Role of Dietary Factors on DNA Methylation Levels of TNF-Alpha Gene and Proteome Profiles in Obese Men
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Anthropometric Measures
2.3. Assessment of Dietary Intake
2.4. Biochemical Analysis
2.5. DNA Methylation Analysis
2.6. Protein Quantitation and Identification by Liquid Chromatography–Mass Spectrometry (LC-MS/MS)
2.7. Statistical Analysis
3. Results
3.1. Characteristics of the Participants
3.2. Associations between DNA Methylation of TNF-α and Metabolic Components and Dietary Factors
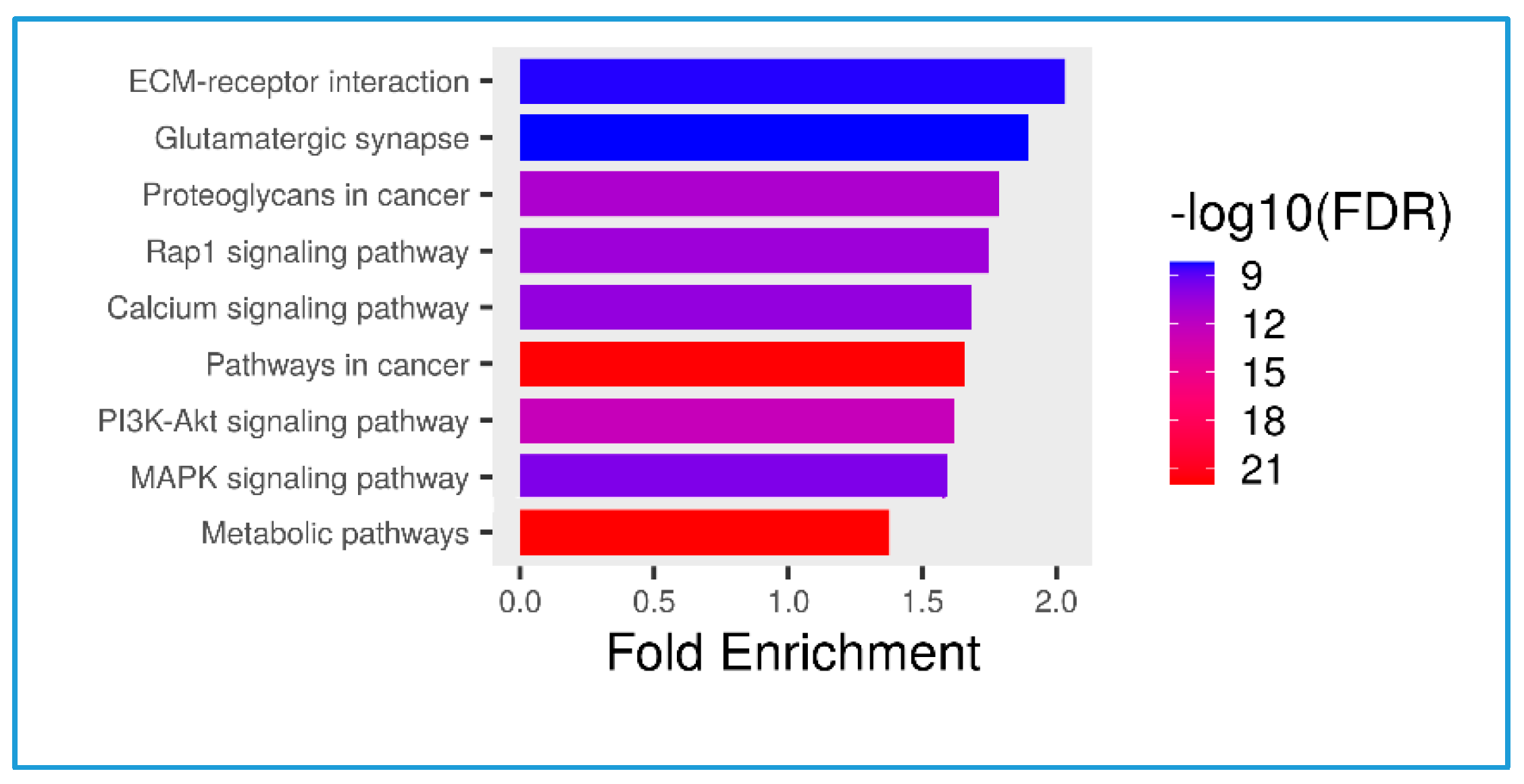
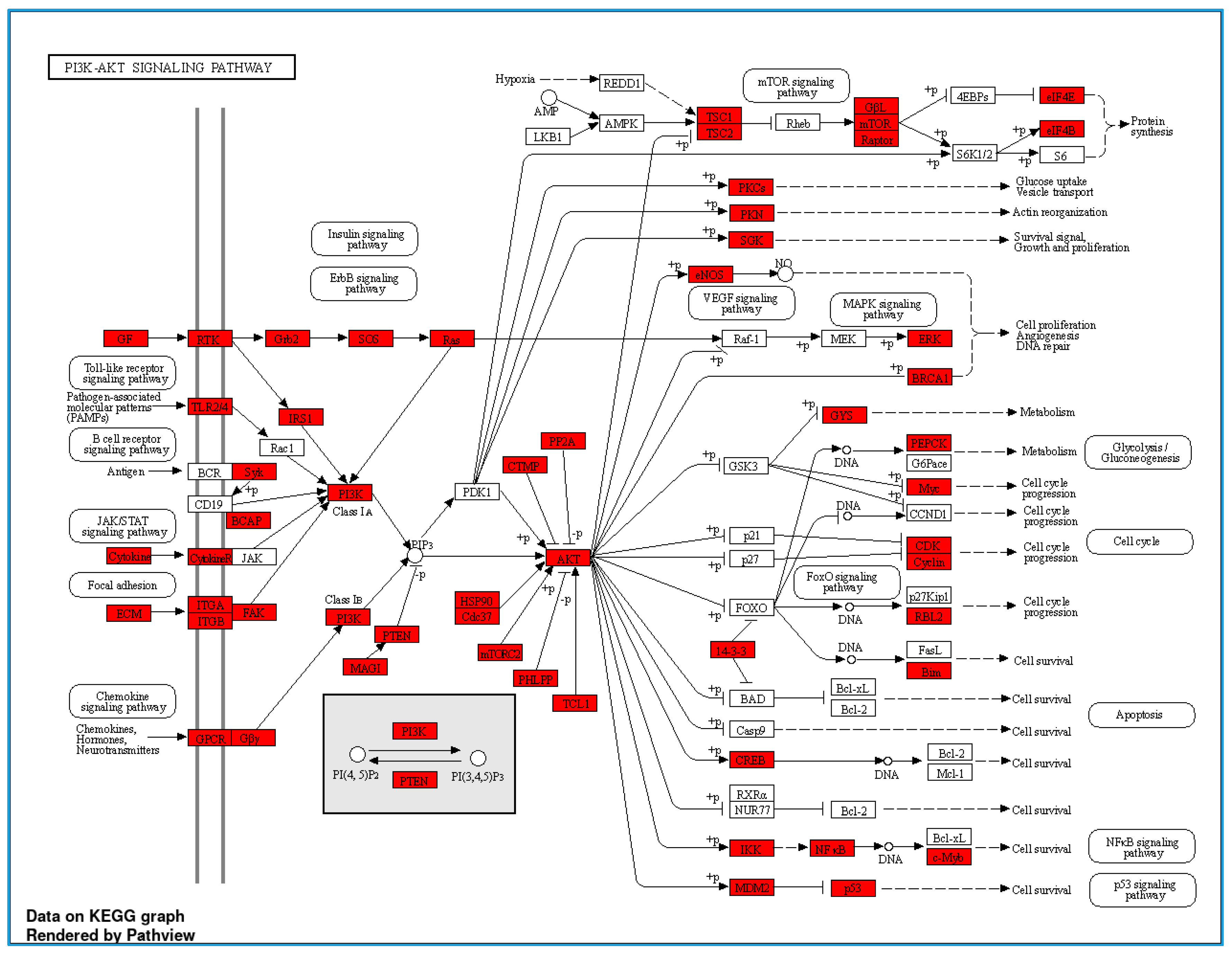
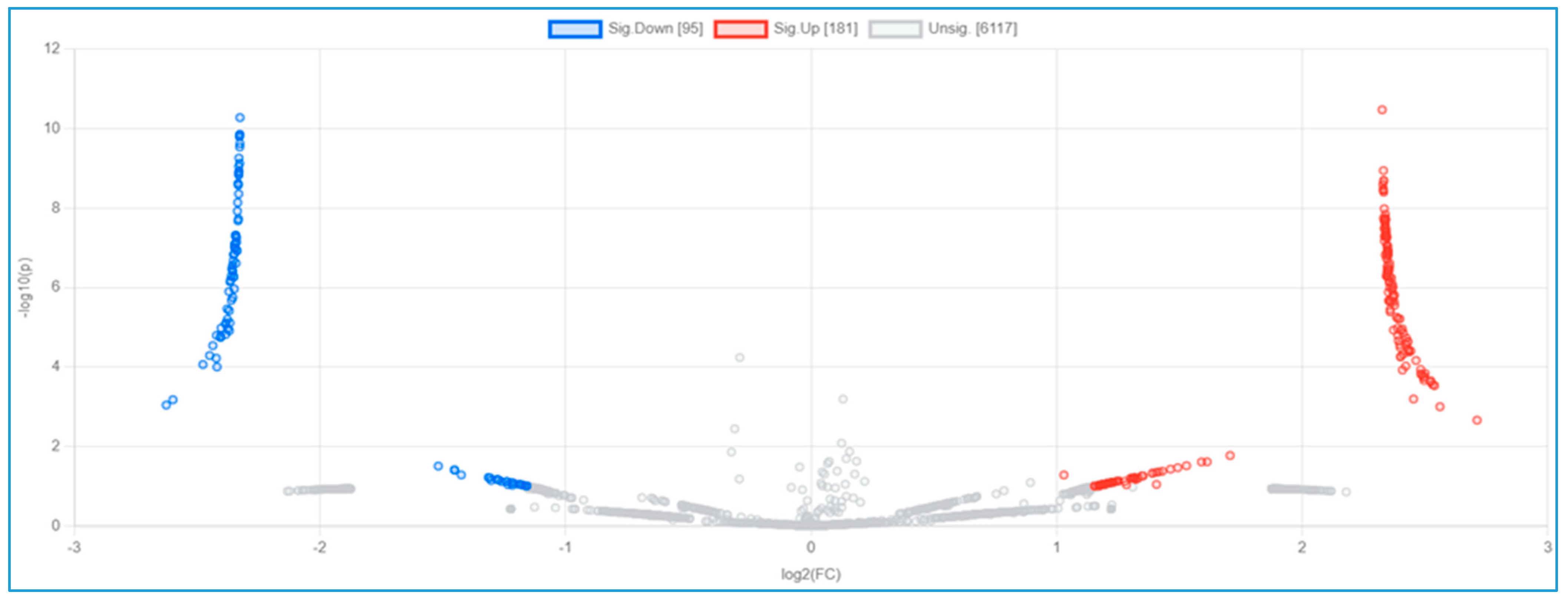
3.3. Serum Proteomic Analysis, Differential Protein Expression, and Potential Mechanisms Related to Identified Proteins in the Non-Obese and Obese Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chew, N.W.S.; Ng, C.H.; Tan, D.J.H.; Kong, G.; Lin, C.; Chin, Y.H.; Lim, W.H.; Huang, D.Q.; Quek, J.; Fu, C.E.; et al. The global burden of metabolic disease: Data from 2000 to 2019. Cell Metab. 2023, 35, 414–428.e3. [Google Scholar] [CrossRef]
- Samblas, M.; Milagro, F.I.; Martínez, A. DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics 2019, 14, 421–444. [Google Scholar] [CrossRef]
- Lomba, A.; Martínez, J.A.; García-Díaz, D.F.; Paternain, L.; Marti, A.; Campión, J.; Milagro, F.I. Weight gain induced by an isocaloric pair-fed high fat diet: A nutriepigenetic study on FASN and NDUFB6 gene promoters. Mol. Genet. Metab. 2010, 101, 273–278. [Google Scholar] [CrossRef]
- Ellulu, M.S.; Patimah, I.; Khaza’ai, H.; Rahmat, A.; Abed, Y. Obesity and inflammation: The linking mechanism and the complications. Arch. Med. Sci. 2017, 13, 851–863. [Google Scholar] [CrossRef]
- Tzanavari, T.; Giannogonas, P.; Karalis, K.P. TNF-alpha and obesity. Curr. Dir. Autoimmun. 2010, 11, 145–156. [Google Scholar] [PubMed]
- Sethi, J.K.; Hotamisligil, G.S. Metabolic Messengers: Tumour necrosis factor. Nat. Metab. 2021, 3, 1302–1312. [Google Scholar] [CrossRef]
- Ramseyer, V.D.; Garvin, J.L. Tumor necrosis factor-α: Regulation of renal function and blood pressure. Am. J. Physiol. Renal Physiol. 2013, 304, F1231–F1242. [Google Scholar] [CrossRef]
- Bollati, V.; Favero, C.; Albetti, B.; Tarantini, L.; Moroni, A.; Byun, H.M.; Motta, V.; Conti, D.M.; Tirelli, A.S.; Vigna, L.; et al. Nutrients intake is associated with DNA methylation of candidate inflammatory genes in a population of obese subjects. Nutrients 2014, 6, 4625–4639. [Google Scholar] [CrossRef]
- Campión, J.; Milagro, F.I.; Goyenechea, E.; Martínez, J.A. TNF-alpha promoter methylation as a predictive biomarker for weight-loss response. Obesity 2009, 17, 1293–1297. [Google Scholar] [CrossRef]
- Aleksandrova, K.; Egea Rodrigues, C.; Floegel, A.; Ahrens, W. Omics biomarkers in obesity: Novel etiological insights and targets for precision prevention. Curr. Obes. Rep. 2020, 9, 219–230. [Google Scholar] [CrossRef]
- Issaq, H.J.; Xiao, Z.; Veenstra, T.D. Serum and plasma proteomics. Chem. Rev. 2007, 107, 3601–3620. [Google Scholar] [CrossRef]
- Cominetti, O.; Galindo, A.N.; Corthésy, J.; Valsesia, A.; Irincheeva, I.; Kussmann, M.; Saris, W.H.M.; Astrup, A.; McPherson, R.; Harper, M.-E.; et al. Obesity shows preserved plasma proteome in large independent clinical cohorts. Sci. Rep. 2018, 8, 16981. [Google Scholar] [CrossRef] [PubMed]
- Zaghlool, S.B.; Kühnel, B.; Elhadad, M.A.; Kader, S.; Halama, A.; Thareja, G.; Engelke, R.; Sarwath, H.; Al-Dous, E.K.; Mohamoud, Y.A.; et al. Epigenetics meets proteomics in an epigenome-wide association study with circulating blood plasma protein traits. Nat. Commun. 2020, 11, 15. [Google Scholar] [CrossRef]
- Lau, F.C.; Bagchi, M.; Sen, C.; Roy, S.; Bagchi, D. Nutrigenomic analysis of diet-gene interactions on functional supplements for weight management. Curr. Genom. 2008, 9, 239–251. [Google Scholar] [CrossRef][Green Version]
- Huang, P.L. A comprehensive definition for metabolic syndrome. Dis. Model. Mech. 2009, 2, 231–237. [Google Scholar] [CrossRef]
- Ng, C. Stratification of BMI categories among older adults within and across countries. Public Health Nutr. 2019, 23, 254–263. [Google Scholar] [CrossRef]
- Kerdsaeng, N.; Roytrakul, S.; Chanprasertyothin, S.; Charernwat, P.; Chansirikarnjana, S.; Sritara, P.; Sirivarasai, J. Serum glycoproteomics and identification of potential mechanisms underlying Alzheimer’s disease. Behav. Neurol. 2021, 2021, 1434076. [Google Scholar] [CrossRef]
- Tansakul, N.; Rattanasrisomporn, J.; Roytrakul, S. Proteomics analysis of serum protein patterns in duck during aflatoxin B1 exposure. Vet. World 2019, 12, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Hermsdorff, H.H.; Mansego, M.L.; Campión, J.; Milagro, F.I.; Zulet, M.A.; Martínez, J.A. TNF-alpha promoter methylation in peripheral white blood cells: Relationship with circulating TNFα, truncal fat and n-6 PUFA intake in young women. Cytokine 2013, 64, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Lim, U.; Song, M.A. Dietary and lifestyle factors of DNA methylation. Methods Mol. Biol. 2012, 863, 359–376. [Google Scholar]
- Cӑtoi, A.F.; Pârvu, A.E.; Andreicuț, A.D.; Mironiuc, A.; Crӑciun, A.; Cӑtoi, C.; Pop, I.D. Metabolically healthy versus unhealthy morbidly obese: Chronic inflammation, nitro-oxidative stress, and insulin resistance. Nutrients 2018, 10, 1199. [Google Scholar] [CrossRef]
- Mazor, R.; Itzhaki, O.; Sela, S.; Yagil, Y.; Cohen-Mazor, M.; Yagil, C.; Kristal, B. Tumor necrosis factor-alpha: A possible priming agent for the polymorphonuclear leukocyte-reduced nicotinamide-adenine dinucleotide phosphate oxidase in hypertension. Hypertension 2010, 55, 353–362. [Google Scholar] [CrossRef]
- Akash, M.S.H.; Rehman, K.; Liaqat, A. Tumor necrosis factor-alpha: Role in development of insulin resistance and pathogenesis of type 2 diabetes mellitus. J. Cell. Biochem. 2018, 119, 105–110. [Google Scholar] [CrossRef]
- Du, X.; Liu, M.; Tai, W.; Yu, H.; Hao, X.; Loor, J.J.; Jiang, Q.; Fang, Z.; Gao, X.; Fan, M.; et al. Tumor necrosis factor-α promotes lipolysis and reduces insulin sensitivity by activating nuclear factor kappa B and c-Jun N-terminal kinase in primary bovine adipocytes. J. Dairy Sci. 2022, 105, 8426–8438. [Google Scholar] [CrossRef]
- Schwedhelm, C.; Pischon, T.; Rohrmann, S.; Himmerich, H.; Linseisen, J.; Nimptsch, K. Plasma inflammation markers of the tumor necrosis factor pathway but not C-reactive protein are associated with processed meat and unprocessed red meat consumption in Bavarian adults. J. Nutr. 2017, 147, 78–85. [Google Scholar] [CrossRef]
- Sajadi Hezaveh, Z.; Sikaroudi, M.K.; Vafa, M.; Clayton, Z.S.; Soltani, S. Effect of egg consumption on inflammatory markers: A systematic review and meta-analysis of randomized controlled clinical trials. J. Sci. Food Agric. 2019, 99, 6663–6670. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, M.N.; Valenzuela, F.; Robles, A.E.; Artalejo, E.; Aguilar, D.; Andersen, C.J.; Valdez, H.; Fernandez, M.L. One egg per day improves inflammation when compared to an oatmeal-based breakfast without increasing other cardiometabolic risk factors in diabetic patients. Nutrients 2015, 7, 3449–3463. [Google Scholar] [CrossRef] [PubMed]
- Gariballa, S.; Al-Bluwi, G.S.M.; Yasin, J. Increased fruit and vegetable consumption mitigates oxidative damage and associated inflammatory response in obese subjects independent of body weight change. Nutrients 2023, 15, 1638. [Google Scholar] [CrossRef] [PubMed]
- Hermsdorff, H.H.; Zulet, M.A.; Puchau, B.; Martínez, J.A. Fruit and vegetable consumption and proinflammatory gene expression from peripheral blood mononuclear cells in young adults: A translational study. Nutr. Metab. 2010, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Quetglas-Llabrés, M.M.; Monserrat-Mesquida, M.; Bouzas, C.; Mateos, D.; Ugarriza, L.; Gómez, C.; Tur, J.A.; Sureda, A. Oxidative stress and inflammatory biomarkers are related to high intake of ultra-processed food in old adults with metabolic syndrome. Antioxidants 2023, 12, 1532. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Barchitta, M. How dietary factors affect DNA methylation: Lesson from epidemiological studies. Medicina 2020, 56, 374. [Google Scholar] [CrossRef]
- Clemente-Suárez, V.J.; Martín-Rodríguez, A.; Redondo-Flórez, L.; López-Mora, C.; Yáñez-Sepúlveda, R.; Tornero-Aguilera, J.F. New insights and potential therapeutic interventions in metabolic diseases. Int. J. Mol. Sci. 2023, 24, 10672. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984–2009. [Google Scholar] [CrossRef] [PubMed]
- Savova, M.S.; Mihaylova, L.V.; Tews, D.; Wabitsch, M.; Georgiev, M.I. Targeting PI3K/AKT signaling pathway in obesity. Biomed. Pharmacother. 2023, 159, 114244. [Google Scholar] [CrossRef] [PubMed]
- Ihanus, E.; Uotila, L.M.; Toivanen, A.; Varis, M.; Gahmberg, C.G. Red-cell ICAM-4 is a ligand for the monocyte/macrophage integrin CD11c/CD18: Characterization of the binding sites on ICAM-4. Blood 2007, 109, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.L.; Foster-Schubert, K.E.; Makar, K.W.; Kratz, M.; Hagman, D.; Schur, E.A.; Habermann, N.; Horton, M.; Abbenhardt, C.; Kuan, L.Y.; et al. Gene expression changes in adipose tissue with diet- and/or exercise-induced weight loss. Cancer Prev. Res. 2013, 6, 217–231. [Google Scholar] [CrossRef]
- Pomerantz, J.L.; Denny, E.M.; Baltimore, D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 2002, 21, 5184–5194. [Google Scholar] [CrossRef]
- Hildebrandt, X.; Ibrahim, M.; Peltzer, N. Cell death and inflammation during obesity: “Know my methods, WAT(son)”. Cell Death Differ. 2023, 30, 279–292. [Google Scholar] [CrossRef]
- Pan, C.; Lei, Z.; Wang, S.; Wang, X.; Wei, D.; Cai, X.; Luoreng, Z.; Wang, l.; Ma, Y. Genome-wide identification of cyclin-dependent kinase (CDK) genes affecting adipocyte differentiation in cattle. BMC Genom. 2021, 22, 532. [Google Scholar] [CrossRef]
- Cianfanelli, V.; Nazio, F.; Cecconi, F. Connecting autophagy: AMBRA1 and its network of regulation. Mol. Cell. Oncol. 2015, 2, e970059. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lyu, Y.; Li, J.; Wang, X. AMBRA1 and its role as a target for anticancer therapy. Front. Oncol. 2022, 12, 946086. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liu, F.; Li, M.; He, J.; Huang, J.; Rao, D.C.; Hixson, J.E.; Gu, C.; Kelly, T.N.; Chen, S.; et al. Associations of variants in the CACNA1A and CACNA1C genes with longitudinal blood pressure changes and hypertension incidence: The GenSalt Study. Am. J. Hypertens. 2016, 29, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Newton-Cheh, C.; Chasman, D.I.; Ehret, G.B.; Johnson, T.; Rose, L.; Rice, K.; Verwoert, G.; Launer, L.J.C.; Gudnason, V.; et al. Cohorts for heart and aging research in genomic epidemiology consortium; Global BPgen Consortium; Women’s Genome Health Study. Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension 2011, 57, 903–910. [Google Scholar] [CrossRef]
- Deng, T.; Lyon, C.J.; Minze, J.; Lin, J.; Zou, J.; Liu, J.Z.; Ren, Y.; Yin, Z.; Hamilton, D.J.; Reardon, P.R.; et al. Class II major histocompatibility complex plays an essential role in obesity-induced adipose inflammation. Cell Metab. 2013, 17, 411–422. [Google Scholar] [CrossRef]
| Characteristics | Total (N = 233) | Non-Obese Group (N = 100) | Obese Group (N = 133) |
|---|---|---|---|
| Age (years) | 57.70 ± 1.41 | 57.68 ± 1.41 | 57.71 ± 1.42 |
| BMI (kg/m2) | 25.46 ± 4.30 | 21.47 ± 1.03 | 28.46 ± 3.27 a |
| Waist circumference (cm.) | 92.9 ± 10.87 | 83.38 ± 5.01 | 100.07 ± 8.29 a |
| SBP (mmHg) | 138.04 ± 16.90 | 133.70 ± 16.20 | 141.30 ± 16.72 a |
| DBP (mmHg) | 79.00 ± 9.45 | 76.21 ± 9.02 | 81.10 ± 9.25 a |
| Smoking, n (%) | |||
| 109 (46.8%) | 52 (52.0%) | 57 (42.9%) |
| 31 (13.3%) | 16 (16.0%) | 15 (11.3%) |
| 93 (39.9%) | 32 (32.0%) | 61 (45.9%) |
| Alcohol consumption, n (%) | |||
| 50 (21.5%) | 24 (24.0%) | 26 (19.5%) |
| 125 (53.6%) | 51 (51.0%) | 74 (55.6%) |
| 58 (24.9%) | 25 (25.0%) | 33 (24.9%) |
| FPG (mg/dL) | 103.06 ± 21.34 | 94.25 ± 11.89 | 109.69 ± 24.32 a |
| HbA1c (%) | 5.98 ± 0.88 | 5.58 ± 0.511 | 6.29 ± 0.98 a |
| TC (mg/dL) | 199.14 ± 43.13 | 201.94 ± 40.81 | 194.03 ± 44.26 |
| TG (mg/dL) | 156.03 ± 108.16 | 126.80 ± 65.49 | 169.30 ± 79.50 a |
| LDL-C (mg/dL) | 131.30 ± 40.23 | 123.10 ± 41.39 | 136.89 ± 38.12 a |
| HDL-C (mg/dL) | 52.39 ± 13.16 | 57.04 ± 13.37 | 48.89 ± 11.90 a |
| ALT (U/L) | 27.24 ± 14.83 | 23.58 ± 13.32 | 30.07 ± 15.35 a |
| AST (U/L) | 25.35 ± 12.39 | 25.02 ± 14.98 | 25.60 ± 10.08 |
| BUN (mg/dL) | 13.57 ± 3.23 | 13.47 ± 3.50 | 13.64 ± 3.03 |
| Creatinine (mg/dL) | 1.01 ± 0.16 | 0.98 ± 0.13 | 1.04 ± 0.18 |
| Study Group | DNA Methylation of TNFα | |||||
|---|---|---|---|---|---|---|
| Position 1 | Position 2 | Position 3 | Position 4 | Total | ||
| Non-obese group (N = 100) | 9.37 ± 2.21 | 8.14 ± 1.96 | 17.46 ± 3.89 | 23.16 ± 4.01 | 63.47 ± 8.99 | |
| Obese group (N = 133) | 8.14 ± 2.01 a | 12.35 ± 3.12 a | 16.12 ± 4.08 a | 21.53 ± 3.97 a | 58.11 ± 10.22 a | |
| Metabolic syndrome-components | ||||||
| Waist circumference | ||||||
| Non-obese group | <90 cm (N = 62) | 10.36 ± 2.24 | 13.20 ± 2.08 | 16.35 ± 3.58 | 21.32 ± 4.69 | 62.36 ± 8.74 |
| ≥90 cm (N = 38) | 9.33 ± 5.69 | 14.59 ± 3.14 | 17.02 ± 5.04 | 23.04 ± 5.07 | 61.99 ± 9.63 | |
| Obese group | <90 cm (N = 49) | 8.99 ± 2.32 | 13.01 ± 4.02 | 17.25 ± 3.08 | 23.44 ± 3.99 | 62.48 ± 8.69 |
| ≥90 cm (N = 84) | 7.61 ± 2.14 | 11.05 ± 3.57 a | 15.37 ± 3.64 a | 21.09 ± 4.04 a | 57.96 ± 10.58 a | |
| HDL-cholesterol | ||||||
| Non-obese group | <40 mg/dL (N = 83) | 9.25 ± 3.95 | 13.33 ± 3.63 | 17.39 ± 4.16 | 22.91 ± 4.69 | 62.89 ± 12.62 |
| ≥40 mg/dL (N = 10) | 11.03 ± 2.85 | 15.21 ± 2.88 | 18.44 ± 4.57 | 26.42 ± 2.17 | 71.12 ± 7.35 | |
| Obese group | <40 mg/dL (N = 103) | 8.01 ± 1.85 | 12.22 ± 2.71 | 16.03 ± 3.21 | 21.75 ± 4.26 | 58.02 ± 10.22 |
| ≥40 mg/dL (N = 30) | 8.58 ± 3.02 | 12.72 ± 3.49 | 16.40 ± 3.48 | 20.69 ± 4.67 | 58.41 ± 11.06 | |
| Triglyceride | ||||||
| Non-obese group | <150 mg/dL (N = 73) | 9.47 ± 4.12 | 13.35 ± 3.67 | 17.74 ± 4.27 | 23.11 ± 4.66 | 63.39 ± 12.95 |
| ≥150 mg/dL (N = 26) | 9.24 ± 3.28 | 13.91 ± 3.43 | 15.46 ± 3.76 a | 20.19 ± 4.69 a | 61.07 ± 11.06 a | |
| Obese group | <150 mg/dL (N = 71) | 8.52 ± 2.47 | 12.97 ± 3.04 | 16.46 ± 3.52 | 22.45 ± 4.84 | 60.41 ± 11.45 |
| ≥150 mg/dL (N = 62) | 7.70 ± 1.67 a | 11.59 ± 2.54 a | 15.72 ± 2.92 | 20.43 ± 3.48 a | 55.46 ± 8.32 a | |
| High blood pressure (SBP ≥ 130 or DBP ≥ 85 mmHg | ||||||
| Non-obese group | No (N = 45) | 8.58 ± 3.66 | 13.41 ± 3.92 | 17.36 ± 3.88 | 22.96 ± 4.82 | 62.33 ± 11.98 |
| Yes (N = 55) | 10.02 ± 4.00 | 13.50 ± 3.35 | 17.55 ± 4.43 | 23.3 ± 4.52 | 64.39 ± 12.90 | |
| Obese group | No (N = 30) | 8.20 ± 2.18 | 12.33 ± 2.80 | 16.26 ± 3.22 | 21.63 ± 4.36 | 58.44 ± 10.01 |
| Yes (N = 103) | 7.92 ± 2.15 | 12.32 ± 3.25 | 15.64 ± 3.41 | 19.46 ± 4.43 | 55.34 ± 11.67 a | |
| Fasting plasma glucose | ||||||
| Non-obese group | <110 mg/dL (N = 85) | 9.29 ± 3.89 | 13.39 ± 3.54 | 17.74 ± 4.25 | 23.03 ± 4.65 | 63.18 ± 12.39 |
| ≥110 mg/dL (N = 15) | 10.95 ± 4.14 | 14.72 ± 5.03 | 17.70 ± 2.64 | 25.49 ± 4.07 | 64.88 ± 14.40 | |
| Obese group | <110 mg/dL (N = 85) | 8.09 ± 2.24 | 14.97 ± 2.91 | 16.17 ± 3.33 | 24.47 ± 4.45 | 61.36 ± 10.62 |
| ≥110 mg/dL (N = 48) | 8.21 ± 2.04 | 12.21 ± 2.89 a | 16.02 ± 3.15 | 21.79 ± 4.24 a | 58.25 ± 10.02 a | |
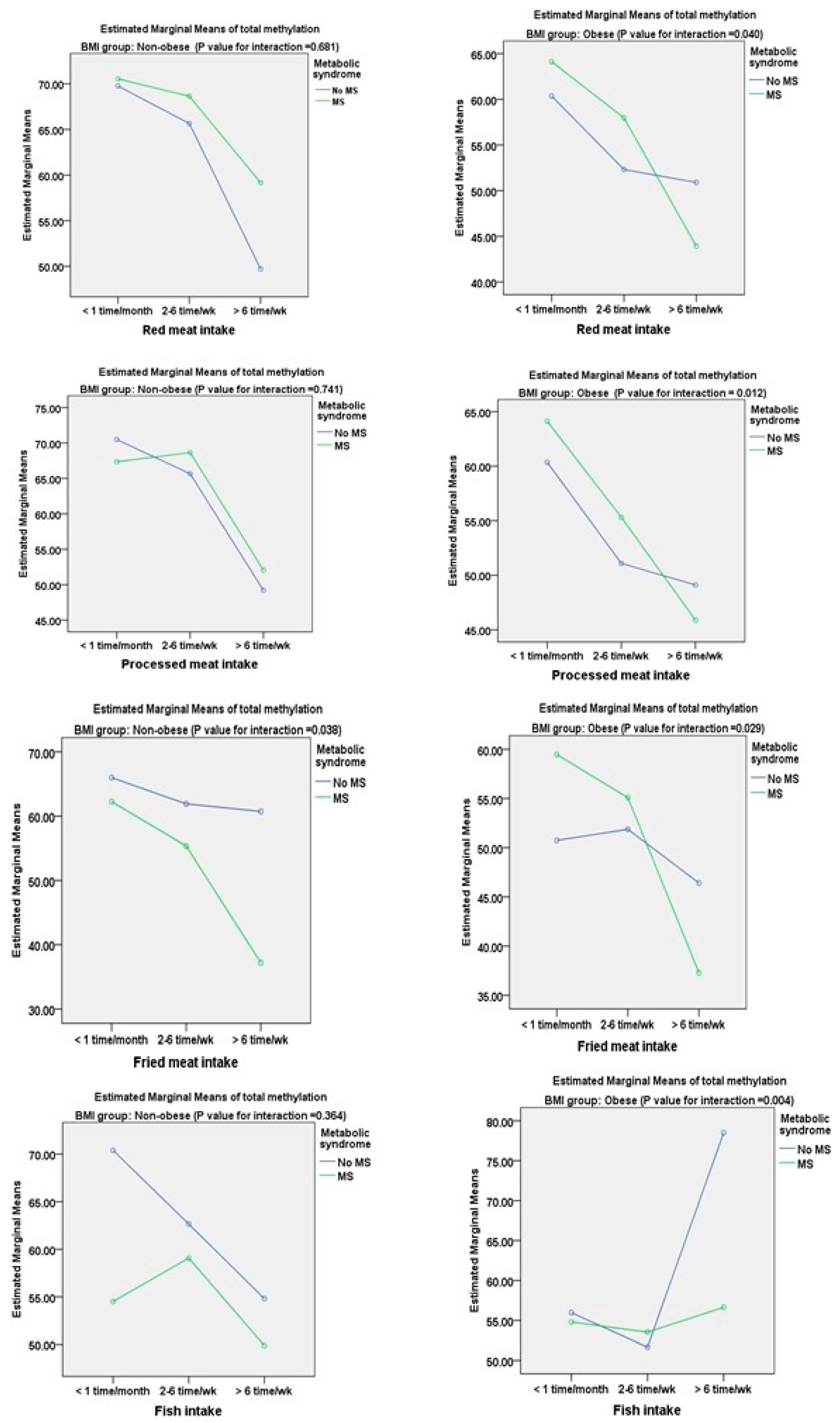
| Dietary Variables | Univariate Regression Analysis | Multivariate Regression Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| Non-Obese Group (N = 100) | Obese Group (N = 133) | Non-Obese Group (N = 100) | Obese Group (N = 133) | |||||
| β | p-Value * | β | p-Value * | β | p-Value * | β | p-Value * | |
| Frequency of red meat per week | ||||||||
| 0–3 times | 0 | (<0.0001) | 0 | (0.005) | 0 | (0.057) | 0 | (0.024) |
| 4–6 times | −3.98 | <0.0001 | −4.68 | 0.361 | −1.21 | 0.071 | −2.36 | 0.124 |
| >6 times | −12.05 | <0.0001 | −11.14 | <0.0001 | −3.65 | 0.084 | −10.25 | <0.0001 |
| Frequency of process meat per week | ||||||||
| 0–3 times | 0 | (0.034) | 0 | (0.027) | 0 | (0.014) | 0 | (0.039) |
| 4–6 times | −4.66 | 0.116 | −2.29 | 0.417 | −3.09 | 0.119 | −1.47 | 0.097 |
| >6 times | −7.61 | <0.0001 | −3.84 | 0.038 | −5.23 | 0.001 | −4.96 | 0.003 |
| Frequency of fried meat per week | ||||||||
| 0–3 times | 0 | (0.025) | 0 | (0.039) | 0 | (0.127) | 0 | (0.029) |
| 4–6 times | −1.64 | 0.458 | −2.81 | 0.660 | −0.98 | 0.233 | −1.29 | 0.068 |
| >6 times | −12.79 | 0.004 | −11.16 | 0.006 | −1.07 | 0.057 | −8.47 | 0.001 |
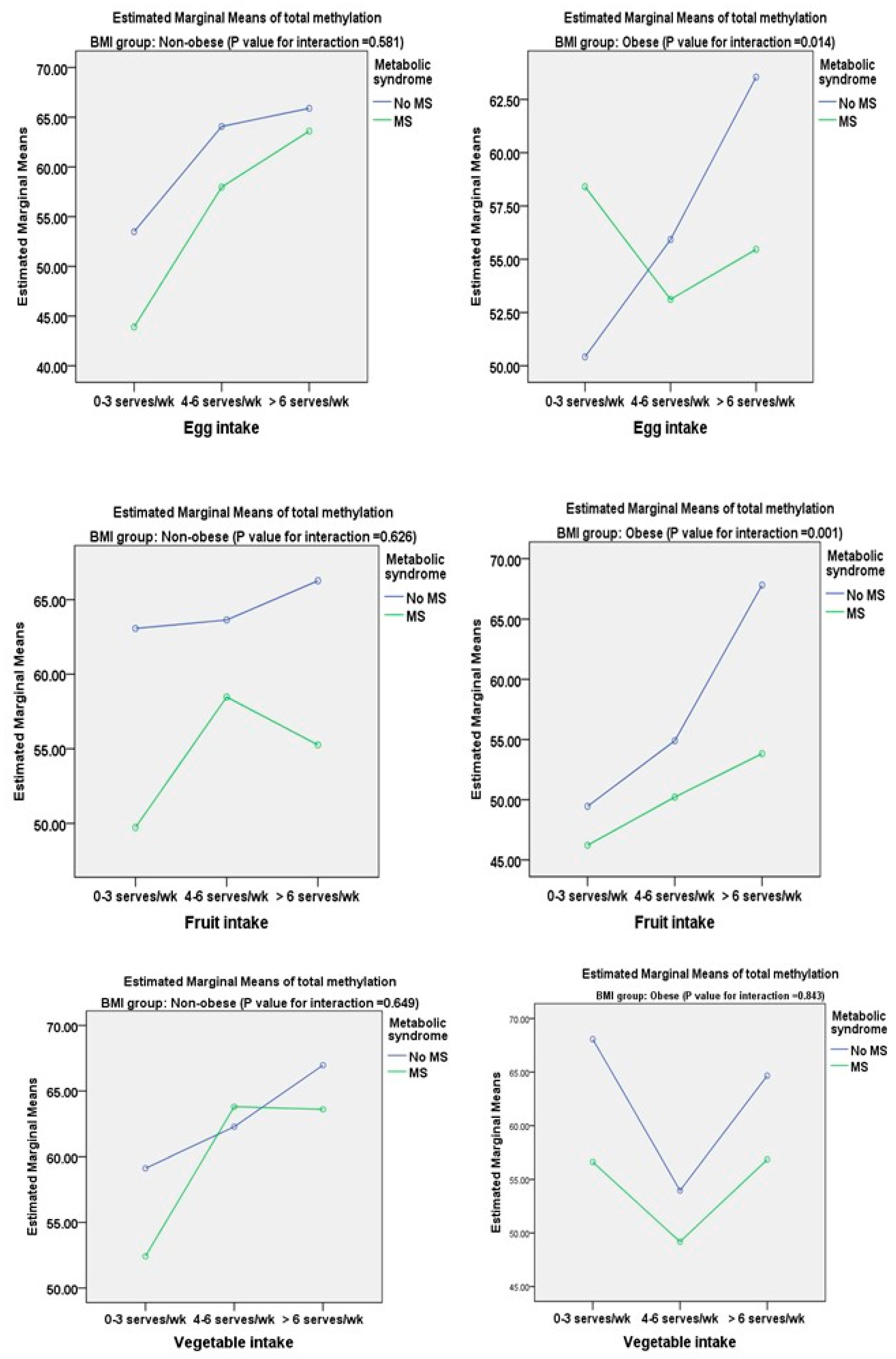
| Frequency of fish per week | ||||||||
| 0–3 times | 0 | (0.124) | 0 | (0.218) | 0 | (0.214) | 0 | (0.067) |
| 4–6 times | −2.43 | 0.362 | −0.85 | 0.737 | 1.96 | 0.198 | 1.14 | 0.314 |
| >6 times | 9.43 | 0.039 | 2.63 | 0.410 | 3.58 | 0.207 | 1.39 | 0.219 |
| Frequency of egg per week | ||||||||
| 0–3 servings | 0 | (0.207) | 0 | (0.011) | 0 | (0.127) | 0 | (0.041) |
| 4–6 servings | 1.18 | 0.865 | 7.66 | 0.002 | 0.95 | 0.141 | 3.04 | 0.053 |
| >6 servings | 4.68 | 0.478 | 6.78 | 0.012 | 2.14 | 0.097 | 5.99 | 0.041 |
| Frequency of fruits per week | ||||||||
| 0–3 servings | 0 | (0.008) | 0 | (0.019) | 0 | (0.039) | 0 | (0.017) |
| 4–6 servings | 4.34 | 0.062 | 8.57 | 0.001 | 2.36 | 0.612 | 6.32 | 0.001 |
| >6 servings | 7.17 | 0.014 | 9.32 | <0.0001 | 4.96 | 0.027 | 7.18 | <0.0001 |
| Frequency of vegetables per week | ||||||||
| 0–3 servings | 0 | (0.011) | 0 | (0.007) | 0 | (0.041) | 0 | (0.022) |
| 4–6 servings | 4.91 | 0.103 | 2.50 | 0.378 | 1.98 | 0.078 | 2.09 | 0.244 |
| >6 servings | 5.07 | 0.042 | 11.24 | <0.0001 | 4.67 | 0.033 | 8.04 | 0.002 |
| Protein ID | Protein Names | Gene Names | Gene Ontology (Biological Process) | log2 (FC) | p-Value |
|---|---|---|---|---|---|
| U5U6J5 | Intercellular adhesion molecule 4 | ICAM4 | Cell adhesion [GO:0007155] | 2.711 | 0.0022 |
| B7Z339 | Phosphatase and actin regulator | 2.560 | 0.0010 | ||
| Q8NEZ4 | Histone-lysine N-methyltransferase 2C | KMT2C | Methylation [GO:0032259]; positive regulation of transcription by RNA polymerase II [GO:0045944]; | 2.538 | 0.0003 |
| Q96RT7 | Gamma-tubulin complex component 6 (GCP-6) | TUBGCP6 | Cytoplasmic microtubule organization [GO:0031122] | 2.531 | 0.0003 |
| Q9BXL7 | Caspase recruitment domain-containing protein 11 | CARD11 | Positive regulation of canonical NF-kappa b signal transduction; regulation of apoptotic process | 2.523 | 0.0002 |
| K7EML2 | RNA binding motif protein 42 | RBM42 | 2.522 | 0.0002 | |
| B1APH4 | Putative zinc finger protein 487 | ZNF487 | Negative regulation of transcription by RNA polymerase II | 2.500 | 0.0001 |
| A0A1C7CYW9 | Tubulin tyrosine ligase-like 8 | TTLL8 | Protein modification process | 2.500 | 0.0002 |
| F8WDP7 | Cyclin-dependent kinase 15 | CDK15 | 2.496 | 0.0002 | |
| Q96M60 | Protein FAM227B | FAM227B | 2.492 | 0.0002 | |
| Q9P2K1 | Coiled-coil and C2 domain-containing protein 2A | CC2D2A | Axoneme assembly; smoothened signaling pathway | −2.6249 | 0.0009 |
| A0A075B6T1 | Autophagy and beclin 1 regulator 1 | AMBRA1 | −2.5975 | 0.0007 | |
| A0A590UJK2 | Voltage-dependent P/Q-type calcium channel subunit alpha | CACNA1A | Regulation of monoatomic ion transmembrane transport | −2.4755 | 0.0001 |
| Q7L0X2 | Glutamate-rich protein 6 | ERICH6 | −2.4482 | 0.0001 | |
| Q8N523 | Tuftelin-interacting protein 11 | TFIP11 | Biomineral tissue development | −2.4352 | 0.0000 |
| A5YKK5 | KIAA0232 | KIAA0232 | −2.4214 | 0.0001 | |
| G3V533 | Spectrin repeat-containing nuclear envelope family member 3 | SYNE3 | −2.4202 | 0.0000 | |
| A0A2H4G345 | MHC class II antigen | HLA-DQB1 | Antigen processing and presentation immune response | −2.4183 | 0.0001 |
| D6RGG8 | Nicotinamide nucleotide adenylyltransferase 3 | NMNAT3 | Biosynthetic process | −2.4039 | 0.0000 |
| A0A3B3IRX3 | Mediator of RNA polymerase II transcription subunit 13 | MED13L | Regulation of transcription by RNA polymerase II | −2.4033 | 0.0000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boonrong, C.; Roytrakul, S.; Shantavasinkul, P.C.; Sritara, P.; Sirivarasai, J. Role of Dietary Factors on DNA Methylation Levels of TNF-Alpha Gene and Proteome Profiles in Obese Men. Nutrients 2024, 16, 877. https://doi.org/10.3390/nu16060877
Boonrong C, Roytrakul S, Shantavasinkul PC, Sritara P, Sirivarasai J. Role of Dietary Factors on DNA Methylation Levels of TNF-Alpha Gene and Proteome Profiles in Obese Men. Nutrients. 2024; 16(6):877. https://doi.org/10.3390/nu16060877
Chicago/Turabian StyleBoonrong, Chayanisa, Sittiruk Roytrakul, Prapimporn Chattranukulchai Shantavasinkul, Piyamitr Sritara, and Jintana Sirivarasai. 2024. "Role of Dietary Factors on DNA Methylation Levels of TNF-Alpha Gene and Proteome Profiles in Obese Men" Nutrients 16, no. 6: 877. https://doi.org/10.3390/nu16060877
APA StyleBoonrong, C., Roytrakul, S., Shantavasinkul, P. C., Sritara, P., & Sirivarasai, J. (2024). Role of Dietary Factors on DNA Methylation Levels of TNF-Alpha Gene and Proteome Profiles in Obese Men. Nutrients, 16(6), 877. https://doi.org/10.3390/nu16060877







