The Relationship between Histological Composition and Metabolic Profile in Breast Tumors and Peritumoral Tissue Determined with 1H HR-MAS NMR Spectroscopy
Abstract
Simple Summary
Abstract
1. Introduction
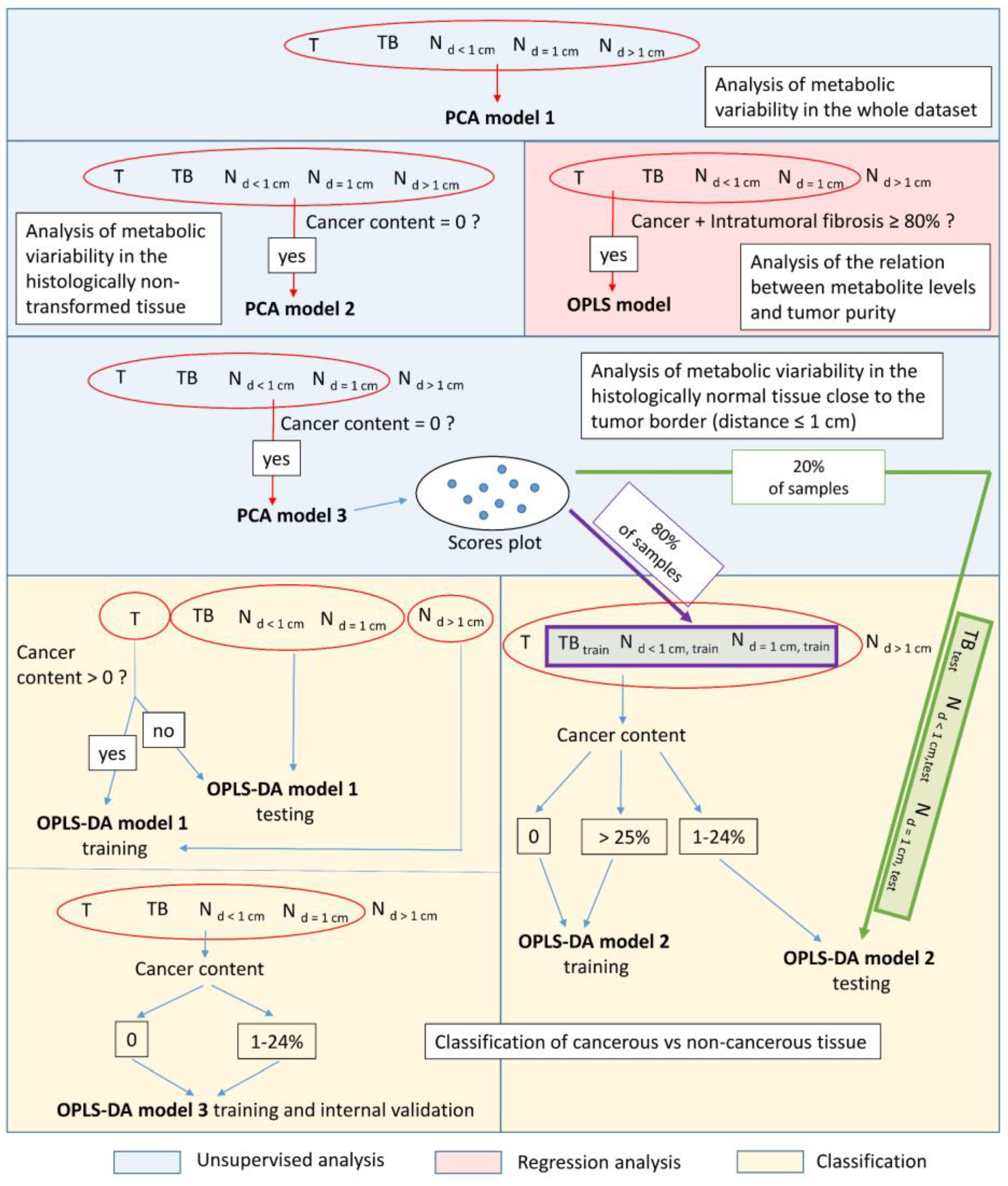
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. 1H HR-MAS NMR
2.3. Data Preprocessing
2.4. Multivariate Data Analysis
2.4.1. Unsupervised Principal Component Analysis (PCA)
2.4.2. Supervised Discrimination of Cancerous vs. Histologically Normal Tissue
2.4.3. Orthogonal Partial Least Squares Regression (OPLS) Modeling of the Relation between the Cancer Content and the Spectral Integrals
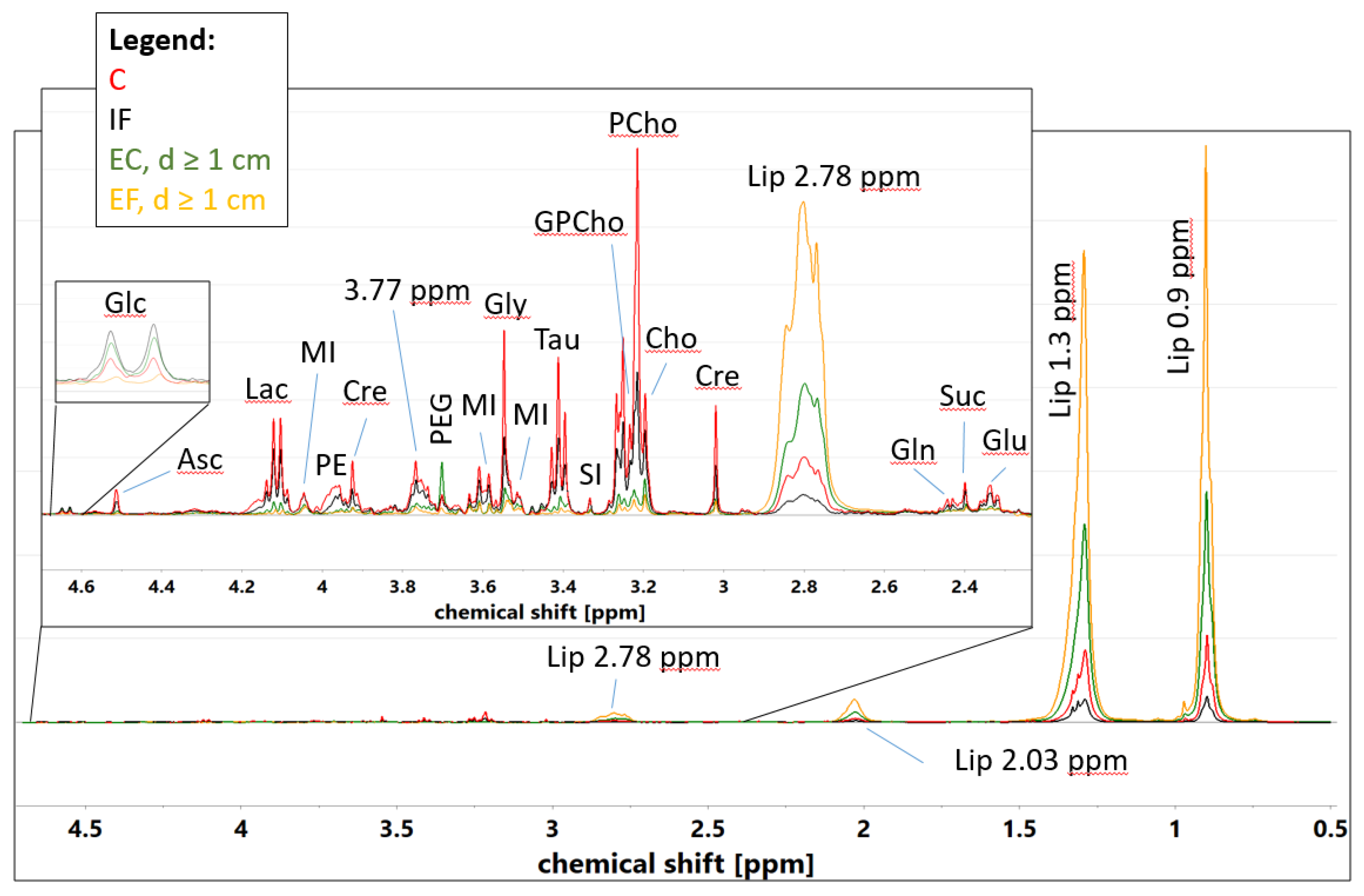
2.5. Comparison of the Metabolic Content of Cancer, Intratumoral Fibrotic Stroma and Extratumoral Connective Tissue–A Univariate Analysis
3. Results
3.1. The Results of the Post-1H HR-MAS NMR Histopathology
3.2. Multivariate Analysis
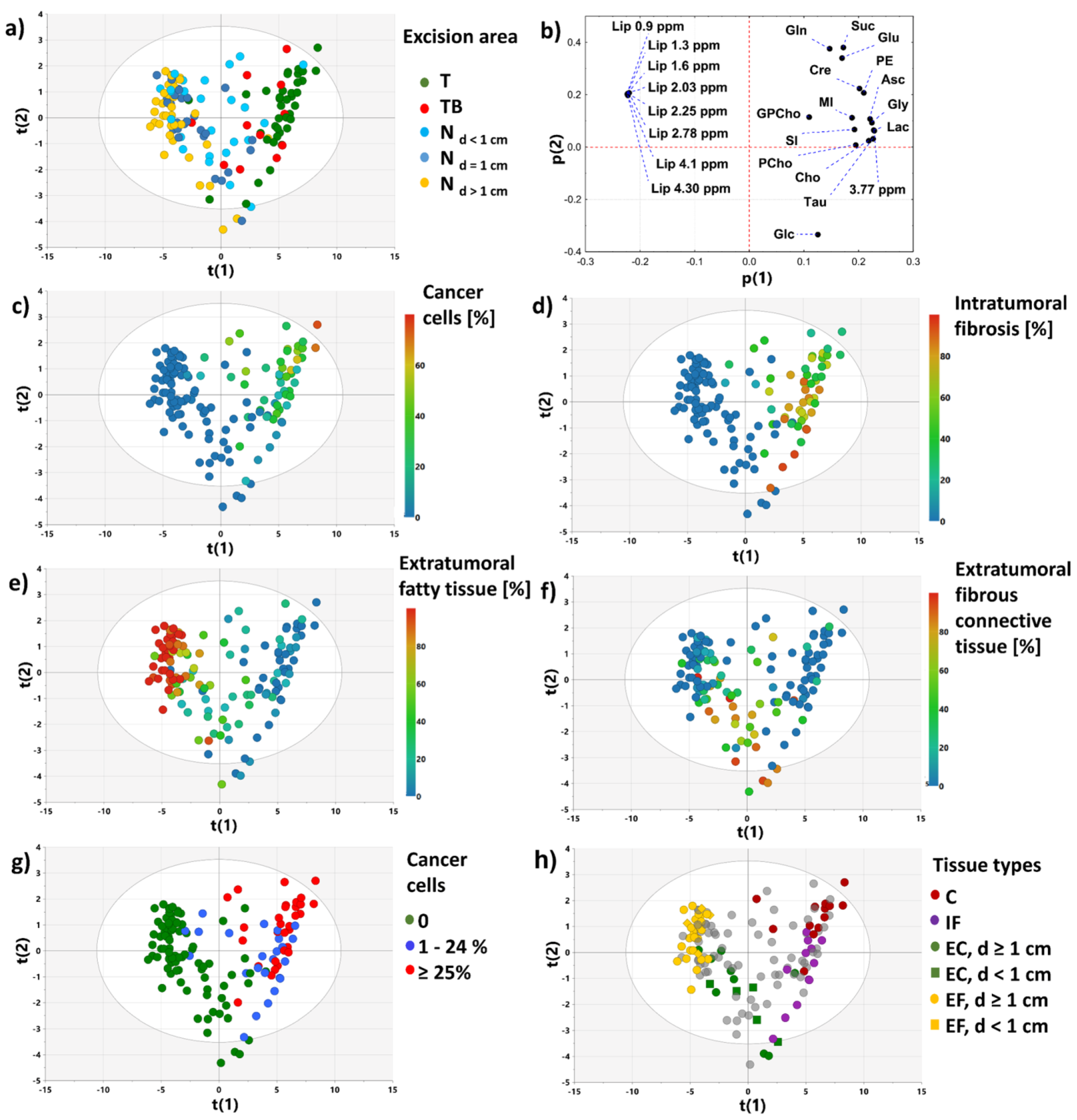
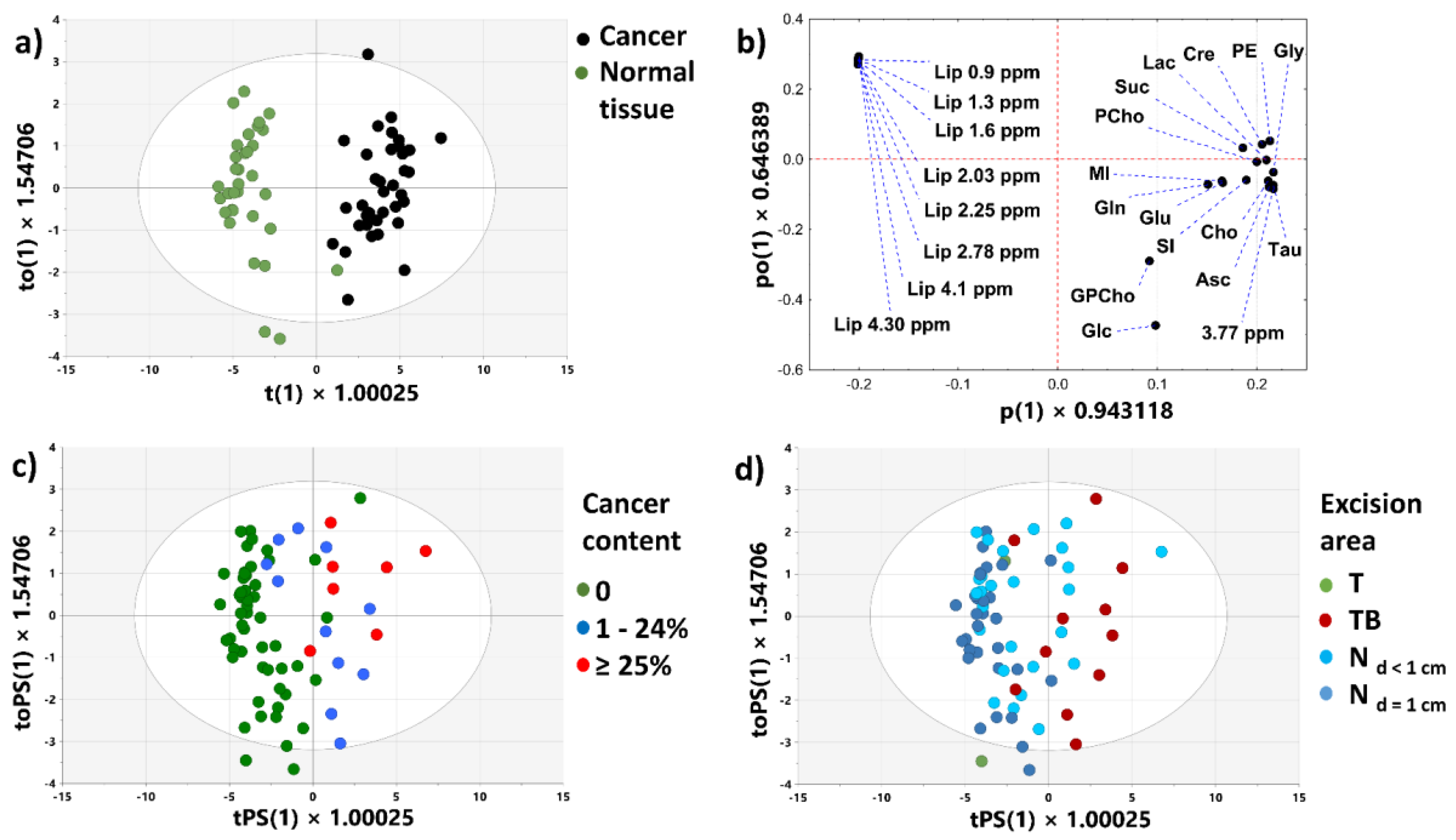
3.2.1. PCA Model 1
3.2.2. PCA Models 2 and 3
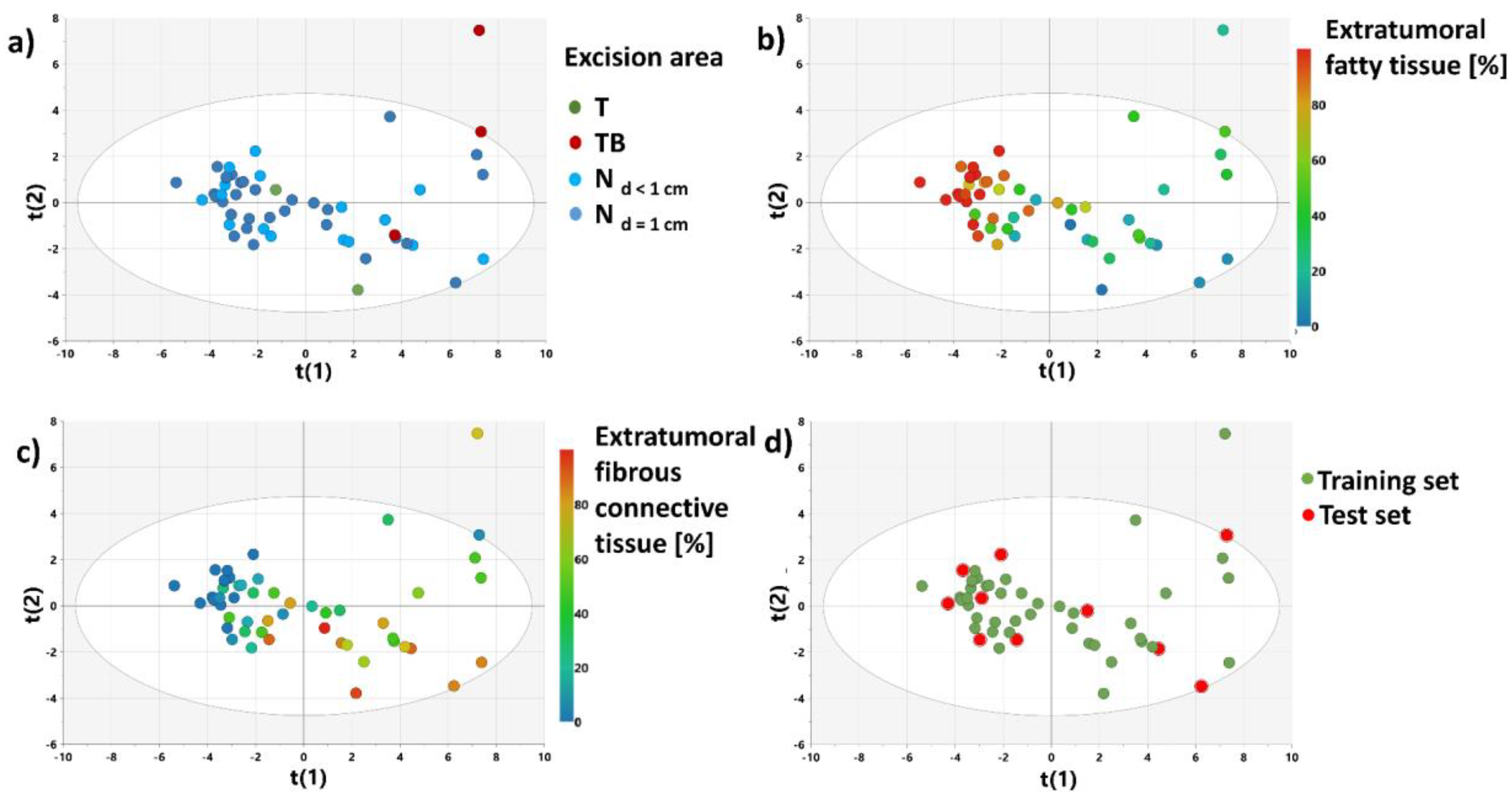
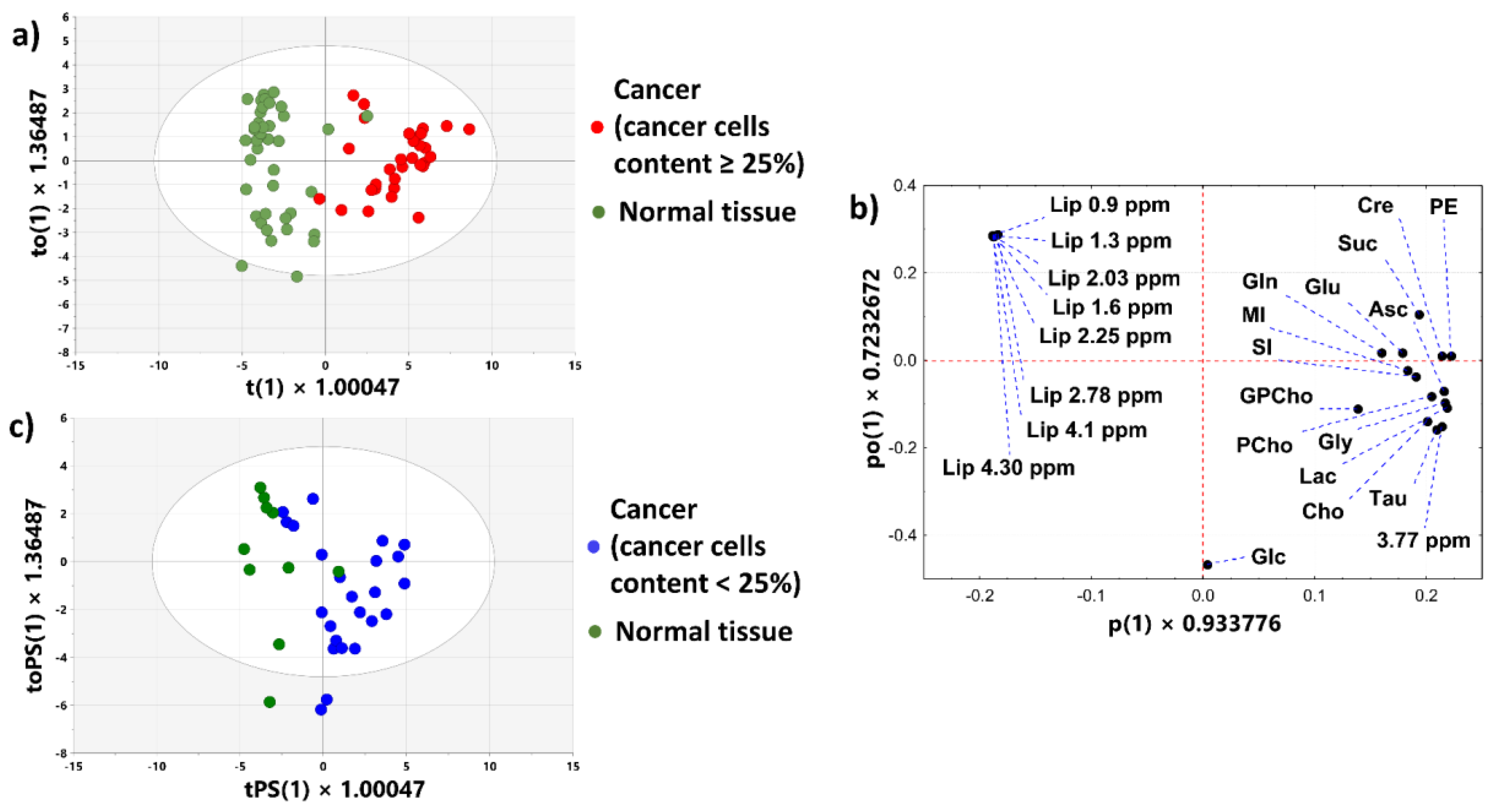
3.2.3. OPLS-DA Model 1
3.2.4. OPLS-DA Model 2
3.2.5. OPLS-DA Model 3
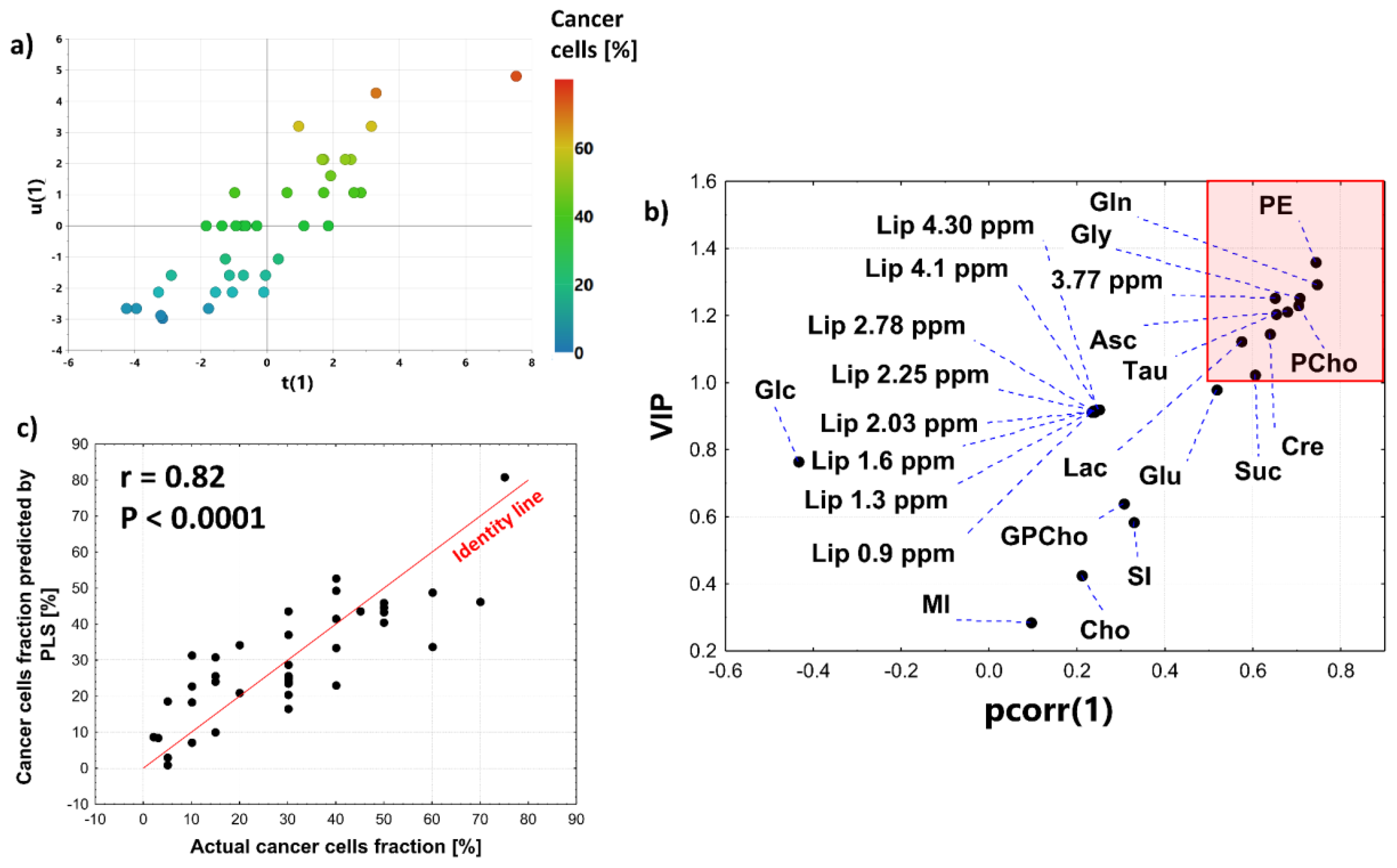
3.2.6. OPLS Model
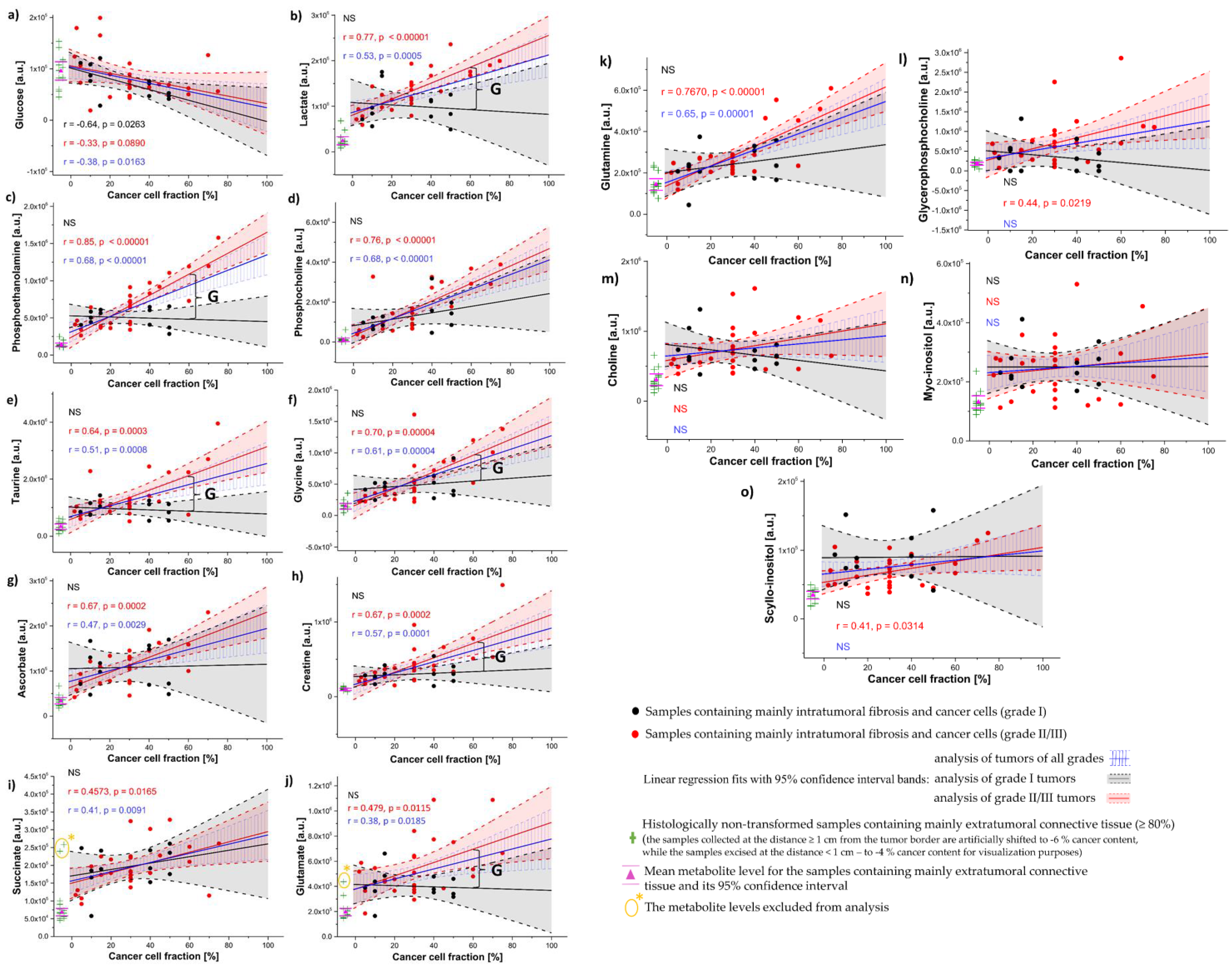
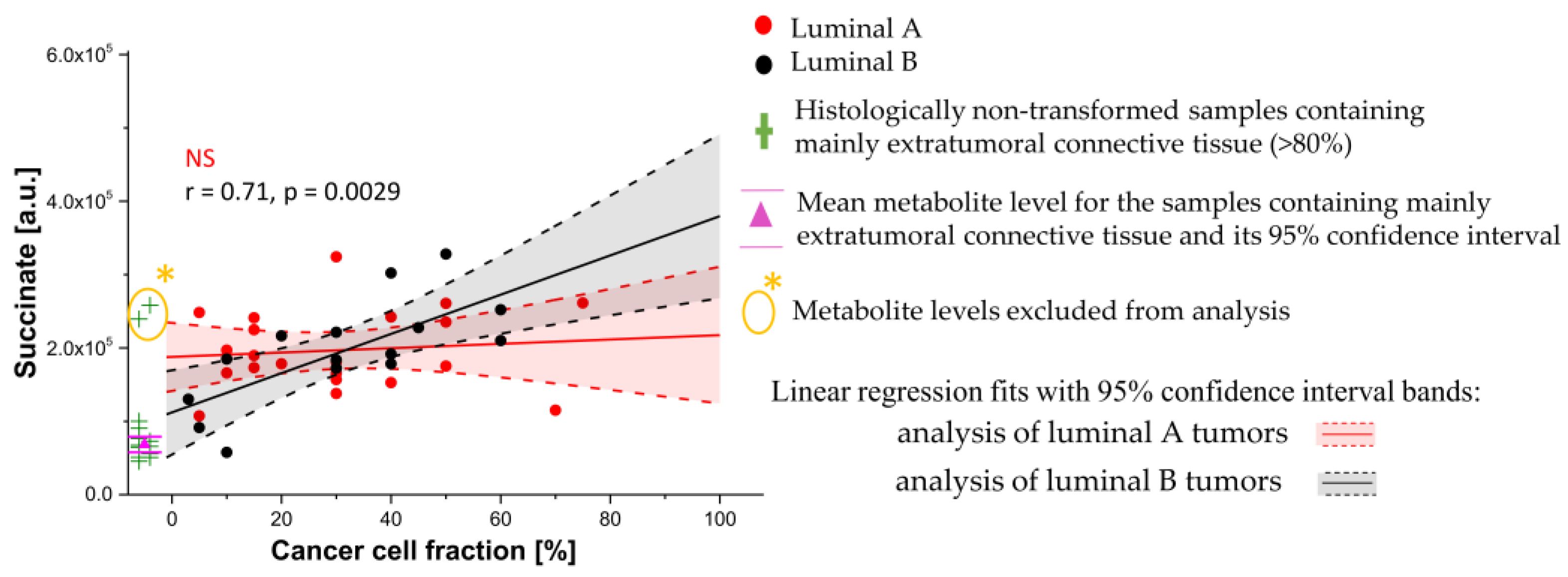
3.3. Univariate Linear Regression Analysis
4. Discussion
5. Conclusions
- 1H HR-MAS NMR spectroscopy coupled with a multivariate OPLS-DA analysis permitted classification of the cancerous vs. non-cancerous tissues with an accuracy of 87% (sensitivity of 72.2%, specificity of 92.3%);
- The correlation coefficients between the metabolite levels and cancer cell fraction were found to be grade dependent;
- The comparison of the tumor purity adjusted levels of the evaluated compounds revealed increased lactate, phosphoethanolamine, taurine, glycine, creatine and glutamate in the grade II/III tumors in comparison to the grade I ones;
- The analysis of the metabolic composition of intratumoral fibrosis (determined using a linear regression approach) revealed both shared and unique metabolic features observed in the breast tumors of different histological grades (I vs. II/III) in reference to the extratumoral fibrous connective tissue. The shared features include the lactate, glutamate and succinate accumulation within the fibrotic compartment, whereas the increased creatine, phosphoethanolamine, taurine and glycine levels were observed in the stroma of the grade I tumors only;
- The levels of ascorbate, choline, myo-inositol, and scyllo-inositol were found to be increased in the fibrotic tissue in the grade I tumors and trended towards an increase in the higher-grade (II/III) ones;
- Although the analysis of the steady-state metabolite levels does not allow for the unambiguous interpretation of the cells interaction within the tumor microenvironment, the results of our study contribute to an understanding of breast tumor molecular heterogeneity which is becoming increasingly important in the era of the introduction of metabolic profiling directly into the surgical theatre.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Fancellu, A.; Sanna, V.; Sedda, M.L.; Delrio, D.; Cottu, P.; Spanu, A.; Giuliani, G.; Conti, M.; Piras, R.; Crivelli, P.; et al. Benefits of Organized Mammographic Screening Programs in Women Aged 50 to 69 years: A Surgical Perspective. Clin. Breast Cancer 2019, 19, e637–e642. [Google Scholar] [CrossRef] [PubMed]
- James, T.A.; Wade, J.E.; Sprague, B.L. The impact of mammographic screening on the surgical management of breast cancer. J. Surg. Oncol. 2016, 113, 496–500. [Google Scholar] [CrossRef]
- Cleveland Clinic. Breast Cancer Recurrence: Symptoms & Treatment. Available online: https://my.clevelandclinic.org/health/diseases/8328-breast-cancer-recurrence (accessed on 10 January 2023).
- Nguyen, D.V.; Kim, S.W.; Oh, Y.T.; Noh, O.K.; Jung, Y.; Chun, M.; Yoon, D.S. Local Recurrence in Young Women with Breast Cancer: Breast Conserving Therapy vs. Mastectomy Alone. Cancers 2021, 13, 2150. [Google Scholar] [CrossRef]
- Gadaleta, E.; Thorn, G.J.; Ross-Adams, H.; Jones, L.J.; Chelala, C. Field cancerization in breast cancer. J. Pathol. 2022, 257, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Lebya, K.; Garcia-Smith, R.; Swaminathan, R.; Jones, A.; Russell, J.; Joste, N.; Bisoffi, M.; Trujillo, K. Towards a personalized surgical margin for breast conserving surgery-Implications of field cancerization in local recurrence. J. Surg. Oncol. 2017, 115, 109–115. [Google Scholar] [CrossRef]
- Li, J.J.; Tsang, J.Y.; Tse, G.M. Tumor Microenvironment in Breast Cancer-Updates on Therapeutic Implications and Pathologic Assessment. Cancers 2021, 13, 4233. [Google Scholar] [CrossRef]
- Casey, T.; Bond, J.; Tighe, S.; Hunter, T.; Lintault, L.; Patel, O.; Eneman, J.; Crocker, A.; White, J.; Tessitore, J.; et al. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res. Treat. 2009, 114, 47–62. [Google Scholar] [CrossRef]
- Soysal, S.D.; Tzankov, A.; Muenst, S.E. Role of the Tumor Microenvironment in Breast Cancer. Pathobiology 2015, 82, 142–152. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. WHO Classification of Tumours. Breast Tumours; International Agency for Research on Cancer: Lyon, France, 2019. [Google Scholar]
- Hagenaars, S.C.; Vangangelt, K.M.H.; Van Pelt, G.W.; Karancsi, Z.; Tollenaar, R.A.E.M.; Green, A.R.; Rakha, E.A.; Kulka, J.; Mesker, W.E. Standardization of the tumor-stroma ratio scoring method for breast cancer research. Breast Cancer Res. Treat. 2022, 193, 545–553. [Google Scholar] [CrossRef]
- Xu, Q.; Yuan, J.P.; Chen, Y.Y.; Zhang, H.Y.; Wang, L.W.; Xiong, B. Prognostic Significance of the Tumor-Stromal Ratio in Invasive Breast Cancer and a Proposal of a New Ts-TNM Staging System. J. Oncol. 2020, 2020, 9050631. [Google Scholar] [CrossRef]
- Vangangelt, K.M.H.; van Pelt, G.W.; Engels, C.C.; Putter, H.; Liefers, G.J.; Smit, V.T.H.B.M.; Tollenaar, R.A.E.M.; Kuppen, P.J.K.; Mesker, W.E. Prognostic value of tumor-stroma ratio combined with the immune status of tumors in invasive breast carcinoma. Breast Cancer Res. Treat. 2018, 168, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Karta, J.; Bossicard, Y.; Kotzamanis, K.; Dolznig, H.; Letellier, E. Mapping the Metabolic Networks of Tumor Cells and Cancer-Associated Fibroblasts. Cells 2021, 10, 304. [Google Scholar] [CrossRef]
- Becker, L.M.; O’Connell, J.T.; Vo, A.P.; Cain, M.P.; Tampe, D.; Bizarro, L.; Sugimoto, H.; McGow, A.K.; Asara, J.M.; Lovisa, S.; et al. Epigenetic Reprogramming of Cancer-Associated Fibroblasts Deregulates Glucose Metabolism and Facilitates Progression of Breast Cancer. Cell Rep. 2020, 31, 107701. [Google Scholar] [CrossRef]
- Selvarajah, B.; Azuelos, I.; Anastasiou, D.; Chambers, R.C. Fibrometabolism-An emerging therapeutic frontier in pulmonary fibrosis. Sci. Signal. 2021, 14, eaay1027. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.; O’Reilly, S. The emerging role of metabolism in fibrosis. Trends Endocrinol. Metab. 2021, 32, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Vandergrift, L.A.; Decelle, E.A.; Kurth, J.; Wu, S.; Fuss, T.L.; DeFeo, E.M.; Halpern, E.F.; Taupitz, M.; McDougal, W.S.; Olumi, A.F.; et al. Metabolomic Prediction of Human Prostate Cancer Aggressiveness: Magnetic Resonance Spectroscopy of Histologically Benign Tissue. Sci. Rep. 2018, 8, 4997. [Google Scholar] [CrossRef]
- Schult, T.A.; Lauer, M.J.; Berker, Y.; Cardoso, M.R.; Vandergrift, L.A.; Habbel, P.; Nowak, J.; Taupitz, M.; Aryee, M.; Mino-Kenudson, M.A.; et al. Screening human lung cancer with predictive models of serum magnetic resonance spectroscopy metabolomics. Proc. Natl. Acad. Sci. USA 2021, 118, e2110633118. [Google Scholar] [CrossRef]
- Li, T.; Deng, P. Nuclear Magnetic Resonance technique in tumor metabolism. Genes Dis. 2017, 4, 28–36. [Google Scholar] [CrossRef]
- Guenther, S.; Muirhead, L.J.; Speller, A.V.; Golf, O.; Strittmatter, N.; Ramakrishnan, R.; Goldin, R.D.; Jones, E.; Veselkov, K.; Nicholson, J.; et al. Spatially resolved metabolic phenotyping of breast cancer by desorption electrospray ionization mass spectrometry. Cancer Res. 2015, 75, 1828–1837. [Google Scholar] [CrossRef]
- Calligaris, D.; Caragacianu, D.; Liu, X.; Norton, I.; Thompson, C.J.; Richardson, A.L.; Golshan, M.; Easterling, M.L.; Santagata, S.; Dillon, D.A.; et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc. Natl. Acad. Sci. USA 2014, 111, 15184–15189. [Google Scholar] [CrossRef] [PubMed]
- Vaysse, P.M.; Kooreman, L.F.S.; Engelen, S.M.E.; Kremer, B.; Olde Damink, S.W.M.; Heeren, R.M.A.; Smidt, M.L.; Porta Siegel, T. Stromal vapors for real-time molecular guidance of breast-conserving surgery. Sci. Rep. 2020, 10, 20109. [Google Scholar] [CrossRef]
- Bathen, T.F.; Geurts, B.; Sitter, B.; Fjøsne, H.E.; Lundgren, S.; Buydens, L.M.; Gribbestad, I.S.; Postma, G.; Giskeødegård, G.F. Feasibility of MR metabolomics for immediate analysis of resection margins during breast cancer surgery. PLoS ONE 2013, 8, e61578. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Baek, H.M.; Kim, S.; Kim, M.J.; Youk, J.H.; Moon, H.J.; Kim, E.K.; Han, K.H.; Kim, D.H.; Kim, S.I.; et al. HR-MAS MR spectroscopy of breast cancer tissue obtained with core needle biopsy: Correlation with prognostic factors. PLoS ONE 2012, 7, e51712. [Google Scholar] [CrossRef]
- Sitter, B.; Bathen, T.F.; Singstad, T.E.; Fjøsne, H.E.; Lundgren, S.; Halgunset, J.; Gribbestad, I.S. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010, 23, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Euceda, L.R.; Haukaas, T.H.; Giskeødegård, G.F.; Vettukattil, R.; Engel, J.; Silwal-Pandit, L.; Lundgren, S.; Borgen, E.; Garred, Ø.; Postma, G.; et al. Evaluation of metabolomic changes during neoadjuvant chemotherapy combined with bevacizumab in breast cancer using MR spectroscopy. Metabolomics 2017, 13, 80. [Google Scholar] [CrossRef]
- Euceda, L.R.; Hill, D.K.; Stokke, E.; Hatem, R.; El Botty, R.; Bièche, I.; Marangoni, E.; Bathen, T.F.; Moestue, S.A. Metabolic Response to Everolimus in Patient-Derived Triple-Negative Breast Cancer Xenografts. J. Proteome Res. 2017, 16, 1868–1879. [Google Scholar] [CrossRef]
- Cao, M.D.; Sitter, B.; Bathen, T.F.; Bofin, A.; Lønning, P.E.; Lundgren, S.; Gribbestad, I.S. Predicting long-term survival and treatment response in breast cancer patients receiving neoadjuvant chemotherapy by MR metabolic profiling. NMR Biomed. 2012, 25, 369–378. [Google Scholar] [CrossRef]
- Gogiashvili, M.; Horsch, S.; Marchan, R.; Gianmoena, K.; Cadenas, C.; Tanner, B.; Naumann, S.; Ersova, D.; Lippek, F.; Rahnenführer, J.; et al. Impact of intratumoral heterogeneity of breast cancer tissue on quantitative metabolomics using high-resolution magic angle spinning 1 H NMR spectroscopy. NMR Biomed. 2018, 31, e3862. [Google Scholar] [CrossRef]
- Park, V.Y.; Yoon, D.; Koo, J.S.; Kim, E.K.; Kim, S.I.; Choi, J.S.; Park, S.; Park, H.S.; Kim, S.; Kim, M.J. Intratumoral Agreement of High-Resolution Magic Angle Spinning Magnetic Resonance Spectroscopic Profiles in the Metabolic Characterization of Breast Cancer. Medicine 2016, 95, e3398. [Google Scholar] [CrossRef]
- Choi, J.S.; Yoon, D.; Han, K.; Koo, J.S.; Kim, S.; Kim, M.J. Impact of intratumoral heterogeneity on the metabolic profiling of breast cancer tissue using high-resolution magic angle spinning magnetic resonance spectroscopy. NMR Biomed. 2022, 35, e4682. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.J. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Van den Eynden, G.G.; Colpaert, C.G.; Couvelard, A.; Pezzella, F.; Dirix, L.Y.; Vermeulen, P.B.; Van Marck, E.A.; Hasebe, T. A fibrotic focus is a prognostic factor and a surrogate marker for hypoxia and (lymph)angiogenesis in breast cancer: Review of the literature and proposal on the criteria of evaluation. Histopathology 2007, 51, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Human Metabolome Database (HMDB). Available online: https://hmdb.ca (accessed on 10 February 2023).
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Hänel, L.; Kwiatkowski, M.; Heikaus, L.; Schlüter, H. Mass spectrometry-based intraoperative tumor diagnostics. Future Sci. OA 2019, 5, FSO373. [Google Scholar] [CrossRef]
- Gribbestad, I.S.; Petersen, S.B.; Fjøsne, H.E.; Kvinnsland, S.; Krane, J. 1H NMR spectroscopic characterization of perchloric acid extracts from breast carcinomas and non-involved breast tissue. NMR Biomed. 1994, 7, 181–194. [Google Scholar] [CrossRef]
- Sitter, B.; Lundgren, S.; Bathen, T.F.; Halgunset, J.; Fjosne, H.E.; Gribbestad, I.S. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR Biomed. 2006, 19, 30–40. [Google Scholar] [CrossRef]
- Haukaas, T.H.; Euceda, L.R.; Giskeødegård, G.F.; Lamichhane, S.; Krohn, M.; Jernström, S.; Aure, M.R.; Lingjærde, O.C.; Schlichting, E.; Garred, Ø.; et al. Metabolic clusters of breast cancer in relation to gene- and protein expression subtypes. Cancer Metab. 2016, 4, 12. [Google Scholar] [CrossRef]
- Gogiashvili, M.; Nowacki, J.; Hergenröder, R.; Hengstler, J.G.; Lambert, J.; Edlund, K. HR-MAS NMR Based Quantitative Metabolomics in Breast Cancer. Metabolites 2019, 9, 19. [Google Scholar] [CrossRef]
- Borgan, E.; Sitter, B.; Lingjærde, O.C.; Johnsen, H.; Lundgren, S.; Bathen, T.F.; Sørlie, T.; Børresen-Dale, A.L.; Gribbestad, I.S. Merging transcriptomics and metabolomics—Advances in breast cancer profiling. BMC Cancer 2010, 10, 628. [Google Scholar] [CrossRef]
- Rohatgi, N.; Ghoshdastider, U.; Baruah, P.; Kulshrestha, T.; Skanderup, A.J. A pan-cancer metabolic atlas of the tumor microenvironment. Cell. Rep. 2022, 39, 110800. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells. Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Kim, D.H.; Jung, W.H.; Koo, J.S. Metabolic interaction between cancer cells and stromal cells according to breast cancer molecular subtype. Breast Cancer Res. 2013, 15, R78. [Google Scholar] [CrossRef]
- Ravazoula, P.; Batistatou, A.; Aletra, C.; Ladopoulos, J.; Kourounis, G.; Tzigounis, B. Immunohistochemical expression of glucose transporter Glut1 and cyclin D1 in breast carcinomas with negative lymph nodes. Eur. J. Gynaecol. Oncol. 2003, 24, 544–546. [Google Scholar]
- Krzeslak, A.; Wojcik-Krowiranda, K.; Forma, E.; Jozwiak, P.; Romanowicz, H.; Bienkiewicz, A.; Brys, M. Expression of GLUT1 and GLUT3 glucose transporters in endometrial and breast cancers. Pathol. Oncol. Res. 2012, 18, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Yin, K. Positive correlation between expression level of mitochondrial serine hydroxymethyltransferase and breast cancer grade. Onco. Targets Ther. 2015, 8, 1069–1074. [Google Scholar] [CrossRef]
- Kim, S.K.; Jung, W.H.; Koo, J.S. Differential expression of enzymes associated with serine/glycine metabolism in different breast cancer subtypes. PLoS ONE 2014, 9, e101004. [Google Scholar] [CrossRef]
- Moestue, S.A.; Borgan, E.; Huuse, E.M.; Lindholm, E.M.; Sitter, B.; Børresen-Dale, A.L.; Engebraaten, O.; Maelandsmo, G.M.; Gribbestad, I.S. Distinct choline metabolic profiles are associated with differences in gene expression for basal-like and luminal-like breast cancer xenograft models. BMC Cancer 2010, 10, 433. [Google Scholar] [CrossRef]
- Vignoli, A.; Risi, E.; McCartney, A.; Migliaccio, I.; Moretti, E.; Malorni, L.; Luchinat, C.; Biganzoli, L.; Tenori, L. Precision Oncology via NMR-Based Metabolomics: A Review on Breast Cancer. Int. J. Mol. Sci. 2021, 22, 4687. [Google Scholar] [CrossRef]
- Ramírez de Molina, A.; Gutiérrez, R.; Ramos, M.A.; Silva, J.M.; Silva, J.; Bonilla, F.; Sánchez, J.J.; Lacal, J.C. Increased choline kinase activity in human breast carcinomas: Clinical evidence for a potential novel antitumor strategy. Oncogene 2002, 21, 4317–4322. [Google Scholar] [CrossRef]
- Beckonert, O.; Monnerjahn, J.; Bonk, U.; Leibfritz, D. Visualizing metabolic changes in breast-cancer tissue using 1H-NMR spectroscopy and self-organizing maps. NMR Biomed. 2003, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.L.; Chang, I.W.; Smith, B.L.; Gonzalez, R.G. Evaluating human breast ductal carcinomas with high-resolution magic-angle spinning proton magnetic resonance spectroscopy. J. Magn. Reson. 1998, 135, 194–202. [Google Scholar] [CrossRef]
- Shah, T.; Krishnamachary, B.; Wildes, F.; Wijnen, J.P.; Glunde, K.; Bhujwalla, Z.M. Molecular causes of elevated phosphoethanolamine in breast and pancreatic cancer cells. NMR Biomed. 2018, 31, e3936. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Shimamura, T.; Saito, K.; Hasegawa, Y.; Ishii, N.; Nishida, M.; Ando, R.; Kondo, A.; Anwar, M.; Tsuchida, R.; et al. Phosphoethanolamine Accumulation Protects Cancer Cells under Glutamine Starvation through Downregulation of PCYT2. Cell Rep. 2019, 29, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Kung, H.N.; Marks, J.R.; Chi, J.T. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 2011, 7, e1002229. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.E.; Park, H.K.; Choi, H.J.; Lee, H.B.; Lee, H.J.; Lee, H.; Yu, E.S.; Son, W.C. Expression of the glutamine metabolism-related proteins glutaminase 1 and glutamate dehydrogenase in canine mammary tumours. Vet. Comp. Oncol. 2018, 16, 239–245. [Google Scholar] [CrossRef]
- Masisi, B.K.; El Ansari, R.; Alfarsi, L.; Craze, M.L.; Jewa, N.; Oldfield, A.; Cheung, H.; Toss, M.; Rakha, E.A.; Green, A.R. The Biological and Clinical Significance of Glutaminase in Luminal Breast Cancer. Cancers 2021, 13, 3963. [Google Scholar] [CrossRef]
- Li, Q.; Liu, M.; Sun, Y.; Jin, T.; Zhu, P.; Wan, X.; Hou, Y.; Tu, G. SLC6A8-mediated intracellular creatine accumulation enhances hypoxic breast cancer cell survival via ameliorating oxidative stress. J. Exp. Clin. Cancer Res. 2021, 40, 168. [Google Scholar] [CrossRef]
- Zhang, L.; Bu, P. The two sides of creatine in cancer. Trends Cell Biol. 2022, 32, 380–390. [Google Scholar] [CrossRef]
- Pavlides, S.; Tsirigos, A.; Migneco, G.; Whitaker-Menezes, D.; Chiavarina, B.; Flomenberg, N.; Frank, P.G.; Casimiro, M.C.; Wang, C.; Pestell, R.G.; et al. The autophagic tumor stroma model of cancer: Role of oxidative stress and ketone production in fueling tumor cell metabolism. Cell Cycle 2010, 9, 3485–3505. [Google Scholar] [CrossRef]
- Li, F.; Simon, M.C. Cancer Cells Don’t Live Alone: Metabolic Communication within Tumor Microenvironments. Dev. Cell 2020, 54, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Bardella, C.; Pollard, P.J.; Tomlinson, I. SDH mutations in cancer. Biochim. Biophys. Acta 2011, 1807, 1432–1443. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, D.H.; Jung, W.H.; Koo, J.S. Succinate dehydrogenase expression in breast cancer. Springerplus 2013, 2, 299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, Y.; Shi, Z.; Liu, J.; Sun, P.; Hou, X.; Zhang, J.; Zhao, S.; Zhou, B.P.; Mi, J. Metabolic reprogramming of cancer-associated fibroblasts by IDH3α downregulation. Cell Rep. 2015, 10, 1335–1348. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.S.; Chan, S.H.; Chang, H.T.; Li, G.C.; Tu, Y.T.; Tseng, H.H.; Fu, T.Y.; Chang, H.Y.; Liou, H.H.; Ger, L.P.; et al. Isocitrate dehydrogenase 1-snail axis dysfunction significantly correlates with breast cancer prognosis and regulates cell invasion ability. Breast Cancer Res. 2018, 20, 25. [Google Scholar] [CrossRef]
- Minemura, H.; Takagi, K.; Sato, A.; Yamaguchi, M.; Hayashi, C.; Miki, Y.; Harada-Shoji, N.; Miyashita, M.; Sasano, H.; Suzuki, T. Isoforms of IDH in breast carcinoma: IDH2 as a potent prognostic factor associated with proliferation in estrogen-receptor positive cases. Breast Cancer 2021, 28, 915–926. [Google Scholar] [CrossRef]
- Xie, N.; Tan, Z.; Banerjee, S.; Cui, H.; Ge, J.; Liu, R.M.; Bernard, K.; Thannickal, V.J.; Liu, G. Glycolytic Reprogramming in Myofibroblast Differentiation and Lung Fibrosis. Am. J. Respir. Crit. Care Med. 2015, 192, 1462–1474. [Google Scholar] [CrossRef]
- Bernard, K.; Logsdon, N.J.; Benavides, G.A.; Sanders, Y.; Zhang, J.; Darley-Usmar, V.M.; Thannickal, V.J. Glutaminolysis is required for transforming growth factor-β1-induced myofibroblast differentiation and activation. J. Biol. Chem. 2018, 293, 1218–1228. [Google Scholar] [CrossRef]
- Fazzari, J.; Lin, H.; Murphy, C.; Ungard, R.; Singh, G. Inhibitors of glutamate release from breast cancer cells; new targets for cancer-induced bone-pain. Sci. Rep. 2015, 5, 8380. [Google Scholar] [CrossRef]
- Ko, Y.H.; Lin, Z.; Flomenberg, N.; Pestell, R.G.; Howell, A.; Sotgia, F.; Lisanti, M.P.; Martinez-Outschoorn, U.E. Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: Implications for preventing chemotherapy resistance. Cancer Biol. Ther. 2011, 12, 1085–1097. [Google Scholar] [CrossRef]
- Bertero, T.; Oldham, W.M.; Grasset, E.M.; Bourget, I.; Boulter, E.; Pisano, S.; Hofman, P.; Bellvert, F.; Meneguzzi, G.; Bulavin, D.V.; et al. Tumor-Stroma Mechanics Coordinate Amino Acid Availability to Sustain Tumor Growth and Malignancy. Cell Metab. 2019, 29, 124–140. [Google Scholar] [CrossRef] [PubMed]

| Age (Median (Range)) | 50 (47–78) Years |
|---|---|
| Tumor type | Invasive carcinoma–32 |
| Invasive lobular carcinoma–7 | |
| Grade | G1–11 |
| G2–21 | |
| G3–6 | |
| Missing data–1 | |
| T-classification | T1–22 |
| T2–17 | |
| N-classification | N0–39 (100%) |
| Receptor status | ER positive–36 (92.3%) |
| PR positive–32 (82.1%) | |
| HER2 positive–4 (10.3%) | |
| Subtype | Luminal A–22 |
| Luminal B (HER2 negative/HER2 positive)–11/3 | |
| HER2–1 | |
| TNBC–2 |
| Excision Area | Total Number of Samples/Number of Samples Containing Cancer Cells | Statistics | Tumor Tissue [%] | Cancer Cells [%] | Intratumoral Fibrosis [%] | Extratumoral Fibrous Connective Tissue [%] | Extratumoral Fatty Tissue [%] | Glandular Tissue [%] | Immune Cells [%] | Necrosis [%] |
|---|---|---|---|---|---|---|---|---|---|---|
| T | 38/36 | Median | 100 | 30 | 70 | 0 | 0 | 0 | 0 | 0 |
| Minimum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Maximum | 100 | 75 | 95 | 95 | 45 | 20 | 50 | 30 | ||
| 1st quartile | 95 | 10 | 40 | 0 | 0 | 0 | 0 | 0 | ||
| 3rd quartile | 100 | 40 | 80 | 0 | 5 | 0 | 0 | 0 | ||
| TB | 11/8 | Median | 60 | 3 | 30 | 5 | 20 | 0 | 5 | 0 |
| Minimum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Maximum | 100 | 30 | 92 | 75 | 60 | 15 | 25 | 20 | ||
| 1st quartile | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 3rd quartile | 100 | 25 | 85 | 40 | 50 | 0 | 5 | 0 | ||
| Nd<1cm | 26/9 | Median | 0 | 0 | 0 | 7.5 | 30 | 0 | 0 | 0 |
| Minimum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Maximum | 100 | 50 | 80 | 90 | 100 | 20 | 5 | 0 | ||
| 1st quartile | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | ||
| 3rd quartile | 50 | 20 | 25 | 60 | 90 | 2 | 0 | 0 | ||
| Nd=1cm | 30/1 | Median | 0 | 0 | 0 | 22.5 | 75 | 0 | 0 | 0 |
| Minimum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Maximum | 5 | 5 | 0 | 100 | 100 | 20 | 5 | 0 | ||
| 1st quartile | 0 | 0 | 0 | 0 | 40 | 0 | 0 | 0 | ||
| 3rd quartile | 0 | 0 | 0 | 50 | 95 | 10 | 0 | 0 | ||
| Nd>1cm | 35/0 | Median | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 |
| Minimum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Maximum | 0 | 0 | 0 | 100 | 100 | 10 | 0 | 0 | ||
| 1st quartile | 0 | 0 | 0 | 0 | 60 | 0 | 0 | 0 | ||
| 3rd quartile | 0 | 0 | 0 | 40 | 100 | 0 | 0 | 0 |
| Tissue Type | Localization/Number of Samples | Inclusion Criteria |
|---|---|---|
| Cancer (C) | T/12 Nd<1cm/1 | Cancer cells ≥ 40%, Fatty tissue ≤ 10%, Glandular tissue ≤ 10% |
| Intratumoral fibrotic stroma (IF) | T/9 TB/3 | Fibrotic stroma ≥ 80% Cancer cells ≤ 15% |
| Extratumoral fibrous connective tissue at a distance < 1 cm from tumor border (EC, d < 1 cm) | Nd<1cm/5 | Connective tissue ≥ 80% |
| Extratumoral fibrous connective tissue at a distance ≥ 1 cm from tumor border (EC, d ≥ 1 cm) | Nd=1cm/4 Nd>1cm /5 | Connective tissue ≥ 80% |
| Extratumoral fatty tissue at a distance <1 cm from tumor border (EF, d < 1 cm) | Nd<1cm/5 | Fatty tissue = 100% |
| Extratumoral fatty tissue at a distance ≥1 cm from tumor border (EF, d ≥ 1 cm) | Nd=1cm/7 Nd>1cm/18 | Fatty tissue = 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skorupa, A.; Ciszek, M.; Turska-d’Amico, M.; Stobiecka, E.; Chmielik, E.; Szumniak, R.; d’Amico, A.; Boguszewicz, Ł.; Sokół, M. The Relationship between Histological Composition and Metabolic Profile in Breast Tumors and Peritumoral Tissue Determined with 1H HR-MAS NMR Spectroscopy. Cancers 2023, 15, 1283. https://doi.org/10.3390/cancers15041283
Skorupa A, Ciszek M, Turska-d’Amico M, Stobiecka E, Chmielik E, Szumniak R, d’Amico A, Boguszewicz Ł, Sokół M. The Relationship between Histological Composition and Metabolic Profile in Breast Tumors and Peritumoral Tissue Determined with 1H HR-MAS NMR Spectroscopy. Cancers. 2023; 15(4):1283. https://doi.org/10.3390/cancers15041283
Chicago/Turabian StyleSkorupa, Agnieszka, Mateusz Ciszek, Maria Turska-d’Amico, Ewa Stobiecka, Ewa Chmielik, Ryszard Szumniak, Andrea d’Amico, Łukasz Boguszewicz, and Maria Sokół. 2023. "The Relationship between Histological Composition and Metabolic Profile in Breast Tumors and Peritumoral Tissue Determined with 1H HR-MAS NMR Spectroscopy" Cancers 15, no. 4: 1283. https://doi.org/10.3390/cancers15041283
APA StyleSkorupa, A., Ciszek, M., Turska-d’Amico, M., Stobiecka, E., Chmielik, E., Szumniak, R., d’Amico, A., Boguszewicz, Ł., & Sokół, M. (2023). The Relationship between Histological Composition and Metabolic Profile in Breast Tumors and Peritumoral Tissue Determined with 1H HR-MAS NMR Spectroscopy. Cancers, 15(4), 1283. https://doi.org/10.3390/cancers15041283








