Systematic Review of Tumor Segmentation Strategies for Bone Metastases
Abstract
Simple Summary
Abstract
1. Introduction
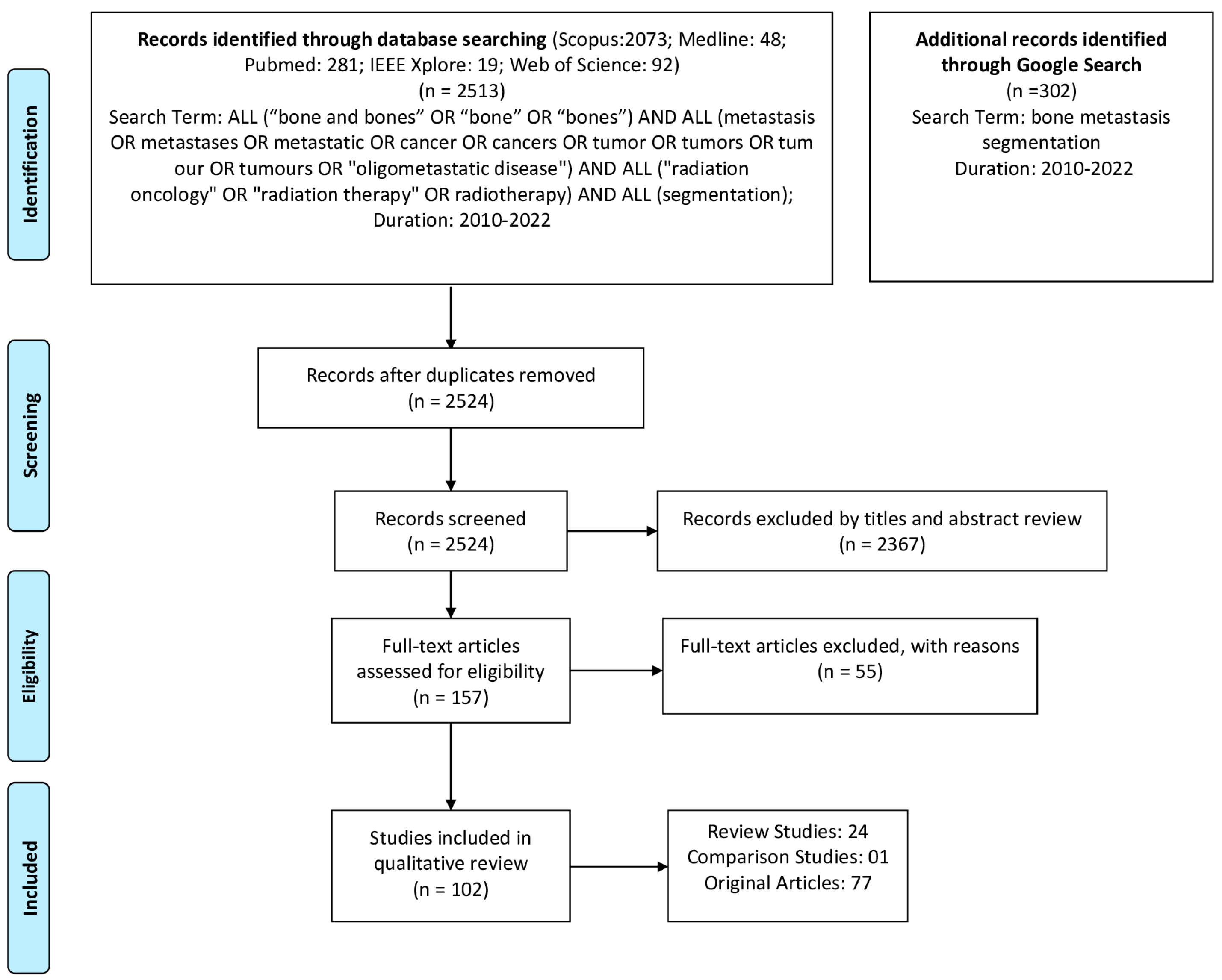
2. Methods
2.1. Literature Search
- “bone metastasis segmentation”.
2.2. Data Extraction
- Enrollment period of the patients;
- Study type: retrospective cohort study or prospective;
- Study population. Extracted the number of scans or images when patient numbers were not provided;
- Training/Validation/testing cohorts;
- Primary tumor and relevant location;
- Imaging modality;
- Methodology;
- Outcome;
- Evaluation Metrics;
- Details of whether the study mentioned the suitability of the approaches for clinical use;
- Country of the Authors.
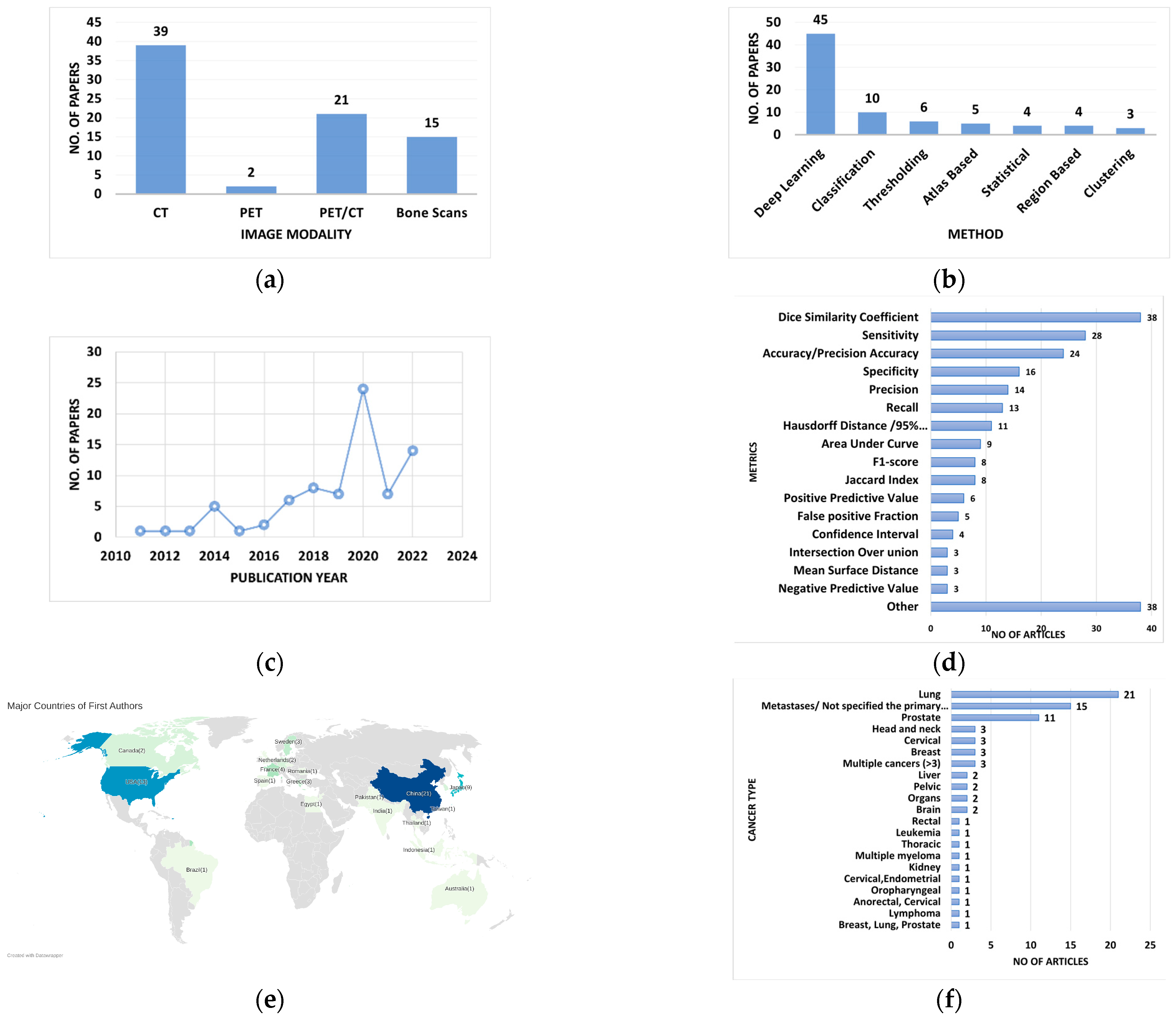
3. Results
4. Discussion
4.1. Deep Learning
4.2. Thresholding
4.3. Clustering/Classification
4.4. Statistical Methods
4.5. Atlas-Based Approaches
4.6. Region-Based Approaches
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Svensson, E.; Christiansen, C.F.; Ulrichsen, S.P.; Rørth, M.R.; Sørensen, H.T. Survival after bone metastasis by primary cancer type: A Danish population-based cohort study. BMJ Open 2017, 7, e016022. [Google Scholar] [CrossRef] [PubMed]
- Chu, G.; Lo, P.; Ramakrishna, B.; Kim, H.; Morris, D.; Goldin, J.; Brown, M. Bone Tumor Segmentation on Bone Scans Using Context Information and Random Forests; Springer International Publishing: Cham, Switzerland, 2014. [Google Scholar]
- Peeters, S.T.H.; Van Limbergen, E.J.; Hendriks, L.E.L.; De Ruysscher, D. Radiation for Oligometastatic Lung Cancer in the Era of Immunotherapy: What Do We (Need to) Know? Cancers 2021, 13, 2132. [Google Scholar] [CrossRef]
- Zeng, K.L.; Tseng, C.L.; Soliman, H.; Weiss, Y.; Sahgal, A.; Myrehaug, S. Stereotactic body radiotherapy (SBRT) for oligometastatic spine metastases: An overview. Front. Oncol. 2019, 9, 337. [Google Scholar] [CrossRef]
- Spencer, K.L.; van der Velden, J.M.; Wong, E.; Seravalli, E.; Sahgal, A.; Chow, E.; Verlaan, J.J.; Verkooijen, H.M.; van der Linden, Y.M. Systematic Review of the Role of Stereotactic Radiotherapy for Bone Metastases. J. Natl. Cancer Inst. 2019, 111, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Loi, M.; Nuyttens, J.J.; Desideri, I.; Greto, D.; Livi, L. Single-fraction radiotherapy (SFRT) for bone metastases: Patient selection and perspectives. Cancer Manag. Res. 2019, 11, 9397–9408. [Google Scholar] [CrossRef] [PubMed]
- Palma, D.A.; Olson, R.; Harrow, S.; Gaede, S.; Louie, A.V.; Haasbeek, C.; Mulroy, L.; Lock, M.; Rodrigues, G.B.; Yaremko, B.P.; et al. Stereotactic Ablative Radiotherapy for the Comprehensive Treatment of Oligometastatic Cancers: Long-Term Results of the SABR-COMET Phase II Randomized Trial. J. Clin. Oncol. 2020, 38, 2830–2838. [Google Scholar] [CrossRef] [PubMed]
- De Ruysscher, D.; Wanders, R.; van Baardwijk, A.; Dingemans, A.M.; Reymen, B.; Houben, R.; Bootsma, G.; Pitz, C.; van Eijsden, L.; Geraedts, W.; et al. Radical treatment of non-small-cell lung cancer patients with synchronous oligometastases: Long-term results of a prospective phase II trial (Nct01282450). J. Thorac. Oncol. 2012, 7, 1547–1555. [Google Scholar] [CrossRef]
- Dercle, L.; Henry, T.; Carré, A.; Paragios, N.; Deutsch, E.; Robert, C. Reinventing radiation therapy with machine learning and imaging bio-markers (radiomics): State-of-the-art, challenges and perspectives. Methods 2020, 188, 44–60. [Google Scholar] [CrossRef]
- Speirs, C.K.; Grigsby, P.W.; Huang, J.; Thorstad, W.L.; Parikh, P.J.; Robinson, C.G.; Bradley, J.D. PET-based radiation therapy planning. PET Clin. 2015, 10, 27–44. [Google Scholar] [CrossRef]
- Lu, W.; Wang, J.; Zhang, H.H. Computerized PET/CT image analysis in the evaluation of tumour response to therapy. Br. J. Radiol. 2015, 88, 20140625. [Google Scholar] [CrossRef]
- Vergalasova, I.; Cai, J. A modern review of the uncertainties in volumetric imaging of respiratory-induced target motion in lung radiotherapy. Med. Phys. 2020, 47, e988–e1008. [Google Scholar] [CrossRef] [PubMed]
- Papandrianos, N.; Papageorgiou, E.; Anagnostis, A.; Papageorgiou, K. Bone metastasis classification using whole body images from prostate cancer patients based on convolutional neural networks application. PLoS ONE 2020, 15, e0237213. [Google Scholar] [CrossRef] [PubMed]
- Foster, B.; Bagci, U.; Mansoor, A.; Xu, Z.; Mollura, D.J. A review on segmentation of positron emission tomography images. Comput. Biol. Med. 2014, 50, 76–96. [Google Scholar] [CrossRef]
- Takahashi, M.E.S.; Mosci, C.; Souza, E.M.; Brunetto, S.Q.; de Souza, C.; Pericole, F.V.; Lorand-Metze, I.; Ramos, C.D. Computed tomography-based skeletal segmentation for quantitative PET metrics of bone involvement in multiple myeloma. Nucl. Med. Commun. 2020, 41, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Bach Cuadra, M.; Favre, J.; Omoumi, P. Quantification in Musculoskeletal Imaging Using Computational Analysis and Machine Learning: Segmentation and Radiomics. Semin. Musculoskelet. Radiol. 2020, 24, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, V.; Nicolini, S.; Caroli, P.; Nanni, C.; Massaro, A.; Marzola, M.C.; Rubello, D.; Fanti, S. PET/CT imaging in different types of lung cancer: An overview. Eur. J. Radiol. 2012, 81, 988–1001. [Google Scholar] [CrossRef]
- Carvalho, L.E.; Sobieranski, A.C.; von Wangenheim, A. 3D Segmentation Algorithms for Computerized Tomographic Imaging: A Systematic Literature Review. J. Digit. Imaging 2018, 31, 799–850. [Google Scholar] [CrossRef]
- Domingues, I.; Pereira, G.; Martins, P.; Duarte, H.; Santos, J.; Abreu, P.H. Using deep learning techniques in medical imaging: A systematic review of applications on CT and PET. Artif. Intell. Rev. 2020, 53, 4093–4160. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- Mansoor, A.; Bagci, U.; Foster, B.; Xu, Z.; Papadakis, G.Z.; Folio, L.R.; Udupa, J.K.; Mollura, D.J. Segmentation and Image Analysis of Abnormal Lungs at CT: Current Approaches, Challenges, and Future Trends. Radiographics 2015, 35, 1056–1076. [Google Scholar] [CrossRef]
- Punn, N.S.; Agarwal, S. Modality specific U-Net variants for biomedical image segmentation: A survey. Artif. Intell. Rev. 2022, 55, 5845–5889. [Google Scholar] [CrossRef] [PubMed]
- Saba, T. Recent advancement in cancer detection using machine learning: Systematic survey of decades, comparisons and challenges. J. Infect. Public Health 2020, 13, 1274–1289. [Google Scholar] [CrossRef] [PubMed]
- Trevor Hastie, J.F.; Tibshirani, R. The Elements of Statistical Learning; Springer: New York, NY, USA, 2001. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Into Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, Z.; Li, Y.; Chen, Z.; Lu, P.; Wang, W.; Liu, W.; Yu, L. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res. 2017, 7, 11. [Google Scholar] [CrossRef]
- Yousefirizi, F.; Pierre, D.; Amyar, A.; Ruan, S.; Saboury, B.; Rahmim, A. AI-Based Detection, Classification and Prediction/Prognosis in Medical Imaging: Towards Radiophenomics. PET Clin. 2022, 17, 183–212. [Google Scholar] [CrossRef]
- Zhang, Z.; Sejdić, E. Radiological images and machine learning: Trends, perspectives, and prospects. Comput. Biol. Med. 2019, 108, 354–370. [Google Scholar] [CrossRef]
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.M.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019, 46, e1–e36. [Google Scholar] [CrossRef]
- Samarasinghe, G.; Jameson, M.; Vinod, S.; Field, M.; Dowling, J.; Sowmya, A.; Holloway, L. Deep learning for segmentation in radiation therapy planning: A review. J. Med. Imaging Radiat. Oncol. 2021, 65, 578–595. [Google Scholar] [CrossRef]
- Faiella, E.; Santucci, D.; Calabrese, A.; Russo, F.; Vadalà, G.; Zobel, B.B.; Soda, P.; Iannello, G.; de Felice, C.; Denaro, V. Artificial Intelligence in Bone Metastases: An MRI and CT Imaging Review. Int. J. Environ. Res. Public Health 2022, 19, 1880. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Wang, R.; Lei, T.; Cui, R.; Zhang, B.; Meng, H.; Nandi, A.K. Medical image segmentation using deep learning: A survey. IET Image Process. 2022, 16, 1243–1267. [Google Scholar] [CrossRef]
- MacManus, M.; Everitt, S. Treatment Planning for Radiation Therapy. PET Clin. 2018, 13, 43–57. [Google Scholar] [CrossRef]
- Yang, W.C.; Hsu, F.M.; Yang, P.C. Precision radiotherapy for non-small cell lung cancer. J. Biomed. Sci. 2020, 27, 82. [Google Scholar] [CrossRef] [PubMed]
- Orcajo-Rincon, J.; Muñoz-Langa, J.; Sepúlveda-Sánchez, J.M.; Fernández-Pérez, G.C.; Martínez, M.; Noriega-Álvarez, E.; Sanz-Viedma, S.; Vilanova, J.C.; Luna, A. Review of imaging techniques for evaluating morphological and functional responses to the treatment of bone metastases in prostate and breast cancer. Clin. Transl. Oncol. 2022, 24, 1290–1310. [Google Scholar] [CrossRef] [PubMed]
- Chmelik, J.; Jakubicek, R.; Walek, P.; Jan, J.; Ourednicek, P.; Lambert, L.; Amadori, E.; Gavelli, G. Deep convolutional neural network-based segmentation and classification of difficult to define metastatic spinal lesions in 3D CT data. Med. Image Anal. 2018, 49, 76–88. [Google Scholar] [CrossRef]
- Elfarra, F.-G.; Calin, M.A.; Parasca, S.V. Computer-aided detection of bone metastasis in bone scintigraphy images using parallelepiped classification method. Ann. Nucl. Med. 2019, 33, 866–874. [Google Scholar] [CrossRef]
- Guo, Y.; Lin, Q.; Zhao, S.; Li, T.; Cao, Y.; Man, Z.; Zeng, X. Automated detection of lung cancer-caused metastasis by classifying scintigraphic images using convolutional neural network with residual connection and hybrid attention mechanism. Insights Into Imaging 2022, 13, 24. [Google Scholar] [CrossRef]
- Hammes, J.; Täger, P.; Drzezga, A. EBONI: A Tool for Automated Quantification of Bone Metastasis Load in PSMA PET/CT. J. Nucl. Med. 2018, 59, 1070–1075. [Google Scholar] [CrossRef]
- Han, S.; Oh, J.S.; Lee, J.J. Diagnostic performance of deep learning models for detecting bone metastasis on whole-body bone scan in prostate cancer. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 585–595. [Google Scholar] [CrossRef]
- Hinzpeter, R.; Baumann, L.; Guggenberger, R.; Huellner, M.; Alkadhi, H.; Baessler, B. Radiomics for detecting prostate cancer bone metastases invisible in CT: A proof-of-concept study. Eur. Radiol. 2022, 32, 1823–1832. [Google Scholar] [CrossRef]
- Li, T.; Lin, Q.; Guo, Y.; Zhao, S.; Zeng, X.; Man, Z.; Cao, Y.; Hu, Y. Automated detection of skeletal metastasis of lung cancer with bone scans using convolutional nuclear network. Phys. Med. Biol. 2022, 67, 015004. [Google Scholar] [CrossRef]
- Lin, Q.; Luo, M.; Gao, R.; Li, T.; Zhengxing, M.; Cao, Y.; Wang, H. Deep learning based automatic segmentation of metastasis hotspots in thorax bone SPECT images. PLoS ONE 2020, 15, e0243253. [Google Scholar] [CrossRef] [PubMed]
- Moreau, N.; Rousseau, C.; Fourcade, C.; Santini, G.; Brennan, A.; Ferrer, L.; Lacombe, M.; Guillerminet, C.; Colombié, M.; Jézéquel, P.; et al. Automatic segmentation of metastatic breast cancer lesions on18f-fdg pet/ct longitudinal acquisitions for treatment response assessment. Cancers 2022, 14, 101. [Google Scholar] [CrossRef] [PubMed]
- Moreau, N.; Rousseau, C.; Fourcade, C.; Santini, G.; Ferrer, L.; Lacombe, M.; Guillerminet, C.; Campone, M.; Colombié, M.; Rubeaux, M.; et al. Deep learning approaches for bone and bone lesion segmentation on 18FDG PET/CT imaging in the context of metastatic breast cancer. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020. [Google Scholar]
- Papandrianos, N.; Papageorgiou, E.; Anagnostis, A. Development of Convolutional Neural Networks to identify bone metastasis for prostate cancer patients in bone scintigraphy. Ann. Nucl. Med. 2020, 34, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Rachmawati, E.; Sumarna, F.R.; Jondri; Kartamihardja, A.H.S.; Achmad, A.; Shintawati, R. Bone Scan Image Segmentation based on Active Shape Model for Cancer Metastasis Detection. In Proceedings of the 2020 8th International Conference on Information and Communication Technology (ICoICT), Yogyakarta, Indonesia, 24–26 June 2020. [Google Scholar]
- Sato, S.; Lu, H.; Kim, H.; Murakami, S.; Ueno, M.; Terasawa, T.; Aoki, T. Enhancement of Bone Metastasis from CT Images Based on Salient Region Feature Registration. In Proceedings of the 2018 18th International Conference on Control, Automation and Systems (ICCAS), PyeongChang, Republic of Korea, 17–20 October 2018. [Google Scholar]
- Song, Y.; Lu, H.; Kim, H.; Murakami, S.; Ueno, M.; Terasawa, T.; Aoki, T. Segmentation of Bone Metastasis in CT Images Based on Modified HED. In Proceedings of the 2019 19th International Conference on Control, Automation and Systems (ICCAS 2019), Institute of Control, Robotics and Systems—ICROS, ICC Jeju, Jeju, Republic of Korea, 11 October–18 October 2019; pp. 812–815. [Google Scholar]
- Wiese, T.; Burns, J.; Jianhua, Y.; Summers, R.M. Computer-aided detection of sclerotic bone metastases in the spine using watershed algorithm and support vector machines. In Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 March–2 April 2011; pp. 152–155. [Google Scholar]
- Zhang, J.; Huang, M.; Deng, T.; Cao, Y.; Lin, Q. Bone metastasis segmentation based on Improved U-NET algorithm. J. Phys. Conf. Ser. 2021, 1848, 012027. [Google Scholar] [CrossRef]
- Hsieh, T.-C.; Liao, C.-W.; Lai, Y.-C.; Law, K.-M.; Chan, P.-K.; Kao, C.-H. Detection of Bone Metastases on Bone Scans through Image Classification with Contrastive Learning. J. Pers. Med. 2021, 11, 1248. [Google Scholar] [CrossRef]
- Liu, S.; Feng, M.; Qiao, T.; Cai, H.; Xu, K.; Yu, X.; Jiang, W.; Lv, Z.; Wang, Y.; Li, D. Deep Learning for the Automatic Diagnosis and Analysis of Bone Metastasis on Bone Scintigrams. Cancer Manag. Res. 2022, 14, 51–65. [Google Scholar] [CrossRef]
- AbuBaker, A.; Ghadi, Y. A novel CAD system to automatically detect cancerous lung nodules using wavelet transform and SVM. Int. J. Electr. Comput. Eng. 2020, 10, 4745–4751. [Google Scholar] [CrossRef]
- Apiparakoon, T.; Rakratchatakul, N.; Chantadisai, M.; Vutrapongwatana, U.; Kingpetch, K.; Sirisalipoch, S.; Rakvongthai, Y.; Chaiwatanarat, T.; Chuangsuwanich, E. MaligNet: Semisupervised Learning for Bone Lesion Instance Segmentation Using Bone Scintigraphy. IEEE Access 2020, 8, 27047–27066. [Google Scholar] [CrossRef]
- Biswas, B.; Ghosh, S.K.; Ghosh, A. A novel CT image segmentation algorithm using PCNN and Sobolev gradient methods in GPU frameworks. Pattern Anal. Appl. 2020, 23, 837–854. [Google Scholar] [CrossRef]
- Borrelli, P.; Góngora, J.L.L.; Kaboteh, R.; Ulén, J.; Enqvist, O.; Trägårdh, E.; Edenbrandt, L. Freely available convolutional neural network-based quantification of PET/CT lesions is associated with survival in patients with lung cancer. EJNMMI Phys. 2022, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Buckless, C.; Yeh, K.J.; Torriani, M. Automated detection and segmentation of sclerotic spinal lesions on body CTs using a deep convolutional neural network. Skelet. Radiol. 2022, 51, 391–399. [Google Scholar] [CrossRef] [PubMed]
- da Cruz, L.B.; Júnior, D.A.D.; Diniz, J.O.B.; Silva, A.C.; de Almeida, J.D.S.; de Paiva, A.C.; Gattass, M. Kidney tumor segmentation from computed tomography images using DeepLabv3+ 2.5D model. Expert Syst. Appl. 2022, 192, 116270. [Google Scholar] [CrossRef]
- Elsayed, O.; Mahar, K.; Kholief, M.; Khater, H.A. Automatic detection of the pulmonary nodules from CT images. In Proceedings of the 2015 SAI Intelligent Systems Conference (IntelliSys), London, UK, 10–11 November 2015. [Google Scholar]
- Guo, Y.; Feng, Y.; Sun, J.; Zhang, N.; Lin, W.; Sa, Y.; Wang, P. Automatic Lung Tumor Segmentation on PET/CT Images Using Fuzzy Markov Random Field Model. Comput. Math. Methods Med. 2014, 2014, 401201. [Google Scholar] [CrossRef] [PubMed]
- Hussain, L.; Rathore, S.; Abbasi, A.A.; Saeed, S. Automated Lung Cancer Detection Based on Multimodal Features Extracting Strategy Using Machine Learning Techniques; SPIE Medical Imaging: San Diego, CA, USA, 2019; Volume 10948. [Google Scholar]
- Kim, N.; Chang, J.S.; Kim, Y.B.; Kim, J.S. Atlas-based auto-segmentation for postoperative radiotherapy planning in endometrial and cervical cancers. Radiat. Oncol. 2020, 15, 106. [Google Scholar] [CrossRef]
- Li, L.; Zhao, X.; Lu, W.; Tan, S. Deep learning for variational multimodality tumor segmentation in PET/CT. Neurocomputing 2020, 392, 277–295. [Google Scholar] [CrossRef]
- Lu, Y.; Lin, J.; Chen, S.; He, H.; Cai, Y. Automatic Tumor Segmentation by Means of Deep Convolutional U-Net with Pre-Trained Encoder in PET Images. IEEE Access 2020, 8, 113636–113648. [Google Scholar] [CrossRef]
- Markel, D.; Caldwell, C.; Alasti, H.; Soliman, H.; Ung, Y.; Lee, J.; Sun, A. Automatic Segmentation of Lung Carcinoma Using 3D Texture Features in 18-FDG PET/CT. Int. J. Mol. Imaging 2013, 2013, 980769. [Google Scholar] [CrossRef]
- Moussallem, M.; Valette, P.J.; Traverse-Glehen, A.; Houzard, C.; Jegou, C.; Giammarile, F. New strategy for automatic tumor segmentation by adaptive thresholding on PET/CT images. J. Appl. Clin. Med. Phys. 2012, 13, 3875. [Google Scholar] [CrossRef]
- Naqiuddin, M.; Sofia, N.N.; Isa, I.S.; Sulaiman, S.N.; Karim, N.K.A.; Shuaib, I.L. Lesion demarcation of CT-scan images using image processing technique. In Proceedings of the 2018 8th IEEE International Conference on Control System, Computing and Engineering (ICCSCE), Penang, Malaysia, 23–25 November 2018. [Google Scholar]
- Perk, T.; Chen, S.; Harmon, S.; Lin, C.; Bradshaw, T.; Perlman, S.; Liu, G.; Jeraj, R. A statistically optimized regional thresholding method (SORT) for bone lesion detection in 18F-NaF PET/CT imaging. Phys. Med. Biol. 2018, 63, 225018. [Google Scholar] [CrossRef]
- Protonotarios, N.E.; Katsamenis, I.; Sykiotis, S.; Dikaios, N.; Kastis, G.A.; Chatziioannou, S.N.; Metaxas, M.; Doulamis, N.; Doulamis, A. A few-shot U-Net deep learning model for lung cancer lesion segmentation via PET/CT imaging. Biomed. Phys. Eng. Express 2022, 8, 025019. [Google Scholar] [CrossRef]
- Rao, C.; Pai, S.; Hadzic, I.; Zhovannik, I.; Bontempi, D.; Dekker, A.; Teuwen, J.; Traverso, A. Oropharyngeal Tumour Segmentation Using Ensemble 3D PET-CT Fusion Networks for the HECKTOR Challenge; Springer International Publishing: Cham, Switzerland, 2021. [Google Scholar]
- Sarker, P.; Shuvo, M.M.H.; Hossain, Z.; Hasan, S. Segmentation and classification of lung tumor from 3D CT image using K-means clustering algorithm. In Proceedings of the 2017 4th International Conference on Advances in Electrical Engineering (ICAEE), Dhaka, Bangladesh, 28–30 September 2017. [Google Scholar]
- Tian, H.; Xiang, D.; Zhu, W.; Shi, F.; Chen, X. Fully convolutional network with sparse feature-maps composition for automatic lung tumor segmentation from PET images. SPIE Med. Imaging 2020, 11313, 1131310. [Google Scholar]
- Xue, Z.; Li, P.; Zhang, L.; Lu, X.; Zhu, G.; Shen, P.; Shah, S.A.A.; Bennamoun, M. Multi-Modal Co-Learning for Liver Lesion Segmentation on PET-CT Images. IEEE Trans. Med. Imaging 2021, 40, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Xiang, D.; Yu, F.; Chen, X. Lung tumor segmentation based on the multi-scale template matching and region growing. SPIE Med. Imaging 2018, 10578, 105782Q. [Google Scholar]
- Zhang, Y.; He, S.; Wa, S.; Zong, Z.; Lin, J.; Fan, D.; Fu, J.; Lv, C. Symmetry GAN Detection Network: An Automatic One-Stage High-Accuracy Detection Network for Various Types of Lesions on CT Images. Symmetry 2022, 14, 234. [Google Scholar] [CrossRef]
- Zhao, X.; Li, L.; Lu, W.; Tan, S. Tumor co-segmentation in PET/CT using multi-modality fully convolutional neural network. Phys. Med. Biol. 2019, 64, 015011. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Luna, L.P.; Chung, H.W.; Rowe, S.P.; Du, Y.; Solnes, L.B.; Frey, E.C. Learning fuzzy clustering for SPECT/CT segmentation via convolutional neural networks. Med. Phys. 2021, 48, 3860–3877. [Google Scholar] [CrossRef]
- Yousefirizi, F.; Rahmim, A. GAN-Based Bi-Modal Segmentation Using Mumford-Shah Loss: Application to Head and Neck Tumors in PET-CT Images; Springer International Publishing: Cham, Switzerland, 2021. [Google Scholar]
- Dong, R.; Lu, H.; Kim, H.; Aoki, T.; Zhao, Y.; Zhao, Y. An Interactive Technique of Fast Vertebral Segmentation for Computed Tomography Images with Bone Metastasis. In Proceedings of the 2nd International Conference on Biomedical Signal and Image Processing, Kitakyushu, Japan, 23–25 August 2017. [Google Scholar]
- Fränzle, A.; Sumkauskaite, M.; Hillengass, J.; Bäuerle, T.; Bendl, R. Fully automated shape model positioning for bone segmentation in whole-body CT scans. J. Phys. Conf. Ser. 2014, 489, 012029. [Google Scholar] [CrossRef]
- Hanaoka, S.; Masutani, Y.; Nemoto, M.; Nomura, Y.; Miki, S.; Yoshikawa, T.; Hayashi, N.; Ohtomo, K.; Shimizu, A. Landmark-guided diffeomorphic demons algorithm and its application to automatic segmentation of the whole spine and pelvis in CT images. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 413–430. [Google Scholar] [CrossRef]
- Hu, Q.; de F. Souza, L.F.; Holanda, G.B.; Alves, S.S.A.; dos S. Silva, F.H.; Han, T.; Rebouças Filho, P.P. An effective approach for CT lung segmentation using mask region-based convolutional neural networks. Artif. Intell. Med. 2020, 103, 101792. [Google Scholar] [CrossRef]
- Lindgren Belal, S.; Sadik, M.; Kaboteh, R.; Enqvist, O.; Ulén, J.; Poulsen, M.H.; Simonsen, J.; Høilund-Carlsen, P.F.; Edenbrandt, L.; Trägårdh, E. Deep learning for segmentation of 49 selected bones in CT scans: First step in automated PET/CT-based 3D quantification of skeletal metastases. Eur. J. Radiol. 2019, 113, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Nishio, M.; Yakami, M.; Nakagomi, K.; Togashi, K. Bone segmentation on whole-body CT using convolutional neural network with novel data augmentation techniques. Comput. Biol. Med. 2020, 121, 103767. [Google Scholar] [CrossRef]
- Polan, D.F.; Brady, S.L.; Kaufman, R.A. Tissue segmentation of computed tomography images using a Random Forest algorithm: A feasibility study. Phys. Med. Biol. 2016, 61, 6553–6569. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-España, S.; Domingo, J.; Díaz-Parra, A.; Dura, E.; D’Ocón-Alcañiz, V.; Arana, E.; Moratal, D. Automatic segmentation of the spine by means of a probabilistic atlas with a special focus on ribs suppression. Med. Phys. 2017, 44, 4695–4707. [Google Scholar] [CrossRef] [PubMed]
- Arends, S.R.S.; Savenije, M.H.F.; Eppinga, W.S.C.; van der Velden, J.M.; van den Berg, C.A.T.; Verhoeff, J.J.C. Clinical utility of convolutional neural networks for treatment planning in radiotherapy for spinal metastases. Phys. Imaging Radiat. Oncol. 2022, 21, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Bernard, M.E.; Hunter, T.; Chen, Q. Improving accuracy and robustness of deep convolutional neural network based thoracic OAR segmentation. Phys. Med. Biol. 2020, 65, 07NT01. [Google Scholar] [CrossRef] [PubMed]
- Fritscher, K.D.; Peroni, M.; Zaffino, P.; Spadea, M.F.; Schubert, R.; Sharp, G. Automatic segmentation of head and neck CT images for radiotherapy treatment planning using multiple atlases, statistical appearance models, and geodesic active contours. Med. Phys. 2014, 41, 051910. [Google Scholar] [CrossRef] [PubMed]
- Ibragimov, B.; Toesca, D.A.S.; Chang, D.T.; Yuan, Y.; Koong, A.C.; Xing, L.; Vogelius, I.R. Deep learning for identification of critical regions associated with toxicities after liver stereotactic body radiation therapy. Med. Phys. 2020, 47, 3721–3731. [Google Scholar] [CrossRef]
- Lin, X.W.; Li, N.; Qi, Q. Organs-At-Risk Segmentation in Medical Imaging Based on the U-Net with Residual and Attention Mechanisms. In Proceedings of the 2021 IEEE 23rd Int Conf on High Performance Computing & Communications; 7th Int Conf on Data Science & Systems; 19th Int Conf on Smart City; 7th Int Conf on Dependability in Sensor, Cloud & Big Data Systems & Application (HPCC/DSS/SmartCity/DependSys), Haikou, China, 20–22 December 2021. [Google Scholar]
- Liu, Z.K.; Liu, X.; Xiao, B.; Wang, S.B.; Miao, Z.; Sun, Y.L.; Zhang, F.Q. Segmentation of organs-at-risk in cervical cancer CT images with a convolutional neural network. Phys. Med. 2020, 69, 184–191. [Google Scholar] [CrossRef]
- Nemoto, T.; Futakami, N.; Yagi, M.; Kumabe, A.; Takeda, A.; Kunieda, E.; Shigematsu, N. Efficacy evaluation of 2D, 3D U-Net semantic segmentation and atlas-based segmentation of normal lungs excluding the trachea and main bronchi. J. Radiat. Res. 2020, 61, 257–264. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Havlicek, J.P.; Chakrabarty, J.H.; Duong, Q.; Vesely, S.K. Towards automatic 3D bone marrow segmentation. In Proceedings of the 2016 IEEE Southwest Symposium on Image Analysis and Interpretation (SSIAI), Santa Fe, NM, USA, 6–8 March 2016. [Google Scholar]
- Yusufaly, T.; Miller, A.; Medina-Palomo, A.; Williamson, C.W.; Nguyen, H.; Lowenstein, J.; Leath, C.A., III; Xiao, Y.; Moore, K.L.; Moxley, K.M.; et al. A Multi-atlas Approach for Active Bone Marrow Sparing Radiation Therapy: Implementation in the NRG-GY006 Trial. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, 1240–1247. [Google Scholar] [CrossRef]
- Xiong, X.; Smith, B.J.; Graves, S.A.; Sunderland, J.J.; Graham, M.M.; Gross, B.A.; Buatti, J.M.; Beichel, R.R. Quantification of uptake in pelvis F-18 FLT PET-CT images using a 3D localization and segmentation CNN. Med. Phys. 2022, 49, 1585–1598. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, T.; Futakami, N.; Yagi, M.; Kunieda, E.; Akiba, T.; Takeda, A.; Shigematsu, N. Simple low-cost approaches to semantic segmentation in radiation therapy planning for prostate cancer using deep learning with non-contrast planning CT images. Phys. Med. 2020, 78, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, M.; Teramoto, A.; Ota, S.; Toyama, H.; Fujita, H. Automated segmentation and detection of increased uptake regions in bone scintigraphy using SPECT/CT images. Ann. Nucl. Med. 2018, 32, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Slattery, A. Validating an image segmentation program devised for staging lymphoma. Australas. Phys. Eng. Sci. Med. 2017, 40, 799–809. [Google Scholar] [CrossRef]
- Martínez, F.; Romero, E.; Dréan, G.; Simon, A.; Haigron, P.; de Crevoisier, R.; Acosta, O. Segmentation of pelvic structures for planning CT using a geometrical shape model tuned by a multi-scale edge detector. Phys. Med. Biol. 2014, 59, 1471–1484. [Google Scholar] [CrossRef]
- Ninomiya, K.; Arimura, H.; Sasahara, M.; Hirose, T.; Ohga, S.; Umezu, Y.; Honda, H.; Sasaki, T. Bayesian delineation framework of clinical target volumes for prostate cancer radiotherapy using an anatomical-features-based machine learning technique. In Medical Imaging 2018: Image-Guided Procedures, Robotic Interventions, and Modeling; Fei, B., Webster, R.J., Eds.; SPIE-Int. Soc. Optical Engineering: Bellingham, WA, USA, 2018. [Google Scholar]
- Men, K.; Dai, J.; Li, Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med. Phys. 2017, 44, 6377–6389. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, Z.; Wang, Z.; Wang, X.; Hu, D.; Ma, P.; Ma, C.; Wei, W.; Li, X.; Xue, X.; et al. Three-dimensional deep neural network for automatic delineation of cervical cancer in planning computed tomography images. J. Appl. Clin. Med. Phys 2022, 23, e13566. [Google Scholar] [CrossRef]
- Sartor, H.; Minarik, D.; Enqvist, O.; Ulén, J.; Wittrup, A.; Bjurberg, M.; Trägårdh, E. Auto-segmentations by convolutional neural network in cervical and anorectal cancer with clinical structure sets as the ground truth. Clin. Transl. Radiat. Oncol. 2020, 25, 37–45. [Google Scholar] [CrossRef]
- Papandrianos, N.; Papageorgiou, E.; Anagnostis, A.; Feleki, A. A deep-learning approach for diagnosis of metastatic breast cancer in bones from whole-body scans. Appl. Sci. 2020, 10, 997. [Google Scholar] [CrossRef]
- Pi, Y.; Zhao, Z.; Xiang, Y.; Li, Y.; Cai, H.; Yi, Z. Automated diagnosis of bone metastasis based on multi-view bone scans using attention-augmented deep neural networks. Med. Image Anal. 2020, 65, 101784. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Dong, D.; Chen, B.; Fang, M.; Cheng, Y.; Gan, Y.; Zhang, R.; Zhang, L.; Zang, Y.; Liu, Z.; et al. Diagnosis of Distant Metastasis of Lung Cancer: Based on Clinical and Radiomic Features. Transl. Oncol. 2018, 11, 31–36. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, G.; Cheng, J.; Song, S.; Zhang, Y.; Shi, L.Q. Diagnostic classification of solitary pulmonary nodules using support vector machine model based on 2-[18F]fluoro-2-deoxy-D-glucose PET/computed tomography texture features. Nucl. Med. Commun. 2020, 41, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Lou, B.; Doken, S.; Zhuang, T.; Wingerter, D.; Gidwani, M.; Mistry, N.; Ladic, L.; Kamen, A.; Abazeed, M.E. An image-based deep learning framework for individualising radiotherapy dose: A retrospective analysis of outcome prediction. Lancet Digit. Health 2019, 1, e136–e147. [Google Scholar] [CrossRef]
- Mao, X.; Pineau, J.; Keyes, R.; Enger, S.A. RapidBrachyDL: Rapid Radiation Dose Calculations in Brachytherapy Via Deep Learning. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, 802–812. [Google Scholar] [CrossRef]
- LabelMe. LabelMe Annotation Tool. 2022. Available online: http://labelme2.csail.mit.edu/Release3.0/ (accessed on 12 July 2022).
- Alzubaidi, L.; Al-Amidie, M.; Al-Asadi, A.; Humaidi, A.J.; Al-Shamma, O.; Fadhel, M.A.; Zhang, J.; Santamaría, J.; Duan, Y. Novel Transfer Learning Approach for Medical Imaging with Limited Labeled Data. Cancers 2021, 13, 1590. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019. [Google Scholar]
- Ge, Z.; Liu, S.; Wang, F.; Li, Z.; Sun, J. Yolox: Exceeding yolo series in 2021. arXiv 2021, arXiv:2107.08430. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Mah, P.; Reeves, T.E.; McDavid, W.D. Deriving Hounsfield units using grey levels in cone beam computed tomography. Dentomaxillofac. Radiol. 2010, 39, 323–335. [Google Scholar] [CrossRef]
- Phan, A.-C.; Vo, V.-Q.; Phan, T.-C. A Hounsfield value-based approach for automatic recognition of brain haemorrhage. J. Inf. Telecommun. 2019, 3, 196–209. [Google Scholar] [CrossRef]
- Sheen, H.; Shin, H.-B.; Kim, J.Y. Comparison of radiomics prediction models for lung metastases according to four semiautomatic segmentation methods in soft-tissue sarcomas of the extremities. J. Korean Phys. Soc. 2022, 80, 247–256. [Google Scholar] [CrossRef]
- Horikoshi, H.; Kikuchi, A.; Onoguchi, M.; Sjöstrand, K.; Edenbrandt, L. Computer-aided diagnosis system for bone scintigrams from Japanese patients: Importance of training database. Ann. Nucl. Med. 2012, 26, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Alarifi, A.; Alwadain, A. Computer-aided cancer classification system using a hybrid level-set image segmentation. Meas. J. Int. Meas. Confed. 2019, 148, 106864. [Google Scholar] [CrossRef]
| Area of the Study | Purpose of the Study | Reference | No of Papers |
|---|---|---|---|
| Reviews/Comparison of methods | Computerized PET/CT Image Analysis in the Evaluation of Tumor | [11] | 1 |
| Machine learning techniques in medical imaging | [19,20,22,27,28,29,33] | 7 | |
| Segmentation methods for Radiology image (s) | [14,16,18,21,23] | 5 | |
| Radiation therapy treatments for metastases | [4,5,6] | 3 | |
| Radiation therapy and planning | [9,10,12,34,35] | 5 | |
| Metastases Segmentation | [26] | 1 | |
| Imaging Techniques | [17,36] | 2 | |
| Radiomics in medical imaging | [25] | 1 | |
| Segmentation | Metastases | [37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54] | 18 |
| Tumor | [2,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80] | 27 | |
| Organ(s)/Organs-at-Risk (OARs) | [81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102] | 22 | |
| Target Volume/OARs + Target Volume | [103,104,105,106] | 4 | |
| Classification | Metastases | [13,107,108,109] | 4 |
| Tumor | [110,111] | 2 | |
| Total | 102 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paranavithana, I.R.; Stirling, D.; Ros, M.; Field, M. Systematic Review of Tumor Segmentation Strategies for Bone Metastases. Cancers 2023, 15, 1750. https://doi.org/10.3390/cancers15061750
Paranavithana IR, Stirling D, Ros M, Field M. Systematic Review of Tumor Segmentation Strategies for Bone Metastases. Cancers. 2023; 15(6):1750. https://doi.org/10.3390/cancers15061750
Chicago/Turabian StyleParanavithana, Iromi R., David Stirling, Montserrat Ros, and Matthew Field. 2023. "Systematic Review of Tumor Segmentation Strategies for Bone Metastases" Cancers 15, no. 6: 1750. https://doi.org/10.3390/cancers15061750
APA StyleParanavithana, I. R., Stirling, D., Ros, M., & Field, M. (2023). Systematic Review of Tumor Segmentation Strategies for Bone Metastases. Cancers, 15(6), 1750. https://doi.org/10.3390/cancers15061750






