Deciphering the Interplay between Genetic Risk Scores and Lifestyle Factors on Individual Obesity Predisposition
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Anthropometric and Lifestyle Variables
2.3. Genetic Risk Score (GRSBMI)
2.4. Statistical Analysis
3. Results
3.1. Participants
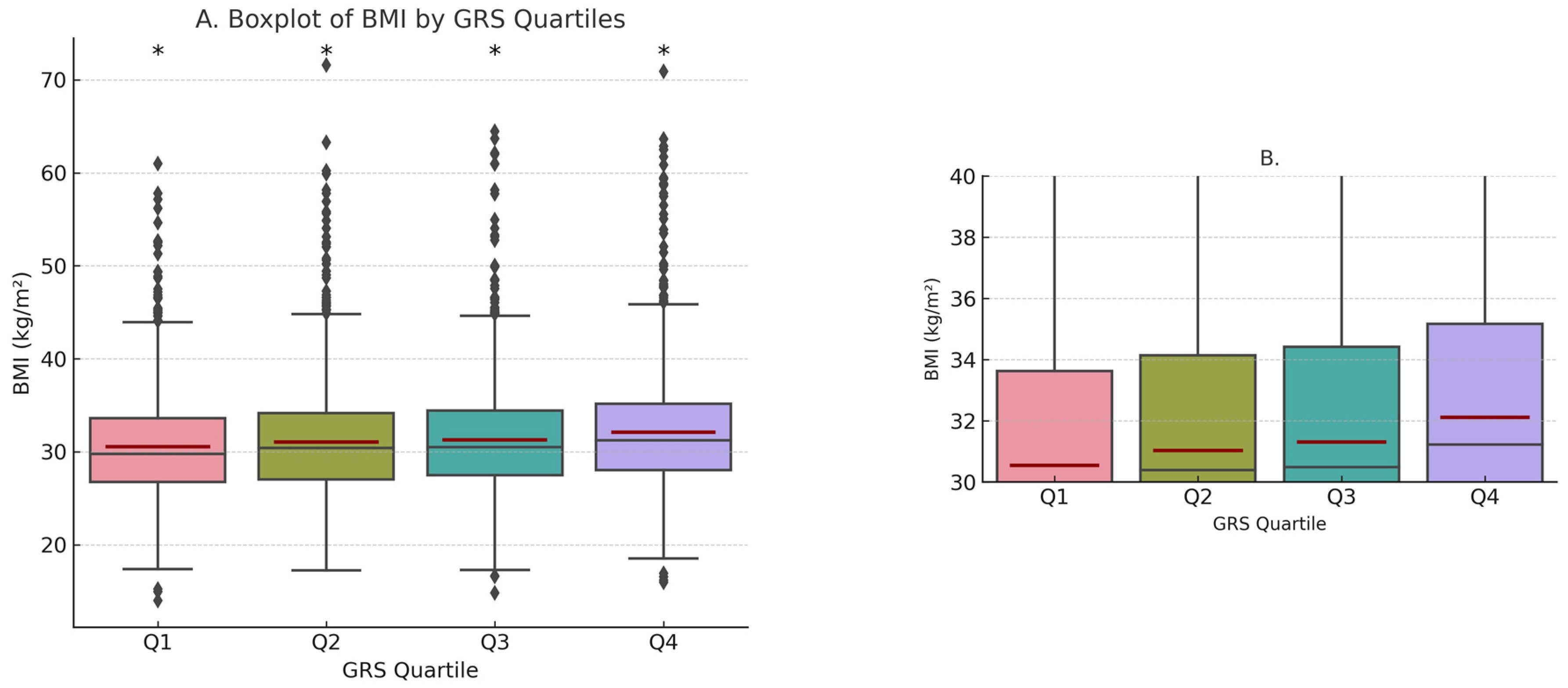
3.2. GRSBMI and Obesity
3.3. GRSBMI and Lifestyle Variables
3.3.1. Physical Activity
3.3.2. Eating Habits Score
3.3.3. Sugar-Sweetened Beverages
3.3.4. Wine
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hruby, A.; Hu, F.B. The Epidemiology of Obesity: A big picture. Pharmacoeconomics 2015, 33, 673–689. [Google Scholar] [CrossRef]
- GBD 2015 Obesity Collaborators; Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; et al. Health effects of overweight and obesity in 195 countries over 25 years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [PubMed]
- Purnell, J.Q. Definitions, classification, and pidemiology of Obesity. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. Available online: http://www.ncbi.nlm.nih.gov/books/NBK279167/ (accessed on 20 October 2023).
- Walter, N.A.R.; McWeeney, S.K.; Peters, S.T.; Belknap, J.K.; Hitzemann, R.; Buck, K.J. Single-nucleotide polymorphism masking. Alcohol. Res. Health 2008, 31, 270–271. [Google Scholar] [PubMed]
- Goodarzi, M.O. Genetics of obesity: What genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol. 2018, 6, 223–236. [Google Scholar] [CrossRef]
- Li, S.; Zhao, J.H.; Luan, J.; Luben, R.N.; Rodwell, S.A.; Khaw, K.-T.; Ong, K.K.; Wareham, N.J.; Loos, R.J.F. Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am. J. Clin. Nutr. 2010, 91, 184–190. [Google Scholar] [CrossRef]
- Sufianov, A.; Beilerli, A.; Kudriashov, V.; Ilyasova, T.; Liang, Y.; Mukhamedzyanov, A.; Bessonova, M.; Mashkin, A.; Beylerli, O. The role of long non-coding RNAs in the development of adipose cells. Noncoding RNA Res. 2023, 8, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Speliotes, E.K.; Willer, C.J.; Berndt, S.I.; Monda, K.L.; Thorleifsson, G.; Jackson, A.U.; Allen, H.L.; Lindgren, C.M.; Luan, J.; Mägi, R.; et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010, 42, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Thorleifsson, G.; Walters, G.B.; Gudbjartsson, D.F.; Steinthorsdottir, V.; Sulem, P.; Helgadottir, A.; Styrkarsdottir, U.; Gretarsdottir, S.; Thorlacius, S.; Jonsdottir, I.; et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009, 41, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M.; Vassos, E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020, 12, 44. [Google Scholar] [CrossRef]
- Popejoy, A.B.; Fullerton, S.M. Genomics is failing on diversity. Nature 2016, 538, 161–164. [Google Scholar] [CrossRef]
- Wardle, J.; Carnell, S.; Haworth, C.M.; Plomin, R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am. J. Clin. Nutr. 2008, 87, 298–404. [Google Scholar] [CrossRef] [PubMed]
- Nicolaidis, S. Environment and obesity. Metabolism 2019, 100S, 153942. [Google Scholar] [CrossRef] [PubMed]
- Heianza, Y.; Zhou, T.; Sun, D.; Hu, F.B.; Qi, L. Healthful plant-based dietary patterns, genetic risk of obesity, and cardiovascular risk in the UK biobank study. Clin. Nutr. 2021, 40, 4694–4701. [Google Scholar] [CrossRef] [PubMed]
- Michałowska, J.; Miller-Kasprzak, E.; Seraszek-Jaros, A.; Mostowska, A.; Bogdański, P. The link between three single nucleotide variants of the GIPR gene and metabolic health. Genes 2022, 13, 1534. [Google Scholar] [CrossRef] [PubMed]
- Belsky, D.W.; Moffitt, T.E.; Sugden, K.; Williams, B.; Houts, R.; McCarthy, J.; Caspi, A. Development and evaluation of a genetic risk score for obesity. Biodemogr. Soc. Biol. 2013, 59, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Seral-Cortes, M.; Sabroso-Lasa, S.; De Miguel-Etayo, P.; Gonzalez-Gross, M.; Gesteiro, E.; Molina-Hidalgo, C.; De Henauw, S.; Gottrand, F.; Mavrogianni, C.; Manios, Y.; et al. Development of a genetic risk score to predict the risk of overweight and obesity in European adolescents from the HELENA study. Sci. Rep. 2021, 11, 3067. [Google Scholar] [CrossRef] [PubMed]
- Damavandi, N.; Soleymaniniya, A.; Zadegan, S.B.; Aref, M.H.S.; Zeinali, S. Development of a genetic risk score for obesity predisposition evaluation. Mol. Genet. Genom. 2022, 297, 1495–1503. [Google Scholar] [CrossRef]
- Hung, C.-F.; Breen, G.; Czamara, D.; Corre, T.; Wolf, C.; Kloiber, S.; Bergmann, S.; Craddock, N.; Gill, M.; Holsboer, F.; et al. A genetic risk score combining 32 SNPs is associated with body mass index and improves obesity prediction in people with major depressive disorder. BMC Med. 2015, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Bhatti, A.A. Predicting anthropometric and metabolic traits with a genetic risk score for obesity in a sample of Pakistanis. Sci. Rep. 2021, 11, 8320. [Google Scholar] [CrossRef]
- Sag, S.J.M.; Mueller, S.; Wallner, S.; Strack, C.; Hubauer, U.; Mohr, M.; Zeller, J.; Loew, T.; Rehli, M.; Wimmer, J.; et al. A multilocus genetic risk score for obesity: Association with BMI and metabolic alterations in a cohort with severe obesity. Medicine 2023, 102, e34597. [Google Scholar] [CrossRef]
- Ahmad, S.; Rukh, G.; Varga, T.V.; Ali, A.; Kurbasic, A.; Shungin, D.; Ericson, U.; Koivula, R.W.; Chu, A.Y.; Rose, L.M.; et al. Gene × physical activity interactions in obesity: Combined analysis of 111,421 individuals of European ancestry. PLoS Genet. 2013, 9, e1003607. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, J.H.; Luan, J.; Ekelund, U.; Luben, R.N.; Khaw, K.-T.; Wareham, N.J.; Loos, R.J.F. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010, 7, e1000332. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Loos, R.J.F.; Lu, L.; Zong, G.; Gan, W.; Ye, X.; Sun, L.; Li, H.; Lin, X. Associations of genetic risk score with obesity and related traits and the modifying effect of physical activity in a Chinese Han population. PLoS ONE 2014, 9, e91442. [Google Scholar] [CrossRef] [PubMed]
- Bull, F.C.; Al-Ansari, S.S.; Biddle, S.; Borodulin, K.; Buman, M.P.; Cardon, G.; Carty, C.; Chaput, J.-P.; Chastin, S.; Chou, R.; et al. World Health Organization 2020 guidelines on physical activity and sedentary behaviour. Br. J. Sports Med. 2020, 54, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Herle, M.; Smith, A.D.; Kininmonth, A.; Llewellyn, C. The Role of Eating Behaviours in Genetic Susceptibility to Obesity. Curr. Obes. Rep. 2020, 9, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.; Drapeau, V.; Tremblay, A.; Provencher, V.; Bouchard, C.; Pérusse, L. The role of eating behavior traits in mediating genetic susceptibility to obesity. Am. J. Clin. Nutr. 2018, 108, 445–452. [Google Scholar] [CrossRef]
- Begum, S.; Hinton, E.C.; Toumpakari, Z.; Frayling, T.M.; Howe, L.; Johnson, L.; Lawrence, N. Mediation and moderation of genetic risk of obesity through eating behaviours in two UK cohorts. Int. J. Epidemiol. 2023, 52, 1926–1938. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.S.; Hu, F.B. The role of sugar-sweetened beverages in the global epidemics of obesity and chronic diseases. Nat. Rev. Endocrinol. 2022, 18, 205–218. [Google Scholar] [CrossRef]
- Brunkwall, L.; Chen, Y.; Hindy, G.; Rukh, G.; Ericson, U.; Barroso, I.; Johansson, I.; Franks, P.W.; Orho-Melander, M.; Renström, F. Sugar-sweetened beverage consumption and genetic predisposition to obesity in 2 Swedish cohorts12. Am. J. Clin. Nutr. 2016, 104, 809–815. [Google Scholar] [CrossRef]
- Qi, Q.; Chu, A.Y.; Kang, J.H.; Jensen, M.K.; Curhan, G.C.; Pasquale, L.R.; Ridker, P.M.; Hunter, D.J.; Willett, W.C.; Rimm, E.B.; et al. Sugar-sweetened beverages and genetic risk of obesity. N. Engl. J. Med. 2012, 367, 1387–1396. [Google Scholar] [CrossRef]
- Inan-Eroglu, E.; Powell, L.; Hamer, M.; O’Donovan, G.; Duncan, M.J.; Stamatakis, E. Is there a link between different types of alcoholic drinks and obesity? An analysis of 280,183 UK biobank participants. Int. J. Environ. Res. Public Health 2020, 17, 5178. [Google Scholar] [CrossRef] [PubMed]
- Thomson, C.A.; Wertheim, B.C.; Hingle, M.; Wang, L.; Neuhouser, M.L.; Gong, Z.; Garcia, L.; Stefanick, M.L.; Manson, J.E. Alcohol consumption and body weight change in postmenopausal women: Results from the Women’s Health Initiative. Int. J. Obes. 2012, 36, 1158–1164. [Google Scholar] [CrossRef] [PubMed]
- Flechtner-Mors, M.; Biesalski, H.K.; Jenkinson, C.P.; Adler, G.; Ditschuneit, H.H. Effects of moderate consumption of white wine on weight loss in overweight and obese subjects. Int. J. Obes. Relat. Metab. Disord. 2004, 28, 1420–1426. [Google Scholar] [CrossRef] [PubMed]
- Jéquier, E. Alcohol intake and body weight: A paradox. Am. J. Clin. Nutr. 1999, 69, 173–174. [Google Scholar] [PubMed]
- Parsamanesh, N.; Asghari, A.; Sardari, S.; Tasbandi, A.; Jamialahmadi, T.; Xu, S.; Sahebkar, A. Resveratrol and endothelial function: A literature review. Pharmacol. Res. 2021, 170, 105725. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liang, X.; Yang, Q.; Fu, X.; Rogers, C.J.; Zhu, M.; Rodgers, B.D.; Jiang, Q.; Dodson, M.V.; Du, M. Resveratrol induces brown-like adipocyte formation in white fat through activation of AMP-activated protein kinase (AMPK) α1. Int. J. Obes. 2015, 39, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Timmers, S.; Konings, E.; Bilet, L.; Houtkooper, R.H.; van de Weijer, T.; Goossens, G.H.; Hoeks, J.; van der Krieken, S.; Ryu, D.; Kersten, S.; et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011, 14, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Zagotta, I.; Dimova, E.Y.; Debatin, K.-M.; Wabitsch, M.; Kietzmann, T.; Fischer-Posovszky, P. Obesity and inflammation: Reduced cytokine expression due to resveratrol in a human in vitro model of inflamed adipose tissue. Front. Pharmacol. 2015, 6, 79. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Gomez, Y.; Mattison, J.A.; Pearson, K.J.; Martin-Montalvo, A.; Palacios, H.H.; Sossong, A.M.; Ward, T.M.; Younts, C.M.; Lewis, K.; Allard, J.S.; et al. Resveratrol improves adipose insulin signaling and reduces the inflammatory response in adipose tissue of rhesus monkeys on a high-fat, high-sugar diet. Cell Metab. 2013, 18, 533–545. [Google Scholar] [CrossRef]
- Chermon, D.; Birk, R. Drinking habits and physical activity interact and attenuate obesity predisposition of TMEM18 polymorphisms carriers. Nutrients 2023, 15, 266. [Google Scholar] [CrossRef]
- Chermon, D.; Birk, R. FTO common obesity SNPs interact with actionable environmental factors: Physical activity, sugar-sweetened beverages and wine consumption. Nutrients 2022, 14, 4202. [Google Scholar] [CrossRef] [PubMed]
| Character | All (n = 5824) | BMI ≥ 30 (n = 3173) | BMI < 30 (n = 2651) | p-Value |
|---|---|---|---|---|
| Age (years) | 55.78 ± 15.3 | 55.91 ± 15.4 | 55.63 ± 15.2 | 0.3 |
| Height (cm) | 166.42 ± 9.15 | 166.52 ± 9.4 | 166.29 ± 8.84 | 0.55 |
| Weight (kg) | 86.83 ± 19.6 | 98.13 ± 17.9 | 73.31 ± 11.2 | <0.0001 |
| BMI (kg/m2) | 31.24 ± 6.06 | 35.28 ± 5.06 | 26.41 ± 2.66 | <0.0001 |
| Sex (female) | 4050 (69.54) | 1919 (72.39) | 2131 (67.16) | <0.001 |
| T2DM (n, %) | 449 (7.7) | 285 (8.98) | 164 (6.19) | <0.0001 |
| EHS score | 11.60 ± 7.55 | 12.33 ± 7.68 | 10.73 ± 7.3 | <0.0001 |
| PA ≥ 90 (n, %) | 1162 (19.95) | 473 (14.9) | 689 (26) | <0.0001 |
| SSB consumers (n, %) | 684 (11.74) | 439 (13.84) | 245 (9.24) | <0.0001 |
| Wine consumers * (n, %) | 1398 (24) | 669 (21.08) | 729 (27.5) | <0.0001 |
| GRSBMI Quartile | Mean BMI—Active (±SD) | Mean BMI—Inactive (±SD) | BMI Difference (Active vs. Inactive) (95% CI) | Obesity OR (Active vs. Inactive within Q) (95% CI) | Mean BMI between GRSBMI Quartiles (Inactive) ** |
|---|---|---|---|---|---|
| Q1 (n = 1465) | 29.0 ± 5.23 | 30.9 ± 5.85 | −1.9 (−2.56–(−1.2)) | 0.56 (0.43–0.72) | - |
| Q2 (n = 1456) | 28.9 ± 4.73 | 31.5 ± 6.17 | −2.6 (−3.25–(−1.95)) | 0.44 (0.34–0.57) | NS |
| Q3 (n = 1455) | 29.3 ± 5.06 | 31.8 ± 6.09 | −2.5 (−3.15–(−1.78)) | 0.51 (0.40–0.67) | 0.003 a |
| Q4 (n = 1448) | 30.2 ± 5.66 | 32.6 ± 6.43 | −2.4 (−3.14–(−1.64)) | 0.48 (0.37–0.63) | <0.001 a <0.0001 b <0.005 c |
| GRSBMI Quartile (Q) | Mean BMI—EHS ≥ Median (±SD) | Mean BMI—EHS < Median (±SD) | BMI Difference (EHS ≥ Median vs. EHS < Median) (95% CI) | EHS ≥ Median vs. EHS < Median within Q OR (95% CI) * | BMI EHS ≥ between across GRSBMI Quartiles ** |
|---|---|---|---|---|---|
| Q1 (n = 1465) | 31.02 ± 5.78 | 30.05 ± 5.74 | +0.97 (0.38–1.56) | 1.42 (1.15–1.75) | NS |
| Q2 (n = 1456) | 31.56 ± 6.11 | 30.44 ± 5.83 | +1.12 (0.51–1.73) | 1.20 (0.97–1.48) | 0.02 b |
| Q3 (n = 1455) | 32.02 ± 6.02 | 30.57 ± 5.86 | +1.45 (0.84–2.06) | 1.51 (1.21–1.86) | 0.008 a |
| Q4 (n = 1448) | 32.51 ± 6.37 | 31.63 ± 6.31 | +0.88 (0.23–1.55) | 1.36 (1.09–1.69) | <0.0001 a |
| GRSBMI Quartile (Q) | Mean BMI—SSB Consumers (±SD) | Mean BMI-Non-SSB Consumers (±SD) | BMI Difference (SSB vs. Non-SSB) (95% CI) | (SSB vs. Non-SSB within Q) OR (95% CI) * | Mean BMI between GRS Quartiles (SSB Consumer) ** |
|---|---|---|---|---|---|
| Q1 (n = 1465) | 32.30 ± 5.66 | 30.36 ± 5.77 | +1.63 (0.69–2.57) | 1.46 (1.05–2.04) | NS |
| Q2 (n = 1456) | 32.4 ± 6.63 | 30.85 ± 5.89 | +1.55 (0.49–2.58) | 1.48 (1.07–2.05) | NS |
| Q3 (n = 1455) | 32.78 ± 5.92 | 31.09 ± 5.97 | +1.69 (0.77–2.64) | 1.88 (1.36–2.63) | NS |
| Q4 (n = 1448) | 33.87 ± 6.91 | 31.88 ± 6.25 | +1.92 (0.78–2.98) | 1.49 (1.05–2.1) | 0.007 a |
| GRSBMI Quartile (Q) | Mean BMI—Wine Drinkers (±SD) | Mean BMI—Non-Drinkers (±SD) | BMI Difference (Drinkers vs. Non-Drinkers) (95% CI) | (Drinkers vs. Non-Drinkers within Q) OR (95% CI) * | Mean BMI between GRSBMI Quartiles (Non-Drinkers) ** |
|---|---|---|---|---|---|
| Q1 (n = 1424) | 29.85 ± 5.79 | 30.86 ± 5.77 | −1.01 (−1.7–(−0.31)) | 0.61 (0.48–0.78) | - |
| Q2 (n = 1423) | 30.18 ± 4.85 | 31.46 ± 6.32 | −1.28 (−1.91–(−0.66)) | 0.67 (0.53–0.86) | NS |
| Q3 (n = 1416) | 30.58 ± 5.32 | 31.62 ± 6.16 | −1.04 (−1.73–(−0.36)) | 0.71 (0.55–0.91) | 0.03 a |
| Q4 (n = 1415) | 31.05 ± 5.35 | 32.56 ± 6.62 | −1.51 (−2.2–(−0.83)) | 0.65 (0.51–0.83) | <0.0001 a <0.0001 b 0.005 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chermon, D.; Birk, R. Deciphering the Interplay between Genetic Risk Scores and Lifestyle Factors on Individual Obesity Predisposition. Nutrients 2024, 16, 1296. https://doi.org/10.3390/nu16091296
Chermon D, Birk R. Deciphering the Interplay between Genetic Risk Scores and Lifestyle Factors on Individual Obesity Predisposition. Nutrients. 2024; 16(9):1296. https://doi.org/10.3390/nu16091296
Chicago/Turabian StyleChermon, Danyel, and Ruth Birk. 2024. "Deciphering the Interplay between Genetic Risk Scores and Lifestyle Factors on Individual Obesity Predisposition" Nutrients 16, no. 9: 1296. https://doi.org/10.3390/nu16091296
APA StyleChermon, D., & Birk, R. (2024). Deciphering the Interplay between Genetic Risk Scores and Lifestyle Factors on Individual Obesity Predisposition. Nutrients, 16(9), 1296. https://doi.org/10.3390/nu16091296







