The Position of Circulating Tumor DNA in the Clinical Management of Colorectal Cancer
Abstract
Simple Summary
Abstract
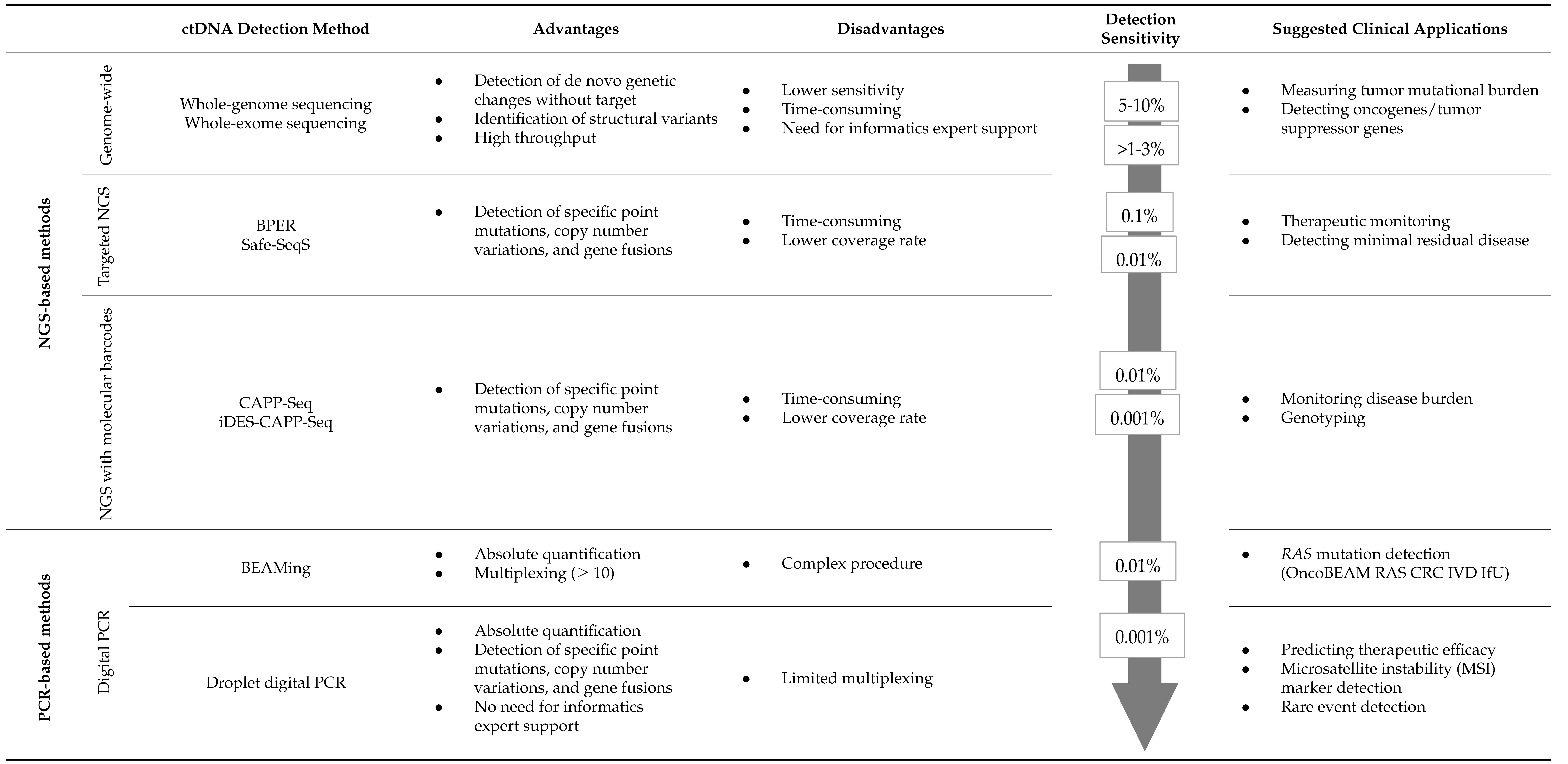
1. Circulating Tumor DNA: Detection
1.1. Pre-Analytical Conditions
1.2. Analytical Conditions
1.3. Delay of Sampling
Delay between Surgery and ctDNA Sampling
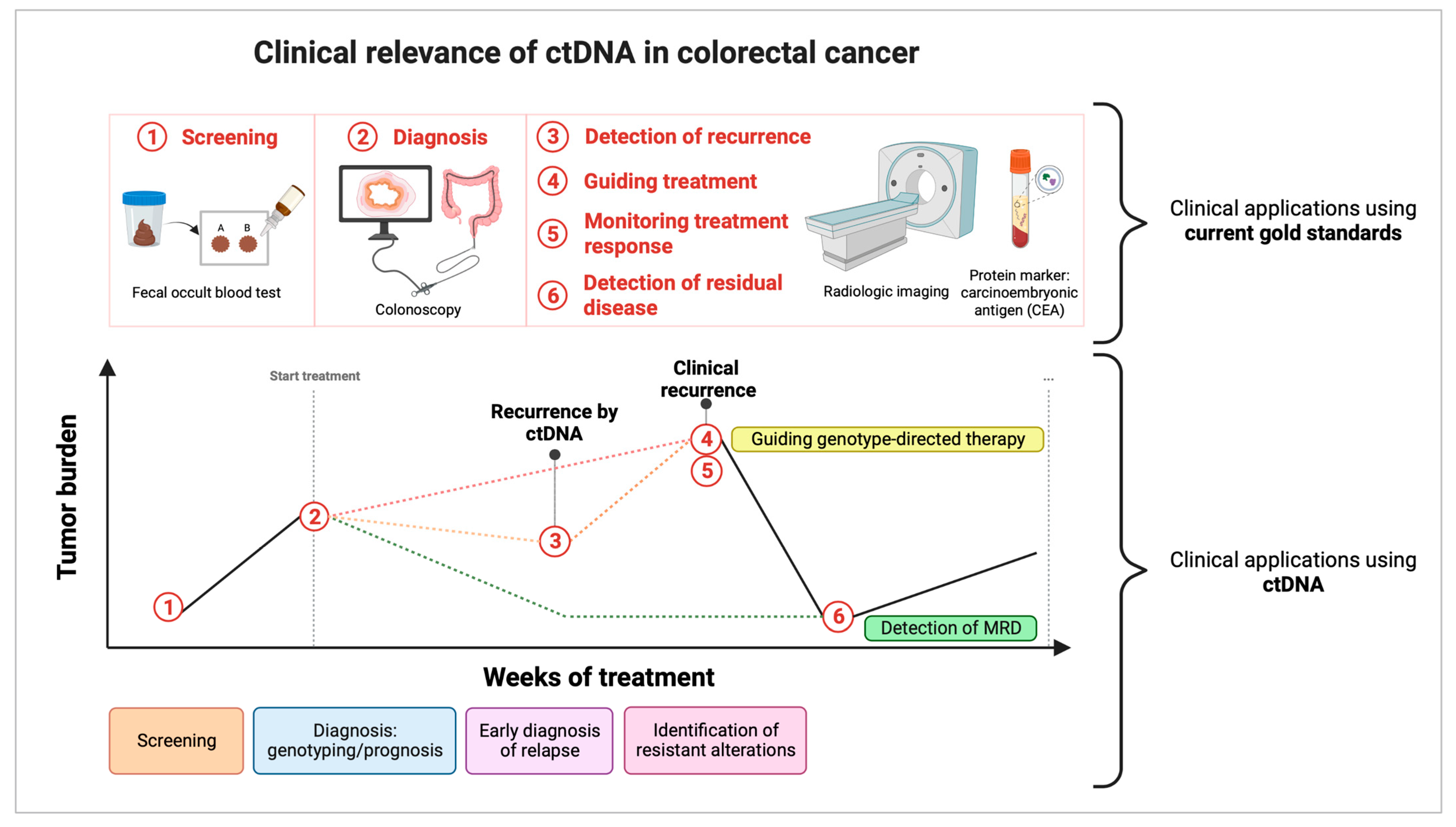
2. ctDNA: Clinical Applications
2.1. ctDNA for Early Cancer Detection: Screening
Screening
2.2. The Value of ctDNA Detection at Diagnosis
2.3. ctDNA as a Prognostic Biomarker in CRC Stage I–III: Detection of Minimal Residual Disease
2.3.1. Detection of MRD/Recurrence after Surgery
2.3.2. ctDNA Clearance after Treatment
- In stages I–III CRC, Reinert et al. observed that 84/94 (89.4%) patients became ctDNA negative and 10/94 (10.6%) patients became ctDNA positive after surgery.
- In stages I–III, Reinert et al. observed 30% of patients who cleared ctDNA after ACT and stayed disease free throughout the study [8].
- In stage II, Tie et al. observed that post-operative ctDNA+ remained negative after ACT in three out of six patients [64].
2.3.3. Value of ctDNA in the Prediction of Relapse before Conventional Imaging Techniques
2.4. ctDNA as a Quantitative Monitoring Tool in Predicting Response to Treatment (Stage IV)
2.4.1. ctDNA Concentration during Treatment
2.4.2. ctDNA Predicts Response to Targeted Therapy
2.5. ctDNA as a Tool for Guiding Treatment
| Name of Study and Country | Recruitment Status | Patient Population | Sample Size | ctDNA Detection Method | Intervention | Primary Objective |
|---|---|---|---|---|---|---|
| Cetuximab Rechallenge in irinotecan-pre-treated mCRC, KRAS, NRAS, and BRAF Wild-type Treated in 1st Line With Anti-EGFR Therapy (CRICKET) (NCT02296203) Italy [71] | Active, not recruiting | KRAS, NRAS, and BRAF wild-type, irinotecan-resistant mCRC patients who have progressed after an initial response to a first-line cetuximab-containing therapy | 28 | ddPCR | Cetuximab and irinotecan (single-arm trial) | Overall response rate |
| Rechallenge With Panitumumab Driven by RAS Dynamic of Resistance (CHRONOS) (NCT03227926) Italy [83] | Active, not recruiting | RAS wild-type mCRC patients who have progressed on first-line anti-EGFR therapy and whose RAS mutation load has decreased over 50% compared to the mutation load at the time of progression on first-line anti-EGFR therapy | 129 | ddPCR | Panitumumab monotherapy (single-arm trial) | Overall response rate |
| Circulating Tumor DNA Based Decision for Adjuvant Treatment in Stage II Colon Cancer based on ctDNA (CIRCULATE-PRODIGE 70-trial) (NCT04120701) France [87] | Recruiting | Stage II colon cancer patients who underwent curative-intent surgery | 1980 | ddPCR | ctDNA-positive randomized into two arms (1) Control arm: observation (2) Experimental arm: adjuvant mFOLFOX6 ctDNA-negative: surveillance | 3-year disease-free survival |
|
Following Therapy Response Through Liquid Biopsy in Metastatic Colorectal Cancer Patients (FOLICOLOR) Belgium | Recruiting | Unresectable, metastatic colorectal cancer patients receiving first-line treatment (pembrolizumab, panitumumab, or FOLFOX/FOLFIRI with(out) targeted therapy) | 336 | ddPCR | Control arm: Treatment decisions are guided by radiographic evaluation Experimental arm: Treatment decisions are guided by ctDNA | Primary: To determine the proportion of patients in which PD can be detected earlier in ctDNA than with conventional CT imaging Secondary: - PFS - 3-year overall survival |
| Circulating Tumour DNA Analysis Informing Adjuvant Chemotherapy in Stage II Colon Cancer (DYNAMIC) (ACTRN12615000381583) Australia [88] | Closed | Stage II colon cancer patients who underwent curative-intent surgery | 455 | Safe-SeqS | Control arm: All decisions were based on conventional clinicopathological criteria) Experimental arm: ctDNA informed (ctDNA positive: adjuvant chemotherapy; ctDNA negative: no adjuvant chemotherapy) | 2-year recurrence-free survival |
| Circulating Tumor DNA Analysis Informing Adjuvant Chemotherapy in Stage III Colon Cancer (DYNAMIC-III) (ACTRN12617001566325) Australia [89] | Recruiting | Stage III colon cancer patients who underwent curative-intent surgery | 1000 | Safe-SeqS | Control arm: standard of care treatment Experimental arm: ctDNA informed (ctDNA negative: therapy de-escalation; ctDNA positive: therapy escalation) | 3-year recurrence-free survival |
| Tracking Mutations in Cell Free Tumor DNA to Predict Relapse in Early Colorectal Cancer (TRACC) (NCT04050345), United Kingdom [90] | Recruiting | High-risk stage II and III patients with CRC who have measurable ctDNA pre-operatively and underwent R0 resection | 1000 | ddPCR | Control arm: standard of care ACT after surgery Experimental arm: ctDNA-guided ACT (ctDNA-negative: therapy de-escalation of ACT for 3 months with single Cape, or no chemotherapy; ctDNA positive: 3 months CapOx) | 1. The incidence of pre-operatively detectable ctDNA in stage II and III CRC patients 2. The correlation between post-operatively detectable ctDNA and DFS |
| Circulating Tumor DNA Analysis to Optimize the Operative and Postoperative Treatment for Patients With Colorectal Cancer—Intervention Tial 2 (IMPROVE-IT2) (NCT04084249) Denmark [91] | Recruiting | Stage I and II patients with colon cancer who underwent surgery | 254 | ddPCR Targeted error correction sequencing (TEC-Seq) [52] | Control arm: surveillance according to current Danish Guidelines with CT-scans at 12- and 36-months post-operative and colonoscopy every 5 years until age 75 Experimental arm: ctDNA-guided surveillance every 4 months postoperatively. (1) ctDNA-positive: patients undergo a whole-body FDG-PET/CT scan and colonoscopy. (2) ctDNA-negative: high-intensive radiological surveillance with FDG-PET/CT-scan every 3 months until recurrence detection or 21 months have passed | Fraction of patients with relapse receiving curative-intended resection or local treatment |
| Circulating Tumor DNA Testing in Predicting Treatment for Patients With Stage IIA Colon Cancer After Surgery (COBRA) (NCT0406810 US [92] | Recruiting | Patients whose stage II colon cancer has been resected and who have no traditional high-risk features | 1408 | LUNAR™ (Guardant Health Inc.) | Control arm: Standard of care, observation Experimental arm: Prospective testing for ctDNA. (1) ctDNA-positive: treatment with 6 months of adjuvant (FOLFOX) chemotherapy (2) ctDNA-negative: active surveillance | 1. Clearance of ctDNA with adjuvant chemotherapy 2. Recurrence-free survival for ctDNA-positive patients treated with or without adjuvant chemotherapy |
| Circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for CRC (CIRCULATE-Japan, consists of the 3 trials (GALAXY, VEGA & ALTAIR)), Japan [93] | ||||||
|
Genetic Alterations and clinical record in radically resected colorectal cancer revealed by Liquid biopsy and whole eXome analYsis (GALAXY) (UMIN000039205) | Recruiting | Stage II high-risk and stage III low-risk CRC patients who have recurrence after initial registration and are eligible for radical surgical resection | 2500 | Signatera™ (Natera Inc.) | Observational study Based on ctDNA results in this study, patients can be enrolled in investigator-initiated phase III trials, either the VEGA (if ctDNA-negative) or the ALTAIL (if ctDNA-positive) trial (see below). | 1. Disease-free survival 2. Sensitivity and specificity of ctDNA for the presence of lymph node metastases in additional colorectal resections |
|
Study to Compare CAPOX Therapy as Post-operative Adjuvant Chemotherapy with Surgery Alone in Patients with Completely Resected Circulating Tumor DNA-negative High-risk Stage II and Low-risk Stage III Colon Cancer (VEGA) (jRCT1031200006) | Recruiting | Colon cancer patients that have a negative ctDNA status at week 4 after surgery in the GALAXY study | 1240 | Signatera™ (Natera Inc.) | Control arm: Observation Experimental arm: CapOx therapy for 3 months | Disease-free survival |
|
Initial Attack on Latent Metastasis Using TAS-102 for ctDNA Identified Colorectal Cancer Patients After Curative Resection (ALTAIR) (NCT04457297) | Recruiting | CRC patients that have a positive ctDNA status within the previous 3 months at any time after surgery in the GALAXY study, and no obvious relapse on CT-scan | 240 | Signatera™ (Natera Inc.) | Control arm: Placebo Experimental arm: 6 months of oral trifluridine/tipiracil (FTD/TPI) | Disease-free survival |
3. Rectal Cancer: The Current State of Management
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’Homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Fettke, H.; Kwan, E.M.; Azad, A.A. Cell-free DNA in cancer: Current insights. Cell. Oncol. 2019, 42, 13–28. [Google Scholar] [CrossRef]
- Moati, E.; Taly, V.; Didelot, A.; Perkins, G.; Blons, H.; Taieb, J.; Laurent-Puig, P.; Zaanan, A. Role of circulating tumor DNA in the management of patients with colorectal cancer. Clin. Res. Hepatol. Gastroenterol. 2018, 42, 396–402. [Google Scholar] [CrossRef]
- Ershova, E.; Sergeeva, V.; Klimenko, M.; Avetisova, K.; Klimenko, P.; Kostyuk, E.; Veiko, N.; Veiko, R.; Izevskaya, V.; Kutsev, S.; et al. Circulating cell-free DNA concentration and DNase I activity of peripheral blood plasma change in case of pregnancy with intrauterine growth restriction compared to normal pregnancy. Biomed. Rep. 2017, 7, 319–324. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.T.V.; Pantel, K. Circulating Tumor Cells and Circulating nucleic acids in oncology. Tietz Textbook of Clinical Chemistry and Molecular Diagnostics, 7th ed. in progress.
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.X.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients with Stages I to III Colorectal Cancer. JAMA Oncol. 2019, 5, 1124–1131. [Google Scholar] [CrossRef]
- Elazezy, M.; Joosse, S.A. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput. Struct. Biotechnol. 2018, 16, 370–378. [Google Scholar] [CrossRef]
- Barták, B.K.; Kalmár, A.; Galamb, O.; Wichmann, B.; Nagy, Z.B.; Tulassay, Z.; Dank, M.; Igaz, P.; Molnár, B. Blood Collection and Cell-Free DNA Isolation Methods Influence the Sensitivity of Liquid Biopsy Analysis for Colorectal Cancer Detection. Pathol. Oncol. Res. 2019, 25, 915–923. [Google Scholar] [CrossRef]
- Van Ginkel, J.H.; van den Broek, D.A.; van Kuik, J.; Linders, D.; de Weger, R.; Willems, S.M.; Huibers, M.M.H. Preanalytical blood sample workup for cell-free DNA analysis using Droplet Digital PCR for future molecular cancer diagnostics. Cancer Med. 2017, 6, 2297–2307. [Google Scholar] [CrossRef]
- Markus, H.; Contente-Cuomo, T.; Farooq, M.; Liang, W.S.; Borad, M.J.; Sivakumar, S.; Gollins, S.; Tran, N.L.; Dhruv, H.D.; Berens, M.E.; et al. Evaluation of pre-analytical factors affecting plasma DNA analysis. Sci. Rep. 2018, 8, 7375. [Google Scholar] [CrossRef] [PubMed]
- Gerber, T.; Taschner-Mandl, S.; Saloberger-Sindhöringer, L.; Popitsch, N.; Heitzer, E.; Witt, V.; Geyeregger, R.; Hutter, C.; Schwentner, R.; Ambros, I.M.; et al. Assessment of Pre-Analytical Sample Handling Conditions for Comprehensive Liquid Biopsy Analysis. J. Mol. Diagn. 2020, 22, 1070–1086. [Google Scholar] [CrossRef] [PubMed]
- Poulet, G. Université de Paris, UMR-S1138, CNRS SNC5096, Équipe labélisée Ligue Nationale Contre le Cancer; Centre de Recherche des Cordeliers: Paris, France, 2022; to be submitted. [Google Scholar]
- Lianidou, E. Detection and relevance of epigenetic markers on ctDNA: Recent advances and future outlook. Mol. Oncol. 2021, 15, 1683–1700. [Google Scholar] [CrossRef] [PubMed]
- Connors, D.; Allen, J.; Alvarez, J.D.; Boyle, J.; Cristofanilli, M.; Hiller, C.; Keating, S.; Kelloff, G.; Leiman, L.; McCormack, R.; et al. International liquid biopsy standardization alliance white paper. Crit. Rev. Oncol. Hematol. 2020, 156, 103112. [Google Scholar] [CrossRef] [PubMed]
- Wojas-Krawczyk, K.; Kalinka-Warzocha, E.; Reszka, K.; Nicos, M.; Szumilo, J.; Mandziuk, S.; Szczepaniak, K.; Kupnicka, D.; Lewandowski, R.; Milanowski, J.; et al. Analysis of KRAS, NRAS, BRAF, and PIK3CA mutations could predict metastases in colorectal cancer: A preliminary study. Adv. Clin. Exp. Med. 2019, 28, 67–73. [Google Scholar] [CrossRef]
- Vidal, J.; Muinelo, L.; Dalmases, A.; Jones, F.; Edelstein, D.; Iglesias, M.; Orrillo, M.; Abalo, A.; Rodriguez, C.; Brozos, E.; et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann. Oncol. 2017, 28, 1325–1332. [Google Scholar] [CrossRef]
- Madic, J.; Jovelet, C.; Lopez, J.; André, B.; Fatien, J.; Miran, I.; Honoré, A.; Mezquita, L.; Besse, B.; Lacroix, L.; et al. EGFR C797S, EGFR T790M and EGFR sensitizing mutations in non-small cell lung cancer revealed by six-color crystal digital PCR. Oncotarget 2018, 9, 37393–37406. [Google Scholar] [CrossRef]
- Madic, J.; Jovelet, C.; Dehri, I.; Mallory, A.C. 6-Color Crystal Digital PCR(TM) for the High-Plex Detection of EGFR Mutations in Non-Small Cell Lung Cancer. Methods Mol. Biol. 2021, 2279, 127–144. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Boeckx, N.; Op de Beeck, K.; Beyens, M.; Deschoolmeester, V.; Hermans, C.; De Clercq, P.; Garrigou, S.; Normand, C.; Monsaert, E.; Papadimitriou, K.; et al. Mutation and Methylation Analysis of Circulating Tumor DNA Can Be Used for Follow-up of Metastatic Colorectal Cancer Patients. Clin. Color. Cancer 2018, 17, e369–e379. [Google Scholar] [CrossRef]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 552–558. [Google Scholar] [CrossRef]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef]
- Pecuchet, N.; Rozenholc, Y.; Zonta, E.; Pietrasz, D.; Didelot, A.; Combe, P.; Gibault, L.; Bachet, J.B.; Taly, V.; Fabre, E.; et al. Analysis of Base-Position Error Rate of Next-Generation Sequencing to Detect Tumor Mutations in Circulating DNA. Clin. Chem. 2016, 62, 1492–1503. [Google Scholar] [CrossRef]
- Manier, S.; Park, J.; Capelletti, M.; Bustoros, M.; Freeman, S.S.; Ha, G.; Rhoades, J.; Liu, C.J.; Huynh, D.; Reed, S.C.; et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat. Commun. 2018, 9, 1691. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Belic, J.; Gutschi, S.; Quehenberger, F.; Fischereder, K.; Benezeder, T.; Auer, M.; Pischler, C.; Mannweiler, S.; et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013, 5, 30. [Google Scholar] [CrossRef]
- Vymetalkova, V.; Cervena, K.; Bartu, L.; Vodicka, P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 3356. [Google Scholar] [CrossRef]
- Fleming, C.A.; O’Leary, D.P.; Wang, J.; Redmond, H.P. Association of Observed Perioperative Cell-Free DNA Dynamics with Early Recurrence in Patients with Colon Cancer. JAMA Surg. 2020, 155, 168–170. [Google Scholar] [CrossRef]
- Henriksen, T.V.; Reinert, T.; Christensen, E.; Sethi, H.; Birkenkamp-Demtröder, K.; Gögenur, M.; Gögenur, I.; Zimmermann, B.G.; Dyrskjøt, L.; Andersen, C.L. The effect of surgical trauma on circulating free DNA levels in cancer patients-implications for studies of circulating tumor DNA. Mol. Oncol. 2020, 14, 1670–1679. [Google Scholar] [CrossRef]
- Scholer, L.V.; Reinert, T.; Orntoft, M.B.W.; Kassentoft, C.G.; Arnadottir, S.S.; Vang, S.; Nordentoft, I.; Knudsen, M.; Lamy, P.; Andreasen, D.; et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin. Cancer Res. 2017, 23, 5437–5445. [Google Scholar] [CrossRef]
- Vatandoost, N.; Ghanbari, J.; Mojaver, M.; Avan, A.; Ghayour-Mobarhan, M.; Nedaeinia, R.; Salehi, R. Early detection of colorectal cancer: From conventional methods to novel biomarkers. J. Cancer Res. Clin. Oncol. 2016, 142, 341–351. [Google Scholar] [CrossRef]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- Bretthauer, M. Colorectal cancer screening. J. Intern. Med. 2011, 270, 87–98. [Google Scholar] [CrossRef]
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis. Primers 2015, 1, 15065. [Google Scholar] [CrossRef]
- Petit, J.; Carroll, G.; Gould, T.; Pockney, P.; Dun, M.; Scott, R.J. Cell-Free DNA as a Diagnostic Blood-Based Biomarker for Colorectal Cancer: A Systematic Review. J. Surg. Res. 2019, 236, 184–197. [Google Scholar] [CrossRef]
- Lamb, Y.N.; Dhillon, S. Epi proColon((R)) 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol. Diagn. Ther. 2017, 21, 225–232. [Google Scholar] [CrossRef]
- Rasmussen, S.L.; Krarup, H.B.; Sunesen, K.G.; Johansen, M.B.; Stender, M.T.; Pedersen, I.S.; Madsen, P.H.; Thorlacius-Ussing, O. Hypermethylated DNA, a circulating biomarker for colorectal cancer detection. PLoS ONE 2017, 12, e0180809. [Google Scholar] [CrossRef]
- Rahier, J.F.; Druez, A.; Faugeras, L.; Martinet, J.P.; Gehenot, M.; Josseaux, E.; Herzog, M.; Micallef, J.; George, F.; Delos, M.; et al. Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer. Clin. Epigenetics 2017, 9, 1–7. [Google Scholar] [CrossRef]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castaños-Vélez, E.; Blumenstein, B.A.; Rösch, T.; Osborn, N.; et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014, 63, 317–325. [Google Scholar] [CrossRef]
- Sun, G.P.; Meng, J.; Duan, H.; Zhang, D.W.; Tang, Y.X. Diagnostic Assessment of septin9 DNA Methylation for Colorectal Cancer Using Blood Detection: A Meta-Analysis. Pathol. Oncol. Res. 2019, 25, 1525–1534. [Google Scholar] [CrossRef]
- Adler, A.; Geiger, S.; Keil, A.; Bias, H.; Schatz, P.; deVos, T.; Dhein, J.; Zimmermann, M.; Tauber, R.; Wiedenmann, B. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol. 2014, 14, 183. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Lin, J.S.; Perdue, L.A.; Henrikson, N.B.; Bean, S.I.; Blasi, P.R. Screening for Colorectal Cancer: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2021, 325, 1978–1998. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631. [Google Scholar] [CrossRef]
- Bi, F.; Wang, Q.; Dong, Q.; Wang, Y.; Zhang, L.; Zhang, J. Circulating tumor DNA in colorectal cancer: Opportunities and challenges. Am. J. Transl. Res. 2020, 12, 1044–1055. [Google Scholar]
- Marcuello, M.; Vymetalkova, V.; Neves, R.P.L.; Duran-Sanchon, S.; Vedeld, H.M.; Tham, E.; van Dalum, G.; Flugen, G.; Garcia-Barberan, V.; Fijneman, R.J.; et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Asp. Med. 2019, 69, 107–122. [Google Scholar] [CrossRef]
- Wang, X.; Shi, X.Q.; Zeng, P.W.; Mo, F.M.; Chen, Z.H. Circulating cell free DNA as the diagnostic marker for colorectal cancer: A systematic review and meta-analysis. Oncotarget 2018, 9, 24514–24524. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019, 5, 1710–1717. [Google Scholar] [CrossRef]
- Benhaim, L.; Bouché, O.; Normand, C.; Didelot, A.; Mulot, C.; Le Corre, D.; Garrigou, S.; Djadi-Prat, J.; Wang-Renault, S.-F.; Perez-Toralla, K.; et al. Circulating tumor DNA is a prognostic marker of tumor recurrence in stage II and III colorectal cancer: Multicentric, prospective cohort study (ALGECOLS). Eur. J. Cancer 2021, 159, 24–33. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef]
- Symonds, E.L.; Pedersen, S.K.; Murray, D.H.; Jedi, M.; Byrne, S.E.; Rabbitt, P.; Baker, R.T.; Bastin, D.; Young, G.P. Circulating tumour DNA for monitoring colorectal cancer—A prospective cohort study to assess relationship to tissue methylation, cancer characteristics and surgical resection. Clin. Epigenetics 2018, 10, 63. [Google Scholar] [CrossRef]
- Tarazona, N.; Gimeno-Valiente, F.; Gambardella, V.; Zuñiga, S.; Rentero-Garrido, P.; Huerta, M.; Roselló, S.; Martinez-Ciarpaglini, C.; Carbonell-Asins, J.A.; Carrasco, F.; et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann. Oncol. 2019, 30, 1804–1812. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yokoyama, S.; Matsuda, K.; Tamura, K.; Mitani, Y.; Iwamoto, H.; Mizumoto, Y.; Murakami, D.; Kitahata, Y.; Yamaue, H. Preoperative detection of KRAS mutated circulating tumor DNA is an independent risk factor for recurrence in colorectal cancer. Sci. Rep. 2021, 11, 441. [Google Scholar] [CrossRef]
- Chen, G.; Peng, J.; Xiao, Q.; Wu, H.X.; Wu, X.; Wang, F.; Li, L.; Ding, P.; Zhao, Q.; Li, Y.; et al. Postoperative circulating tumor DNA as markers of recurrence risk in stages II to III colorectal cancer. J. Hematol. Oncol. 2021, 14, 80. [Google Scholar] [CrossRef]
- Bregni, G.; Pretta, A.; Senti, C.; Acedo Reina, E.; Vandeputte, C.; Trevisi, E.; Gkolfakis, P.; Kehagias, P.; Deleporte, A.; Van Laethem, J.L.; et al. Circulating DNA in the neoadjuvant setting of early stage colon cancer. Acta Oncol. 2022, 61, 1223–1229. [Google Scholar] [CrossRef]
- Norcic, G. Liquid Biopsy in Colorectal Cancer—Current Status and Potential Clinical Applications. Micromachines 2018, 9, 300. [Google Scholar] [CrossRef]
- Basnet, S.; Zhang, Z.-y.; Liao, W.-q.; Li, S.-h.; Li, P.-s.; Ge, H.-y. The Prognostic Value of Circulating Cell-Free DNA in Colorectal Cancer: A Meta-Analysis. J. Cancer 2016, 7, 1105–1113. [Google Scholar] [CrossRef]
- Manca, P.; Corallo, S.; Lonardi, S.; Fucà, G.; Busico, A.; Leone, A.G.; Corti, F.; Antoniotti, C.; Procaccio, L.; Smiroldo, V.; et al. Variant allele frequency in baseline circulating tumour DNA to measure tumour burden and to stratify outcomes in patients with RAS wild-type metastatic colorectal cancer: A translational objective of the Valentino study. Br. J. Cancer 2021, 126, 449–455. [Google Scholar] [CrossRef]
- Garlan, F.; Laurent-Puig, P.; Sefrioui, D.; Siauve, N.; Didelot, A.; Sarafan-Vasseur, N.; Michel, P.; Perkins, G.; Mulot, C.; Blons, H.; et al. Early Evaluation of Circulating Tumor DNA as Marker of Therapeutic Efficacy in Metastatic Colorectal Cancer Patients (PLACOL Study). Clin. Cancer Res. 2017, 23, 5416–5425. [Google Scholar] [CrossRef]
- Osterman, E.; Glimelius, B. Recurrence Risk After Up-to-Date Colon Cancer Staging, Surgery, and Pathology: Analysis of the Entire Swedish Population. Dis. Colon Rectum 2018, 61, 1016–1025. [Google Scholar] [CrossRef]
- Pachman, D.R.; Qin, R.; Seisler, D.K.; Smith, E.M.L.; Beutler, A.S.; Ta, L.E.; Lafky, J.M.; Wagner-Johnston, N.D.; Ruddy, K.J.; Dakhil, S.; et al. Clinical Course of Oxaliplatin-Induced Neuropathy: Results from the Randomized Phase III Trial N08CB (Alliance). J. Clin. Oncol. 2015, 33, 3416–3422. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra392. [Google Scholar] [CrossRef] [PubMed]
- Li, L.R.; Zhou, W.H.; Li, C.; Li, P.S.; Gong, Y.H.; Guan, Y.F. Analysis of circulating tumor DNA to monitor disease status in colorectal cancer after surgery. J. Clin. Oncol. 2018, 36, e15583. [Google Scholar] [CrossRef]
- Taieb, J.; Taly, V.; Henriques, J.; Bourreau, C.; Mineur, L.; Bennouna, J.; Desrame, J.; Louvet, C.; Lepere, C.; Mabro, M.; et al. Prognostic Value and Relation with Adjuvant Treatment Duration of ctDNA in Stage III Colon Cancer: A Post Hoc Analysis of the PRODIGE-GERCOR IDEA-France Trial. Clin. Cancer Res. 2021, 27, 5638. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.; Wang, Y.X.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; Cho, J.H.; Faragher, I.; McKendrick, J.J.; et al. Serial circulating tumor DNA (ctDNA) analysis as a prognostic marker and a real-time indicator of adjuvant chemotherapy (CT) efficacy in stage III colon cancer (CC). J. Clin. Oncol. 2018, 36, 3516. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Cohen, J.D.; Kinde, I.; Ptak, J.; Popoli, M.; Schaefer, J.; Silliman, N.; Dobbyn, L.; Tie, J.; et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol. 2019, 5, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Reinert, T.; Scholer, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.R.; Nordentoft, I.; Lamy, P.; Kannerup, A.S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef]
- Henriksen, T.V.; Tarazona, N.; Frydendahl, A.; Reinert, T.; Gimeno-Valiente, F.; Carbonell-Asins, J.A.; Sharma, S.; Renner, D.; Hafez, D.; Roda, D.; et al. Circulating Tumor DNA in Stage III Colorectal Cancer, beyond Minimal Residual Disease Detection, toward Assessment of Adjuvant Therapy Efficacy and Clinical Behavior of Recurrences. Clin. Cancer Res. 2021, 28, 507–517. [Google Scholar] [CrossRef]
- Cremolini, C.; Rossini, D.; Dell’Aquila, E.; Lonardi, S.; Conca, E.; Del Re, M.; Busico, A.; Pietrantonio, F.; Danesi, R.; Aprile, G.; et al. Rechallenge for Patients with RAS and BRAF Wild-Type Metastatic Colorectal Cancer with Acquired Resistance to First-line Cetuximab and Irinotecan: A Phase 2 Single-Arm Clinical Trial. JAMA Oncol. 2019, 5, 343–350. [Google Scholar] [CrossRef]
- Siravegna, G.; Sartore-Bianchi, A.; Nagy, R.J.; Raghav, K.; Odegaard, J.I.; Lanman, R.B.; Trusolino, L.; Marsoni, S.; Siena, S.; Bardelli, A. Plasma HER2 (ERBB2) Copy Number Predicts Response to HER2-targeted Therapy in Metastatic Colorectal Cancer. Clin. Cancer Res. 2019, 25, 3046–3053. [Google Scholar] [CrossRef]
- Herbst, A.; Vdovin, N.; Gacesa, S.; Philipp, A.; Nagel, D.; Holdt, L.M.; op den Winkel, M.; Heinemann, V.; Stieber, P.; Graeven, U.; et al. Methylated free-circulating HPP1 DNA is an early response marker in patients with metastatic colorectal cancer. Int. J. Cancer 2017, 140, 2134–2144. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Barault, L.; Amatu, A.; Siravegna, G.; Ponzetti, A.; Moran, S.; Cassingena, A.; Mussolin, B.; Falcomata, C.; Binder, A.M.; Cristiano, C.; et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 2018, 67, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Murray, D.; Symonds, E.L.; Pedersen, S.K.; Young, G.P. Detection of ctDNA biomarkers post resection and relationship to recurrent disease in colorectal cancer patients. J. Clin. Oncol. 2018, 36, e15616. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Thierry, A.R.; El Messaoudi, S.; Mollevi, C.; Raoul, J.L.; Guimbaud, R.; Pezet, D.; Artru, P.; Assenat, E.; Borg, C.; Mathonnet, M.; et al. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann. Oncol. 2017, 28, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Knebel, F.H.; Bettoni, F.; da Fonseca, L.G.; Camargo, A.A.; Sabbaga, J.; Jardim, D.L. Circulating Tumor DNA Detection in the Management of Anti-EGFR Therapy for Advanced Colorectal Cancer. Front. Oncol. 2019, 9, 170. [Google Scholar] [CrossRef]
- Van Emburgh, B.O.; Arena, S.; Siravegna, G.; Lazzari, L.; Crisafulli, G.; Corti, G.; Mussolin, B.; Baldi, F.; Buscarino, M.; Bartolini, A.; et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat. Commun. 2016, 7, 13665. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef]
- Raghav, K.P.S.; Overman, M.J.; Yu, R.; Meric-Bernstam, F.; Menter, D.; Kee, B.K.; Muranyi, A.; Singh, S.; Routbort, M.; Chen, K.; et al. HER2 amplification as a negative predictive biomarker for anti-epidermal growth factor receptor antibody therapy in metastatic colorectal cancer. J. Clin. Oncol. 2016, 34, 3517. [Google Scholar] [CrossRef]
- Sartore-Bianchi, A.; Pietrantonio, F.; Lonardi, S.; Mussolin, B.; Rua, F.; Fenocchio, E.; Amatu, A.; Corallo, S.; Manai, C.; Tosi, F.; et al. Phase II study of anti-EGFR rechallenge therapy with panitumumab driven by circulating tumor DNA molecular selection in metastatic colorectal cancer: The CHRONOS trial. J. Clin. Oncol. 2021, 39, 3506. [Google Scholar] [CrossRef]
- Turner, N.C.; Kingston, B.; Kilburn, L.S.; Kernaghan, S.; Wardley, A.M.; Macpherson, I.R.; Baird, R.D.; Roylance, R.; Stephens, P.; Oikonomidou, O.; et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): A multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020, 21, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Di Leo, A.; Johnston, S.; Lee, K.S.; Ciruelos, E.; Lønning, P.E.; Janni, W.; O’Regan, R.; Mouret-Reynier, M.-A.; Kalev, D.; Egle, D.; et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018, 19, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non–Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef]
- Taïeb, J.; Benhaim, L.; Laurent Puig, P.; Le Malicot, K.; Emile, J.F.; Geillon, F.; Tougeron, D.; Manfredi, S.; Chauvenet, M.; Taly, V.; et al. Decision for adjuvant treatment in stage II colon cancer based on circulating tumor DNA:The CIRCULATE-PRODIGE 70 trial. Dig. Liver Dis. 2020, 52, 730–733. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef]
- Tie, J.; Lo, S.N.; Cohen, J.D.; Wang, Y.; Wong, R.; Shapiro, J.; Harris, S.; Khattak, A.; Burge, M.; Vogelstein, B.; et al. Circulating tumour DNA analysis informing adjuvant chemotherapy in stage III colon cancer: A multicentre phase II/III randomised controlled study (DYNAMIC-III). Aust. N. Z. Clin. Regist. 2017. [Google Scholar]
- Anandappa, G.; Starling, N.; Peckitt, C.; Bryant, A.; Begum, R.; Carter, P.; Hatt, S.; Khakoo, S.S.; Turner, A.; Kidd, S.; et al. TRACC: Tracking mutations in cell-free DNA to predict relapse in early colorectal cancer—A randomized study of circulating tumour DNA (ctDNA) guided adjuvant chemotherapy versus standard of care chemotherapy after curative surgery in patients with high risk stage II or stage III colorectal cancer (CRC). J. Clin. Oncol. 2020, 38, TPS4120. [Google Scholar] [CrossRef]
- Nors, J.; Henriksen, T.V.; Gotschalck, K.A.; Juul, T.; Søgaard, J.; Iversen, L.H.; Andersen, C.L. IMPROVE-IT2: Implementing noninvasive circulating tumor DNA analysis to optimize the operative and postoperative treatment for patients with colorectal cancer—Intervention trial 2. Study protocol. Acta Oncol. 2020, 59, 336–341. [Google Scholar] [CrossRef]
- Morris, V.K.; Yothers, G.; Kopetz, S.; Jacobs, S.A.; Lucas, P.C.; Iqbal, A.; Boland, P.M.; Deming, D.A.; Scott, A.J.; Lim, H.J.; et al. NRG-GI005 (COBRA): Phase II/III study of circulating tumor DNA as a predictive biomarker in adjuvant chemotherapy in patients with stage II colon cancer. J. Clin. Oncol. 2020, 38, TPS261. [Google Scholar] [CrossRef]
- Taniguchi, H.; Nakamura, Y.; Kotani, D.; Yukami, H.; Mishima, S.; Sawada, K.; Shirasu, H.; Ebi, H.; Yamanaka, T.; Aleshin, A.; et al. CIRCULATE-Japan: Circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021, 112, 2915–2920. [Google Scholar] [CrossRef]
- Sauer, R.; Becker, H.; Hohenberger, W.; Rödel, C.; Wittekind, C.; Fietkau, R.; Martus, P.; Tschmelitsch, J.; Hager, E.; Hess, C.F.; et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N. Engl. J. Med. 2004, 351, 1731–1740. [Google Scholar] [CrossRef]
- Delibegovic, S. Introduction to Total Mesorectal Excision. Med. Arch. 2017, 71, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Garcia-Aguilar, J. Advances and challenges in treatment of locally advanced rectal cancer. J. Clin. Oncol. 2015, 33, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Dattani, M.; Heald, R.J.; Goussous, G.; Broadhurst, J.; Sao Juliao, G.P.; Habr-Gama, A.; Perez, R.O.; Moran, B.J. Oncological and Survival Outcomes in Watch and Wait Patients with a Clinical Complete Response after Neoadjuvant Chemoradiotherapy for Rectal Cancer: A Systematic Review and Pooled Analysis. Ann. Surg. 2018, 268, 955–967. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Aguilar, J.; Patil, S.; Gollub, M.J.; Kim, J.K.; Yuval, J.B.; Thompson, H.M.; Verheij, F.S.; Omer, D.M.; Lee, M.; Dunne, R.F.; et al. Organ Preservation in Patients with Rectal Adenocarcinoma Treated with Total Neoadjuvant Therapy. J. Clin. Oncol. 2022, 40, 2546–2556. [Google Scholar] [CrossRef] [PubMed]
- Van der Valk, M.J.M.; Hilling, D.E.; Bastiaannet, E.; Meershoek-Klein Kranenbarg, E.; Beets, G.L.; Figueiredo, N.L.; Habr-Gama, A.; Perez, R.O.; Renehan, A.G.; van de Velde, C.J.H. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): An international multicentre registry study. Lancet 2018, 391, 2537–2545. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, C.; Lin, G.; Xiao, Y.; Jia, W.; Xiao, G.; Liu, Q.; Wu, B.; Wu, A.; Qiu, H.; et al. Serial Circulating Tumor DNA in Predicting and Monitoring the Effect of Neoadjuvant Chemoradiotherapy in Patients with Rectal Cancer: A Prospective Multicenter Study. Clin. Cancer Res. 2021, 27, 301–310. [Google Scholar] [CrossRef]
- Pazdirek, F.; Minarik, M.; Benesova, L.; Halkova, T.; Belsanova, B.; Macek, M.; Stepanek, L.; Hoch, J. Monitoring of Early Changes of Circulating Tumor DNA in the Plasma of Rectal Cancer Patients Receiving Neoadjuvant Concomitant Chemoradiotherapy: Evaluation for Prognosis and Prediction of Therapeutic Response. Front. Oncol. 2020, 10, 1028. [Google Scholar] [CrossRef]
- Vidal, J.; Casadevall, D.; Bellosillo, B.; Pericay, C.; Garcia-Carbonero, R.; Losa, F.; Layos, L.; Alonso, V.; Capdevila, J.; Gallego, J.; et al. Clinical Impact of Presurgery Circulating Tumor DNA after Total Neoadjuvant Treatment in Locally Advanced Rectal Cancer: A Biomarker Study from the GEMCAD 1402 Trial. Clin. Cancer Res. 2021, 27, 2890–2898. [Google Scholar] [CrossRef]
- Carpinetti, P.; Donnard, E.; Bettoni, F.; Asprino, P.; Koyama, F.; Rozanski, A.; Sabbaga, J.; Habr-Gama, A.; Parmigiani, R.B.; Galante, P.A.F.; et al. The use of personalized biomarkers and liquid biopsies to monitor treatment response and disease recurrence in locally advanced rectal cancer after neoadjuvant chemoradiation. Oncotarget 2015, 6, 38360–38371. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D.; et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2019, 68, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Khakoo, S.; Carter, P.D.; Brown, G.; Valeri, N.; Picchia, S.; Bali, M.A.; Shaikh, R.; Jones, T.; Begum, R.; Rana, I.; et al. MRI Tumor Regression Grade and Circulating Tumor DNA as Complementary Tools to Assess Response and Guide Therapy Adaptation in Rectal Cancer. Clin. Cancer Res. 2020, 26, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, L.; Bao, H.; Fan, X.; Xia, F.; Wan, J.; Shen, L.; Guan, Y.; Bao, H.; Wu, X.; et al. Utility of ctDNA in predicting response to neoadjuvant chemoradiotherapy and prognosis assessment in locally advanced rectal cancer: A prospective cohort study. PLoS Med. 2021, 18, e1003741. [Google Scholar] [CrossRef]
- Liu, W.Y.; Li, Y.F.; Tang, Y.; Song, Q.Q.; Wang, J.J.; Li, N.; Chen, S.L.; Shi, J.M.; Wang, S.L.; Li, Y.X.; et al. Response prediction and risk stratification of patients with rectal cancer after neoadjuvant therapy through an analysis of circulating tumour DNA. Ebiomedicine 2022, 78, 103945. [Google Scholar] [CrossRef]
- McDuff, S.G.R.; Hardiman, K.M.; Ulintz, P.J.; Parikh, A.R.; Zheng, H.; Kim, D.W.; Lennerz, J.K.; Hazar-Rethinam, M.; Van Seventer, E.E.; Fetter, I.J.; et al. Circulating Tumor DNA Predicts Pathologic and Clinical Outcomes Following Neoadjuvant Chemoradiation and Surgery for Patients with Locally Advanced Rectal Cancer. JCO Precis. Oncol. 2021, 5. [Google Scholar] [CrossRef]
- Hofste, L.S.M.; Geerlings, M.J.; von Rhein, D.; Rutten, H.; Westenberg, A.H.; Weiss, M.M.; Gilissen, C.; Hofste, T.; van der Post, R.S.; Klarenbeek, B.R.; et al. Circulating tumor DNA detection after neoadjuvant treatment and surgery predicts recurrence in patients with early-stage and locally advanced rectal cancer. Eur. J. Surg. Oncol. 2023. [Google Scholar] [CrossRef]
- Agostini, M.; Pucciarelli, S.; Enzo, M.V.; Del Bianco, P.; Briarava, M.; Bedin, C.; Maretto, I.; Friso, M.L.; Lonardi, S.; Mescoli, C.; et al. Circulating cell-free DNA: A promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapy. Ann. Surg. Oncol. 2011, 18, 2461–2468. [Google Scholar] [CrossRef]
- Kramer, A.; Schuuring, E.; Vessies, D.C.L.; van der Leest, P.; Geerlings, M.J.; Rozendal, P.; Lanfermeijer, M.; Linders, T.C.; van Kempen, L.C.; Fijneman, R.J.A.; et al. A Micro-Costing Framework for Circulating Tumor DNA Testing in Dutch Clinical Practice. J. Mol. Diagn. 2023, 25, 36–45. [Google Scholar] [CrossRef]
 |
| Sensitivity * | Specificity * | Advantages | Limitations | |||
|---|---|---|---|---|---|---|
| Adenoma | CRC | Adenoma | CRC | |||
| Colonoscopy | 75–95% | 18–100% | 89–94% | 100% |
|
|
| gFOBT | 6–17% | 50–75% | 96–99% | 96–98% |
|
|
| iFOBT/FIT | 23% | 74% | 96% | 94% |
|
|
| ctDNA (Epi proColon) | 22% | 68% | 79% | 79% |
|
|
| Study | Tumor Stage | Rate of ctDNA Detection before Surgery | ctDNA Detection Method | Outcome | |
|---|---|---|---|---|---|
| Tumor burden at diagnosis | [52] | I II III | 50% 89% 90% | TEC-Seq | Patients with increased pre-operative ctDNA had a shorter PFS and OS compared to patients with a lower ctDNA (HR 1.13, 95% CI: 1.03–1.24) |
| [32] | 60% 56% 86% | ddPCR | 8/10 ctDNA+ patients relapsed 6/11 ctDNA+ patients did not relapse | ||
| [54] | 64% | ddPCR | No relation between baseline ctDNA and DFS (HR 0.93, 95% CI: 0.33–2.69) | ||
| [8] | 60% 92% 90% | Multiplex PCR-based NGS | No significant association between ctDNA and the outcome | ||
| [55] | 30% | ddPCR | Pre-operative ctDNA was associated with inferior RFS (HR 2.18, 95% CI: 1.02–4.61) | ||
| [30] | NM | Spectrophotometry (NanoDrop) | Significantly higher cfDNA levels were observed in patients, with early recurrence compared to non-recurrent patients | ||
| [56] | II III | 64% | NGS | Pre-operative ctDNA+ patients had reduced RFS compared with pre-operative ctDNA− patients (HR 5.66; 95% CI: 1.72–18.57) | |
| [57] | 42% | ddPCR | Baseline ctDNA was an independent prognostic factor of DFS (HR 3.35, 95% CI: 1.15–9.77) | ||
| [51] | 25% 30% | ddPCR | The rate of recurrence was 32.7% in ctDNA+ patients and 11.6% in ctDNA− patients (p = 0.001) | ||
| [53] | 64% 74% | Real-time multiplex PCR assay | 12/47 (25.5%) ctDNA+ patients relapsed |
| Study Design | Sample Size | Study Population | ctDNA Detection Method | Timepoint of ctDNA Sampling | Post-Operative ctDNA Detection Rate | Post-Operative Recurrence for ctDNA+ Patients after Surgery | Post-Operative Recurrence for ctDNA− Patients after Surgery | |
|---|---|---|---|---|---|---|---|---|
| Detection of MRD/recurrence after surgery | Prospective cohort study [64] | 230 | Stage II CC | Safe-SeqS | 4–10 weeks after surgery | 8.7% | 79% | 9.8% |
| Prospective cohort study [50] | 96 | Stage II-III CC | Safe-SeqS | 4–10 weeks after surgery | 21% | 42% | NM | |
| Prospective cohort study [32] | 21 | Stage I-III CRC | ddPCR | 1–4 weeks after surgery | 28.5% | 100% | 27% | |
| Cohort study [65] | 63 | Stage II-III CRC | NGS | Within 1 week after surgery | 28.6% | 27.8% | 4.4% | |
| Prospective, multi-center cohort study [8] | 94 | Stage I-III CRC | Multiplex PCR-based NGS (Signatera™) | 4 weeks after surgery | 10.6% | 70% | 11.9% | |
| Prospective study [66] | 1017 | Stage III CC | ddPCR | 35–50 days after surgery | 13.8% | After 2 years: 31.4% | After 2 years: 17.2% | |
| Prospective, multi-center cohort study [51] | 184 | Stage II-III CRC | ddPCR | 1–6 months after surgery | 10.5% | 44.4% | 10.4% | |
| Prospective, cohort study [56] | 240 | Stage II-III CRC | 425-gene NGS panel-based | 3–7 days after surgery | 8.3% | 60% | NM |
| Study Design | Sample Size | Study Population | ctDNA Detection Method | Timepoint of ctDNA Sampling | ctDNA Clearance Rate | Outcome | |
|---|---|---|---|---|---|---|---|
| After surgery | Prospective, multi-center cohort study [51] | 49 | Stage II CRC Stage III CRC | Droplet Digital PCR | Day 5 after surgery | 75% | Recurrence rate ctDNA+: 44.4% ctDNA−: 13.7% |
| Prospective, multi-center study [56] | 240 | Stage II CRC Stage III CRC | NGS | Days 3–7 after surgery | 92% | 2-year RFS: ctDNA+: 39.3% ctDNA−: 89.4% | |
| Prospective, multi-center cohort study [8] | 94 | Stage I CRC Stage II CRC Stage III CRC | Multiplex PCR-based NGS | Day 30 after surgery | 89% | Recurrence rate: ctDNA+: 70% ctDNA−: 11.9% | |
| After adjuvant chemotherapy | Prospective, multi-center cohort study [8] | 10 | Stage I CRC Stage II CRC Stage III CRC | Multiplex PCR-based NGS | After completion of chemotherapy | 30% | NM |
| Prospective cohort study [64] | 6 | Stage II CC | Safe-SeqS | After completion of chemotherapy | 50% | 2-year RFS: ctDNA+: 27% ctDNA−: 82% | |
| Multi-center, cohort study [50] | 95 | Stage III CC | Safe-SeqS | After completion of chemotherapy | 68% | 3-year RFI: ctDNA+: 30% ctDNA−: 77% |
| Study Design | Sample Size | Study Population | ctDNA Detection Method | ctDNA Positivity vs. Recurrence Rate | Frequency of Sampling | Delay of Anticipation | |
|---|---|---|---|---|---|---|---|
| The biological anticipation of radiological recurrence | Cohort study [68] | 58 | Stage I–III CRC | Safe-SeqS | ctDNA+ and recurrence: 100% ctDNA− and recurrence: 0% | Post-surgery: 1 month Follow-up: Every 3–6 months | 3 months |
| Prospective cohort study [32] | 27 | Stage I–III CRC | ddPCR | ctDNA+ and recurrence: 100% ctDNA− and recurrence: 0% | Post-surgery: Days 8 and 30 Follow-up: Every 3 months | 9 months | |
| Prospective cohort study [64] | 178 | Stage II CC | Safe-SeqS | ctDNA+ and recurrence: 78.6% ctDNA− and recurrence: 9.8% | Follow-up: Every 3 months | 167 days (5 months) (IQR, 81–279 days) | |
| Prospective study [69] | 11 | Stage I–IV CRC | ddPCR | ctDNA+ and recurrence: 100% ctDNA− and recurrence: 0% | Post-surgery: Days 8 and 30 Follow-up: Every 3 months | 2–15 months (mean of 10 months) | |
| Prospective, multicenter cohort study [51] | 139 | Stage II–III CRC | ddPCR | ctDNA+ and recurrence: 32.7% ctDNA− and recurrence: 11.6% | Post-surgery: Day 5 Follow-up: Every 3–6 months | 13.1 weeks (IQR, 28 weeks) | |
| Prospective, multicenter study [70] | 160 | Stage III CRC | Multiplex PCR-based NGS | ctDNA+ and recurrence: 96% ctDNA− and recurrence: 3% | Follow-up: Every 3 months | 9.8 months (IQR, 5–12 months) | |
| Prospective, multicenter study [56] | 276 | Stage II–III CRC | NGS | ctDNA+ and recurrence: 76% ctDNA− and recurrence: 4% | Post-surgery: Days 5–8 Follow-up: 6 months after surgery, and then every 3 months | 5.01 months |
| Study Design | Sample Size | Study Population | ctDNA Detection Method | PFS | |
|---|---|---|---|---|---|
| Predicting response to treatment | Prospective, multi-center study [71] | 28 | Stage IV CRC | ddPCR NGS * (Ion AmpliSeq Cancer Hotspot Panel) | ctDNA−: 4.0 months ctDNA+: 1.9 months Hazard ratio, 0.44; 95% CI, 0.18–0.98; p = 0.03) |
| Clinical trial [72] | 29 | Stage IV CRC | Guardant 360 ** assay | ApCN ≥ 25.82: 22.5 weeks ApCN ≤ 25.82: 14.8 weeks Mantel Cox, p = 0.0347 | |
| Prospective study [73] | 467 | Stage IV CRC | Methy-Light | No effect on PFS | |
| Prospective study [74] | 53 | Stage IV CRC | Safe-SeqS | ≥10-fold reduction in ctDNA: 14.7 months ≤10-fold reduction in ctDNA: 8.1 months Hazard ratio, 1.87; 95% CI, 0.62–5.61; p = 0.266 | |
| Prospective (PLACOL) study [61] | 82 | Stage IV CRC | ddPCR | “good ctDNA responder” = ctDNA concentration < 0.1 ng/mL and SlopeΔctDNA ≥ 80%: 8.5 months “bad ctDNA responder” = ctDNA concentration > 0.1 ng/mL and SlopeΔctDNA < 80%: 2.4 months Hazard ratio, 0.19; 95% CI, 0.09–0.40; p < 0.0001 | |
| Prospective study [75] | 45 | Stage IV CRC | dPCR (Methyl-BEAMing) | A negative change in ctDNA is associated with improved PFS |
| Study Design | Sample Size | Study Population | ctDNA Detection Method | ctDNA Positivity | Outcome |
|---|---|---|---|---|---|
| Cohort study [110] | 67 | LARC | Real-time PCR |
| Baseline levels of cfDNA are not associated with tumor response. Post-RCT integrity index is associated with tumor response |
| Prospective cohort study [103] | 4 | LARC | NGS |
| ctDNA concentration can help monitor response to RCT |
| Prospective cohort study [104] | 159 | LARC T3/T4 and/or N+ | Safe-SeqS |
| ctDNA+ after RCT and/or surgery is associated with lower 3-year-RFS (33% vs. 87%) |
| Prospective cohort study [101] | 36 | LARC | BEAMing |
| ctDNA+ at baseline reduced post-operative DFS and OS |
| Prospective cohort study [105] | 47 | LARC | ddPCR |
| Patients with ctDNA+ during RCT, after RCT, and after surgery have lower RFS |
| Cohort study [100] | 104 | Rectal cancer T4 or N1b-3 | NGS |
|
|
| Prospective cohort [106] | 119 | LARC | NGS |
| ctDNA clearance is associated with tumor regression grade |
| Prospective study (phase II trial) [102] | 71 | LARC | NGS |
| Pre-operative ctDNA+ is significantly associated with shorter DFS and OS |
| Prospective cohort study [108] | 29 | LARC | NGS |
| Patients with post-operative ctDNA+ experience poor RFS compared to ctDNA− patients |
| Prospective cohort study [107] | 60 | LARC | Agnostic and tumor-informed assays |
| ctDNA+ after RCT is associated with lower RFS |
| Cohort study [109] | 51 | LARC | NGS |
| Patients with post-neoadjuvant treatment and post-operative ctDNA+ experienced poorer RFS than ctDNA− patients |
| Name of Study and Country | Recruitment Status | Patient Population | Sample Size | ctDNA Detection Method | Intervention | Primary Objective |
|---|---|---|---|---|---|---|
| Circulating Tumor DNA-guided Neoadjuvant Treatment Strategy for Locally Advanced Rectal Cancer (CINTS-R) (NCT05601505) China | Recruiting | Patients with rectal adenocarcinoma who have not received any treatment yet | 465 | NGS | Control arm: Traditional neoadjuvant chemoradiotherapy Experimental arm: Randomization based on ctDNA results:
| Disease-related treatment failure (DrTF) |
| Establishing a ctDNA Biomarker to Improve Organ Preserving Strategies in Patients With Rectal Cancer (ctTRAC) (NCT05081024) USA | Recruiting | Patients with stage II–III rectal adenocarcinoma | 50 | Signatera™ (Natera Inc., Austin, TX, USA) | Observational | Complete clinical response (cCR) |
| Application of Circulating Tumor DNA Test in the Diagnosis and Treatment of Patients with Advanced Rectal Cancer (NCT03615170) China | Recruiting | Patients with locally advanced rectal cancer, who need to receive neoadjuvant radiotherapy and radical operation | 200 | Not mentioned | Observational | Disease-free survival |
| Systemic Neoadjuvant and Adjuvant Control by Precision Medicine in Rectal Cancer (SYNCOPE) (NCT04842006) Finland | Recruiting | Patients with rectal adenocarcinoma that require either radiotherapy or long chemoradiotherapy | 93 | Not mentioned | Randomization based on ctDNA results:
| RFS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Abreu, A.R.; Op de Beeck, K.; Laurent-Puig, P.; Taly, V.; Benhaim, L. The Position of Circulating Tumor DNA in the Clinical Management of Colorectal Cancer. Cancers 2023, 15, 1284. https://doi.org/10.3390/cancers15041284
de Abreu AR, Op de Beeck K, Laurent-Puig P, Taly V, Benhaim L. The Position of Circulating Tumor DNA in the Clinical Management of Colorectal Cancer. Cancers. 2023; 15(4):1284. https://doi.org/10.3390/cancers15041284
Chicago/Turabian Stylede Abreu, Ana Regina, Ken Op de Beeck, Pierre Laurent-Puig, Valerie Taly, and Leonor Benhaim. 2023. "The Position of Circulating Tumor DNA in the Clinical Management of Colorectal Cancer" Cancers 15, no. 4: 1284. https://doi.org/10.3390/cancers15041284
APA Stylede Abreu, A. R., Op de Beeck, K., Laurent-Puig, P., Taly, V., & Benhaim, L. (2023). The Position of Circulating Tumor DNA in the Clinical Management of Colorectal Cancer. Cancers, 15(4), 1284. https://doi.org/10.3390/cancers15041284






