Computer-Aided Diagnosis of Melanoma Subtypes Using Reflectance Confocal Images
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Design and Participants
2.2. RCM Acquisition Procedure and Exclusion Procedure
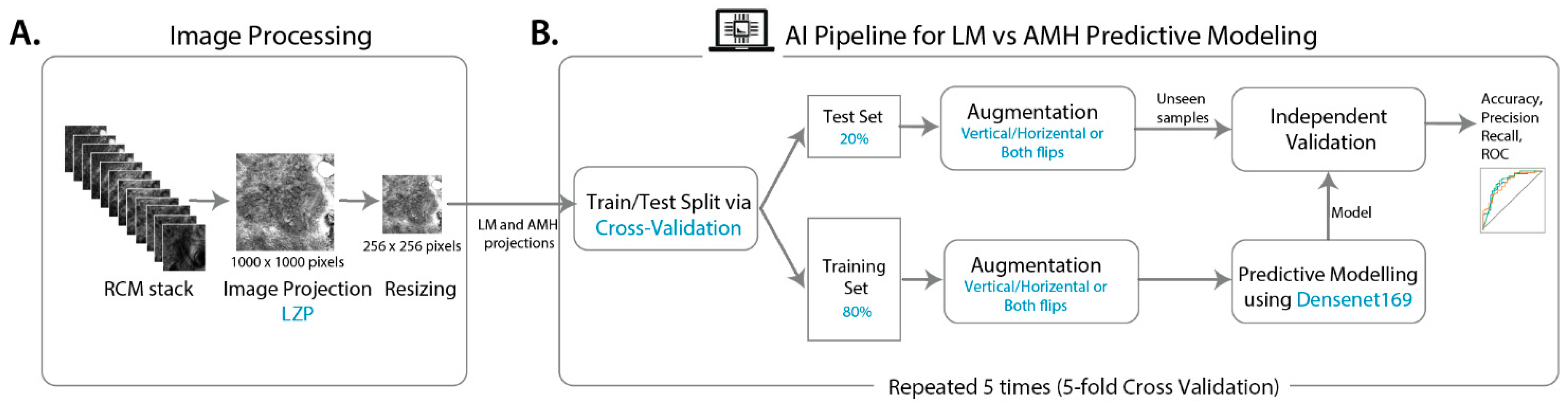
2.3. Image Processing
2.4. Predictive Modelling
2.4.1. Model Development
2.4.2. Model Validation and Performance Metrics
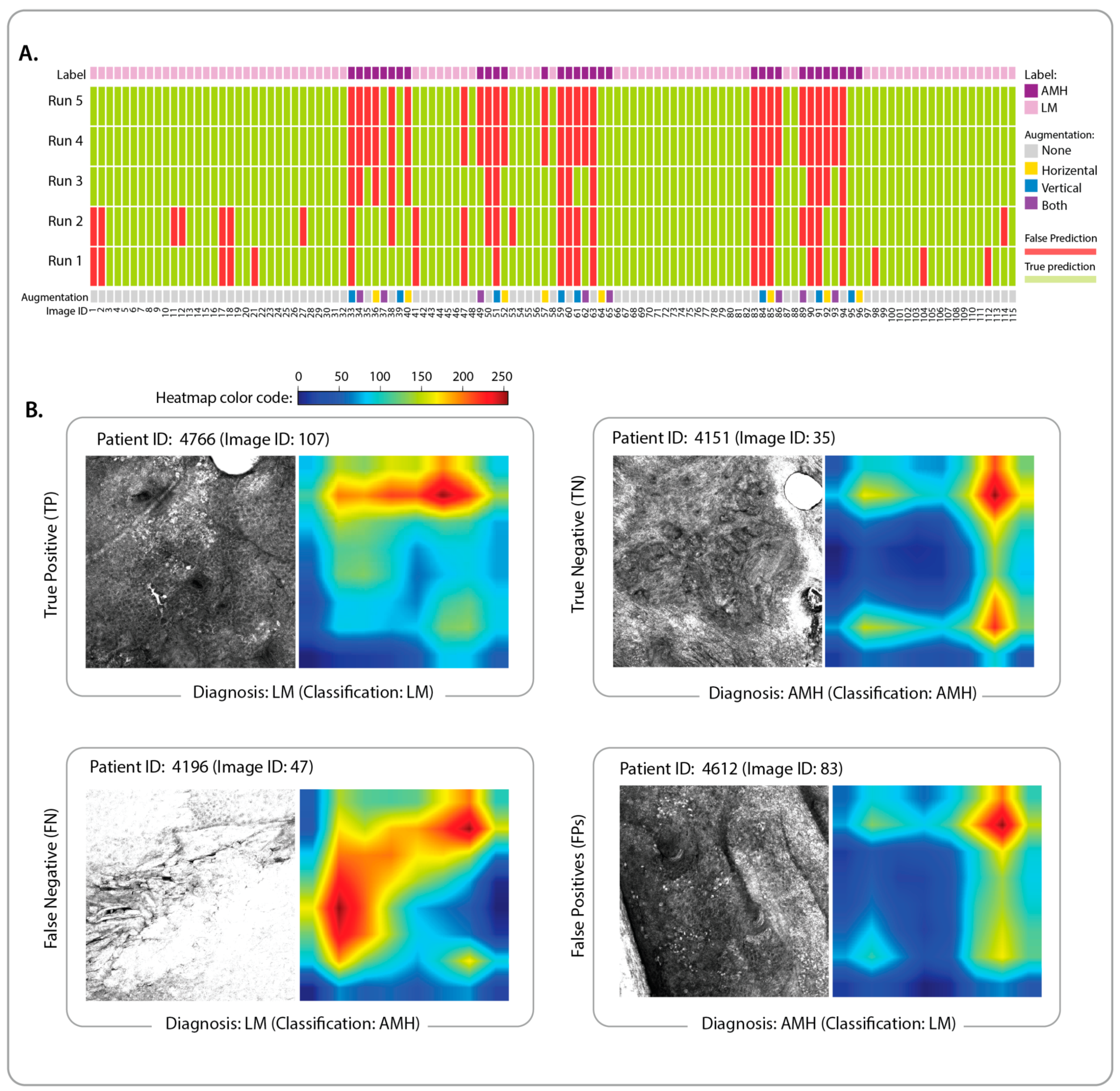
2.5. Prediction Interpretation
2.6. Statistical Analysis
3. Results
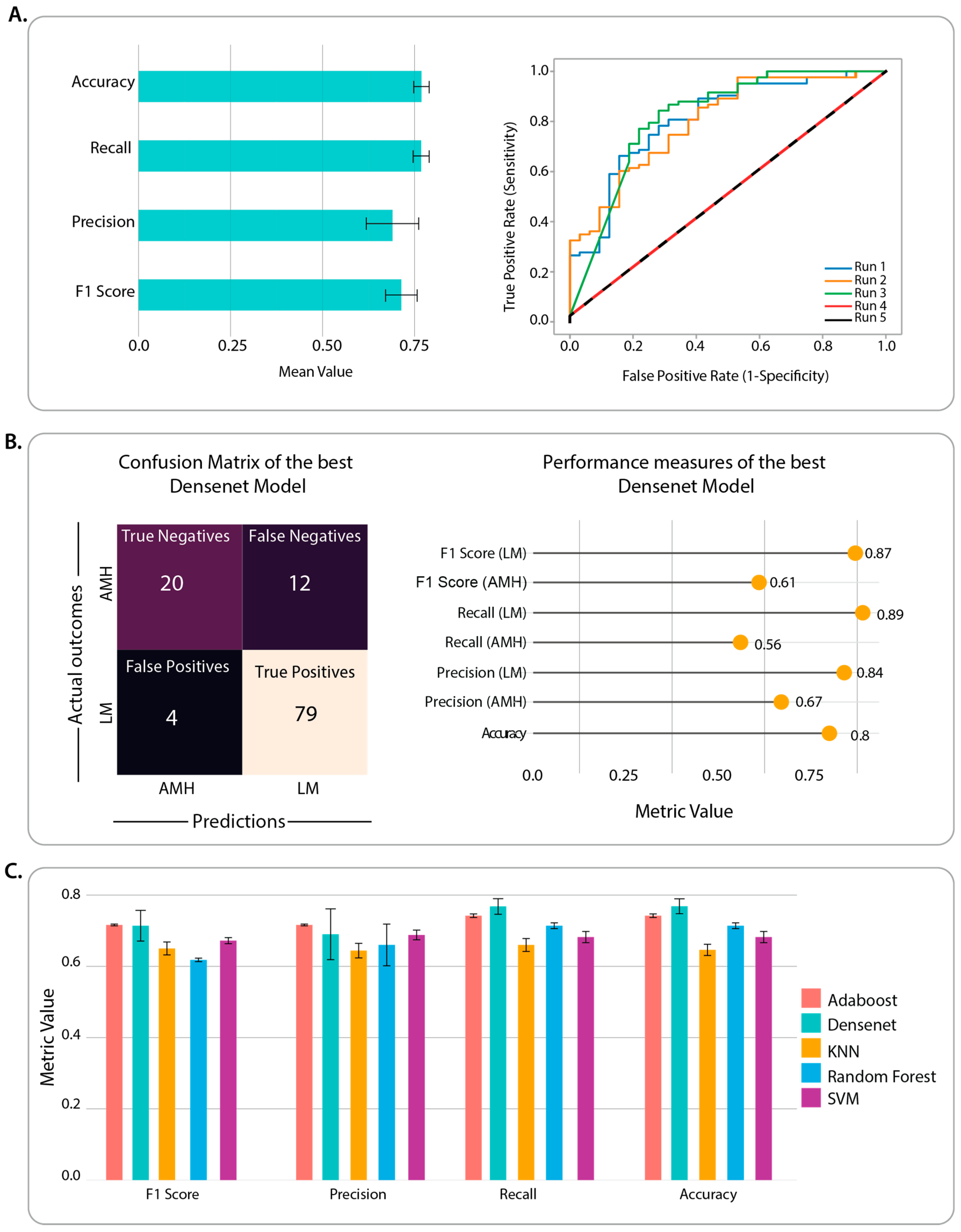
3.1. Benchmarking of CNN Architectures through Classification Performance
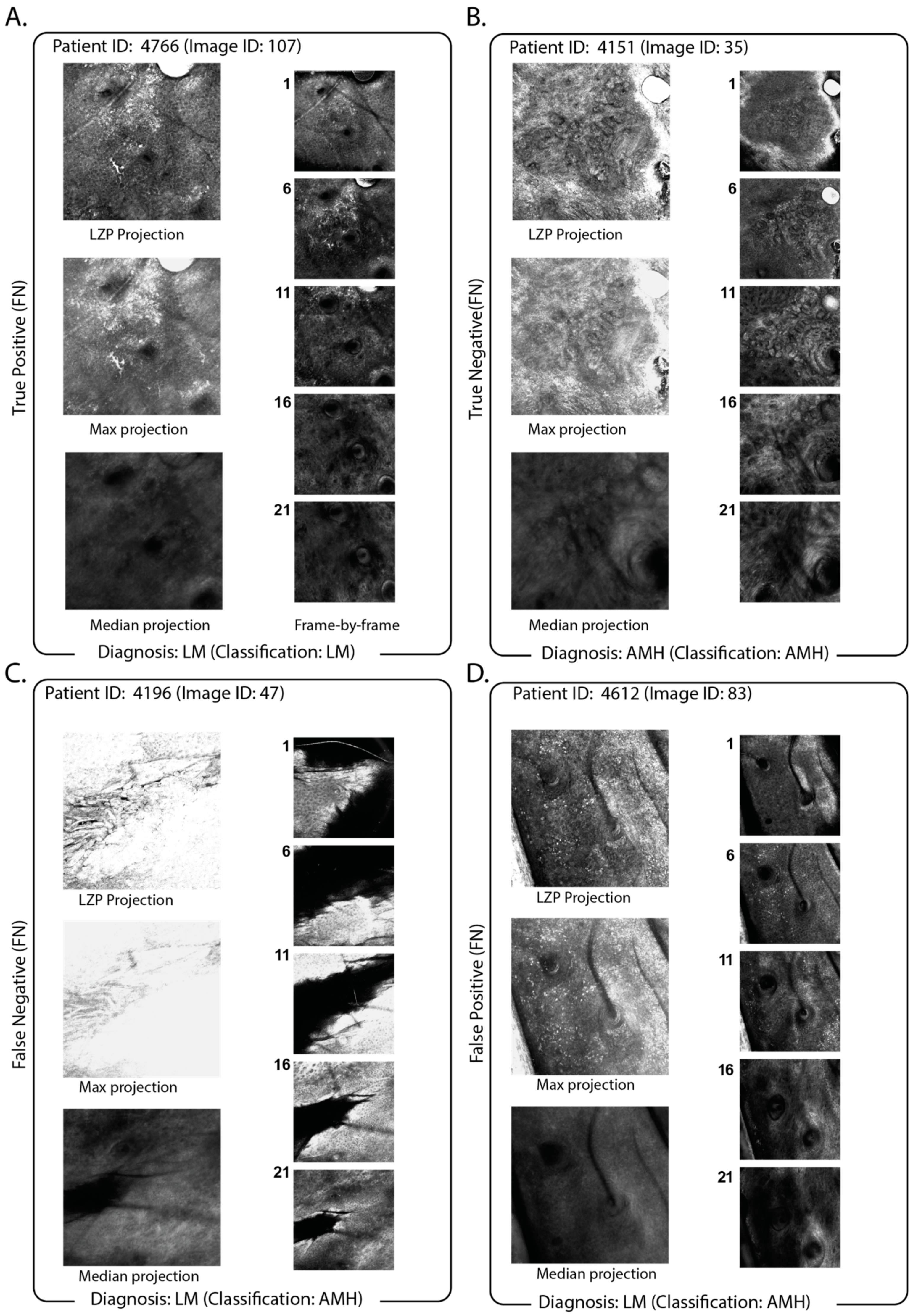
3.2. Identification of Classification Features and Examination of Misclassified Images
3.3. Impact of the Use of Projection in the Classification Pipeline
3.4. Comparison of Projection with Slice-by-Slice Classification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LM | lentigo maligna |
| AIMP | Atypical Intraepidermal Melanocytic Proliferation |
| RCM | reflectance confocal microscopy |
| LZP | local z-projection |
References
- Koller, S.; Gerger, A.; Ahlgrimm-Siess, V.; Weger, W.; Smolle, J.; Hofmann-Wellenhof, R. In vivo reflectance confocal microscopy of erythematosquamous skin diseases. Exp. Dermatol. 2009, 18, 536–540. [Google Scholar] [CrossRef]
- Rocha, L.K.F.L.; Vilain, R.E.; Scolyer, R.A.; Lo, S.N.; Ms, M.D.; Star, P.; Fogarty, G.B.; Hong, A.M.; Guitera, P.; BMedSci, M.R.A.S.; et al. Confocal microscopy, dermoscopy, and histopathology features of atypical intraepidermal melanocytic proliferations associated with evolution to melanoma in situ. Int. J. Dermatol. 2022, 61, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Guitera, P.; Pellacani, G.; Crotty, K.A.; Scolyer, R.A.; Li, L.-X.L.; Bassoli, S.; Vinceti, M.; Rabinovitz, H.; Longo, C.; Menzies, S.W. The Impact of In Vivo Reflectance Confocal Microscopy on the Diagnostic Accuracy of Lentigo Maligna and Equivocal Pigmented and Nonpigmented Macules of the Face. J. Investig. Dermatol. 2010, 130, 2080–2091. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Martín, I.; Moreno, S.; López, E.A.; Hernández-Muñoz, I.; Gallardo, F.; Barranco, C.; Pujol, R.M.; Segura, S. Histopathologic and Immunohistochemical Correlates of Confocal Descriptors in Pigmented Facial Macules on Photodamaged Skin. JAMA Dermatol. 2017, 153, 771–780. [Google Scholar] [CrossRef]
- Bou-Prieto, A.; Sarriera-Lázaro, C.J.; Valentín-Nogueras, S.M.; Sánchez, J.E.; Sánchez, J.L. Defining the Histopathological Term Atypical Intraepidermal Melanocytic Proliferation: A Retrospective Cross-Sectional Study. Am. J. Dermatopathol. 2021, 43, 252–258. [Google Scholar] [CrossRef]
- Elmore, J.G.; Barnhill, R.L.; Elder, D.E.; Longton, G.M.; Pepe, M.S.; Reisch, L.M.; Carney, P.A.; Titus, L.J.; Nelson, H.D.; Onega, T.; et al. Pathologists’ diagnosis of invasive melanoma and melanocytic proliferations: Observer accuracy and reproducibility study. BMJ 2017, 357, j2813. [Google Scholar] [CrossRef]
- Fusano, M.; Gianotti, R.; Bencini, P.L. Reflectance confocal microscopy in atypical intraepidermal melanocytic proliferation: Two cases with dermoscopic and histologic correlation. Ski. Res. Technol. Off. J. Int. Soc. Bioeng. Skin ISBS Int. Soc. Digit. Imaging Skin ISDIS Int. Soc. Skin Imaging ISSI 2020, 26, 773–775. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Chuchvara, N.; Cucalon, J.; Haroon, A.; Rao, B. Evaluating residual melanocytic atypia in a post-excision scar using in vivo reflectance confocal microscopy. Ski. Res. Technol. 2021, 27, 985–987. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Campanella, G.; Navarrete-Dechent, C.; Liopyris, K.; Monnier, J.; Aleissa, S.; Minhas, B.; Scope, A.; Longo, C.; Guitera, P.; Pellacani, G.; et al. Deep Learning for Basal Cell Carcinoma Detection for Reflectance Confocal Microscopy. J. Investig. Dermatol. 2021, 142, 97–103. [Google Scholar] [CrossRef]
- Soenen, A.; Vourc’H, M.; Dréno, B.; Chiavérini, C.; Alkhalifah, A.; Dessomme, B.K.; Roussel, K.; Chambon, S.; Debarbieux, S.; Monnier, J.; et al. Diagnosis of congenital pigmented macules in infants with reflectance confocal microscopy and machine learning. J. Am. Acad. Dermatol. 2021, 85, 1308–1309. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, A.; Kose, K.; Coll-Font, J.; Alessi-Fox, C.; Brooks, D.H.; Dy, J.G.; Rajadhyaksha, M. Skin strata delineation in reflectance confocal microscopy images using recurrent convolutional networks with attention. Sci. Rep. 2021, 11, 12567. [Google Scholar] [CrossRef] [PubMed]
- D’Alonzo, M.; Bozkurt, A.; Alessi-Fox, C.; Gill, M.; Brooks, D.H.; Rajadhyaksha, M.; Kose, K.; Dy, J.G. Semantic segmentation of reflectance confocal microscopy mosaics of pigmented lesions using weak labels. Sci. Rep. 2021, 11, 3679. [Google Scholar] [CrossRef] [PubMed]
- Kose, K.; Bozkurt, A.; Alessi-Fox, C.; Brooks, D.H.; Dy, J.G.; Rajadhyaksha, M.; Gill, M. Utilizing Machine Learning for Image Quality Assessment for Reflectance Confocal Microscopy. J. Investig. Dermatol. 2020, 140, 1214–1222. [Google Scholar] [CrossRef] [PubMed]
- Kose, K.; Bozkurt, A.; Alessi-Fox, C.; Gill, M.; Longo, C.; Pellacani, G.; Dy, J.G.; Brooks, D.H.; Rajadhyaksha, M. Segmentation of cellular patterns in confocal images of melanocytic lesions in vivo via a multiscale encoder-decoder network (MED-Net). Med. Image Anal. 2021, 67, 101841. [Google Scholar] [CrossRef] [PubMed]
- Wodzinski, M.; Pajak, M.; Skalski, A.; Witkowski, A.; Pellacani, G.; Ludzik, J. Automatic Quality Assessment of Reflectance Confocal Microscopy Mosaics Using Attention-Based Deep Neural Network. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1824–1827. [Google Scholar]
- Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3D Deep Learning on Medical Images: A Review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef] [PubMed]
- Haertter, D.; Wang, X.; Fogerson, S.M.; Ramkumar, N.; Crawford, J.M.; Poss, K.D.; Talia, S.D.; Kiehart, D.P.; Schmidt, C.F. DeepProjection: Rapid and Structure-Specific Projections of Tissue Sheets Embedded in 3D Microscopy Stacks Using Deep Learning. bioRxiv 2021, 11, 468809. [Google Scholar]
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools Prof. Program. 2000, 25, 120–123. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. arXiv 2015, arXiv:151203385. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:14091556. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:160806993. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimisation. arXiv 2017, arXiv:14126980. [Google Scholar]
- Kurbiel, T.; Khaleghian, S. Training of Deep Neural Networks Based on Distance Measures Using RMSProp. arXiv 2017, arXiv:170801911. [Google Scholar]
- Wang, R. AdaBoost for Feature Selection, Classification and Its Relation with SVM, A Review. Phys. Procedia 2012, 25, 800–807. [Google Scholar] [CrossRef]
- Kohavi, R. A Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence; Morgan Kaufman Publishers Inc.: San Francisco, CA, USA, 1995; pp. 1137–1143. [Google Scholar]
- Hoo, Z.H.; Candlish, J.; Teare, D. What is an ROC curve? Emerg. Med. J. 2017, 34, 357–359. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Malciu, A.M.; Lupu, M.; Voiculescu, V.M. Artificial Intelligence-Based Approaches to Reflectance Confocal Microscopy Image Analysis in Dermatology. J. Clin. Med. 2022, 11, 429. [Google Scholar] [CrossRef]
- Zorgui, S.; Chaabene, S.; Bouaziz, B.; Batatia, H.; Chaari, L. A Convolutional Neural Network for Lentigo Diagnosis. In Proceedings of the Impact of Digital Technologies on Public Health in Developed and Developing Countries, Hammamet, Tunisia, 24–26 June 2020; Jmaiel, M., Mokhtari, M., Abdulrazak, B., Aloulou, H., Kallel, S., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 89–99. [Google Scholar]
- Kaur, P.; Dana, K.J.; Cula, G.O.; Mack, M.C. Hybrid Deep Learning for Reflectance Confocal Microscopy Skin Images. In Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 1466–1471. [Google Scholar]
- Kassem, M.; Hosny, K.; Damaševičius, R.; Eltoukhy, M. Machine Learning and Deep Learning Methods for Skin Lesion Classification and Diagnosis: A Systematic Review. Diagnostics 2021, 11, 1390. [Google Scholar] [CrossRef]
- Guo, S.; Yang, Z. Multi-Channel-ResNet: An integration framework towards skin lesion analysis. Inform. Med. Unlocked 2018, 12, 67–74. [Google Scholar] [CrossRef]
- Sarkar, R.; Chatterjee, C.C.; Hazra, A. Diagnosis of melanoma from dermoscopic images using a deep depthwise separable residual convolutional network. IET Image Process. 2019, 13, 2130–2142. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Xia, Y.; Shen, C. Attention Residual Learning for Skin Lesion Classification. IEEE Trans. Med. Imaging 2019, 38, 2092–2103. [Google Scholar] [CrossRef] [PubMed]
- Rundo, F.; Conoci, S.; Banna, G.L.; Ortis, A.; Stanco, F.; Battiato, S. Evaluation of Levenberg–Marquardt neural networks and stacked autoencoders clustering for skin lesion analysis, screening and follow-up. IET Comput. Vis. 2018, 12, 957–962. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Ellinger, I.; Ecker, R.; Pitiot, A.; Wang, C. Fusing fine-tuned deep features for skin lesion classification. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc. 2019, 71, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Parslow, A.; Cardona, A.; Bryson-Richardson, R.J. Sample Drift Correction Following 4D Confocal Time-lapse Imaging. J. Vis. Exp. JoVE 2014, 86, e51086. [Google Scholar] [CrossRef]
- Zunair, H.; Rahman, A.; Mohammed, N.; Cohen, J.P. Uniformizing Techniques to Process CT Scans with 3D CNNs for Tuberculosis Prediction. In Proceedings of the Predictive Intelligence in Medicine: Third International Workshop, PRIME 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, 8 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 156–168. [Google Scholar]
- Yang, C.; Rangarajan, A.; Ranka, S. Visual Explanations from Deep 3D Convolutional Neural Networks for Alzheimer’s Disease Classification. AMIA Annu. Symp. Proc. 2018, 2018, 1571–1580. [Google Scholar] [PubMed]
- Kang, H.Y.; Bahadoran, P.; Ortonne, J.-P. Reflectance confocal microscopy for pigmentary disorders. Exp. Dermatol. 2010, 19, 233–239. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandal, A.; Priyam, S.; Chan, H.H.; Gouveia, B.M.; Guitera, P.; Song, Y.; Baker, M.A.B.; Vafaee, F. Computer-Aided Diagnosis of Melanoma Subtypes Using Reflectance Confocal Images. Cancers 2023, 15, 1428. https://doi.org/10.3390/cancers15051428
Mandal A, Priyam S, Chan HH, Gouveia BM, Guitera P, Song Y, Baker MAB, Vafaee F. Computer-Aided Diagnosis of Melanoma Subtypes Using Reflectance Confocal Images. Cancers. 2023; 15(5):1428. https://doi.org/10.3390/cancers15051428
Chicago/Turabian StyleMandal, Ankita, Siddhaant Priyam, Hsien Herbert Chan, Bruna Melhoranse Gouveia, Pascale Guitera, Yang Song, Matthew Arthur Barrington Baker, and Fatemeh Vafaee. 2023. "Computer-Aided Diagnosis of Melanoma Subtypes Using Reflectance Confocal Images" Cancers 15, no. 5: 1428. https://doi.org/10.3390/cancers15051428
APA StyleMandal, A., Priyam, S., Chan, H. H., Gouveia, B. M., Guitera, P., Song, Y., Baker, M. A. B., & Vafaee, F. (2023). Computer-Aided Diagnosis of Melanoma Subtypes Using Reflectance Confocal Images. Cancers, 15(5), 1428. https://doi.org/10.3390/cancers15051428







