Build Deep Neural Network Models to Detect Common Edible Nuts from Photos and Estimate Nutrient Portfolio
Abstract
1. Introduction
2. Methods
2.1. Data
2.2. Model
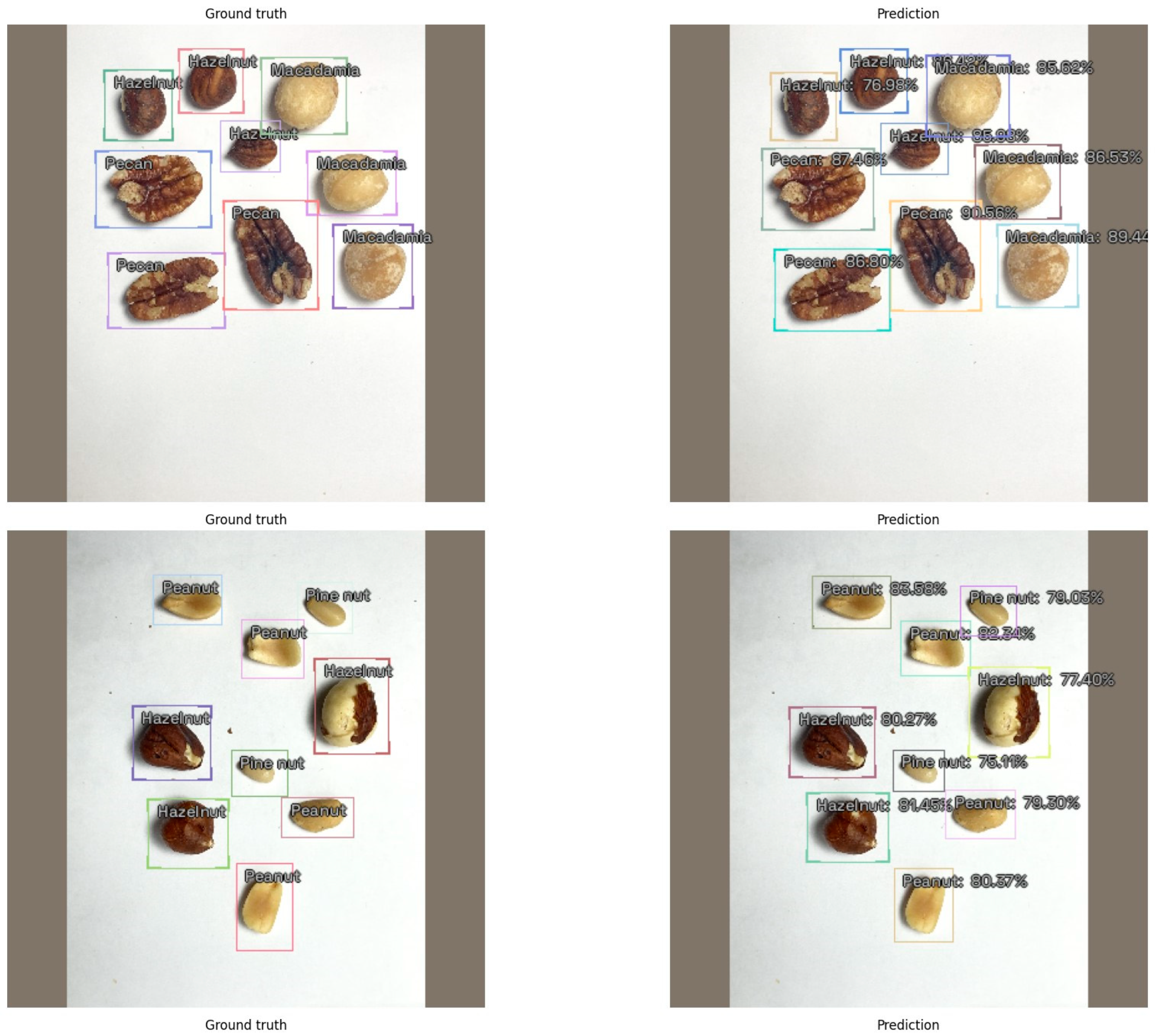
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ros, E. Health benefits of nut consumption. Nutrients 2010, 2, 652–682. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.G.M.; Schincaglia, R.M.; Pimentel, G.D.; Mota, J.F. Nuts and Human Health Outcomes: A Systematic Review. Nutrients 2017, 9, 1311. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.K.; Alasalvar, C.; Bolling, B.W.; Shahidi, F. Nuts and their co-products: The impact of processing (roasting) on phenolics, bioavailability, and health benefits—A comprehensive review. J. Funct. Foods 2016, 26, 88–122. [Google Scholar] [CrossRef]
- McGuire, S. US Department of Agriculture and US Department of Health and Human Services, Dietary Guidelines for Americans, 2010. Washington, DC: US Government Printing Office, January 2011. Adv. Nutr. 2011, 2, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Aune, D.; Keum, N.; Giovannucci, E.; Fadnes, L.T.; Boffetta, P.; Greenwood, D.C.; Tonstad, S.; Vatten, L.J.; Riboli, E.; Norat, T. Nut consumption and risk of cardiovascular disease, total cancer, all-cause and cause-specific mortality: A systematic review and dose-response meta-analysis of prospective studies. BMC Med. 2016, 14, 207. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Keogh, J.; Clifton, P. Benefits of nut consumption on insulin resistance and cardiovascular risk factors: Multiple potential mechanisms of actions. Nutrients 2017, 9, 1271. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Yuan, S.; Jin, Y.; Lu, J. Nut consumption and risk of metabolic syndrome and overweight/obesity: A meta-analysis of prospective cohort studies and randomized trials. Nutr. Metab. 2018, 15, 46. [Google Scholar] [CrossRef] [PubMed]
- Barnett, I.; Edwards, D. Mobile Phones for Real-Time Nutrition Surveillance: Approaches, Challenges and Opportunities for Data Presentation and Dissemination. IDS Evidence Report 75, Brighton: IDS. Available online: https://opendocs.ids.ac.uk/opendocs/handle/20.500.12413/4020 (accessed on 15 September 2022).
- Ferrara, G.; Kim, J.; Lin, S.; Hua, J.; Seto, E. A focused review of smartphone diet-tracking apps: Usability, functionality, coherence with behavior change theory, and comparative validity of nutrient intake and energy estimates. JMIR mHealth uHealth 2019, 7, e9232. [Google Scholar] [CrossRef] [PubMed]
- Zečević, M.; Mijatović, D.; Koklič, M.K.; Žabkar, V.; Gidaković, P. User Perspectives of Diet-Tracking Apps: Reviews Content Analysis and Topic Modeling. J. Med. Internet Res. 2021, 23, e25160. [Google Scholar] [CrossRef]
- West, J.H.; Belvedere, L.M.; Andreasen, R.; Frandsen, C.; Hall, P.C.; Crookston, B.T. Controlling your “app” etite: How diet and nutrition-related mobile apps lead to behavior change. JMIR mHealth uHealth 2017, 5, e95. [Google Scholar] [CrossRef]
- Limketkai, B.N.; Mauldin, K.; Manitius, N.; Jalilian, L.; Salonen, B.R. The Age of Artificial Intelligence: Use of Digital Technology in Clinical Nutrition. Curr. Surg. Rep. 2021, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Abiodun, O.I.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Mohamed, N.A.; Arshad, H. State-of-the-art in artificial neural network applications: A survey. Heliyon 2018, 4, e00938. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Côté, M.; Lamarche, B. Artificial intelligence in nutrition research: Perspectives on current and future applications. Appl. Physiol. Nutr. Metab. 2021, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gemming, L.; Utter, J.; Ni Mhurchu, C. Image-assisted dietary assessment: A systematic review of the evidence. J. Acad. Nutr. Diet. 2015, 115, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Francois, C. Deep Learning with Python; Manning Publications: Shelter Island, NY, USA, 2017. [Google Scholar]
- Dheir, I.M.; Mettleq, A.S.A.; Elsharif, A.A. Nuts Types Classification Using Deep learning. Int. J. Acad. Inf. Syst. Res. 2020, 3, 12–17. [Google Scholar]
- An, R.; Perez-Cruet, J.; Wang, J. We got nuts! use deep neural networks to classify images of common edible nuts. Nutr. Health 2022. Online ahead of print. [Google Scholar] [CrossRef]
- Dutta, A.; Zisserman, A. The VIA Annotation Software for Images, Audio and Video. In Proceedings of the 27th ACM International Conference on Multimedia, Nice, France, 21–25 October 2019; pp. 2276–2279. [Google Scholar] [CrossRef]
- IceVision. IceVision: An Agnostic Computer Vision Framework. Available online: https://pypi.org/project/icevision/ (accessed on 1 March 2024).
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst. 2015, 28, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. arXiv 2017, arXiv:1708.02002. [Google Scholar]
- Ultralytics. YOLOv5 in PyTorch > ONNX > CoreML > TFLite. 2020. Available online: https://github.com/ultralytics/yolov5 (accessed on 1 March 2024).
- Tan, M.; Pang, R.; Le, Q.V. Efficientdet: Scalable and efficient object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2020), Washington, DC, USA, 14–19 June 2020; pp. 10781–10790. Available online: https://ar5iv.org/abs/1911.09070 (accessed on 1 March 2024).
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Smith, L.N. Cyclical learning rates for training neural networks. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision, WACV 2017, Santa Rosa, CA, USA, 24–31 March 2017; pp. 464–472. [Google Scholar] [CrossRef]
- Henderson, P.; Ferrari, V. End-to-end training of object class detectors for mean average precision. In Proceedings of the 13th Asian Conference on Computer Vision, Taibei, Taiwan, 20–24 November 2016; Springer: Berlin, Germany, 2016; pp. 198–213. Available online: https://ar5iv.labs.arxiv.org/html/1607.03476 (accessed on 1 March 2024).
- Haytowitz, D.; Ahuja, J.; Wu, X.; Khan, M.; Somanchi, M.; Nickle, M.; Nguyen, Q.; Roseland, J.; Williams, J.; Patterson, K. USDA National Nutrient Database for Standard Reference, Legacy. In USDA National Nutrient Database for Standard Reference; The United States Department of Agriculture: Washington, DC, USA, 2018. Available online: https://www.ars.usda.gov/ARSUserFiles/80400525/Data/SR-Legacy/SR-Legacy_Doc.pdf (accessed on 1 March 2024).
- Anthony, L.F.W.; Kanding, B.; Selvan, R. Carbontracker: Tracking and predicting the carbon footprint of training deep learning models. arXiv 2020, arXiv:2007.03051. [Google Scholar]
- Ferranti, E.P.; Dunbar, S.B.; Higgins, M.; Dai, J.; Ziegler, T.R.; Frediani, J.; Reilly, C.; Brigham, K.L. Psychosocial factors associated with diet quality in a working adult population. Res. Nurs. Health 2013, 36, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Grossniklaus, D.A.; Dunbar, S.B.; Tohill, B.C.; Gary, R.; Higgins, M.K.; Frediani, J.K. Psychological factors are important correlates of dietary pattern in overweight adults. J. Cardiovasc. Nurs. 2010, 25, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Baranowski, T.; Cullen, K.W.; Baranowski, J. Psychosocial correlates of dietary intake: Advancing dietary intervention. Annu. Rev. Nutr. 1999, 19, 17–40. [Google Scholar] [CrossRef] [PubMed]
- McClain, A.D.; Chappuis, C.; Nguyen-Rodriguez, S.T.; Yaroch, A.L.; Spruijt-Metz, D. Psychosocial correlates of eating behavior in children and adolescents: A review. Int. J. Behav. Nutr. Phys. Act. 2009, 6, 54. [Google Scholar] [CrossRef]
- Veloso, S.M.; Matos, M.G.; Carvalho, M.; Diniz, J.A. Psychosocial factors of different health behaviour patterns in adolescents: Association with overweight and weight control behaviours. J. Obes. 2012, 2012, 852672. [Google Scholar] [CrossRef] [PubMed]
- West, J.H.; Cougar Hall, P.; Arredondo, V.; Berrett, B.; Guerra, B.; Farrell, J. Health Behavior Theories in Diet Apps. J. Consum. Health Internet 2013, 17, 10–24. [Google Scholar] [CrossRef]
- Bohr, A.; Memarzadeh, K. The rise of artificial intelligence in healthcare applications. In Artificial Intelligence in Healthcare; Academic Press: Cambridge, MA, USA, 2020; pp. 25–60. [Google Scholar] [CrossRef]
- Garcia-Ceja, E.; Riegler, M.; Nordgreen, T.; Jakobsen, P.; Oedegaard, K.J.; Tørresen, J. Mental health monitoring with multimodal sensing and machine learning: A survey. Pervasive Mob. Comput. 2018, 51, 1–26. [Google Scholar] [CrossRef]

| Model | Highest mAP Score | Number of Epochs |
|---|---|---|
| Faster R-CNN | 0.7114 | 17 |
| RetinaNet | 0.4905 | 19 |
| YOLOv5 | 0.7596 | 19 |
| EfficientDet | 0.7250 | 13 |
| Model | Predicted Mean | Ground Truth Mean | Proportion of Discrepancy ± Standard Error |
|---|---|---|---|
| Total energy (kcal) | 101.24 | 99.68 | 1.56% |
| Protein (g) | 2.36 | 2.33 | 1.13% |
| Carbohydrate (g) | 4.32 | 4.21 | 2.58% |
| Total fat (g) | 9.02 | 8.90 | 1.37% |
| Saturated fat (g) | 1.42 | 1.40 | 1.18% |
| Fiber (g) | 1.14 | 1.13 | 1.34% |
| Vitamin E (mg) | 0.78 | 0.77 | 1.31% |
| Magnesium (mg) | 36.67 | 36.20 | 1.28% |
| Phosphorus (mg) | 71.59 | 70.86 | 1.02% |
| Copper (mg) | 0.20 | 0.20 | 1.19% |
| Manganese (mg) | 0.42 | 0.41 | 2.19% |
| Selenium (µg) | 85.76 | 85.06 | 0.82% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, R.; Perez-Cruet, J.M.; Wang, X.; Yang, Y. Build Deep Neural Network Models to Detect Common Edible Nuts from Photos and Estimate Nutrient Portfolio. Nutrients 2024, 16, 1294. https://doi.org/10.3390/nu16091294
An R, Perez-Cruet JM, Wang X, Yang Y. Build Deep Neural Network Models to Detect Common Edible Nuts from Photos and Estimate Nutrient Portfolio. Nutrients. 2024; 16(9):1294. https://doi.org/10.3390/nu16091294
Chicago/Turabian StyleAn, Ruopeng, Joshua M. Perez-Cruet, Xi Wang, and Yuyi Yang. 2024. "Build Deep Neural Network Models to Detect Common Edible Nuts from Photos and Estimate Nutrient Portfolio" Nutrients 16, no. 9: 1294. https://doi.org/10.3390/nu16091294
APA StyleAn, R., Perez-Cruet, J. M., Wang, X., & Yang, Y. (2024). Build Deep Neural Network Models to Detect Common Edible Nuts from Photos and Estimate Nutrient Portfolio. Nutrients, 16(9), 1294. https://doi.org/10.3390/nu16091294







