A Handheld Visible Resonance Raman Analyzer Used in Intraoperative Detection of Human Glioma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Glioma Grading Using Machine Learning
4. Analysis and Results
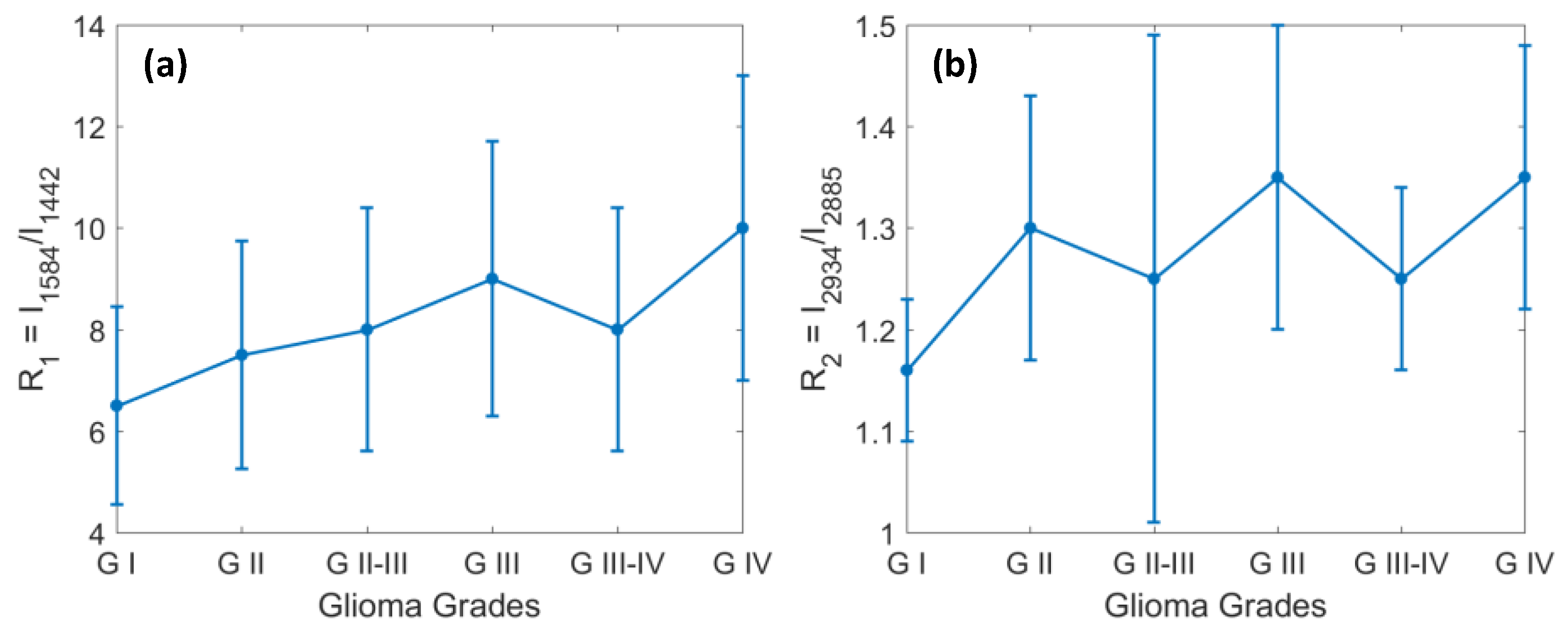
4.1. The Changes in Lipids and Proteins for Identification of Grades 1 through 4 of Glioma
4.2. Identification of Glioma Margin by Carotenoids and the Ratio of Protein to Lipids
4.3. The New VRR Biomarkers of Glioma in the High-Wavenumber Region
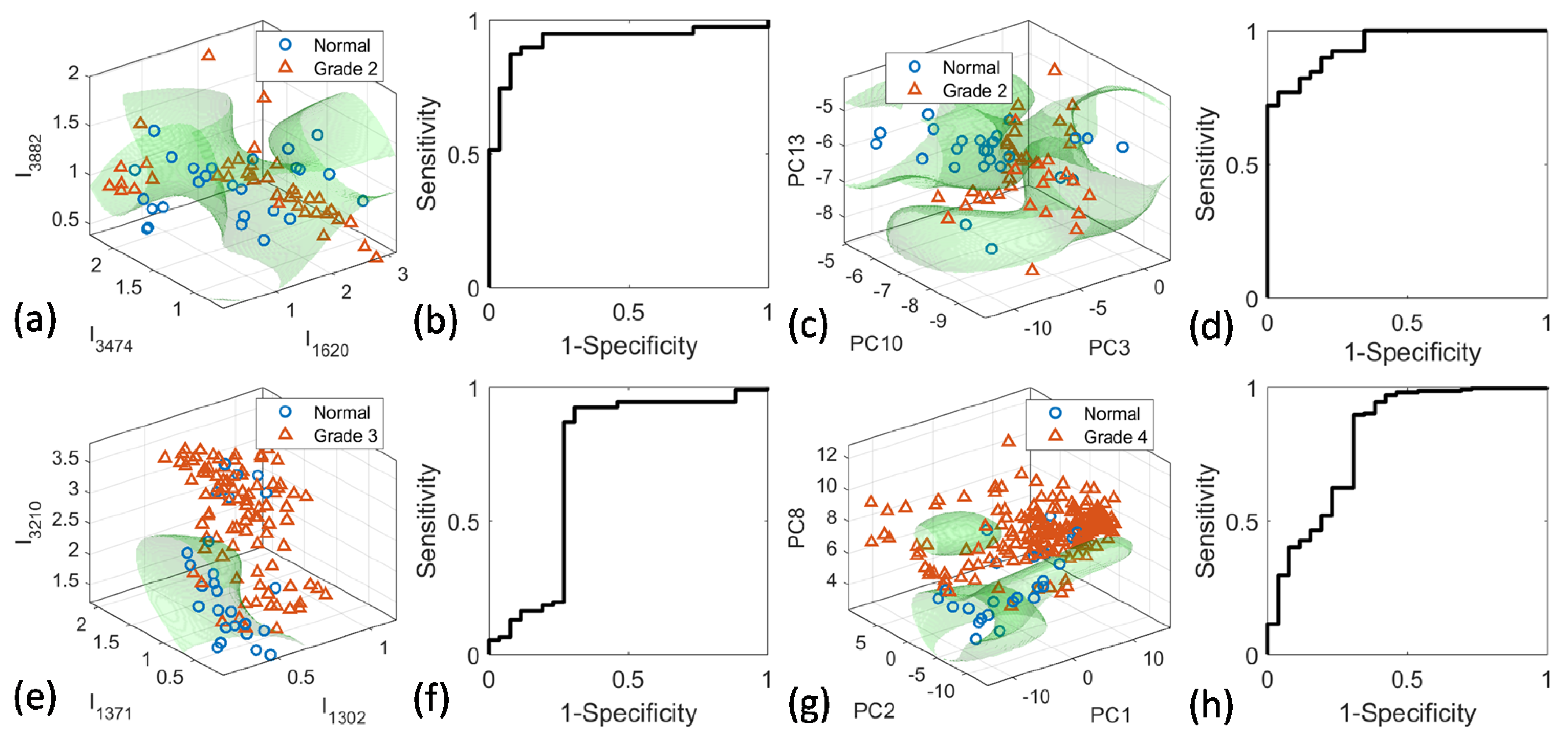
4.4. PCA-SVM and Peak-SVM Analyses
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro-Oncol. 2019, 21, v1–v100. [Google Scholar] [CrossRef]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.J.; Brennan, M.C.; Li, M.; Church, E.W.; Brandmeir, N.J.; Rakszawski, K.L.; Patel, A.S.; Rizk, E.B.; Suki, D.; Sawaya, R.; et al. Association of the extent of resection with survival in glioblastoma: A systematic review and meta-analysis. JAMA Oncol. 2016, 2, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Silantyev, A.S.; Falzone, L.; Libra, M.; Gurina, O.I.; Kardashova, K.S.; Nikolouzakis, T.K.; Nosyrev, A.E.; Sutton, C.W.; Mitsias, P.D.; Tsatsakis, A. Current and future trends on diagnosis and prognosis of glioblastoma: From molecular biology to proteomics. Cells 2019, 8, 863. [Google Scholar] [CrossRef]
- Mair, M.J.; Geurts, M.; van den Bent, M.J.; Berghoff, A.S. A basic review on systemic treatment options in WHO grade II–III gliomas. Cancer Treat. Rev. 2020, 92, 102124. [Google Scholar] [CrossRef]
- Wijnenga, M.M.J.; French, P.J.; Dubbink, H.J.; Dinjens, W.N.M.; Atmodimedjo, P.N.; Kros, J.M.; Smits, M.; Gahrmann, R.; Rutten, G.-J.; Verheul, J.B.; et al. The impact of surgery in molecularly defined low-grade glioma: An integrated clinical, radiological, and molecular analysis. Neuro-Oncol. 2018, 20, 103–112. [Google Scholar] [CrossRef]
- D’Amico, R.S.; Englander, Z.K.; Canoll, P.; Bruce, J.N. Extent of resection in glioma-A review of the cutting edge. World Neurosurg. 2017, 103, 538–549. [Google Scholar] [CrossRef]
- Cahill, D.P. Extent of resection of glioblastoma: A critical evaluation in the molecular era. Neurosurg. Clin. N. Am. 2021, 32, 23–29. [Google Scholar] [CrossRef]
- Parney, I.F.; Berger, M.S. Chapter 15-Principles of brain tumor surgery. In Handbook of Clinical Neurology; Aminoff, M.J., Boller, F., Swaab, D.F., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; Volume 104, pp. 187–213. [Google Scholar]
- Landy, H.J.; Lee, T.T.; Potter, P.; Feun, L.; Markoe, A. Early MRI findings in high grade glioma. J. Neurooneol. 2000, 47, 65–72. [Google Scholar] [CrossRef] [PubMed]
- McGahan, J.P.; Ellis, W.G.; Budenz, R.W.; Walter, J.P.; Boggan, J. Brain gliomas: Sono-graphic characterization. J. Radiol. 1986, 159, 485–492. [Google Scholar] [CrossRef]
- Picca, A.; Berzero, G.; Sanson, M. Current therapeutic approaches to diffuse grade II and III gliomas. Ther. Adv. Neurol. Disord. 2018, 11, 1756285617752039. [Google Scholar] [CrossRef]
- Al-Muslet, N.A.; Ali, E.E.; El Mahal, M. Spectroscopic analysis of bladder cancer tissues using laser Raman spectroscopy. Res. J. Pharm. 2013, 1, 1–11. [Google Scholar]
- Krafft, C.; Sobottka, S.B.; Schackert, G.; Salzer, R. Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors. Analyst 2005, 130, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Matthews, Q.; Jirasek, A.; Lum, J.; Duan, X.; Brolo, A.G. Variability in Raman spectra of single human tumor cell cultured in vitro: Correlation with cell cycle and culture confluency. Appl. Spectrosc. 2010, 64, 871–887. [Google Scholar] [CrossRef] [PubMed]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Abramczyk, H.; Brozek-Pluska, B. New look inside human breast ducts with Raman imaging. Raman candidates as diagnostic markers for breast cancer prognosis: Mammaglobin, palmitic acid and sphingomyelin. Anal. Chim. Acta 2016, 909, 91–100. [Google Scholar] [CrossRef]
- Köhler, M.; Machill, S.; Salzer, R.; Krafft, C. Characterization of lipid extracts from brain tissue and tumors using Raman spectroscopy and mass spectrometry. Anal. Bioanal. Chem. 2009, 393, 1513–1520. [Google Scholar] [CrossRef]
- Koljenovic, S.; Choo-Smith, L.P.; Schut, T.C.B.; Kros, J.M.; van den Berge, H.J.; Puppels, G.J. Discriminating vital tumor from necrotic tissue in human glioblastoma tissue samples by Raman spectroscopy. Lab Investig. 2002, 82, 1265. [Google Scholar] [CrossRef]
- Ji, M.; Orringer, D.A.; Freudiger, C.W.; Ramkissoon, S.; Liu, X.; Lau, D.; Golby, A.J.; Norton, I.; Hayashi, M.; Agar, N.Y.R.; et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci. Transl. Med. 2013, 5, 201ra119. [Google Scholar] [CrossRef]
- Liu, C.-H.; Zhou, Y.; Sun, Y.; Li, J.Y.; Zhou, L.X.; Boydston-White, S.; Masilamani, V.; Zhu, K.; Pu, K.Y.; Alfano, R.R. Resonance Raman and Raman spectroscopy for breast cancer detection. Technol. Cancer Res. Treat. 2013, 12, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Bergholt, M.S.; Zheng, W.; Lin, K.; Ho, K.Y.; Teh, M.; Yeoh, K.G.; So, J.B.; Huang, Z. In vivo diagnosis of esophageal cancer using image-guided Raman endoscopy and biomolecular modeling. Technol. Cancer Res. Treat. 2011, 10, 103–112. [Google Scholar] [CrossRef]
- Kircher, M.F.; Zerda, A.; Jokerst, J.; Zavalet, C.; Kempen, P.J.; Mittr, E.; Pitter, K.; Huang, R.; Campos, C.; Habt, F.; et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat. Med. 2012, 18, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, C.-H.; Sun, Y.; Pu, Y.; Boydston-White, S.; Liu, Y.L.; Alfano, R.R. Human brain cancer studied by resonance Raman spectroscopy. J. Biomed. Opt. 2012, 17, 116021. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P. Raman spectroscopy as a promising noninvasive tool in brain cancer detection. J. Innov. Opt. Health Sci. 2017, 10, 1730012. [Google Scholar] [CrossRef]
- Hollon, T.; Lewis, S.; Freudiger, C.W.; Xie, X.S.; Orringer, D.A. Improving the accuracy of brain tumor surgery via Raman-based technology. Neurosurg. Focus 2016, 40, E9. [Google Scholar] [CrossRef]
- Nothinger, I. Raman spectroscopy cell-based biosensors. Sensors 2007, 7, 1343–1358. [Google Scholar] [CrossRef]
- Jermyn, M.; Mok, K.; Mercier, J.; Desroches, J.; Pichette, J.; Saint-Arnaud, K.; Bernstein, L.; Guiot, M.C.; Petrecca, K.; Leblond, F. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci. Transl. Med. 2015, 7, 274ra19. [Google Scholar] [CrossRef]
- Bergholt, M.S.; Zheng, W.; Ho, K.Y.; Teh, M.; Yeoh, K.G.; Yan, S.J.B.; Shabbir, A.; Huang, Z. Fiberoptic confocal raman spectroscopy for real-time in vivo diagnosis of dysplasia in Barrett’s esophagus. Gastroenterology 2014, 146, 27–32. [Google Scholar] [CrossRef]
- Krafft, C.; Belay, B.; Bergner, N.; Romeike, B.F.; Reichart, R.; Kalff, R.; Popp, J. Advances in optical biopsy—Correlation of malignancy and cell density of primary brain tumors using Raman microspectroscopic imaging. Analyst 2012, 137, 5533–5537. [Google Scholar] [CrossRef]
- Abramczyk, H.; Brozek-Pluskaa, B.; Surmackia, J.; Jablonska-Gajewiczb, J.; Kordek, R. Raman ‘optical biopsy’ of human breast cancer. Progr. Biophys. Molec. Biol. 2012, 108, 74–81. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, C.-H.; Zhou, L.X.; Zhu, K.; Liu, Y.L.; Zhang, L.; Boydston-White, S.; Cheng, G.G.; Pu, Y.; Bidyut, D.; et al. Resonant Raman spectra of grades of human brain glioma tumors reveal the content of tryptophan by the 1588 cm−1 mode. Proc. SPIE 2015, 9318, 931810. [Google Scholar]
- Imiela, A.; Polis, B.; Polis, L.; Abramczyk, H. Novel strategies of Raman imaging for brain tumor research. Oncotarget 2017, 8, 85290–85310. [Google Scholar] [CrossRef]
- Desroches, J.; Jermyn, M.; Mok, K.; Lemieux-Leduc, C.; Mercier, J.; St-Arnaud, K.; Urmey, K.; Guiot, M.-C.; Marple, E.; Petrecca, K.; et al. Characterization of a Raman spectroscopy probe system for intraoperative brain tissue classification. Biomed. Opt. Express 2015, 6, 2380–2397. [Google Scholar] [CrossRef]
- Belykh, E.; Patel, A.A.; Miller, E.J.; Bozkurt, B.; Yağmurlu, K.; Woolf, E.C.; Scheck, A.C.; Eschbacher, J.M.; Nakaji, P.; Preul, M.C. Probe-based three-dimensional confocal laser endomicroscopy of brain tumors: Technical note. Cancer Manag. Res. 2018, 10, 3109–3123. [Google Scholar] [CrossRef] [PubMed]
- Hollon, T.C.; Lewis, S.; Pandian, B.; Niknafs, Y.S.; Garrard, M.R.; Garton, H.; Maher, C.O.; McFadden, K.; Snuderl, M.; Lieberman, A.P.; et al. Rapid intraoperative diagnosis of pediatric brain tumors using stimulated Raman histology. Cancer Res. 2018, 78, 278–289. [Google Scholar] [CrossRef]
- Du, J.; Su, Y.; Qian, C.; Yuan, D.; Miao, K.; Lee, D.; Ng, A.H.C.; Wijker, R.S.; Ribas, A.; Levine, R.D.; et al. Raman-guided subcellular pharmaco-metabolomics for metastatic melanoma cells. Nat. Commun. 2020, 11, 4830. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, C.-H.; Pu, Y.; Wu, B.; Nguyen, T.A.; Cheng, G.; Zhou, L.; Zhu, K.; Chen, J.; Li, Q.; et al. Combined spatial frequency spectroscopy analysis with visible resonance Raman for optical biopsy of human brain metastases of lung cancers. J. Innov. Opt. Health Sci. 2019, 12, 1950010. [Google Scholar] [CrossRef]
- Liu, C.-H.; Boydston-White, S.; Weisberg, A.; Wang, W.; Sordillo, L.A.; Perotte, A.; Tomaselli, V.P.; Sordillo, P.P.; Pei, Z.; Shi, L.; et al. Vulnerable atherosclerotic plaque detection by resonance Raman spectroscopy. J. Biomed. Opt. 2016, 21, 127006. [Google Scholar] [CrossRef] [PubMed]
- Rodionova, N.N.; Allakhverdiev, E.S.; Maksimov, G.V. Study of myelin structure changes during the nerve fibers demyelination. PLoS ONE 2017, 12, e0185170. [Google Scholar] [CrossRef]
- Mehta, K.; Atak, A.; Sahu, A.; Srivastava, S.; Krishna, C.M. An early investigative serum Raman spectroscopy study of meningioma. Analyst 2018, 143, 1916–1923. [Google Scholar] [CrossRef]
- Liu, C.-H.; Boydston-White, S.; Wang, W.B.; Sordillo, L.A.; Shi, L.Y.; Weisberg, A.; Tomaselli, V.P.; Sordillo, P.P.; Alfano, R.R. Optical pathology study of human abdominal aorta tissues using confocal micro resonance Raman spectroscopy. Proc. SPIE 2016, 9703, 97031S. [Google Scholar]
- Zhou, Y.; Liu, C.-H.; Wu, B.; Yu, X.; Cheng, G.; Zhu, K.; Wang, K.; Zhang, C.; Zhao, M.; Zong, R.; et al. Optical biopsy identification and grading of gliomas using label-free visible resonance Raman spectroscopy. J. Biomed. Opt. 2019, 24, 095001. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, C.-H.; Wu, B.; Zhang, C.; Yu, X.; Cheng, G.; Chen, H.; Li, S.; Liang, Q.; Zhang, M.; et al. Invited Article: Molecular biomarkers characterization for human brain glioma grading using visible resonance Raman spectroscopy. APL Photonics 2018, 3, 120802. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, Y.; Wu, B.; Zhang, S.; Zhu, K.; Liu, C.-H.; Yu, X.; Alfano, R.R. Intraoperative detection of human meningioma using a handheld visible resonance Raman analyzer. Lasers Med. Sci. 2021, 37, 1311–1319. [Google Scholar] [CrossRef]
- Riva, M.; Sciortino, T.; Secoli, R.; D’Amico, E.; Moccia, S.; Fernandes, B.; Conti, N.M.; Gay, L.; Rossi, M.; De Momi, E.; et al. Glioma biopsies classification using Raman spectroscopy and machine learning models on fresh tissue samples. Cancers 2021, 13, 1073. [Google Scholar] [CrossRef]
- Zito, G.; Rusciano, G.; Pesce, G.; Dochshanov, A.; Sasso, A. Surface-enhanced Raman imaging of cell membrane by a highly homogeneous and isotropic silver nanostructure. Nanoscale 2015, 7, 8593–8606. [Google Scholar] [CrossRef] [PubMed]
- Schechinger, M.; Marks, H.; Locke, A.; Choudhury, M.; Coté, G. Development of a miRNA surface-enhanced Raman scattering assay using benchtop and handheld Raman systems. J. Biomed. Opt. 2018, 23, 017002. [Google Scholar] [CrossRef]
- Polis, B.; Imiela, A.; Polis, L.; Abramczyk, H. Raman spectroscopy for medulloblastoma. Childs. Nerv. Syst. 2018, 34, 2425–2430. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, S.; Wu, B.; Yu, X.; Cheng, G.; Zhu, K.; Zhao, M.; Zheng, J.; Zhang, M.; Liang, Q.; et al. A portable visible resonance Raman analyzer with a handheld optical fiber probe for in vivo diagnosis of brain glioblastoma multiforme in an animal model. In Proceedings of the Frontiers in Optics + Laser Science APS/DLS, OSA Technical Digest, Washington, DC, USA, 15–19 September 2019; p. JW3A.5. [Google Scholar]
- Zhou, Y.; Zhang, S.; Wu, B.; Yu, X.; Cheng, G.; Zhu, K.; Zhao, M.; Zheng, J.; Zhang, L.; Zhang, M.; et al. Human glioma tumors detection by a portable visible resonance Raman analyzer with a hand-held optical fiber probe. Proc. SPIE 2020, 11236, 1123608. [Google Scholar]
- Di, L.; Eichberg, D.G.; Huang, K.; Shah, A.H.; Jamshidi, A.M.; Luther, E.M.; Lu, V.M.; Komotar, R.J.; Ivan, M.E.; Gultekin, S.H. Stimulated Raman histology for rapid intraoperative diagnosis of gliomas. World Neurosurg. 2021, 150, e135–e143. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.R.; Wen, P.Y.; Lang-Orsini, M.; Chukwueke, U.N. World Health Organization 2021 classification of central nervous system tumors and implications for therapy for adult-type gliomas: A review. JAMA Oncol. 2022, 8, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Stoyanov, G.S.; Lyutfi, E.; Georgieva, R.; Georgiev, R.; Dzhenkov, D.L.; Petkova, L.; Ivanov, B.D.; Kaprelyan, A.; Ghenev, P. Reclassification of glioblastoma multiforme according to the 2021 World Health Organization classification of central nervous system tumors: A single institution report and practical significance. Cureus 2022, 14, e21822. [Google Scholar] [CrossRef]
- Jain, M.; Robinson, B.D.; Wu, B.; Khani, F.; Mukherjee, S. Exploring multiphoton microscopy as a novel tool to differentiate chromophobe renal cell carcinoma from oncocytoma in fixed tissue sections. Arch. Pathol. Lab. Med. 2018, 142, 383–390. [Google Scholar] [CrossRef]
- Krafft, C.; Neudert, L.; Simat, T.; Salzer, R. Near infrared Raman spectra of human brain lipids. Spectrochim. Acta 2005, 61, 1529–1535. [Google Scholar] [CrossRef]
- Abramczyk, H.; Imiela, A. The biochemical, nanomechanical and chemometric signatures of brain cancer. Spectrochim. Acta Mol. Biomol. Spectrosc. 2018, 188, 8–19. [Google Scholar] [CrossRef]
- Ermakov, I.V.; Sharifzadeh, M.; Ermakova, M.; Gellermann, W. Resonance Raman detection of carotenoid antioxidants in living human tissue. J. Biomed. Opt. 2005, 10, 064028. [Google Scholar] [CrossRef]
- Abramczyk, H.; Brozek-Pluska, B.; Surmacki, J.; Jablonska, J.; Kordek, R. The label-free Raman imaging of human breast cancer. J. Mol. Liq. 2011, 164, 123–131. [Google Scholar] [CrossRef]
- Liu, C.-H.; Wu, B.; Sordillo, L.A.; Boydston-White, S.; Sriramoju, V.; Zhang, C.; Beckman, H.; Zhang, L.; Pei, Z.; Shi, L.; et al. A pilot study for distinguishing basal cell carcinoma from normal human skin tissues using visible resonance Raman spectroscopy. J. Cancer Metastasis Treat. 2019, 5, 4. [Google Scholar] [CrossRef]
- Brozek-Pluska, B.; Musial, J.; Kordek, R.; Abramczyk, H. Analysis of human colon by Raman spectroscopy and imaging-Elucidation of biochemical changes in carcinogenesis. Int. J. Mol. Sci. 2019, 20, 3398. [Google Scholar] [CrossRef]
- Johnson, E.J. The role of carotenoids in human health. Nutr. Clin. Care 2002, 5, 56–65. [Google Scholar] [CrossRef]
- Vishwanathan, R.; Kuchan, M.J.; Sen, S.; Johnson, E.J. Lutein and preterm infants with decreased concentrations of brain carotenoids. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 659–665. [Google Scholar] [CrossRef]
- Erdman, J.W.J.; Smith, J.W.; Kuchan, M.J.; Mohn, E.S.; Johnson, E.J.; Rubakhin, S.S.; Wang, L.; Sweedler, J.V.; Neuringer, M. Lutein and Brain Function. Foods 2015, 4, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Nguyen, H.Q.; Huang, Q.R.; Lin, C.K.; Kuo, J.L.; Patwari, G.N. Vibrational spectroscopic signatures of hydrogen bond induced NH stretch-bend Fermi-resonance in amines: The methylamine clusters and other N-H⋯N hydrogen-bonded complexes. J. Chem. Phys. 2020, 153, 194301. [Google Scholar] [CrossRef]
- Salunkhe, S.; Mishra, S.V.; Ghorai, A.; Hole, A.; Chandrani, P.; Dutt, A.; Chilakapati, M.; Dutt, S. Metabolic rewiring in drug resistant cells exhibit higher OXPHOS and fatty acids as preferred major source to cellular energetics. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148300. [Google Scholar] [CrossRef] [PubMed]
- Moroni, F. Tryptophan metabolism and brain function: Focus on kynurenine and other indole metabolites. Eur. J. Pharmacol. 1999, 375, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.W.; Sparks, R.; Polam, P.; Modi, D.; Douty, B.; Wayland, B.; Glass, B.; Takvorian, A.; Glenn, J.; Zhu, W.; et al. INCB24360 (Epacadostat), a Highly Potent and Selective Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitor for Immuno-oncology. ACS Med. Chem. Lett. 2017, 8, 486–491. [Google Scholar] [CrossRef]
- Ren, H.; Biggs, J.D.; Mukamel, S. Two-dimensional stimulated ultraviolet resonance Raman spectra of tyrosine and tryptophan; a simulation study. J. Raman Spectrosc. 2013, 44, 544–559. [Google Scholar] [CrossRef]
- Rava, R.P.; Spiro, T.G. Selective Enhancement of Tyrosine and Tryptophan Resonance Raman Spectra via Ultraviolet Laser Excitation. J. Am. Chem. Soc. 1984, 106, 4062–4064. [Google Scholar] [CrossRef]
- Platten, M.; Weller, M.; Wick, W. Shaping the glioma immune microenvironment through tryptophan metabolism. CNS Oncol. 2012, 1, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Sahm, F.; Oezen, I.; Opitz, C.A.; Radlwimmer, B.; von Deimling, A.; Ahrendt, T.; Adams, S.; Bode, H.B.; Guillemin, G.J.; Wick, W.; et al. The endogenous tryptophan metabolite and NAD+ precursor quinolinic acid confers resistance of gliomas to oxidative stress. Cancer Res. 2013, 73, 3225–3234. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C. Why tumours eat tryptophan. Nature 2011, 478, 192–194. [Google Scholar] [CrossRef]
- Platten, M.; Wick, W.; Van den Eynde, B.J. Tryptophan catabolism in cancer: Beyond IDO and tryptophan depletion. Cancer Res. 2012, 72, 5435–5440. [Google Scholar] [CrossRef]
- Xue, J.; Pu, Y.; Smith, J.; Gao, X.; Wang, C.; Wu, B. Identifying metastatic ability of prostate cancer cell lines using native fluorescence spectroscopy and machine learning methods. Sci. Rep. 2021, 11, 2282. [Google Scholar] [CrossRef]
- Lai, C.W.; Schwab, M.; Hill, S.C.; Santarpia, J.; Pan, Y.L. Raman scattering and red fluorescence in the photochemical transformation of dry tryptophan particles. Opt. Express 2016, 24, 11654–11667. [Google Scholar] [CrossRef]
- Bartlett, J.S.; Voss, K.J.; Sathyendranath, S.; Vodacek, A. Raman scattering by pure water and seawater. Appl. Opt. 1998, 37, 3324–3332. [Google Scholar] [CrossRef]
- Ryan, C.G.; Clayton, E.; Griffin, W.L.; Sie, S.H.; Cousens, D.R. SNIP, a statistics sensitive background treatment for the quantitative analysis of PIXE spectra in geoscience applications. Nucl. Instrum. Methods Phys. Res. A 1988, B34, 396–402. [Google Scholar] [CrossRef]
- Morháč, M.; Kliman, J.; Matoušek, V.; Veselský, M.; Turzo, I. Background elimination methods for multidimensional coincidence γ-ray spectra. Nucl. Instr. Meth. 1997, A401, 113–132. [Google Scholar] [CrossRef]
- Pirro, V.; Alfaro, C.M.; Jarmusch, A.K.; Hattab, E.M.; Cohen-Gadol, A.A.; Cooks, R.G. Intraoperative assessment of tumor margins during glioma resection by desorption electrospray ionization-mass spectrometry. Proc. Natl. Acad. Sci. USA 2017, 114, 6700–6705. [Google Scholar] [CrossRef] [PubMed]

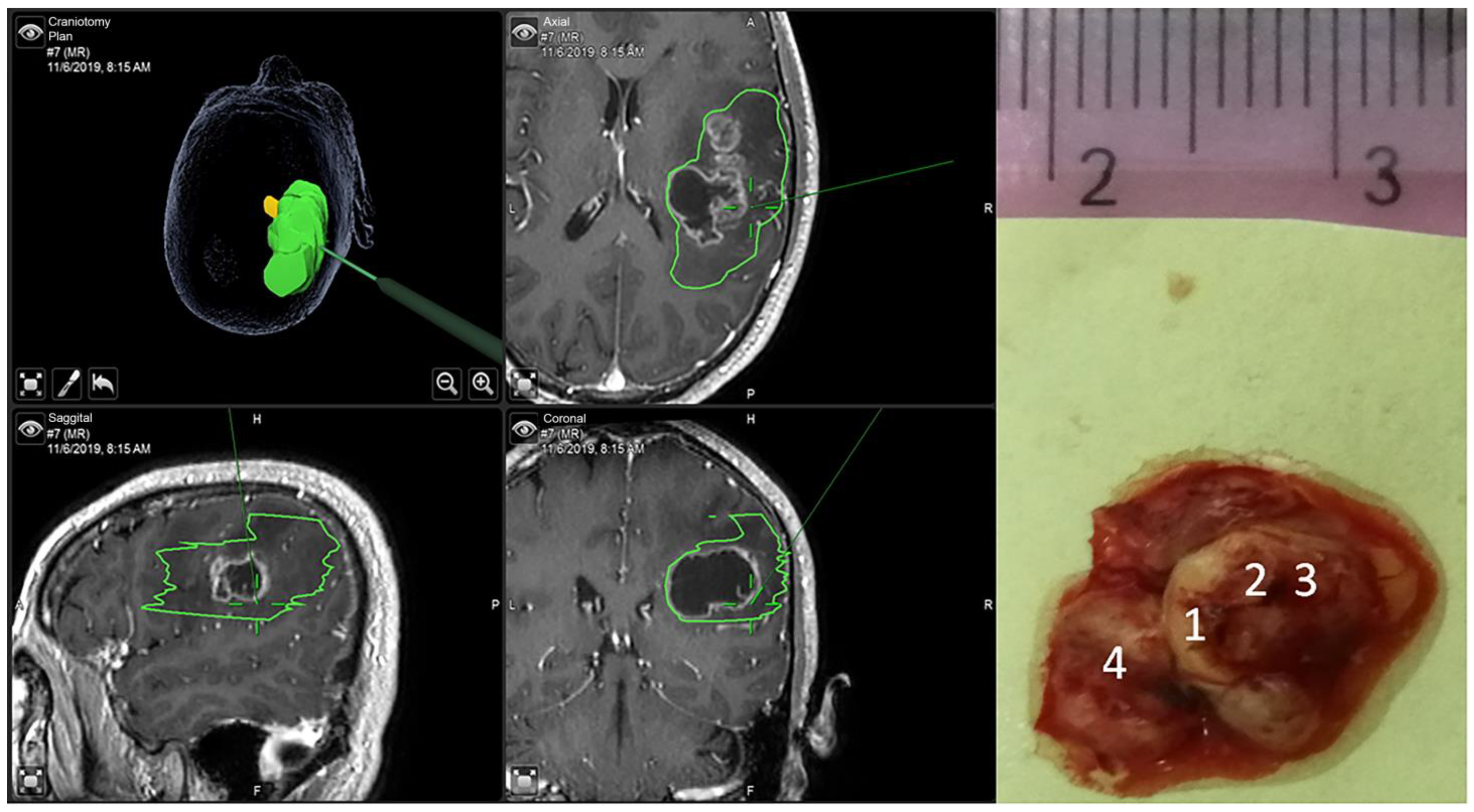

| Tissue Type | No. of Samples | No. of Spectra |
|---|---|---|
| Normal | 7 | 26 |
| Grade 2 | 6 | 39 |
| Grade 3 | 15 | 92 |
| Grade 4 | 31 | 202 |
| Total | 59 | 359 |
| Binary Classes | N vs. C | N vs. G2 | N vs. G3 | N vs. G4 | |
|---|---|---|---|---|---|
| Peak-SVM | Peaks (cm−1) | 1371, 1512, 3002, 3474 | 1620, 3474, 3882 | 1302, 1371, 3210 | 1302, 1371, 1512, 3002 |
| Sensitivity (%) | 99.1 | 94.9 | 92.4 | 98.5 | |
| Specificity (%) | 50.0 | 80.8 | 65.4 | 57.7 | |
| Accuracy (%) | 95.5 | 89.2 | 86.4 | 93.9 | |
| AUROC (%) | 75.5 | 92.4 | 72.7 | 77.3 | |
| PCA-SVM | PCs | 1, 9, 19 | 3, 10, 13 | 1, 5, 9, 24 | 1, 2, 8 |
| Sensitivity (%) | 96.9 | 94.9 | 90.2 | 97.0 | |
| Specificity (%) | 50.0 | 76.9 | 61.5 | 53.8 | |
| Accuracy (%) | 93.1 | 87.7 | 83.9 | 92.1 | |
| AUROC (%) | 74.7 | 94.6 | 81.6 | 80.9 | |
| Multiclass | N vs. G2 vs. G3 vs. G4 | ||||
| Model | Peak-SVM | PCA-SVM | |||
| Peaks (cm−1)/PCs | 1512, 3210, 3497, 3541, 3847 | 1, 4, 10, 14 | |||
| Accuracy | N (%) | 50.0 | 42.3 | ||
| G2 (%) | 56.4 | 43.6 | |||
| G3 (%) | 37.0 | 40.2 | |||
| G4 (%) | 73.8 | 90.1 | |||
| Total (%) | 60.7 | 68.8 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhou, Y.; Wu, B.; Zhang, S.; Zhu, K.; Liu, C.-H.; Yu, X.; Alfano, R.R. A Handheld Visible Resonance Raman Analyzer Used in Intraoperative Detection of Human Glioma. Cancers 2023, 15, 1752. https://doi.org/10.3390/cancers15061752
Zhang L, Zhou Y, Wu B, Zhang S, Zhu K, Liu C-H, Yu X, Alfano RR. A Handheld Visible Resonance Raman Analyzer Used in Intraoperative Detection of Human Glioma. Cancers. 2023; 15(6):1752. https://doi.org/10.3390/cancers15061752
Chicago/Turabian StyleZhang, Liang, Yan Zhou, Binlin Wu, Shengjia Zhang, Ke Zhu, Cheng-Hui Liu, Xinguang Yu, and Robert R. Alfano. 2023. "A Handheld Visible Resonance Raman Analyzer Used in Intraoperative Detection of Human Glioma" Cancers 15, no. 6: 1752. https://doi.org/10.3390/cancers15061752
APA StyleZhang, L., Zhou, Y., Wu, B., Zhang, S., Zhu, K., Liu, C.-H., Yu, X., & Alfano, R. R. (2023). A Handheld Visible Resonance Raman Analyzer Used in Intraoperative Detection of Human Glioma. Cancers, 15(6), 1752. https://doi.org/10.3390/cancers15061752








