Nucleocytoplasmic Shuttling of STATs. A Target for Intervention?
Abstract
:1. Aim and Scope
2. General Mechanisms of Nucleocytoplasmic Transport of Proteins
2.1. The Nuclear Pore Complex
2.2. Importins
2.3. Exportins
2.4. Ran-GTP/GDP Cycle
3. STAT1—Using a Side Track for Nuclear Import
4. STAT3—Acting on Many Stages
4.1. Role of STAT3 in Cancer
4.2. Nuclear Import
4.3. Nuclear Export and Nucleocytoplasmic Shuttling
5. STAT5—Leukemia and More
6. Perspectives for Therapeutic Interventions
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Feldherr, C.M. The uptake and accumulation of proteins by the cell nucleus. Bioessays 1985, 3, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Mohr, D.; Frey, S.; Fischer, T.; Guttler, T.; Gorlich, D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009, 28, 2541–2553. [Google Scholar] [CrossRef] [PubMed]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef]
- Frey, S.; Gorlich, D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 2007, 130, 512–523. [Google Scholar] [CrossRef]
- Lim, R.Y.; Huang, N.P.; Koser, J.; Deng, J.; Lau, K.H.; Schwarz-Herion, K.; Fahrenkrog, B.; Aebi, U. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 2006, 103, 9512–9517. [Google Scholar] [CrossRef]
- Rout, M.P.; Aitchison, J.D.; Magnasco, M.O.; Chait, B.T. Virtual gating and nuclear transport: The hole picture. Trends Cell Biol. 2003, 13, 622–628. [Google Scholar] [CrossRef]
- Schmidt, H.B.; Gorlich, D. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 2016, 41, 46–61. [Google Scholar] [CrossRef]
- Hulsmann, B.B.; Labokha, A.A.; Gorlich, D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 2012, 150, 738–751. [Google Scholar] [CrossRef]
- Radu, A.; Moore, M.S.; Blobel, G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 1995, 81, 215–222. [Google Scholar] [CrossRef]
- Ribbeck, K.; Gorlich, D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001, 20, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Rexach, M.; Blobel, G. Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 1995, 83, 683–692. [Google Scholar] [CrossRef]
- Wente, S.R.; Rout, M.P. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect. Biol. 2010, 2, a000562. [Google Scholar] [CrossRef] [PubMed]
- Aksu, M.; Trakhanov, S.; Vera Rodriguez, A.; Gorlich, D. Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122. J. Cell Biol. 2019, 218, 1839–1852. [Google Scholar] [CrossRef] [PubMed]
- Harel, A.; Forbes, D.J. Importin beta: Conducting a much larger cellular symphony. Mol. Cell 2004, 16, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Yamashita, E.; Yasuhara, N.; Song, J.; Son, S.Y.; Won, Y.H.; Hong, H.R.; Shin, Y.S.; Sekimoto, T.; Park, I.Y.; et al. Structural basis for the selective nuclear import of the C2H2 zinc-finger protein Snail by importin beta. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 1050–1060. [Google Scholar] [CrossRef]
- Yamasaki, H.; Sekimoto, T.; Ohkubo, T.; Douchi, T.; Nagata, Y.; Ozawa, M.; Yoneda, Y. Zinc finger domain of Snail functions as a nuclear localization signal for importin beta-mediated nuclear import pathway. Genes Cells 2005, 10, 455–464. [Google Scholar] [CrossRef]
- Lee, S.J.; Sekimoto, T.; Yamashita, E.; Nagoshi, E.; Nakagawa, A.; Imamoto, N.; Yoshimura, M.; Sakai, H.; Chong, K.T.; Tsukihara, T.; et al. The structure of importin-beta bound to SREBP-2: Nuclear import of a transcription factor. Science 2003, 302, 1571–1575. [Google Scholar] [CrossRef]
- Huber, J.; Cronshagen, U.; Kadokura, M.; Marshallsay, C.; Wada, T.; Sekine, M.; Luhrmann, R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998, 17, 4114–4126. [Google Scholar] [CrossRef]
- Bayliss, R.; Littlewood, T.; Strawn, L.A.; Wente, S.R.; Stewart, M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 2002, 277, 50597–50606. [Google Scholar] [CrossRef]
- Bednenko, J.; Cingolani, G.; Gerace, L. Importin beta contains a COOH-terminal nucleoporin binding region important for nuclear transport. J. Cell Biol. 2003, 162, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Christie, M.; Chang, C.W.; Rona, G.; Smith, K.M.; Stewart, A.G.; Takeda, A.A.; Fontes, M.R.; Stewart, M.; Vertessy, B.G.; Forwood, J.K.; et al. Structural biology and regulation of protein import into the nucleus. J. Mol. Biol. 2016, 428, 2060–2090. [Google Scholar] [CrossRef] [PubMed]
- Pumroy, R.A.; Cingolani, G. Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem. J. 2015, 466, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Kalderon, D.; Richardson, W.D.; Markham, A.F.; Smith, A.E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 1984, 311, 33–38. [Google Scholar] [CrossRef]
- Dingwall, C.; Robbins, J.; Dilworth, S.M.; Roberts, B.; Richardson, W.D. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 1988, 107, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Kelley, J.B.; Talley, A.M.; Spencer, A.; Gioeli, D.; Paschal, B.M. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010, 11, 63. [Google Scholar] [CrossRef]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef]
- Sankhala, R.S.; Lokareddy, R.K.; Begum, S.; Pumroy, R.A.; Gillilan, R.E.; Cingolani, G. Three-dimensional context rather than NLS amino acid sequence determines importin alpha subtype specificity for RCC1. Nat. Commun. 2017, 8, 979. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Matsumura, N.; Takashima, H.; Miyamoto-Sato, E.; Tomita, M.; Yanagawa, H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009, 284, 478–485. [Google Scholar] [CrossRef]
- Cingolani, G.; Petosa, C.; Weis, K.; Muller, C.W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 1999, 399, 221–229. [Google Scholar] [CrossRef]
- Lee, B.J.; Cansizoglu, A.E.; Suel, K.E.; Louis, T.H.; Zhang, Z.; Chook, Y.M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006, 126, 543–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kressler, D.; Bange, G.; Ogawa, Y.; Stjepanovic, G.; Bradatsch, B.; Pratte, D.; Amlacher, S.; Strauss, D.; Yoneda, Y.; Katahira, J.; et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science 2012, 338, 666–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, M.C.; Lin, R.I.; Tarn, W.Y. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 10154–10159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baade, I.; Spillner, C.; Schmitt, K.; Valerius, O.; Kehlenbach, R.H. Extensive identification and in-depth validation of importin 13 cargoes. Mol. Cell. Proteomics 2018, 17, 1337–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunwald, M.; Lazzaretti, D.; Bono, F. Structural basis for the nuclear export activity of Importin13. EMBO J. 2013, 32, 899–913. [Google Scholar] [CrossRef] [Green Version]
- Hanover, J.A.; Love, D.C.; Prinz, W.A. Calmodulin-driven nuclear entry: Trigger for sex determination and terminal differentiation. J. Biol. Chem. 2009, 284, 12593–12597. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Jamieson, C.; Lui, C.; Henderson, B.R. Distinct hydrophobic “patches” in the N- and C-tails of beta-catenin contribute to nuclear transport. Exp. Cell Res. 2016, 348, 132–145. [Google Scholar] [CrossRef]
- Kirli, K.; Karaca, S.; Dehne, H.J.; Samwer, M.; Pan, K.T.; Lenz, C.; Urlaub, H.; Gorlich, D. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. eLife 2015, 4, e11466. [Google Scholar] [CrossRef]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef] [Green Version]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic 2008, 9, 2053–2062. [Google Scholar] [CrossRef]
- Stade, K.; Ford, C.S.; Guthrie, C.; Weis, K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 1997, 90, 1041–1050. [Google Scholar] [CrossRef] [Green Version]
- Andrade, M.A.; Petosa, C.; O’Donoghue, S.I.; Muller, C.W.; Bork, P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001, 309, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, Y. Mechanistic insights from structural analyses of Ran-GTPase-Driven nuclear export of proteins and RNAs. J. Mol. Biol. 2016, 428, 2025–2039. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Hurt, E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007, 8, 761–773. [Google Scholar] [CrossRef]
- Kutay, U.; Bischoff, F.R.; Kostka, S.; Kraft, R.; Gorlich, D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 1997, 90, 1061–1071. [Google Scholar] [CrossRef] [Green Version]
- Lipowsky, G.; Bischoff, F.R.; Schwarzmaier, P.; Kraft, R.; Kostka, S.; Hartmann, E.; Kutay, U.; Gorlich, D. Exportin 4: A mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 2000, 19, 4362–4371. [Google Scholar] [CrossRef] [Green Version]
- Kurisaki, A.; Kurisaki, K.; Kowanetz, M.; Sugino, H.; Yoneda, Y.; Heldin, C.H.; Moustakas, A. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol. Cell. Biol. 2006, 26, 1318–1332. [Google Scholar] [CrossRef] [Green Version]
- Gontan, C.; Guttler, T.; Engelen, E.; Demmers, J.; Fornerod, M.; Grosveld, F.G.; Tibboel, D.; Gorlich, D.; Poot, R.A.; Rottier, R.J. Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J. Cell Biol. 2009, 185, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Mingot, J.M.; Bohnsack, M.T.; Jakle, U.; Gorlich, D. Exportin 7 defines a novel general nuclear export pathway. EMBO J. 2004, 23, 3227–3236. [Google Scholar] [CrossRef] [Green Version]
- Aksu, M.; Pleiner, T.; Karaca, S.; Kappert, C.; Dehne, H.J.; Seibel, K.; Urlaub, H.; Bohnsack, M.T.; Gorlich, D. Xpo7 is a broad-spectrum exportin and a nuclear import receptor. J. Cell Biol. 2018, 217, 2329–2340. [Google Scholar] [CrossRef]
- Nachury, M.V.; Weis, K. The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. USA 1999, 96, 9622–9627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuersten, S.; Ohno, M.; Mattaj, I.W. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 2001, 11, 497–503. [Google Scholar] [CrossRef]
- Macara, I.G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001, 65, 570–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melchior, F.; Gerace, L. Two-way trafficking with Ran. Trends Cell Biol. 1998, 8, 175–179. [Google Scholar] [CrossRef]
- Guttler, T.; Gorlich, D. Ran-dependent nuclear export mediators: A structural perspective. EMBO J. 2011, 30, 3457–3474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frey, S.; Rees, R.; Schunemann, J.; Ng, S.C.; Funfgeld, K.; Huyton, T.; Gorlich, D. Surface properties determining passage rates of proteins through nuclear pores. Cell 2018, 174, 202–217. [Google Scholar] [CrossRef] [Green Version]
- Parker, B.S.; Rautela, J.; Hertzog, P.J. Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 2016, 16, 131–144. [Google Scholar] [CrossRef]
- Meissl, K.; Macho-Maschler, S.; Muller, M.; Strobl, B. The good and the bad faces of STAT1 in solid tumours. Cytokine 2017, 89, 12–20. [Google Scholar] [CrossRef] [Green Version]
- Green, D.S.; Young, H.A.; Valencia, J.C. Current prospects of type II interferon gamma signaling and autoimmunity. J. Biol. Chem. 2017, 292, 13925–13933. [Google Scholar] [CrossRef] [Green Version]
- Sekimoto, T.; Imamoto, N.; Nakajima, K.; Hirano, T.; Yoneda, Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997, 16, 7067–7077. [Google Scholar] [CrossRef] [Green Version]
- Melen, K.; Fagerlund, R.; Franke, J.; Kohler, M.; Kinnunen, L.; Julkunen, I. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 2003, 278, 28193–28200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nardozzi, J.; Wenta, N.; Yasuhara, N.; Vinkemeier, U.; Cingolani, G. Molecular basis for the recognition of phosphorylated STAT1 by importin alpha5. J. Mol. Biol. 2010, 402, 83–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meissner, T.; Krause, E.; Lodige, I.; Vinkemeier, U. Arginine methylation of STAT1: A reassessment. Cell 2004, 119, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, T.; Begitt, A.; Lodige, I.; van Rossum, M.; Vinkemeier, U. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 2002, 21, 344–354. [Google Scholar] [CrossRef] [Green Version]
- Fagerlund, R.; Melen, K.; Kinnunen, L.; Julkunen, I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J. Biol. Chem. 2002, 277, 30072–30078. [Google Scholar] [CrossRef] [Green Version]
- McBride, K.M.; McDonald, C.; Reich, N.C. Nuclear export signal located within theDNA-binding domain of the STAT1transcription factor. EMBO J. 2000, 19, 6196–6206. [Google Scholar] [CrossRef]
- Xu, W.; Edwards, M.R.; Borek, D.M.; Feagins, A.R.; Mittal, A.; Alinger, J.B.; Berry, K.N.; Yen, B.; Hamilton, J.; Brett, T.J.; et al. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 2014, 16, 187–200. [Google Scholar] [CrossRef] [Green Version]
- McBride, K.M.; Banninger, G.; McDonald, C.; Reich, N.C. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 2002, 21, 1754–1763. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Cao, X. Regulation of Stat3 nuclear import by importin alpha5 and importin alpha7 via two different functional sequence elements. Cell. Signal. 2006, 18, 1117–1126. [Google Scholar] [CrossRef]
- Ushijima, R.; Sakaguchi, N.; Kano, A.; Maruyama, A.; Miyamoto, Y.; Sekimoto, T.; Yoneda, Y.; Ogino, K.; Tachibana, T. Extracellular signal-dependent nuclear import of STAT3 is mediated by various importin alphas. Biochem. Biophys. Res. Commun. 2005, 330, 880–886. [Google Scholar] [CrossRef]
- Liu, L.; McBride, K.M.; Reich, N.C. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc. Natl. Acad. Sci. USA 2005, 102, 8150–8155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martincuks, A.; Fahrenkamp, D.; Haan, S.; Herrmann, A.; Kuster, A.; Muller-Newen, G. Dissecting functions of the N-terminal domain and GAS-site recognition in STAT3 nuclear trafficking. Cell. Signal. 2016, 28, 810–825. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.Y.; Reich, N.C. Dynamic trafficking of STAT5 depends on an unconventional nuclear localization signal. J. Cell Sci. 2013, 126, 3333–3343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strehlow, I.; Schindler, C. Amino-terminal signal transducer and activator of transcription (STAT) domains regulate nuclear translocation and STAT deactivation. J. Biol. Chem. 1998, 273, 28049–28056. [Google Scholar] [CrossRef] [Green Version]
- Vogt, M.; Domoszlai, T.; Kleshchanok, D.; Lehmann, S.; Schmitt, A.; Poli, V.; Richtering, W.; Muller-Newen, G. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J. Cell Sci. 2011, 124, 900–909. [Google Scholar] [CrossRef] [Green Version]
- Iyer, J.; Reich, N.C. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. FASEB J. 2008, 22, 391–400. [Google Scholar] [CrossRef]
- Zeng, R.; Aoki, Y.; Yoshida, M.; Arai, K.; Watanabe, S. Stat5B shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J. Immunol. 2002, 168, 4567–4575. [Google Scholar] [CrossRef] [Green Version]
- Begitt, A.; Meyer, T.; van Rossum, M.; Vinkemeier, U. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. USA 2000, 97, 10418–10423. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Schindler, C. Regulation of Stat3 nuclear export. J. Clin. Investig. 2003, 111, 553–559. [Google Scholar] [CrossRef]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E., Jr. Stat3 as an oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef] [Green Version]
- Pilati, C.; Zucman-Rossi, J. Mutations leading to constitutive active gp130/JAK1/STAT3 pathway. Cytokine Growth Factor Rev. 2015, 26, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Aigner, P.; Mizutani, T.; Horvath, J.; Eder, T.; Heber, S.; Lind, K.; Just, V.; Moll, H.P.; Yeroslaviz, A.; Fischer, M.J.M.; et al. STAT3beta is a tumor suppressor in acute myeloid leukemia. Blood Adv. 2019, 3, 1989–2002. [Google Scholar] [CrossRef] [Green Version]
- Pencik, J.; Schlederer, M.; Gruber, W.; Unger, C.; Walker, S.M.; Chalaris, A.; Marie, I.J.; Hassler, M.R.; Javaheri, T.; Aksoy, O.; et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat. Commun. 2015, 6, 7736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagard, R.; Metelev, V.; Souissi, I.; Baran-Marszak, F. STAT3 inhibitors for cancer therapy: Have all roads been explored? JAKSTAT 2013, 2, e22882. [Google Scholar] [CrossRef] [Green Version]
- Meyer, T.; Vinkemeier, U. STAT nuclear translocation: Potential for pharmacological intervention. Expert Opin. Ther. Targets 2007, 11, 1355–1365. [Google Scholar] [CrossRef]
- Kohler, M.; Ansieau, S.; Prehn, S.; Leutz, A.; Haller, H.; Hartmann, E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997, 417, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Kawashima, T.; Bao, Y.C.; Minoshima, Y.; Nomura, Y.; Hatori, T.; Hori, T.; Fukagawa, T.; Fukada, T.; Takahashi, N.; Nosaka, T.; et al. A Rac GTPase-activating protein, MgcRacGAP, is a nuclear localizing signal-containing nuclear chaperone in the activation of STAT transcription factors. Mol. Cell. Biol. 2009, 29, 1796–1813. [Google Scholar] [CrossRef] [Green Version]
- van Adrichem, A.J.; Wennerberg, K. MgcRacGAP inhibition stimulates JAK-dependent STAT3 activity. FEBS Lett. 2015, 589, 3859–3865. [Google Scholar] [CrossRef]
- Martincuks, A.; Andryka, K.; Kuster, A.; Schmitz-Van de Leur, H.; Komorowski, M.; Muller-Newen, G. Nuclear translocation of STAT3 and NF-kappaB are independent of each other but NF-kappaB supports expression and activation of STAT3. Cell. Signal. 2017, 32, 36–47. [Google Scholar] [CrossRef]
- Meyer, T.; Marg, A.; Lemke, P.; Wiesner, B.; Vinkemeier, U. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 2003, 17, 1992–2005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimoto, A.; Yu, Y.; Lu, Z.; Mao, X.; Ren, Z.; Watowich, S.S.; Mills, G.B.; Liao, W.S.; Chen, X.; Bast, R.C., Jr.; et al. A Ras homologue member I directly inhibits signal transducers and activators of transcription 3 translocation and activity in human breast and ovarian cancer cells. Cancer Res. 2005, 65, 6701–6710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohr, A.; Chatain, N.; Domoszlai, T.; Rinis, N.; Sommerauer, M.; Vogt, M.; Muller-Newen, G. Dynamics and non-canonical aspects of JAK/STAT signalling. Eur. J. Cell Biol. 2012, 91, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Pranada, A.L.; Metz, S.; Herrmann, A.; Heinrich, P.C.; Muller-Newen, G. Real time analysis of STAT3 nucleocytoplasmic shuttling. J. Biol. Chem. 2004, 279, 15114–15123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, T.; Gavenis, K.; Vinkemeier, U. Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp. Cell Res. 2002, 272, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Marg, A.; Shan, Y.; Meyer, T.; Meissner, T.; Brandenburg, M.; Vinkemeier, U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 2004, 165, 823–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braunstein, J.; Brutsaert, S.; Olson, R.; Schindler, C. STATs dimerize in the absence of phosphorylation. J. Biol. Chem. 2003, 278, 34133–34140. [Google Scholar] [CrossRef] [Green Version]
- Haan, S.; Kortylewski, M.; Behrmann, I.; Muller-Esterl, W.; Heinrich, P.C.; Schaper, F. Cytoplasmic STAT proteins associate prior to activation. Biochem. J. 2000, 345 Pt 3, 417–421. [Google Scholar] [CrossRef]
- Wenta, N.; Strauss, H.; Meyer, S.; Vinkemeier, U. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc. Natl. Acad. Sci. USA 2008, 105, 9238–9243. [Google Scholar] [CrossRef] [Green Version]
- Domoszlai, T.; Martincuks, A.; Fahrenkamp, D.; Schmitz-Van de Leur, H.; Kuster, A.; Muller-Newen, G. Consequences of the disease-related L78R mutation for dimerization and activity of STAT3. J. Cell Sci. 2014, 127, 1899–1910. [Google Scholar] [CrossRef] [Green Version]
- Majoros, A.; Platanitis, E.; Kernbauer-Holzl, E.; Rosebrock, F.; Muller, M.; Decker, T. Canonical and non-canonical aspects of JAK-STAT signaling: Lessons from interferons for cytokine responses. Front. Immunol. 2017, 8, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingelhofer, B.; Neubauer, H.A.; Valent, P.; Han, X.; Constantinescu, S.N.; Gunning, P.T.; Muller, M.; Moriggl, R. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia 2018, 32, 1713–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferbeyre, G.; Moriggl, R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochim. Biophys. Acta 2011, 1815, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Herrington, J.; Rui, L.; Luo, G.; Yu-Lee, L.Y.; Carter-Su, C. A functional DNA binding domain is required for growth hormone-induced nuclear accumulation of Stat5B. J. Biol. Chem. 1999, 274, 5138–5145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lillemeier, B.F.; Koster, M.; Kerr, I.M. STAT1 from the cell membrane to the DNA. EMBO J. 2001, 20, 2508–2517. [Google Scholar] [CrossRef] [Green Version]
- Park, H.J.; Li, J.; Hannah, R.; Biddie, S.; Leal-Cervantes, A.I.; Kirschner, K.; Flores Santa Cruz, D.; Sexl, V.; Gottgens, B.; Green, A.R. Cytokine-induced megakaryocytic differentiation is regulated by genome-wide loss of a uSTAT transcriptional program. EMBO J. 2016, 35, 580–594. [Google Scholar] [CrossRef]
- Kazansky, A.V.; Kabotyanski, E.B.; Wyszomierski, S.L.; Mancini, M.A.; Rosen, J.M. Differential effects of prolactin and src/abl kinases on the nuclear translocation of STAT5B and STAT5A. J. Biol. Chem. 1999, 274, 22484–22492. [Google Scholar] [CrossRef] [Green Version]
- Harir, N.; Pecquet, C.; Kerenyi, M.; Sonneck, K.; Kovacic, B.; Nyga, R.; Brevet, M.; Dhennin, I.; Gouilleux-Gruart, V.; Beug, H.; et al. Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood 2007, 109, 1678–1686. [Google Scholar] [CrossRef] [Green Version]
- Chatain, N.; Ziegler, P.; Fahrenkamp, D.; Jost, E.; Moriggl, R.; Schmitz-Van de Leur, H.; Muller-Newen, G. Src family kinases mediate cytoplasmic retention of activated STAT5 in BCR-ABL-positive cells. Oncogene 2013, 32, 3587–3597. [Google Scholar] [CrossRef] [Green Version]
- Fahrenkamp, D.; de Leur, H.S.; Kuster, A.; Chatain, N.; Muller-Newen, G. Src family kinases interfere with dimerization of STAT5A through a phosphotyrosine-SH2 domain interaction. Cell Commun. Signal. 2015, 13, 10. [Google Scholar] [CrossRef] [Green Version]
- Berger, A.; Hoelbl-Kovacic, A.; Bourgeais, J.; Hoefling, L.; Warsch, W.; Grundschober, E.; Uras, I.Z.; Menzl, I.; Putz, E.M.; Hoermann, G.; et al. PAK-dependent STAT5 serine phosphorylation is required for BCR-ABL-induced leukemogenesis. Leukemia 2014, 28, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Cagatay, T.; Chook, Y.M. Karyopherins in cancer. Curr. Opin. Cell Biol. 2018, 52, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Mahipal, A.; Malafa, M. Importins and exportins as therapeutic targets in cancer. Pharmacol. Ther. 2016, 164, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Poon, I.K.; Jans, D.A. Regulation of nuclear transport: Central role in development and transformation? Traffic 2005, 6, 173–186. [Google Scholar] [CrossRef]
- Turner, J.G.; Dawson, J.; Cubitt, C.L.; Baz, R.; Sullivan, D.M. Inhibition of CRM1-dependent nuclear export sensitizes malignant cells to cytotoxic and targeted agents. Semin. Cancer Biol. 2014, 27, 62–73. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.G.; Engel, R.; Derderian, J.A.; Jove, R.; Sullivan, D.M. Human topoisomerase IIalpha nuclear export is mediated by two CRM-1-dependent nuclear export signals. J. Cell Sci. 2004, 117, 3061–3071. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.G.; Dawson, J.; Emmons, M.F.; Cubitt, C.L.; Kauffman, M.; Shacham, S.; Hazlehurst, L.A.; Sullivan, D.M. CRM1 inhibition sensitizes drug resistant human myeloma cells to topoisomerase II and proteasome inhibitors both In Vitro and Ex Vivo. J. Cancer 2013, 4, 614–625. [Google Scholar] [CrossRef]
- Hong, A.L.; Tseng, Y.Y.; Cowley, G.S.; Jonas, O.; Cheah, J.H.; Kynnap, B.D.; Doshi, M.B.; Oh, C.; Meyer, S.C.; Church, A.J.; et al. Integrated genetic and pharmacologic interrogation of rare cancers. Nat. Commun. 2016, 7, 11987. [Google Scholar] [CrossRef]
- Kim, J.; McMillan, E.; Kim, H.S.; Venkateswaran, N.; Makkar, G.; Rodriguez-Canales, J.; Villalobos, P.; Neggers, J.E.; Mendiratta, S.; Wei, S.; et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature 2016, 538, 114–117. [Google Scholar] [CrossRef] [Green Version]
- Newlands, E.S.; Rustin, G.J.; Brampton, M.H. Phase I trial of elactocin. Br. J. Cancer 1996, 74, 648–649. [Google Scholar] [CrossRef] [Green Version]
- Stelma, T.; Chi, A.; van der Watt, P.J.; Verrico, A.; Lavia, P.; Leaner, V.D. Targeting nuclear transporters in cancer: Diagnostic, prognostic and therapeutic potential. IUBMB Life 2016, 68, 268–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beg, A.A.; Ruben, S.M.; Scheinman, R.I.; Haskill, S.; Rosen, C.A.; Baldwin, A.S., Jr. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: A mechanism for cytoplasmic retention. Genes Dev. 1992, 6, 1899–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerlund, R.; Kinnunen, L.; Kohler, M.; Julkunen, I.; Melen, K. NF-κB is transported into the nucleus by importin α3 and importin α4. J. Biol. Chem. 2005, 280, 15942–15951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Echeverria, P.C.; Picard, D. Molecular chaperones, essential partners of steroid hormone receptors for activity and mobility. Biochim. Biophys. Acta 2010, 1803, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Verheugd, P.; Forst, A.H.; Milke, L.; Herzog, N.; Feijs, K.L.; Kremmer, E.; Kleine, H.; Luscher, B. Regulation of NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat. Commun. 2013, 4, 1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagstaff, K.M.; Jans, D.A. Importins and beyond: Non-conventional nuclear transport mechanisms. Traffic 2009, 10, 1188–1198. [Google Scholar] [CrossRef]
- Wiechens, N.; Fagotto, F. CRM1- and Ran-independent nuclear export of beta-catenin. Curr. Biol. 2001, 11, 18–27. [Google Scholar] [CrossRef] [Green Version]
- Yokoya, F.; Imamoto, N.; Tachibana, T.; Yoneda, Y. Beta-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell 1999, 10, 1119–1131. [Google Scholar] [CrossRef]
- Asally, M.; Yoneda, Y. Beta-catenin can act as a nuclear import receptor for its partner transcription factor, lymphocyte enhancer factor-1 (lef-1). Exp. Cell Res. 2005, 308, 357–363. [Google Scholar] [CrossRef]
- Brunet, A.; Kanai, F.; Stehn, J.; Xu, J.; Sarbassova, D.; Frangioni, J.V.; Dalal, S.N.; DeCaprio, J.A.; Greenberg, M.E.; Yaffe, M.B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002, 156, 817–828. [Google Scholar] [CrossRef]
- van Hemert, M.J.; Niemantsverdriet, M.; Schmidt, T.; Backendorf, C.; Spaink, H.P. Isoform-specific differences in rapid nucleocytoplasmic shuttling cause distinct subcellular distributions of 14-3-3 sigma and 14-3-3 zeta. J. Cell Sci. 2004, 117, 1411–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthu, K.; Panneerselvam, M.; Topno, N.S.; Jayaraman, M.; Ramadas, K. Structural perspective of ARHI mediated inhibition of STAT3 signaling: An insight into the inactive to active transition of ARHI and its interaction with STAT3 and importinbeta. Cell. Signal. 2015, 27, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, O.A.; Tarasova, N.I.; Zhang, X.; Chasovskikh, S.; Cheema, A.K.; Wang, H.; Brown, M.L.; Dritschilo, A. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc. Natl. Acad. Sci. USA 2013, 110, 1267–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, T.; Yeh, J.E.; Pinello, L.; Jacob, J.; Chakravarthy, S.; Yuan, G.C.; Chopra, R.; Frank, D.A. Impact of the N-Terminal Domain of STAT3 in STAT3-Dependent Transcriptional Activity. Mol. Cell. Biol. 2015, 35, 3284–3300. [Google Scholar] [CrossRef] [Green Version]
- Haspel, R.L.; Darnell, J.E., Jr. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. USA 1999, 96, 10188–10193. [Google Scholar] [CrossRef] [Green Version]
- Aoki, N.; Matsuda, T. A nuclear protein tyrosine phosphatase TC-PTP is a potential negative regulator of the PRL-mediated signaling pathway: Dephosphorylation and deactivation of signal transducer and activator of transcription 5a and 5b by TC-PTP in nucleus. Mol. Endocrinol. 2002, 16, 58–69. [Google Scholar] [CrossRef]
- ten Hoeve, J.; de Jesus Ibarra-Sanchez, M.; Fu, Y.; Zhu, W.; Tremblay, M.; David, M.; Shuai, K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 2002, 22, 5662–5668. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Sekine, Y.; Kashima, K.; Kubota, A.; Sato, N.; Aoki, N.; Matsuda, T. The nuclear isoform of protein-tyrosine phosphatase TC-PTP regulates interleukin-6-mediated signaling pathway through STAT3 dephosphorylation. Biochem. Biophys. Res. Commun. 2002, 297, 811–817. [Google Scholar] [CrossRef] [Green Version]
- Lodige, I.; Marg, A.; Wiesner, B.; Malecova, B.; Oelgeschlager, T.; Vinkemeier, U. Nuclear export determines the cytokine sensitivity of STAT transcription factors. J. Biol. Chem. 2005, 280, 43087–43099. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, A.; Vogt, M.; Monnigmann, M.; Clahsen, T.; Sommer, U.; Haan, S.; Poli, V.; Heinrich, P.C.; Muller-Newen, G. Nucleocytoplasmic shuttling of persistently activated STAT3. J. Cell Sci. 2007, 120, 3249–3261. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Holloway, M.P.; Nguyen, K.; McCauley, D.; Landesman, Y.; Kauffman, M.G.; Shacham, S.; Altura, R.A. XPO1 (CRM1) inhibition represses STAT3 activation to drive a survivin-dependent oncogenic switch in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 675–686. [Google Scholar] [CrossRef] [PubMed]
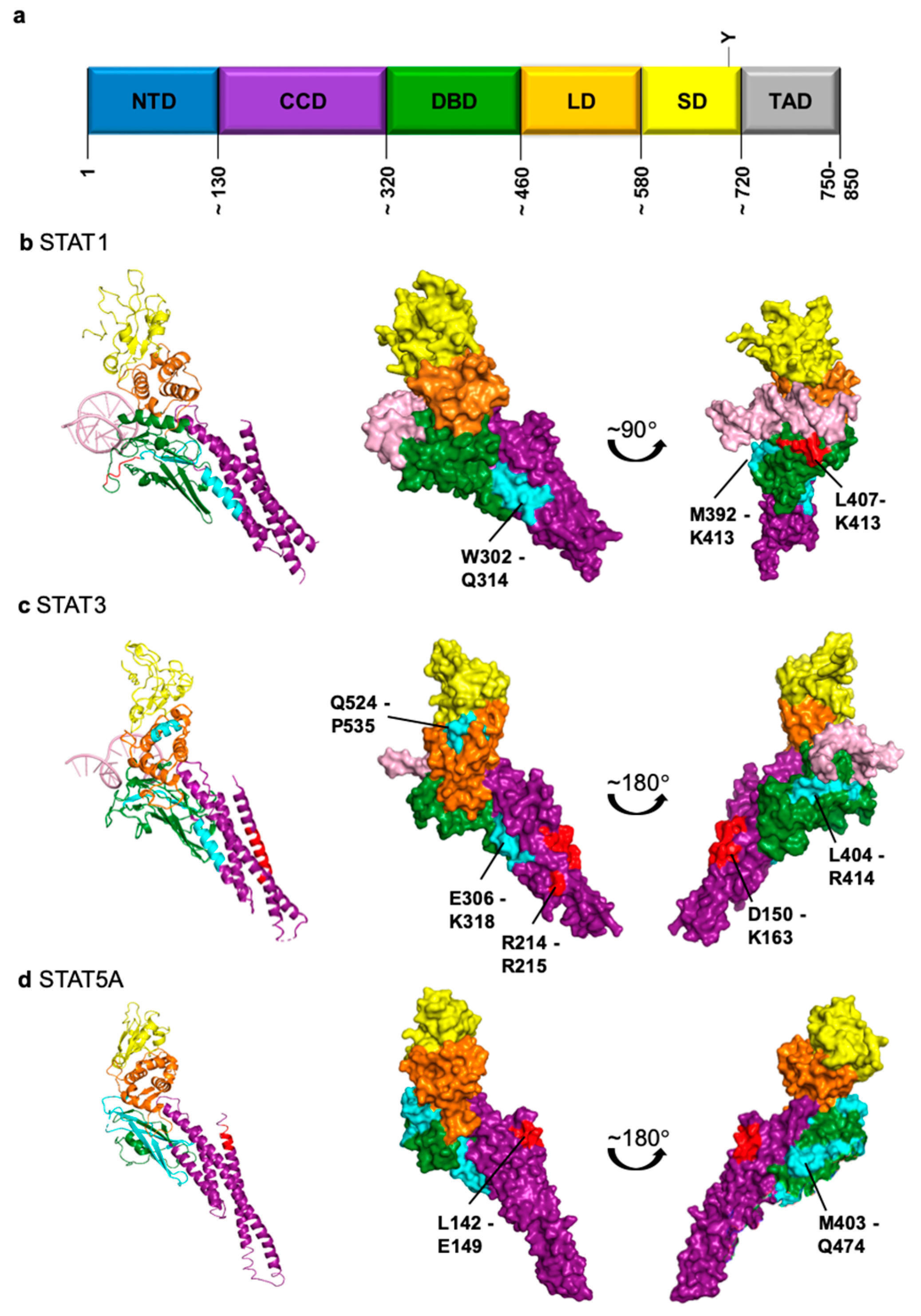
| STAT1 | STAT3 | STAT5A | STAT5B | |
|---|---|---|---|---|
| Importin | α5 [60,68], α7 [69] | α1 [70], α3 [70,71,72], α5 [69,70,72], α6 [71], α7 [69,72] | α3 [73] | n.d. 1 |
| Putative 2 NLS | L407-K413 [64,68] NTD involved [63,74] | D150-K163 [71] R214/R215 [69] NTD involved [75] | L142-E149 [76] intact CCD required [73] | n.d. |
| Exportin | Crm1 [66] | Crm1 [75] | Crm1 (not exclusively) [73] | Crm1 (not exclusively) [77] |
| Putative 2 NES | W302-Q314 [78] M392-K413 [66] | E306-K318 L404-R414 Q524-P535 [79] | L119/L133 M403-Q474 [73] | n.d. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ernst, S.; Müller-Newen, G. Nucleocytoplasmic Shuttling of STATs. A Target for Intervention? Cancers 2019, 11, 1815. https://doi.org/10.3390/cancers11111815
Ernst S, Müller-Newen G. Nucleocytoplasmic Shuttling of STATs. A Target for Intervention? Cancers. 2019; 11(11):1815. https://doi.org/10.3390/cancers11111815
Chicago/Turabian StyleErnst, Sabrina, and Gerhard Müller-Newen. 2019. "Nucleocytoplasmic Shuttling of STATs. A Target for Intervention?" Cancers 11, no. 11: 1815. https://doi.org/10.3390/cancers11111815





