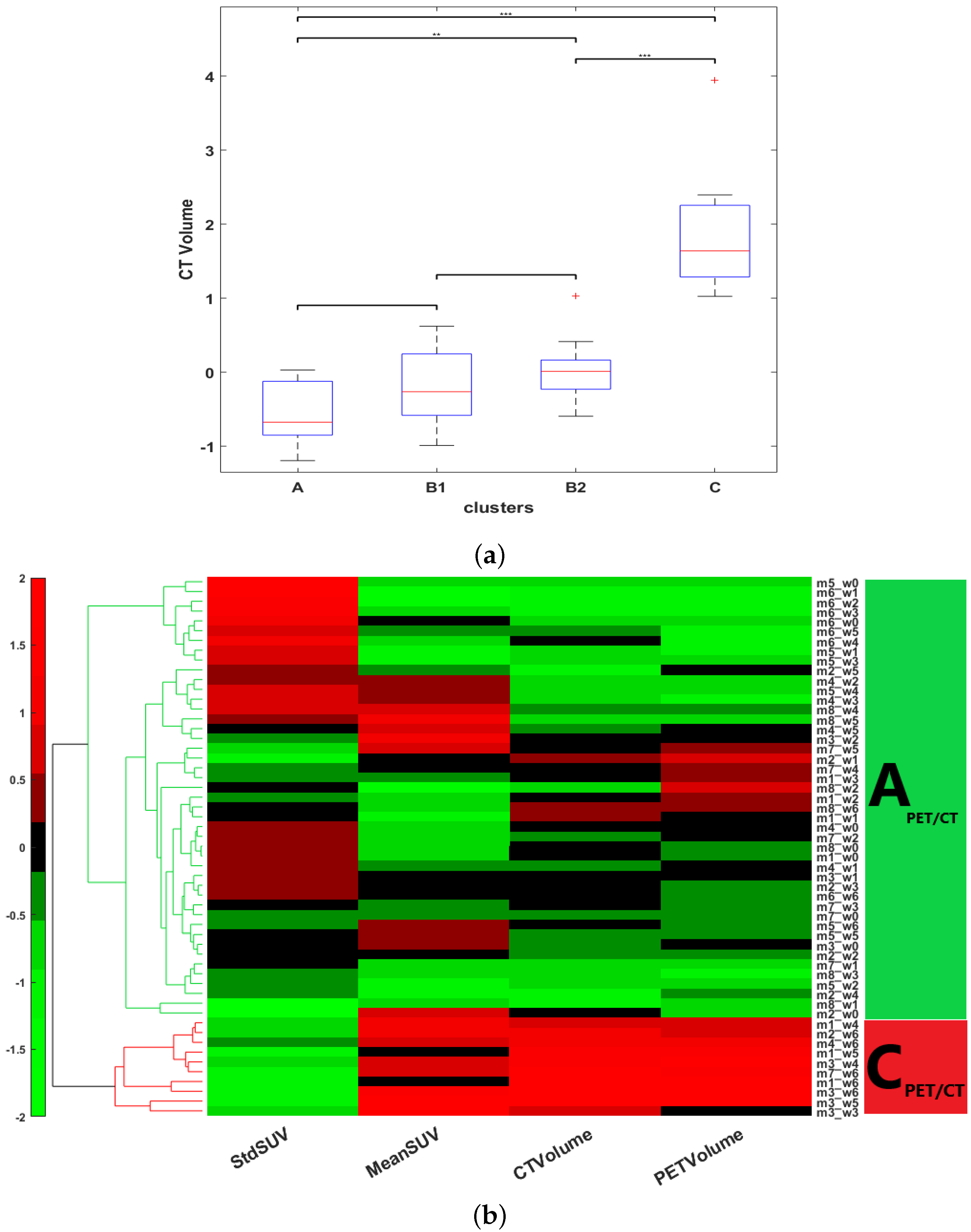
One of the fundamental objectives of our study was the determination of phenotypically representative clusters, each cluster being a representative combination of metabolic, anatomical and vascular features associated with a stage of response to sunitinib. Clusters were determined by the individual response of the subject, independently of the time of treatment by assembling all the longitudinal features extracted. HCA, an unsupervised machine-learning clustering approach [
41], was used to stratify the tumor response by finding common metabolic, anatomical and vascular phenotypic patterns of the image descriptors selected. The HCA was applied on each of the training datasets separately,
and
, in order to determine whether or not the treatment changes the time course of tumor evolution. First, the input data were standardized using the z-score. Then, the interrelationship between individual records was measured by computing the unweighted average Euclidean distance. This was followed by computing the average link as a similarity metric to define the closest pair of clusters. Finally, a heat map with dendrograms was constructed to display the patterns observed and the clusters identified. The length of the dendrogram branches connecting records and features is inversely proportional to the similarity of their profiles. Gap statistics [
42] was applied in order to evaluate the optimal number of clusters, and Welch’s
t-test was applied to identify significantly different clusters [
43]. The outcome of this analysis provided the optimal number of clusters corresponding to a particular phenotype identified for each instance in the data-base. HCA and statistical tests were implemented in MATLAB (version 2021-b) using the
clustergram, ttest2, and evalclusters functions, respectively.
2.5. Supervised Classification: Model Building and Validation
To test the stability of the method, we compared the clustering results applied on an external population ( ) to a classification produced as a generalization of the clustering performed on our initial population (). More precisely, we considered the clusters of the initial population () as classes of a supervised classification algorithm to predict the classes expected in the new population ().
Because our training dataset has an unbalanced number of instances per class, which can undermine the predictability of the models, we performed oversampling through the synthetic minority over-sampling technique (SMOTE), which balances the minority classes [
44]. This technique uses the k-nearest neighbors approach to synthesize new observations based on the existing records. We applied smote using the four nearest neighbors to balance each of the four clusters (A, B1, B2, and C).
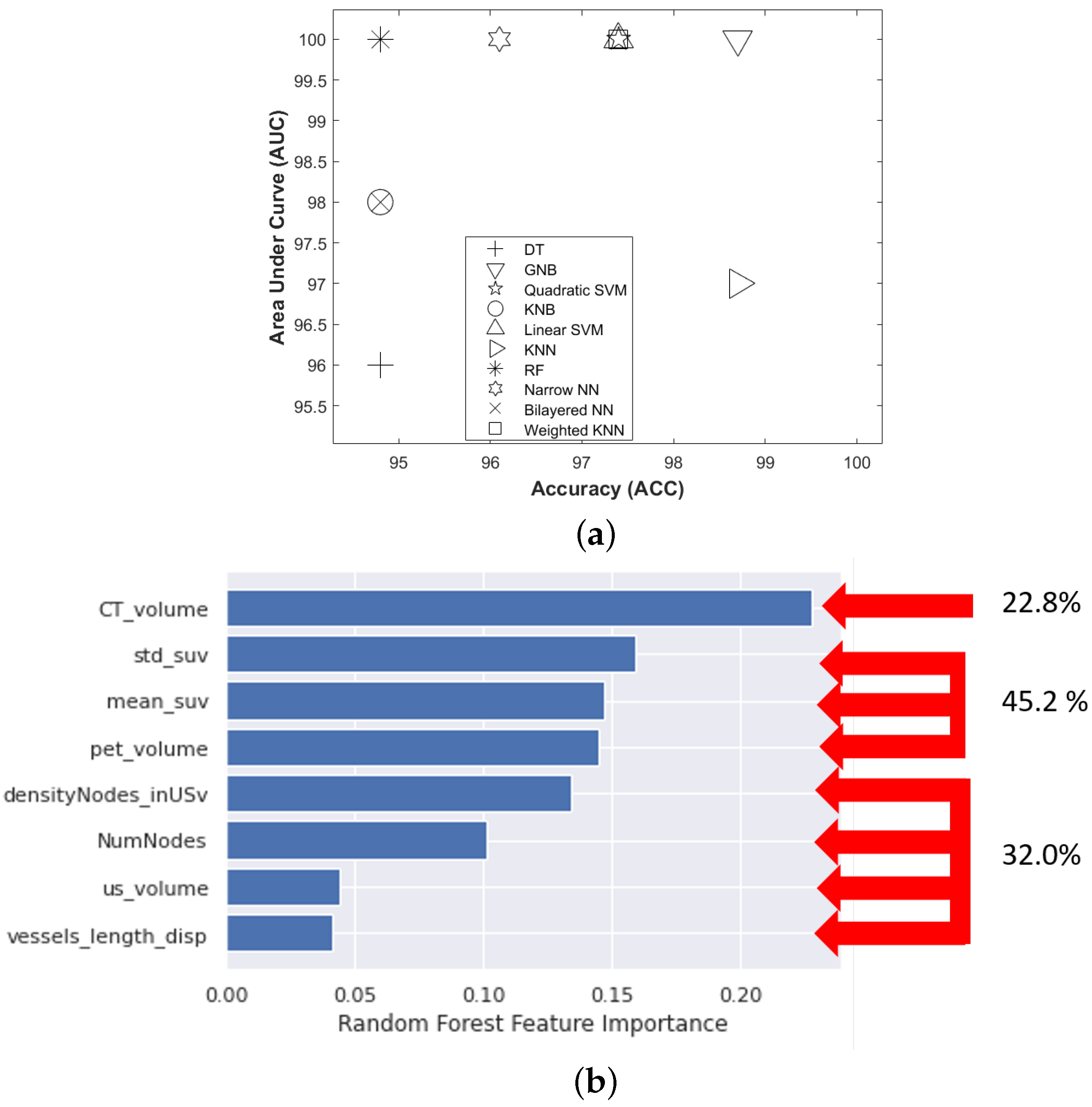
The selected features of our were brought into ten machine learning classifiers, including decision tree (DT), Gaussian naive Bayes (GNB), kernel naive Bayes (KNB), linear support vector machine (Linear SVM), quadratic support vector machine (Quadratic SVM), k-nearest neighbors (KNN), weighted k-nearest neighbors (Weighted KNN), random forest (RF), narrow neural network (Narrow NN), bilayered neural network (Bilayered NN). The best-performing model was selected by comparing the area under the receiver operating characteristic curve (AUC) and accuracy (ACC) values. The control parameters of the best model were further optimized by Bayesian optimization and five-fold cross-validation to evaluate the performance of the classifier. All classifiers were trained and validated using the classification learner application implemented in MATLAB version 2021-b.
In order to check the relative importance of each of the metabolic, vascular, and morphological features in the classification problem, we used the predictor importance attribute associated with the RF model. The predictor importance attribute is an implicit technique performed using the RF model and is evaluated using the Gini impurity criterion index. This index is based on the principle of impurity reduction to provide the power of each feature in the classification [
45].
2.6. Identification of Trajectories of Treatment Responses
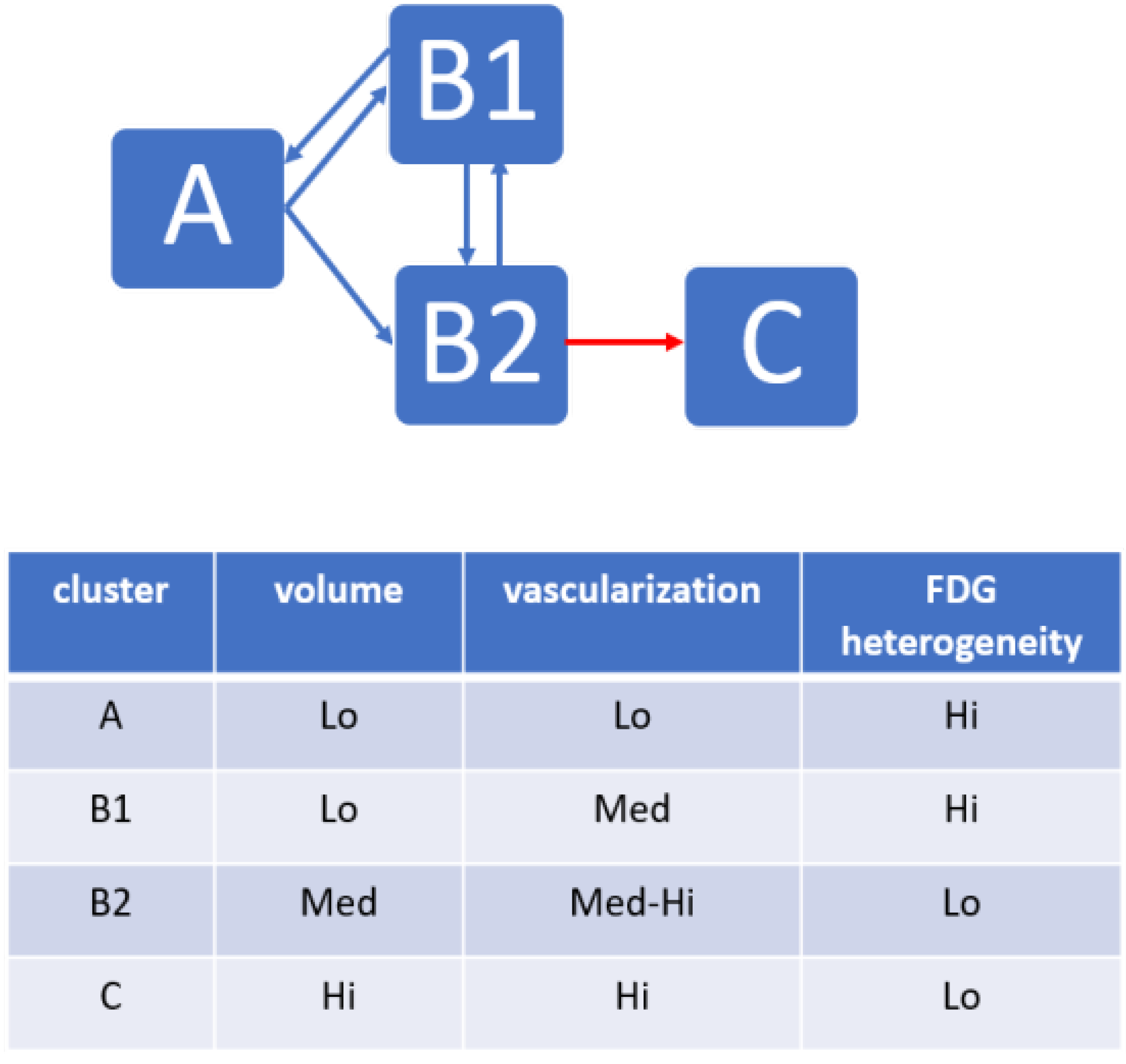
We then tested whether the records assembled within each cluster, corresponding to a tumor state with specific biomarkers, could represent a chronological stage of tumor evolution. By referring back to the time point of each record (the week after the beginning of treatment) in both the CON and SUNI groups, the clusters were ordered chronologically, and a time-dependent trajectory was obtained for each mouse. We applied an test to the states at each of the seven time points of the study (classes obtained from the HCA, considering A = 1, B1 = 2, B2 = 3, and C = 4) to determine if these states indicated temporal stages of treatment response. Finally, the transitional matrix between clusters was analyzed.
4. Discussion
Previous studies used ML to study the correspondence between gene expression and tumor progression [
47,
48], including PGL [
49]. To the best of our knowledge, this is the first application of ML based on HCA and supervised ML algorithms to noninvasive multimodal imaging of PGL. PGL lesions may concern the whole sympathetic and parasympathetic chains from the base of the skull to the pelvis. Germline mutations in one of the SDHx genes are responsible for approximately 20% of cases of PGL and also in some other tumors [
50,
51]. PGL patients carrying SDHx mutations show a higher rate of metastatic disease and a lower rate of survival than non-SDHx PGL patients. Surgery is not without risk and may be impractical for numerous or misplaced lesions. Clinical trials with sunitinib have reported modest results in SDHB mutation carriers [
32,
52].
There is an international consensus on the use of repeated non-invasive imaging for the screening, management and follow-up of PGL patients [
53], as well as for asymptomatic SDHx mutation carriers [
54]. Our results show that unsupervised ML of serial noninvasive and multimodal imaging data can define the phenotypic stages of mouse Sdhb
PGL tumors under anti-angiogenic treatment. The main finding is that, although the records fed to the ML algorithm had not been time stamped for the duration of treatment, unsupervised ML applied to multimodal multiparametric imaging features yielded clusters relevant to disease progression and to the response to sunitinib. In the sham-treated group, all mice switched, generally in less than three weeks, from cluster
, an early stage with small and poorly developed tumors, low vascularization, and heterogeneous FDG uptake, to cluster
, an advanced stage with large tumors, large vessels, high and relatively homogeneous FDG uptake, corresponding to an end-stage cancer disease. In the sunitinib-treated group, a given tumor from a given mouse could, over time, move from one cluster to another, suggesting that the changes from one cluster to another depicted trajectories of tumor evolution related to the response or the escape from treatment. Some sunitinib-treated tumors showed a progression similar to sham-treated tumors, which infers that sunitinib-treated mice entering the advanced-stage
cluster have escaped sunitinib treatment.
Two other clusters,
, and
, representing intermediate tumor stages, were observed only in the sunitinib-treated group, supporting the view that their phenotypes represent the effects of sunitinib on PGL tumors. The first one,
, encompassed small-sized tumors with a significant but moderate level of vascularization and heterogeneity in the distribution of glucose uptake. The second cluster,
, encompassed tumors of moderate volume and vascularization, and low heterogeneity in the distribution of glucose uptake. ML did not identify these two intermediate stages when the vascular features derived from ultrafast ultrasound were removed from the analysis. Therefore, the
and
intermediate stages identified the effect of sunitinib on tumor vascularization, likely by inhibition of vascular endothelial growth factors receptors (VEGFRs), the major pharmacological target of the drug [
55]. Previous studies have documented the relationship between tumor vascular types and the malignancy of PGL or pheochromocytoma, which is the adrenal form of paraganglioma. In a pioneering study, Favier et al. [
56] divided pheochromocytomas into two groups according to their vascular architecture. Tumors with short, straight vascular segments distributed regularly over large areas of tumoral tissue had a vascular density equivalent to that observed in the normal adrenal medulla, while tumors with longer vascular segments of irregular length and a lower density of vessels corresponded to the malignant form. These regular and irregular patterns observed using in vitro stained sections of tumor tissue samples are remarkably similar to the states that we observed here in vivo, A and C [
56]. A few years later, a study attempted to use “Favier’s criteria” of the vascular patterns on histological sections of pheochromocytomas and PGL for the prediction of clinical behavior [
57]. Again, malignancy was associated with an irregular vascular pattern; however, in spite of the correct agreement between observers, sensitivity and specificity were relatively modest and the authors concluded that vascular patterns, although useful, were not sufficient as “stand-alone […] prognostic tool for the distinction between benign and malignant PCC…”. Interestingly, we observed a difference in vascular morphology reminiscent of regular/irregular patterns under sunitinib treatment, tumor vessels being larger in diameter at stage
than at stage
(see
Figure 6b). Therefore, while the analysis of vascularization may by itself not be sufficient, and notwithstanding the fact that the morphology of vessels in fixed tissue may not reflect their in vivo morphology, there is good agreement with changes in vessel morphology and the response to sunitinib, suggesting that the in vivo exploration of vascular morphology may be useful for the management of PGL. In addition, the link between FDG heterogeneity and microvascular density was theorized using a spatiotemporal computational model [
58]. Our present results are in agreement with the authors’ conclusion that “as microvascular densities increase […], the spatiotemporal distribution of total FDG uptake by tumor tissue changes towards a more homogenous distribution [
58]”. Therefore, combined imaging of vascularization and metabolism could be an advantage for the follow-up of PGL patients under treatment.
Interestingly, all of the three mice that pertained to a B cluster ( or ) at baseline ended up in the cluster at the end of the 6-week sunitinib treatment, while only one of the four mice pertaining to the cluster at baseline ended up in the cluster. Although further studies are necessary to determine whether the tumor’s biology prior to the administration of sunitinib could predict future escape from treatment, this may indicate that tumors that have already developed a significant vessel network are less prone to respond to sunitinib therapy. Thus, even though the switch from to was reversible under sunitinib treatment ( to ), increased vascularization and decreased metabolic heterogeneity defining the stage were necessary features for passage to the stage, in other words, for escape from sunitinib treatment. From a cancer biology point of view, this suggests that escape from sunitinib treatment involves both a metabolic and a vascular switch.
From a statistical point of view, the analysis of each record independently without time stamping allows to extraction of information regarding the rates of tumor evolution in a small group of eight mice. This would not have been possible with conventional methods based on time-stamped groups of individuals unless the number of individuals would have been drastically increased. Considering the necessity to reduce the use of animals in research, the unsupervised method for the analysis of multimodal imaging presented here is an attractive alternative for the preclinical exploration of treatments in cancer models.
Moreover, cluster extraction using multiple features could allow gaining a better understanding of the sequence of events underlying drug response. The fact that cancer is a multiform disease with multiple intermingled hallmarks has been extensively documented and reviewed in the classical paper by Hanahan and Weinberg [
59]. Therefore, it is unlikely that assessing only one biomarker, even one that informs on the activity toward the pharmacological target, may be sufficient to assess treatment response, and, even less so, to identify complex escape mechanisms. All in all, our results support the recourse to multimodal imaging with the careful selection of relevant imaging biomarkers, ideally including one or several biomarker(s) of the hallmark targeted by the treatment. In this respect, other tumor variants could also benefit from similar approaches extracting biomarkers specific to the tumor type and/or treatment. Finally, it may also be interesting to apply a radiomics analysis in order to compile mathematically defined image features and determine whether they represent phenotypic states predictive of tumor stage predictive of treatment response.
The main limitation of our study is that it is based on preclinical data. Serial imaging sessions, even non-invasive, are difficult to envision in clinical settings. However, we show that comprehensive longitudinal explorations in a patient-relevant animal model can identify key imaging features leading to sunitinib resistance, and may inspire translational methods for tumor follow-up in patients. ML analysis of multimodal hybrid imaging could offer individual monitoring of the vascular and metabolic states of a tumor, thus providing valuable information for personalized treatment decisions. Our results need to be further validated on prospective cohorts and extended to the clinical situation.