A Cross-Sectional Quantitative Metabolomics Study Evidencing the Metabolic Signature in Six Organs during a 14-Week High-Fat High-Sucrose and Standard Diet in Mice
Abstract
1. Introduction
2. Materials and Methods
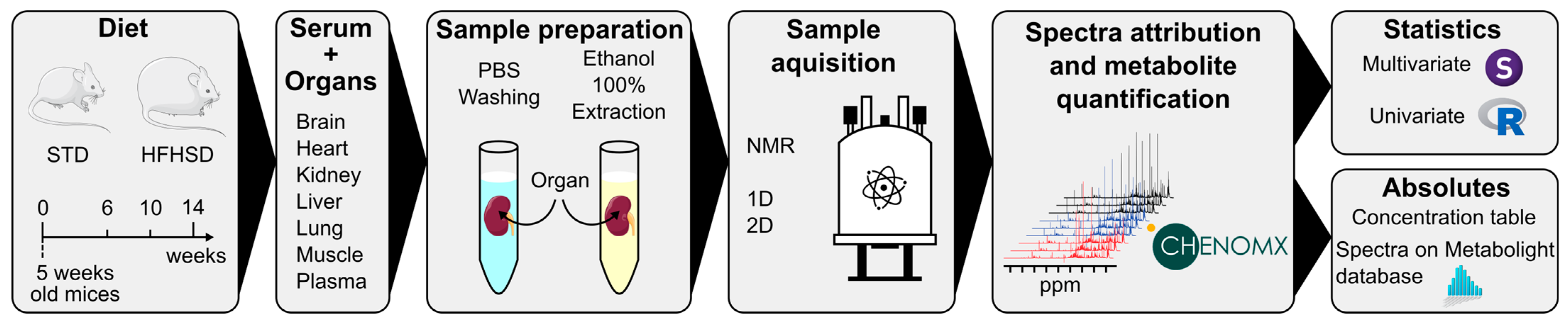
2.1. Animal Protocol and Sample Collection
2.2. Metabolite Extraction and Sample Preparation for NMR Analysis
2.3. NMR Spectra Analysis
2.4. Data Processing
2.5. Statistical Analysis
3. Results
3.1. Experimental Design
3.1.1. Phenotype of the HFHSD-Induced Obese Mice
3.1.2. NMR Determination of Organ and Serum Metabolic Profiles
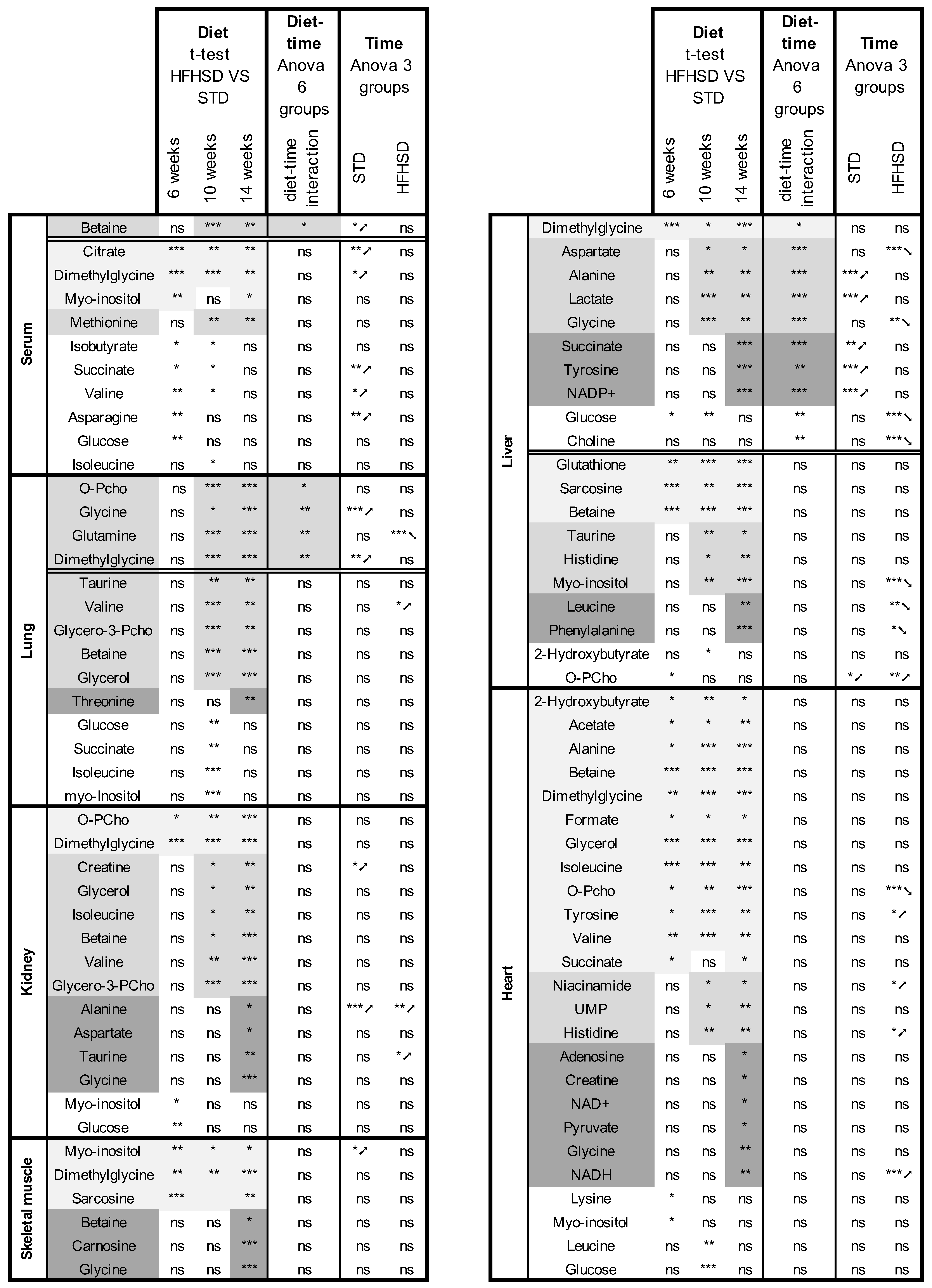
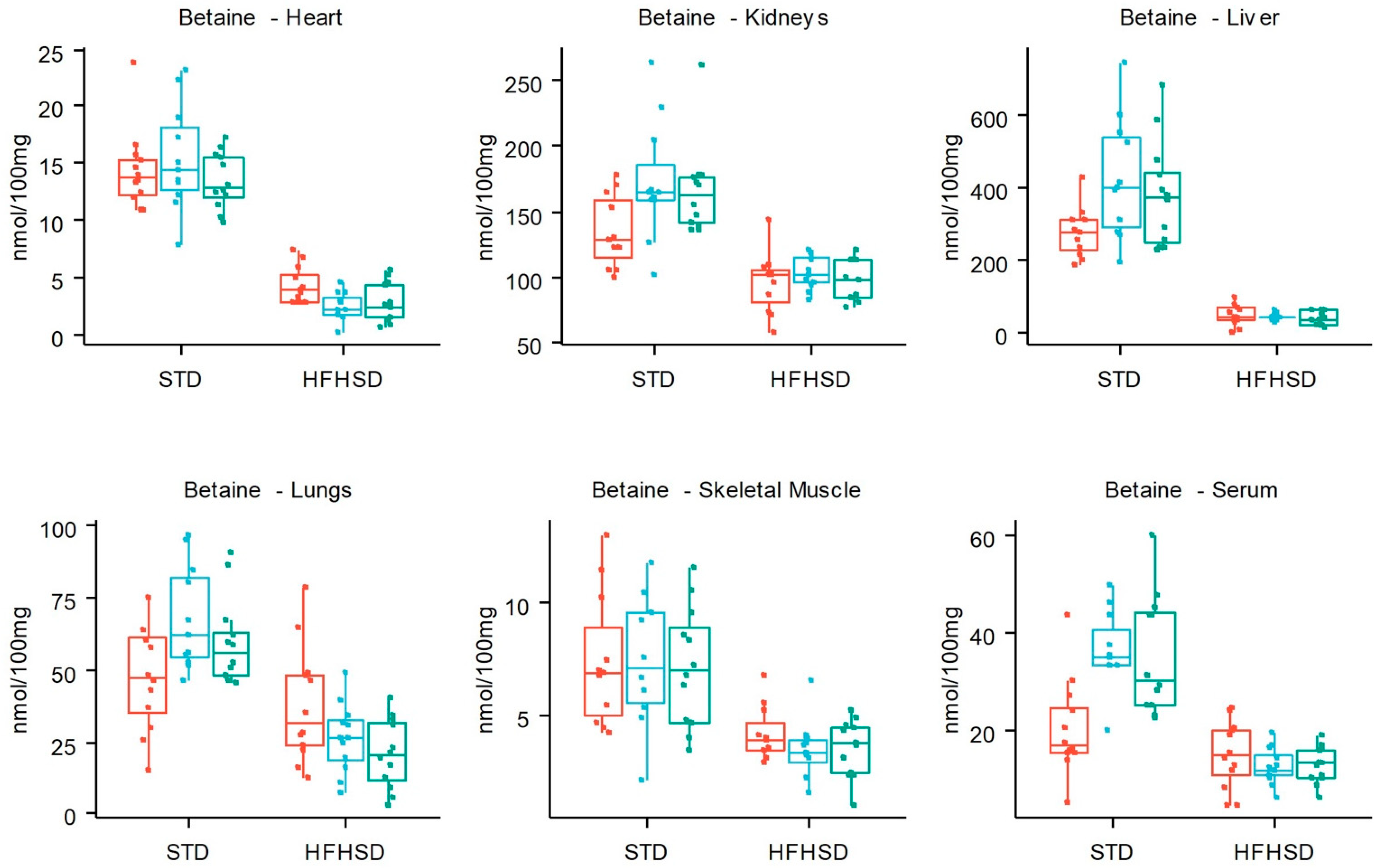
3.2. HFHSD-Induced Metabolic Alterations
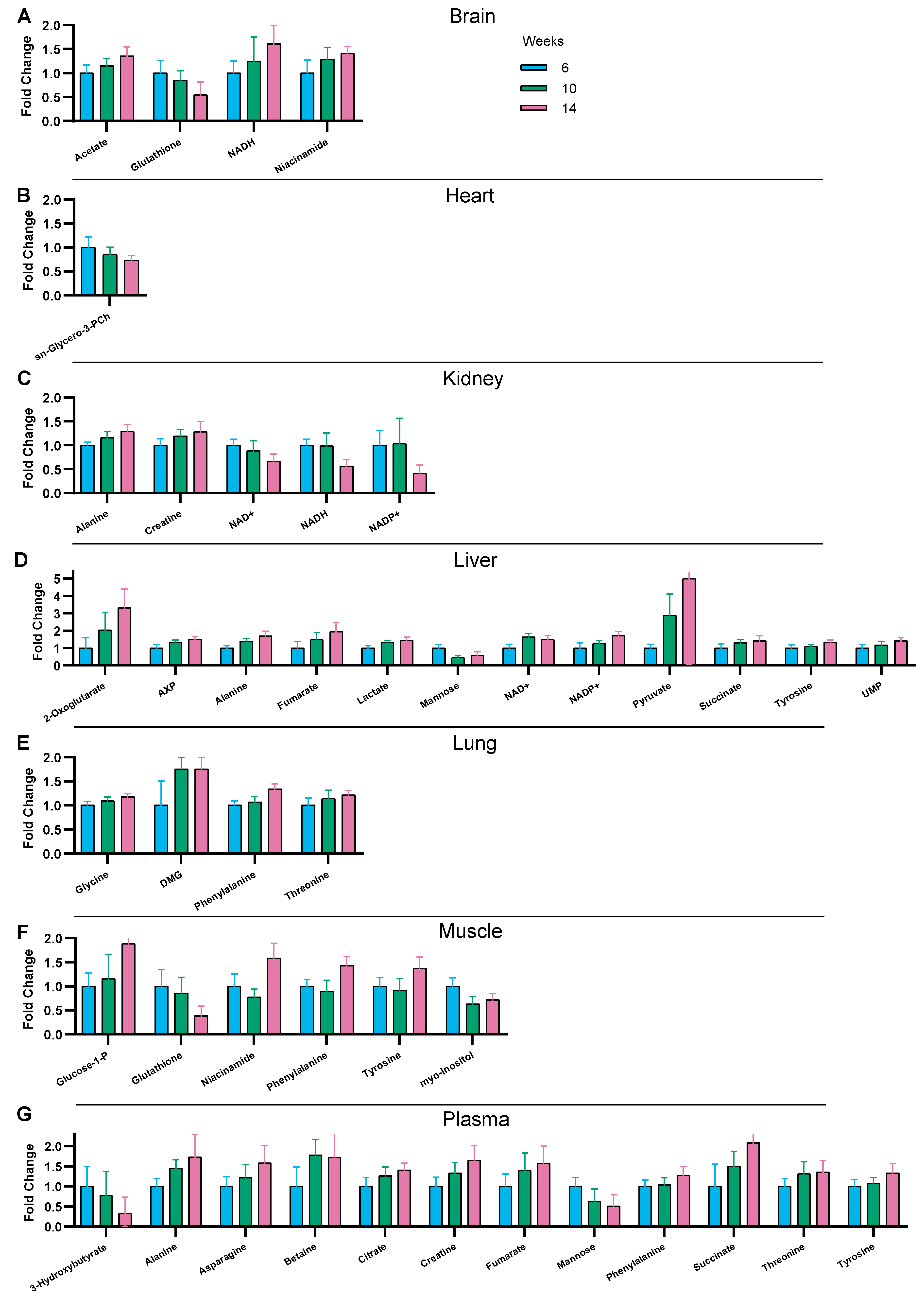
3.3. Time-Related Metabolite Evolution over 14 Weeks of Diet
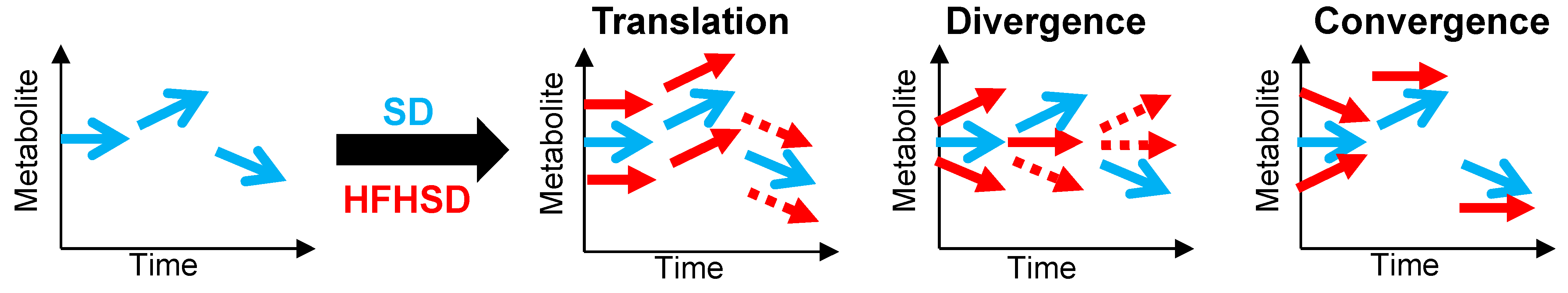
3.4. Interaction between Diet and Time in the Juvenile-to-Adult Transition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- SDGs Sustainable Development Goals. Available online: https://cdn.who.int/media/docs/default-source/gho-documents/world-health-statistic-reports/2023/world-health-statistics-2023_20230519_pdf (accessed on 8 March 2024).
- Virtue, S.; Vidal-Puig, A. Adipose Tissue Expandability, Lipotoxicity and the Metabolic Syndrome—An Allostatic Perspective. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2010, 1801, 338–349. [Google Scholar] [CrossRef]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef]
- Neeland, I.J.; Poirier, P.; Després, J.-P. Cardiovascular and Metabolic Heterogeneity of Obesity. Circulation 2018, 137, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Hazart, J.; Montel, F.; Gentes, E.; Lahaye, C.; Pouget, M.; Farigon, N.; Miolanne, M.; Mulliez, A.; Boirie, Y. Body Mass Trajectory Affects the Long-Term Occurrence of Metabolic Syndrome in Adult Patients with Severe Obesity. Children 2022, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Bautista, R.; Alarcón-Aguilar, F.; Del, C. Escobar-Villanueva, M.; Almanza-Pérez, J.; Merino-Aguilar, H.; Fainstein, M.; López-Diazguerrero, N. Biochemical Alterations during the Obese-Aging Process in Female and Male Monosodium Glutamate (MSG)-Treated Mice. Int. J. Mol. Sci. 2014, 15, 11473–11494. [Google Scholar] [CrossRef] [PubMed]
- Showalter, M.R.; Nonnecke, E.B.; Linderholm, A.L.; Cajka, T.; Sa, M.R.; Lönnerdal, B.; Kenyon, N.J.; Fiehn, O. Obesogenic Diets Alter Metabolism in Mice. PLoS ONE 2018, 13, e0190632. [Google Scholar] [CrossRef]
- Mora-Ortiz, M.; Nuñez Ramos, P.; Oregioni, A.; Claus, S.P. NMR Metabolomics Identifies over 60 Biomarkers Associated with Type II Diabetes Impairment in Db/Db Mice. Metabolomics 2019, 15, 89. [Google Scholar] [CrossRef]
- Pietiläinen, K.H.; Sysi-Aho, M.; Rissanen, A.; Seppänen-Laakso, T.; Yki-Järvinen, H.; Kaprio, J.; Orešič, M. Acquired Obesity Is Associated with Changes in the Serum Lipidomic Profile Independent of Genetic Effects—A Monozygotic Twin Study. PLoS ONE 2007, 2, e218. [Google Scholar] [CrossRef]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A Branched-Chain Amino Acid-Related Metabolic Signature That Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, J.Y.; Kim, O.Y.; Ham, B.M.; Kim, H.-J.; Kwon, D.Y.; Jang, Y.; Lee, J.H. Metabolic Profiling of Plasma in Overweight/Obese and Lean Men Using Ultra Performance Liquid Chromatography and Q-TOF Mass Spectrometry (UPLC−Q-TOF MS). J. Proteome Res. 2010, 9, 4368–4375. [Google Scholar] [CrossRef]
- Zeng, M.; Liang, Y.; Li, H.; Wang, M.; Wang, B.; Chen, X.; Zhou, N.; Cao, D.; Wu, J. Plasma Metabolic Fingerprinting of Childhood Obesity by GC/MS in Conjunction with Multivariate Statistical Analysis. J. Pharm. Biomed. Anal. 2010, 52, 265–272. [Google Scholar] [CrossRef]
- Wang, C.; Feng, R.; Sun, D.; Li, Y.; Bi, X.; Sun, C. Metabolic Profiling of Urine in Young Obese Men Using Ultra Performance Liquid Chromatography and Q-TOF Mass Spectrometry (UPLC/Q-TOF MS). J. Chromatogr. B 2011, 879, 2871–2876. [Google Scholar] [CrossRef] [PubMed]
- Mihalik, S.J.; Michaliszyn, S.F.; de las Heras, J.; Bacha, F.; Lee, S.; Chace, D.H.; DeJesus, V.R.; Vockley, J.; Arslanian, S.A. Metabolomic Profiling of Fatty Acid and Amino Acid Metabolism in Youth with Obesity and Type 2 Diabetes. Diabetes Care 2012, 35, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Morio, B.; Comte, B.; Martin, J.-F.; Chanseaume, E.; Alligier, M.; Junot, C.; Lyan, B.; Boirie, Y.; Vidal, H.; Laville, M.; et al. Metabolomics Reveals Differential Metabolic Adjustments of Normal and Overweight Subjects during Overfeeding. Metabolomics 2015, 11, 920–938. [Google Scholar] [CrossRef]
- Asgharpour, A.; Cazanave, S.C.; Pacana, T.; Seneshaw, M.; Vincent, R.; Banini, B.A.; Kumar, D.P.; Daita, K.; Min, H.-K.; Mirshahi, F.; et al. A Diet-Induced Animal Model of Non-Alcoholic Fatty Liver Disease and Hepatocellular Cancer. J. Hepatol. 2016, 65, 579–588. [Google Scholar] [CrossRef]
- Rautureau, G.J.P.; Morio, B.; Guibert, S.; Lefevre, C.; Perrier, J.; Alves, A.; Chauvin, M.A.; Pinteur, C.; Monet, M.A.; Godet, M.; et al. Dietary Obesity in Mice Is Associated with Lipid Deposition and Metabolic Shifts in the Lungs Sharing Features with the Liver. Sci. Rep. 2021, 11, 8712. [Google Scholar] [CrossRef]
- Rangel-Huerta, O.D.; Pastor-Villaescusa, B.; Gil, A. Are We Close to Defining a Metabolomic Signature of Human Obesity? A Systematic Review of Metabolomics Studies. Metabolomics 2019, 15, 93. [Google Scholar] [CrossRef]
- Surwit, R.S.; Kuhn, C.M.; Cochrane, C.; McCubbin, J.A.; Feinglos, M.N. Diet-Induced Type II Diabetes in C57BL/6J Mice. Diabetes 1988, 37, 1163–1167. [Google Scholar] [CrossRef]
- Abdesselam, I.; Pepino, P.; Troalen, T.; Macia, M.; Ancel, P.; Masi, B.; Fourny, N.; Gaborit, B.; Giannesini, B.; Kober, F.; et al. Time Course of Cardiometabolic Alterations in a High Fat High Sucrose Diet Mice Model and Improvement after GLP-1 Analog Treatment Using Multimodal Cardiovascular Magnetic Resonance. J. Cardiovasc. Magn. Reson. 2015, 17, 95. [Google Scholar] [CrossRef]
- Sinasac, D.S.; Riordan, J.D.; Spiezio, S.H.; Yandell, B.S.; Croniger, C.M.; Nadeau, J.H. Genetic Control of Obesity, Glucose Homeostasis, Dyslipidemia and Fatty Liver in a Mouse Model of Diet-Induced Metabolic Syndrome. Int. J. Obes. 2016, 40, 346–355. [Google Scholar] [CrossRef]
- Buettner, R.; Schölmerich, J.; Bollheimer, L.C. High-Fat Diets: Modeling the Metabolic Disorders of Human Obesity in Rodents*. Obesity 2007, 15, 798–808. [Google Scholar] [CrossRef]
- Lefevre, C.; Chartoire, D.; Ferraz, J.C.; Verdier, T.; Pinteur, C.; Chanon, S.; Pesenti, S.; Vieille-Marchiset, A.; Genestier, L.; Vidal, H.; et al. Obesity Activates Immunomodulating Properties of Mesenchymal Stem Cells in Adipose Tissue with Differences between Localizations. FASEB J. 2021, 35, e21650. [Google Scholar] [CrossRef]
- Jung, Y.-S.; Hyeon, J.-S.; Hwang, G.-S. Software-Assisted Serum Metabolite Quantification Using NMR. Anal. Chim. Acta 2016, 934, 194–202. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for Significance Testing of PLS and OPLS® Models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Blaise, B.J.; Correia, G.D.S.; Haggart, G.A.; Surowiec, I.; Sands, C.; Lewis, M.R.; Pearce, J.T.M.; Trygg, J.; Nicholson, J.K.; Holmes, E.; et al. Statistical Analysis in Metabolic Phenotyping. Nat. Protoc. 2021, 16, 4299–4326. [Google Scholar] [CrossRef]
- Jackson, S.J.; Andrews, N.; Ball, D.; Bellantuono, I.; Gray, J.; Hachoumi, L.; Holmes, A.; Latcham, J.; Petrie, A.; Potter, P.; et al. Does Age Matter? The Impact of Rodent Age on Study Outcomes. Lab. Anim. 2017, 51, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Pann, P.; de Angelis, M.H.; Prehn, C.; Adamski, J. Mouse Age Matters: How Age Affects the Murine Plasma Metabolome. Metabolites 2020, 10, 472. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.; Sahlin, K.; Henriksson, J. Muscle ATP Turnover Rate during Isometric Contraction in Humans. J. Appl. Physiol. 1986, 60, 1839–1842. [Google Scholar] [CrossRef]
- Perrier, J.; Nawrot, M.; Madec, A.-M.; Chikh, K.; Chauvin, M.-A.; Damblon, C.; Sabatier, J.; Thivolet, C.H.; Rieusset, J.; Rautureau, G.J.P.; et al. Human Pancreatic Islets React to Glucolipotoxicity by Secreting Pyruvate and Citrate. Nutrients 2023, 15, 4791. [Google Scholar] [CrossRef] [PubMed]
- Houtkooper, R.H.; Argmann, C.; Houten, S.M.; Cantó, C.; Jeninga, E.H.; Andreux, P.A.; Thomas, C.; Doenlen, R.; Schoonjans, K.; Auwerx, J. The Metabolic Footprint of Aging in Mice. Sci. Rep. 2011, 1, 134. [Google Scholar] [CrossRef]
- Petr, M.A.; Alfaras, I.; Krawcyzk, M.; Bair, W.-N.; Mitchell, S.J.; Morrell, C.H.; Studenski, S.A.; Price, N.L.; Fishbein, K.W.; Spencer, R.G.; et al. A Cross-Sectional Study of Functional and Metabolic Changes during Aging through the Lifespan in Male Mice. eLife 2021, 10, e62952. [Google Scholar] [CrossRef]
- Varshavi, D.; Scott, F.H.; Varshavi, D.; Veeravalli, S.; Phillips, I.R.; Veselkov, K.; Strittmatter, N.; Takats, Z.; Shephard, E.A.; Everett, J.R. Metabolic Biomarkers of Ageing in C57BL/6J Wild-Type and Flavin-Containing Monooxygenase 5 (FMO5)-Knockout Mice. Front. Mol. Biosci. 2018, 5, 28. [Google Scholar] [CrossRef]
- Zhang, F.; Kerbl-Knapp, J.; Akhmetshina, A.; Korbelius, M.; Kuentzel, K.B.; Vujić, N.; Hörl, G.; Paar, M.; Kratky, D.; Steyrer, E.; et al. Tissue-Specific Landscape of Metabolic Dysregulation during Ageing. Biomolecules 2021, 11, 235. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Kerbl-knapp, J.; Zhang, F.; Korbelius, M.; Kuentzel, K.B.; Akhmetshina, A.; Hörl, G.; Paar, M.; Steyrer, E.; Kratky, D. Metabolomic Profiles of Mice Tissues Reveals an Interplay between Aging and Energy Metabolism. Metabolites 2022, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Ji, J.; Rabow, Z.; Shen, T.; Folz, J.; Brydges, C.R.; Fan, S.; Lu, X.; Mehta, S.; Showalter, M.R.; et al. A Metabolome Atlas of the Aging Mouse Brain. Nat. Commun. 2021, 12, 6021. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drevet Mulard, E.; Guibert, S.; Mey, A.; Lefevre, C.; Chauvin, M.-A.; Pinteur, C.; Monet, M.-A.; Godet, M.; Madec, A.-M.; Morio, B.; et al. A Cross-Sectional Quantitative Metabolomics Study Evidencing the Metabolic Signature in Six Organs during a 14-Week High-Fat High-Sucrose and Standard Diet in Mice. Nutrients 2024, 16, 803. https://doi.org/10.3390/nu16060803
Drevet Mulard E, Guibert S, Mey A, Lefevre C, Chauvin M-A, Pinteur C, Monet M-A, Godet M, Madec A-M, Morio B, et al. A Cross-Sectional Quantitative Metabolomics Study Evidencing the Metabolic Signature in Six Organs during a 14-Week High-Fat High-Sucrose and Standard Diet in Mice. Nutrients. 2024; 16(6):803. https://doi.org/10.3390/nu16060803
Chicago/Turabian StyleDrevet Mulard, Eva, Sylvie Guibert, Anne Mey, Camille Lefevre, Marie-Agnès Chauvin, Claudie Pinteur, Marie-Ambre Monet, Murielle Godet, Anne-Marie Madec, Béatrice Morio, and et al. 2024. "A Cross-Sectional Quantitative Metabolomics Study Evidencing the Metabolic Signature in Six Organs during a 14-Week High-Fat High-Sucrose and Standard Diet in Mice" Nutrients 16, no. 6: 803. https://doi.org/10.3390/nu16060803
APA StyleDrevet Mulard, E., Guibert, S., Mey, A., Lefevre, C., Chauvin, M.-A., Pinteur, C., Monet, M.-A., Godet, M., Madec, A.-M., Morio, B., Rieusset, J., Rautureau, G. J. P., & Panthu, B. (2024). A Cross-Sectional Quantitative Metabolomics Study Evidencing the Metabolic Signature in Six Organs during a 14-Week High-Fat High-Sucrose and Standard Diet in Mice. Nutrients, 16(6), 803. https://doi.org/10.3390/nu16060803






