RNA Sequencing Reveals the Inhibitory Effect of High Levels of Arachidonic Acid and Linoleic Acid on C2C12 Differentiation and Myogenic Biomarkers
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Proliferation Assay
2.3. Giemsa Staining and Differentiation Morphology
2.4. qRT-PCR Measurement
2.5. Western Blotting
2.6. RNA-Sequencing and Data Analysis
2.6.1. Analysis Process and Transcriptome Analysis Flow
2.6.2. Correlation between Samples
2.6.3. PCA Analysis
2.7. Statistical Analysis
3. Results
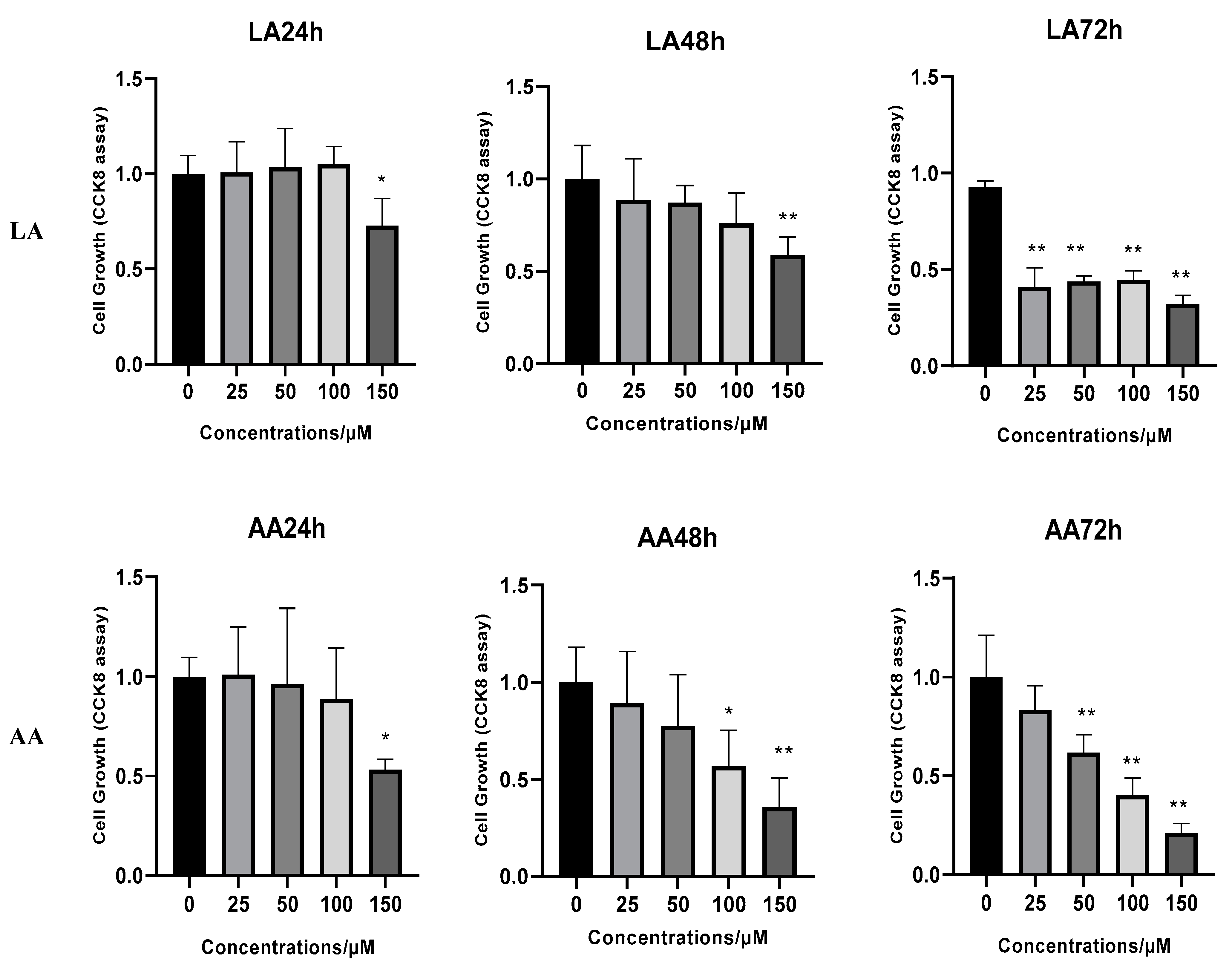
3.1. The Effect of Omega-6 Fatty Acids (LA and AA) on Cell Proliferation
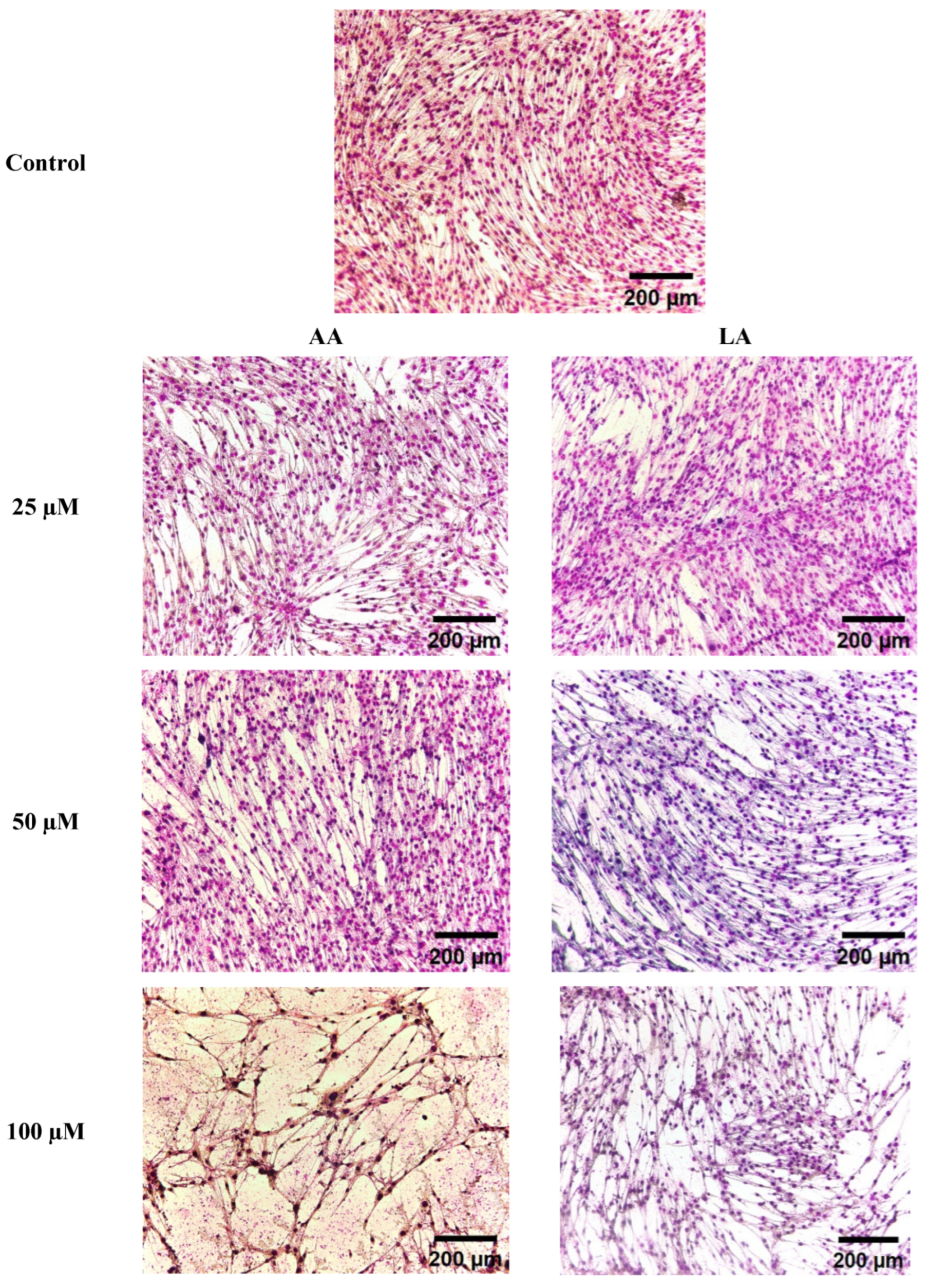
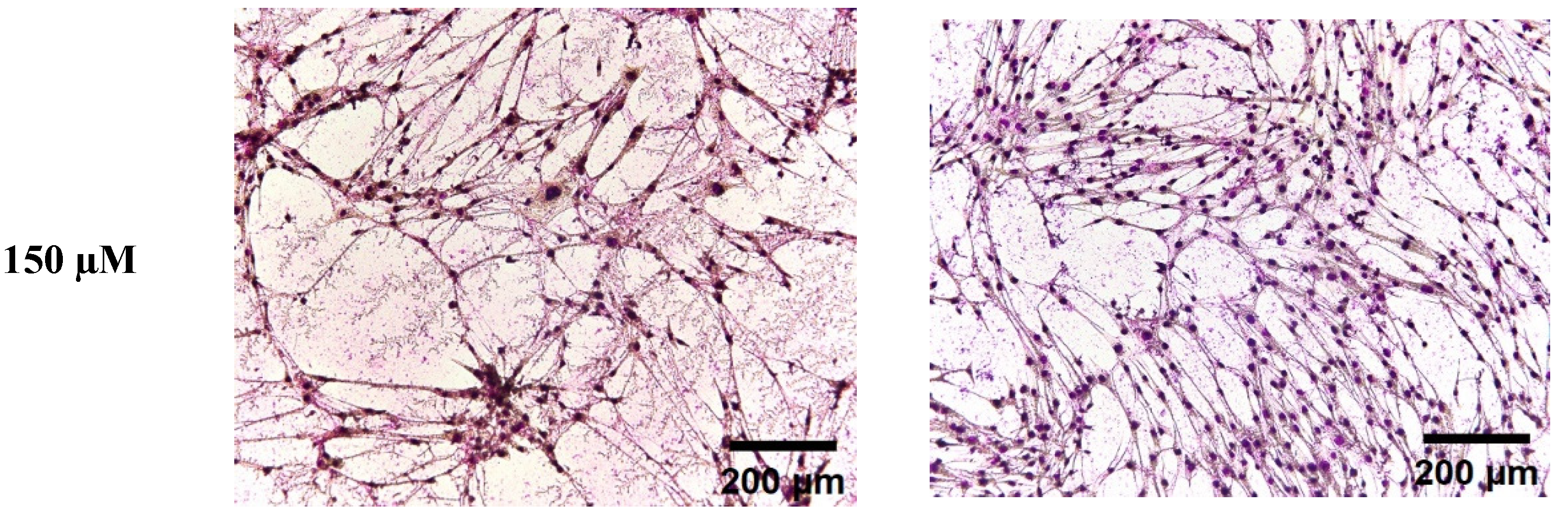
3.2. The Effect of Omega-6 Fatty Acids (LA and AA) on Morphological Changes in C2C12 Myoblasts
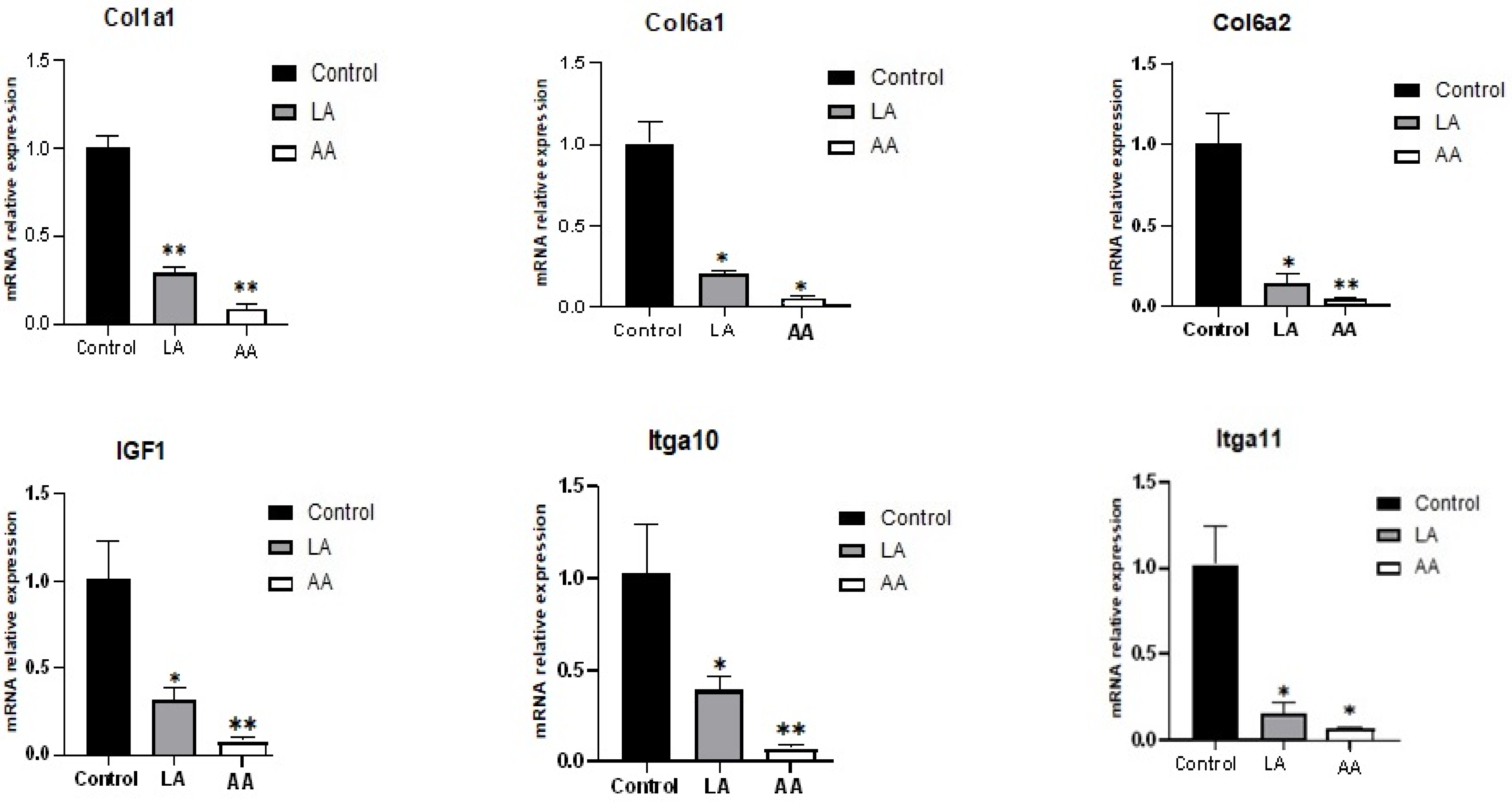
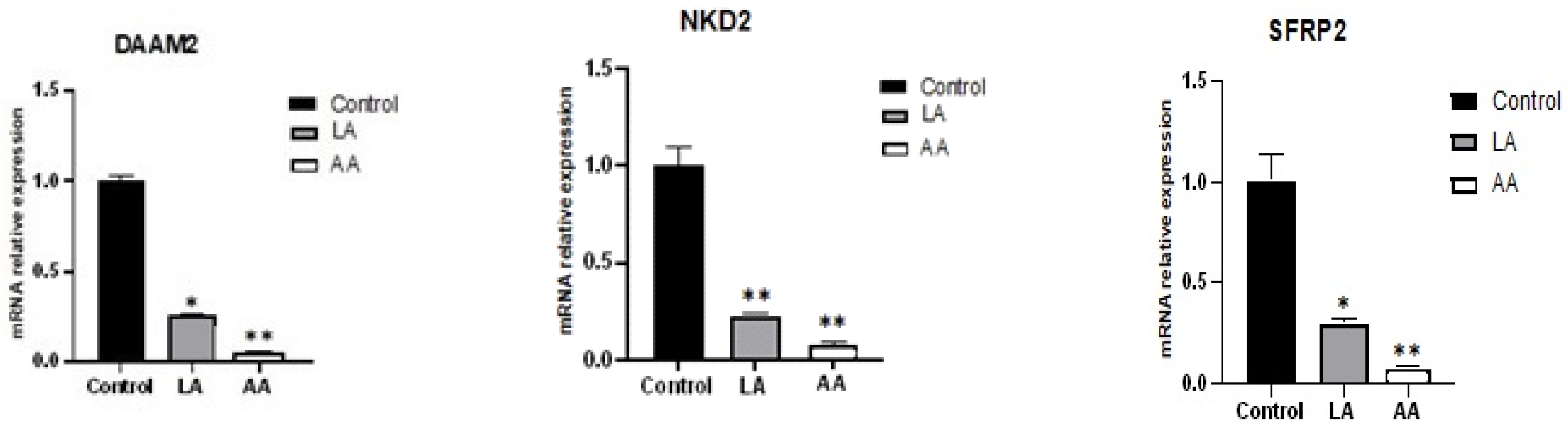
3.3. The Effect of Omega-6 Fatty Acids (LA and AA) on mRNA Expression of Myogenesis Biomarkers in the Wnt Pathway
3.4. RNA Sequencing Results
3.4.1. Data Filtering
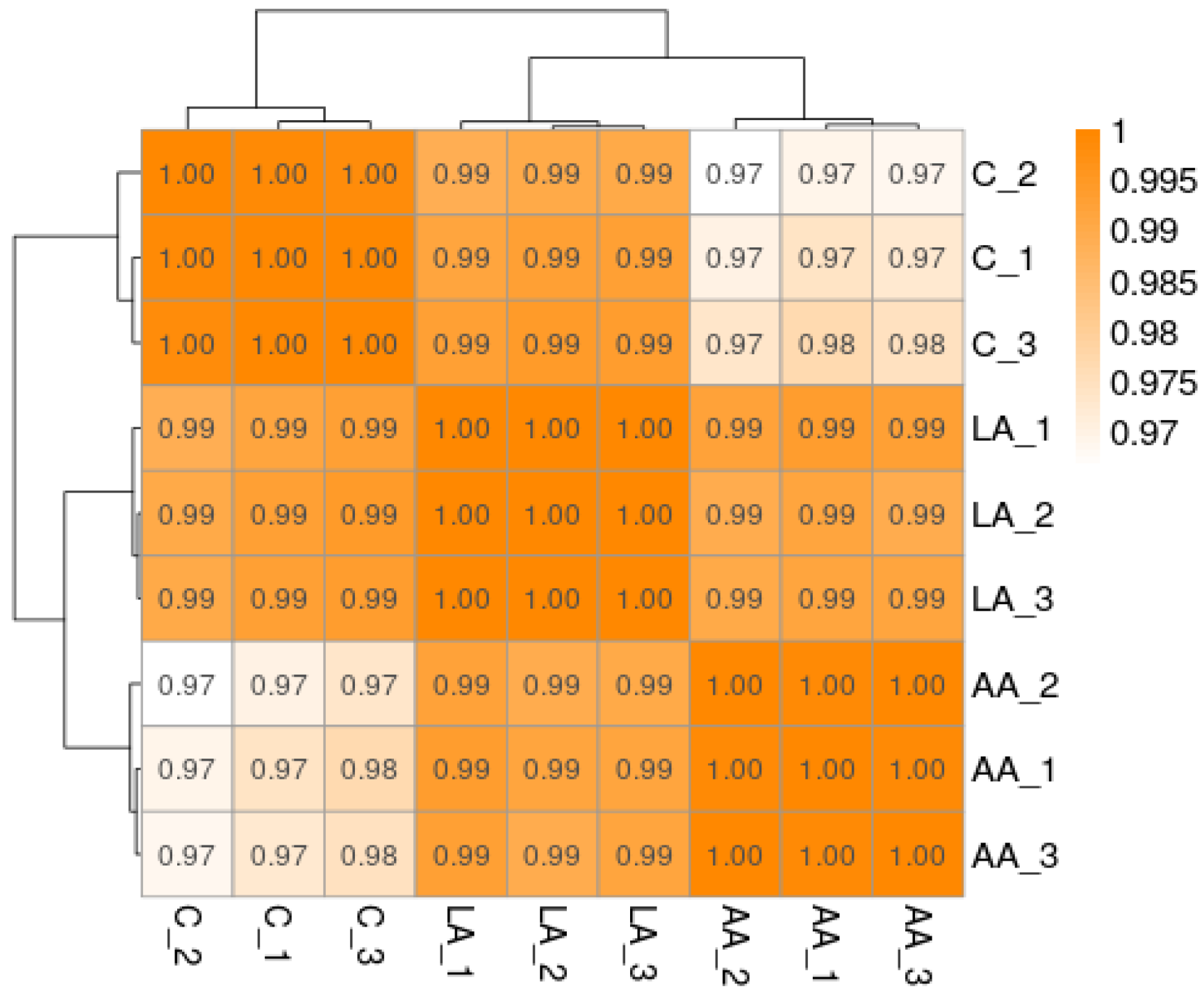
3.4.2. Sample Correlation Test
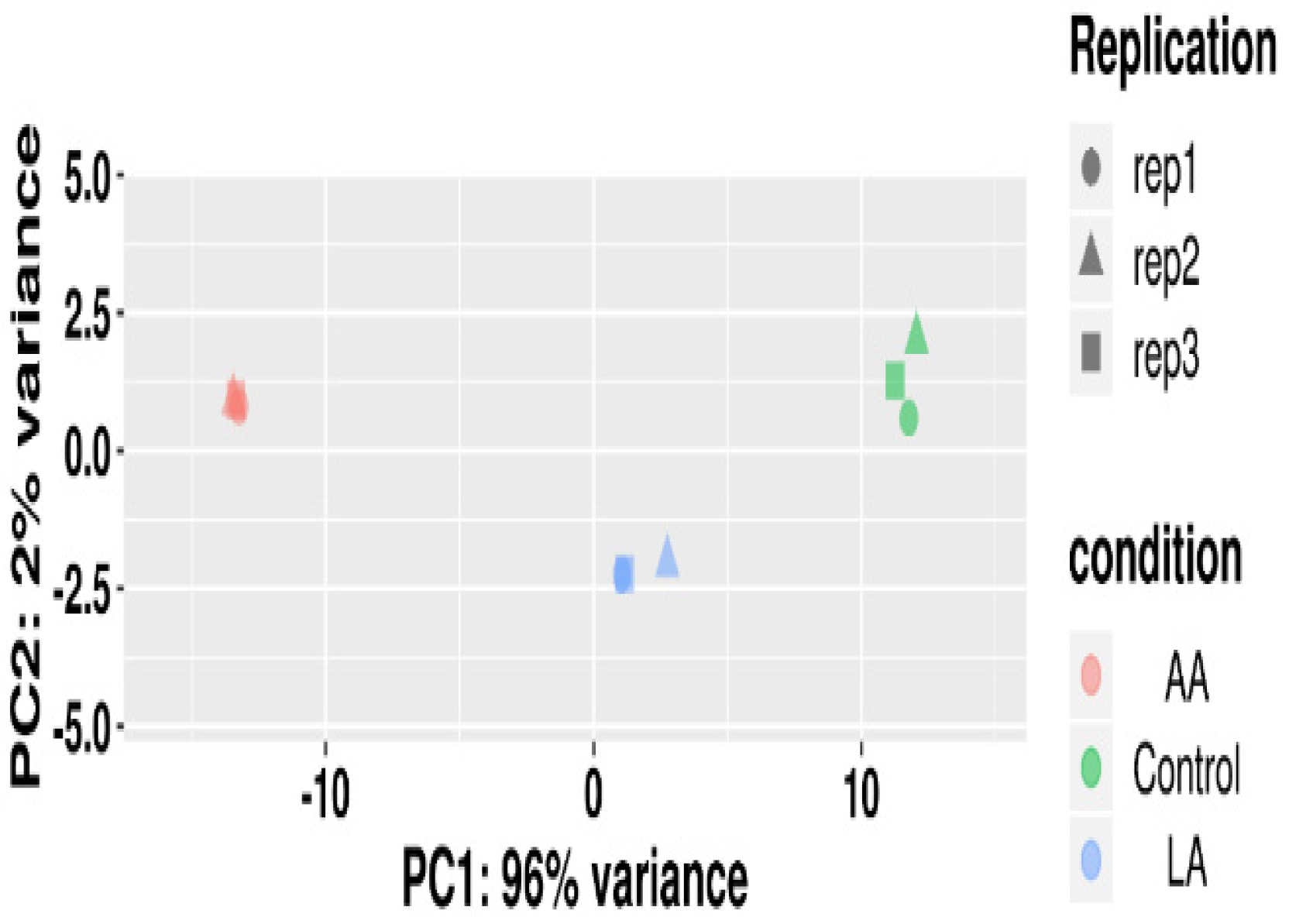
3.4.3. PCA Analysis
3.4.4. Analysis and Comparison of Differential Expression among Multiple Groups
3.4.5. Cluster Analysis
3.4.6. Trend Analysis
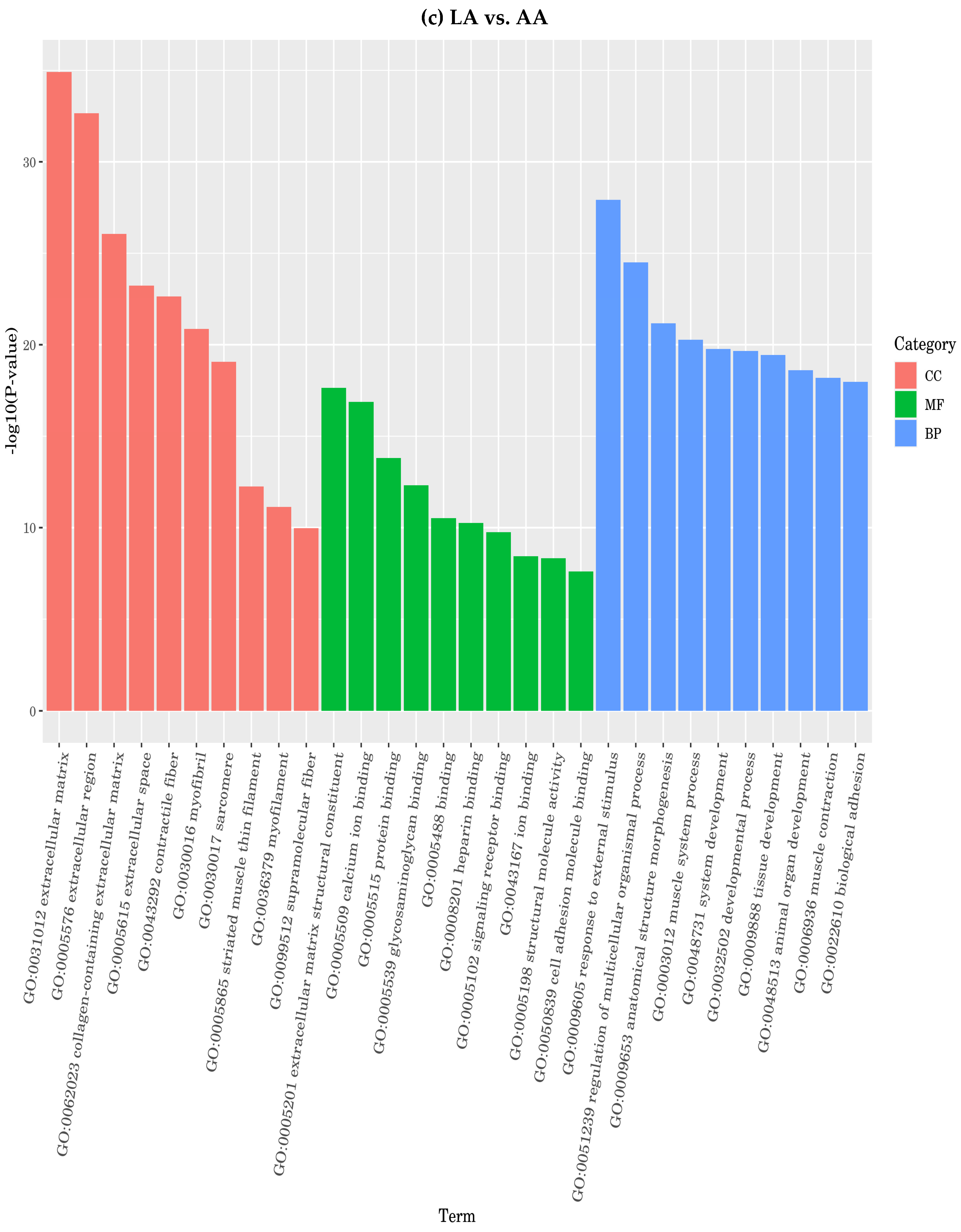
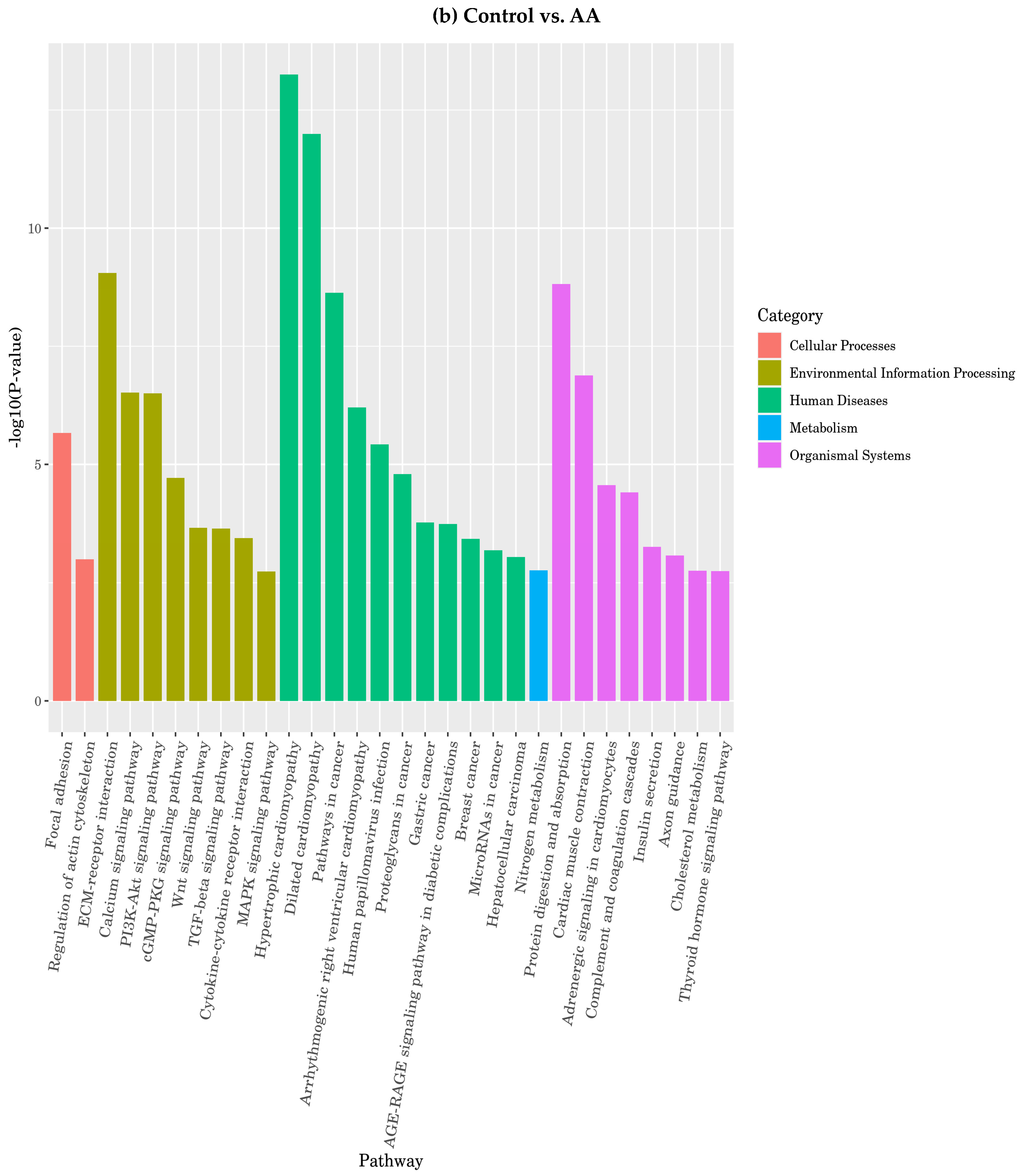
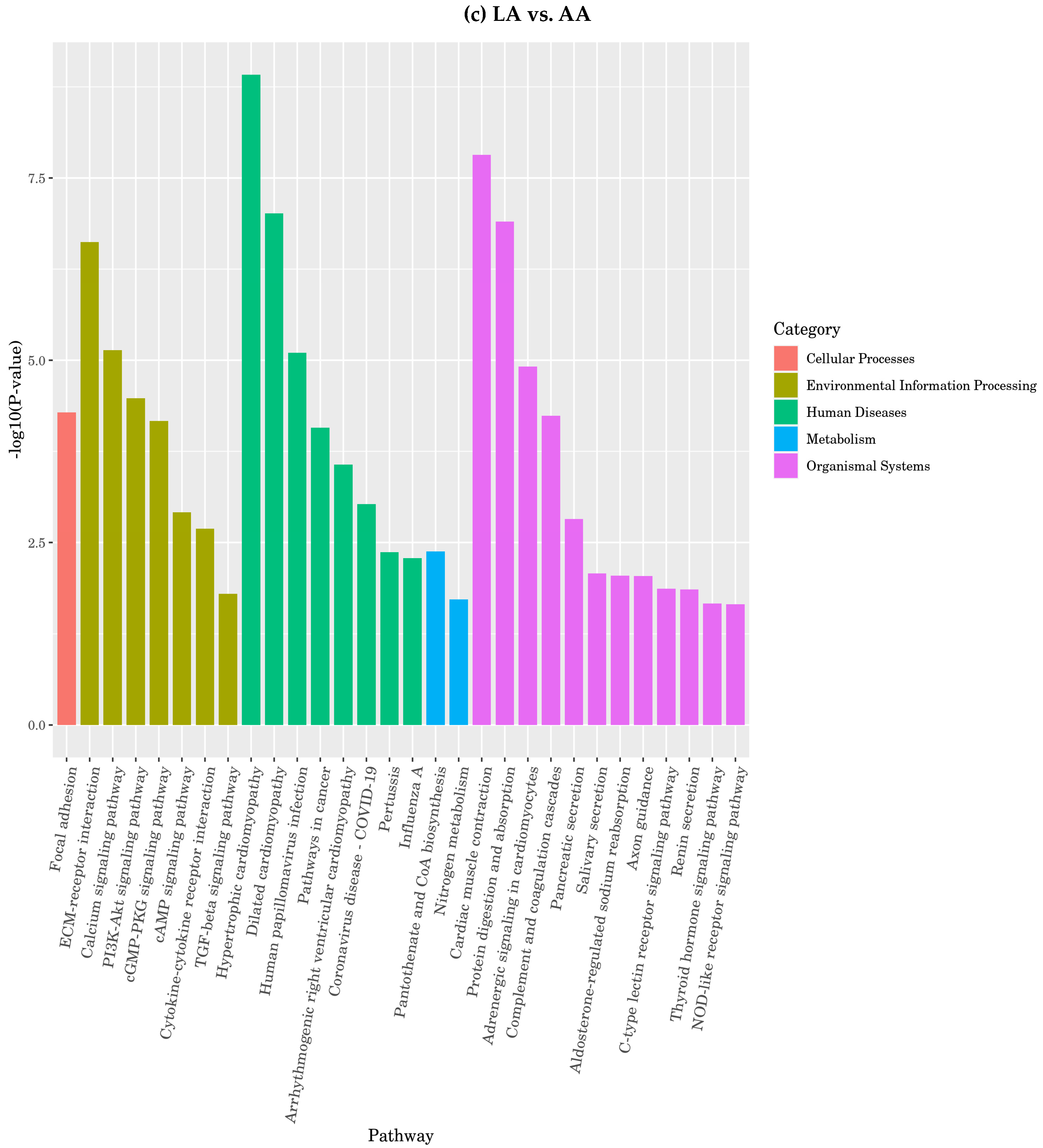
3.4.7. Function of Differentially Expressed Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ogłuszka, M.; Szostak, A.; te Pas, M.F.W.; Poławska, E.; Urbański, P.; Blicharski, T.; Pareek, C.S.; Juszczuk-Kubiak, E.; Dunkelberger, J.R.; Horbańczuk, J.O.; et al. A porcine gluteus medius muscle genome-wide transcriptome analysis: Dietary effects of omega-6 and omega-3 fatty acids on biological mechanisms. Genes Nutr. 2017, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. An Increase in the Omega-6/Omega-3 Fatty Acid Ratio Increases the Risk for Obesity. Nutrients 2016, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- da Paixão, A.O.; Bolin, A.P.; Silvestre, J.G.; Rodrigues, A.C. Palmitic acid impairs myogenesis and alters temporal expression of miR-133a and miR-206 in C2C12 myoblasts. Int. J. Mol. Sci. 2021, 22, 2748. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.; Paton, C.M. Lipid metabolic features of skeletal muscle in pathological and physiological conditions. In Lipid Signaling and Metabolism; Elsevier: Amsterdam, The Netherlands, 2020; pp. 359–383. [Google Scholar]
- Kaur, N.; Chugh, V.; Gupta, A.K. Essential fatty acids as functional components of foods—A review. J. Food Sci. Technol. 2014, 51, 2289–2303. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Sleep, S.L.; Cuffe, J.S.; Holland, O.J.; Perkins, A.V.; Yau, S.Y.; McAinch, A.J.; Hryciw, D.H. Role Of Omega-6 and Omega-3 fatty acids in fetal programming. Clin. Exp. Pharmacol. Physiol. 2020, 47, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Jeukendrup, A.E. Regulation of fat metabolism in skeletal muscle. Ann. N. Y. Acad. Sci. 2002, 967, 217–235. [Google Scholar] [CrossRef] [PubMed]
- Marques, R.; Cooke, R.; Rodrigues, M.; Brandão, A.; Schubach, K.; Lippolis, K.; Moriel, P.; Perry, G.; Lock, A.; Bohnert, D. Effects of supplementing calcium salts of polyunsaturated fatty acids to late-gestating beef cows on performance and physiological responses of the offspring. J. Anim. Sci. 2017, 95, 5347–5357. [Google Scholar] [CrossRef]
- Thomas, D.R. Loss of skeletal muscle mass in aging: Examining the relationship of starvation, sarcopenia and cachexia. Clin. Nutr. 2007, 26, 389–399. [Google Scholar] [CrossRef]
- Chaillou, T.; Lanner, J.T. Regulation of myogenesis and skeletal muscle regeneration: Effects of oxygen levels on satellite cell activity. FASEB J. 2016, 30, 3929–3941. [Google Scholar] [CrossRef]
- Jang, Y.J.; Son, H.J.; Kim, J.-S.; Jung, C.H.; Ahn, J.; Hur, J.; Ha, T.Y. Coffee consumption promotes skeletal muscle hypertrophy and myoblast differentiation. Food Funct. 2018, 9, 1102–1111. [Google Scholar] [CrossRef]
- Kaminski, J.; Lançon, A.; Aires, V.; Limagne, E.; Tili, E.; Michaille, J.-J.; Latruffe, N. Resveratrol initiates differentiation of mouse skeletal muscle-derived C2C12 myoblasts. Biochem. Pharmacol. 2012, 84, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.M.; Klass, M.; Harris, C.; Csete, M. A reducing redox environment promotes C2C12 myogenesis: Implications for regeneration in aged muscle. Cell Biol. Int. 2007, 31, 546–553. [Google Scholar] [CrossRef]
- Beaudry, M.; Hidalgo, M.; Launay, T.; Bello, V.; Darribère, T. Regulation of myogenesis by environmental hypoxia. J. Cell Sci. 2016, 129, 2887–2896. [Google Scholar] [CrossRef] [PubMed]
- Magee, P.; Pearson, S.; Allen, J. The omega-3 fatty acid, eicosapentaenoic acid (EPA), prevents the damaging effects of tumour necrosis factor (TNF)-alpha during murine skeletal muscle cell differentiation. Lipids Health Dis. 2008, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, Z.; Ling, D.; Li, J.; Wang, Y.; Shan, T. The regulatory role of dietary factors in skeletal muscle development, regeneration and function. Crit. Rev. Food Sci. Nutr. 2022, 62, 764–782. [Google Scholar] [CrossRef] [PubMed]
- David, B.; Albert, G.; Juana, A.F.-L.R.; Montserrat, F.; Oscar, P.; Jose, R.-M.; Carme, S.; Manuel, P.; Gemma, L.; Elisenda, C.; et al. Bariatric surgery and LDL cholesterol (BASALTO) trial study protocol: Randomised controlled study evaluating the effect of gastric bypass versus sleeve gastrectomy on high LDL cholesterol. BMJ Open 2020, 10, e037712. [Google Scholar] [CrossRef]
- Koletzko, B.; Brands, B.; Poston, L.; Godfrey, K.; Demmelmair, H. Early nutrition programming of long-term health. Proc. Nutr. Soc. 2012, 71, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Tong, J.; Zhao, J.; Underwood, K.; Zhu, M.; Ford, S.; Nathanielsz, P. Fetal programming of skeletal muscle development in ruminant animals. Anim. Sci. 2010, 88, E51–E60. [Google Scholar] [CrossRef]
- Luo, H.; Zhou, Y.; Hu, X.; Peng, X.; Wei, H.; Peng, J.; Jiang, S. Activation of PPARγ2 by PPARγ1 through a functional PPRE in transdifferentiation of myoblasts to adipocytes induced by EPA. Cell Cycle 2015, 14, 1830–1841. [Google Scholar] [CrossRef]
- Petrany, M.J.; Millay, D.P. Cell fusion: Merging membranes and making muscle. Trends Cell Biol. 2019, 29, 964–973. [Google Scholar] [CrossRef]
- Rahkonen, O.; Su, M.; Hakovirta, H.; Koskivirta, I.; Hormuzdi, S.G.; Vuorio, E.; Bornstein, P.; Penttinen, R. Mice with a deletion in the first intron of the Col1a1 gene develop age-dependent aortic dissection and rupture. Circ. Res. 2004, 94, 83–90. [Google Scholar] [CrossRef]
- Mohammadabadi, M.; Bordbar, F.; Jensen, J.; Du, M.; Guo, W. Key Genes Regulating Skeletal Muscle Development and Growth in Farm Animals. Animals 2021, 11, 835. [Google Scholar] [CrossRef] [PubMed]
- Nesvadbova, M.; Borilova, G. Molecular regulation of skeletal muscle tissue formation and development. Veterinární Medicína 2018, 63, 489–499. [Google Scholar] [CrossRef]
- Theil, P.K.; Sørensen, I.L.; Nissen, P.M.; Oksbjerg, N. Temporal expression of growth factor genes of primary porcine satellite cells during myogenesis. Anim. Sci. J. 2006, 77, 330–337. [Google Scholar] [CrossRef]
- Fahey, A.; Brameld, J.; Parr, T.; Buttery, P. Ontogeny of factors associated with proliferation and differentiation of muscle in the ovine fetus. Anim. Sci. 2005, 83, 2330–2338. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Ogawa, T.; Sato, M.; Tsuchida, N.; Fotovati, A.; Iwamoto, H.; Ikeuchi, Y.; Cassens, R.G.; Ito, T. S-myotrophin promotes the hypertrophy of myotube as insulin-like growth factor-I does. Int. J. Biochem. Cell Biol. 2001, 33, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Hurley, M.S.; Flux, C.; Salter, A.M.; Brameld, J.M. Effects of fatty acids on skeletal muscle cell differentiation in vitro. Br. J. Nutr. 2006, 95, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Hiden, U.; Glitzner, E.; Hartmann, M.; Desoye, G. Insulin and the IGF system in the human placenta of normal and diabetic pregnancies. J. Anat. 2009, 215, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Fowden, A.L. The insulin-like growth factors and feto-placental growth. Placenta 2003, 24, 803–812. [Google Scholar] [CrossRef]
- Roberts, B.M.; Kolb, A.L.; Geddis, A.V.; Naimo, M.A.; Matheny, R.W. The dose–response effects of arachidonic acid on primary human skeletal myoblasts and myotubes. Int. Soc. Sports Nutr. 2023, 20, 2164209. [Google Scholar] [CrossRef]
- Takenaka-Ninagawa, N.; Kim, J.; Zhao, M.; Sato, M.; Jonouchi, T.; Goto, M.; Yoshioka, C.K.B.; Ikeda, R.; Harada, A.; Sato, T.; et al. Collagen-VI supplementation by cell transplantation improves muscle regeneration in Ullrich congenital muscular dystrophy model mice. Stem Cell Res. Ther. 2021, 12, 446. [Google Scholar] [CrossRef]
- Tabib, T.; Huang, M.; Morse, N.; Papazoglou, A.; Behera, R.; Jia, M.; Bulik, M.; Monier, D.E.; Benos, P.V.; Chen, W.; et al. Myofibroblast transcriptome indicates SFRP2hi fibroblast progenitors in systemic sclerosis skin. Nat. Commun. 2021, 12, 4384. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, M.; Gao, P.; Cao, G.; Zhang, W.; Liu, M.; Wang, H.; Qin, B.; Liu, J.; Wang, L. Identification of candidate genes of growth traits in pigs using RNA-sequencing. Ital. J. Anim. Sci. 2019, 18, 279–286. [Google Scholar] [CrossRef]
- Levin, J.M.; Boubaker el Andalousi, R.A.; Dainat, J.; Reyne, Y.; Bacou, F. SFRP2 expression in rabbit myogenic progenitor cells and in adult skeletal muscles. J. Muscle Res. Cell Motil. 2001, 22, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Descamps, S.; Arzouk, H.; Bacou, F.; Bernardi, H.; Fedon, Y.; Gay, S.; Reyne, Y.; Rossano, B.; Levin, J. Inhibition of myoblast differentiation by Sfrp1 and Sfrp2. Cell Tissue Res. 2008, 332, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Aspenström, P.; Richnau, N.; Johansson, A.-S. The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp. Cell Res. 2006, 312, 2180–2194. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tian, Y.; Du, J.; Hu, Z.; Yang, L.; Liu, J.; Gu, L. Dvl2-dependent activation of Daam1 and RhoA regulates Wnt5a-induced breast cancer cell migration. PLoS ONE. 2012, 7, e37823. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, Y.; Pan, M.-H.; Kim, N.-H.; Sun, S.-C.; Cui, X.-S. Daam1 regulates fascin for actin assembly in mouse oocyte meiosis. Cell Cycle 2017, 16, 1350–1356. [Google Scholar] [CrossRef]
- Xiong, H.; Yan, T.; Zhang, W.; Shi, F.; Jiang, X.; Wang, X.; Li, S.; Chen, Y.; Chen, C.; Zhu, Y. miR-613 inhibits cell migration and invasion by downregulating Daam1 in triple-negative breast cancer. Cell. Signal. 2018, 44, 33–42. [Google Scholar] [CrossRef]
- Ajima, R.; Bisson, J.A.; Helt, J.-C.; Nakaya, M.-A.; Habas, R.; Tessarollo, L.; He, X.; Morrisey, E.E.; Yamaguchi, T.P.; Cohen, E.D. DAAM1 and DAAM2 are co-required for myocardial maturation and sarcomere assembly. Dev. Biol. 2015, 408, 126–139. [Google Scholar] [CrossRef]
- Li, H. A Study of Fibroblast-Mediated Contraction in Ocular Scarring: Gene Expression Profiling and the Role of Small GTPases in Matrix Metalloproteinase 1 (MMP1) Regulation. Ph.D. Thesis, UCL (University College London), London, UK, 2017. [Google Scholar]
- Blasbalg, T.L.; Hibbeln, J.R.; Ramsden, C.E.; Majchrzak, S.F.; Rawlings, R.R. Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am. J. Clin. Nutr. 2011, 93, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.; Wall, R.; Fitzgerald, G.; Ross, R.; Stanton, C. Health implications of high dietary omega-6 polyunsaturated fatty acids. Nutr. Metab. 2012, 2012, 539426. [Google Scholar] [CrossRef]
- DiNicolantonio, J.J.; O’Keefe, J. The Importance of Maintaining a Low Omega-6/Omega-3 Ratio for Reducing the Risk of Autoimmune Diseases, Asthma, and Allergies. Mo. Med. 2021, 118, 453–459. [Google Scholar]
- Son, Y.; Lorenz, W.W.; Paton, C.M. Linoleic acid-induced ANGPTL4 inhibits C2C12 skeletal muscle differentiation by suppressing Wnt/β-catenin. J. Nutr. Biochem. 2023, 116, 109324. [Google Scholar] [CrossRef]
- Markworth, J.F.; Cameron-Smith, D. Arachidonic acid supplementation enhances in vitro skeletal muscle cell growth via a COX-2-dependent pathway. Am. J. Physiol.-Cell Physiol. 2013, 304, C56–C67. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-H.; Du, Y.-P.; Wen, J.-T.; Lu, B.-F.; Zhao, Y. snoRNAs: Functions and mechanisms in biological processes, and roles in tumor pathophysiology. Cell Death Discov. 2022, 8, 259. [Google Scholar] [CrossRef]
- Geddis, A.; Kolb, A.L. Arachidonic Acid Impairs Primary Human Skeletal Muscle Myoblasts Proliferation and Differentiation. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]









| Gene Name | Primer Sequences | Accession Number | Product Size (bp) |
|---|---|---|---|
| Itga11 | F 5′ CAGCCTTTGGCCAGGATTCA3′ | NM_176922.6 | 159 |
| R 5′ CCATTGGTTTCCATTGGGGC3′ | |||
| Col6a2 | F 5′CCCTAACAGGAACCTAAACGAA3′ | NM_001347207.1 | 100 |
| R 5′ GGTAGAGTCGGGTCGCATG3′ | |||
| Col6a1 | F 5′ACACTCAACGGGACACGAC3′ | NM_009933.4 | 147 |
| R 5′GCCGACCTTGCGATAAGC3′ | |||
| IGF1 | F 5′GGACCGAGGGGCTTTTAC3′ | NM_001111274.1 | 163 |
| R 5′TAGAGCGGGCTGCTTTTG3′ | |||
| Col1a1 | F 5′AGAGCCTGAGTCAGCAGATTG3′ | NM_007742.4 | 138 |
| R 5′AGCCTTGGTTAGGGTCGA3′ | |||
| Itga10 | F 5′ATCTCTGGCAATGCAAGCTG3′ | NM_001302471.1 | 242 |
| R 5′AAGGTGCTGACCACTGTCAC3′ | |||
| NKD2 | F 5′ACAACCGCCAAGAATGGACAT3′ | NM_001347535.1 | 99 |
| R 5′CCTCGTAGATGGTGTGCATCA3′ | |||
| SFRP2 | F 5′CCCCTGTCTGTCTCGACGA3′ | NM_009144.2 | 131 |
| R 5′GTCGCACTCCAGCATGTCT3′ | |||
| DAAM2 | F 5′TGACCTTCCCGAGATCGACC3′ | NM_001008231.2 | 103 |
| R 5′CTCTGCAAAGCGGACATTGAG3′ | |||
| GAPDH | F 5′AGGTCGGTGTGAACGGATTTG3′ | NM_001289726.2 | 123 |
| R 5′TGTAGACCATGTAGTTGAGGTCA3′ |
| Sample | Clean Reads No. | Clean Data (bp) | Clean Reads% | Clean Data % |
|---|---|---|---|---|
| C_1 | 44,560,604 | 6,728,651,204 | 94.61 | 94.61 |
| C_2 | 41,755,710 | 6,305,112,210 | 94.69 | 94.69 |
| C_3 | 48,940,596 | 7,390,029,996 | 94.63 | 94.63 |
| LA_1 | 42,811,766 | 6,464,576,666 | 94.62 | 94.62 |
| LA_2 | 45,734,926 | 6,905,973,826 | 94.64 | 94.64 |
| LA_3 | 40,923,626 | 6,179,467,526 | 94.58 | 94.58 |
| AA_1 | 44,172,092 | 6,669,985,892 | 94.56 | 94.56 |
| AA_2 | 40,074,740 | 6,051,285,740 | 94.59 | 94.59 |
| AA_3 | 48,299,736 | 7,293,260,136 | 94.58 | 94.58 |
| Comparison | Upregulated | Downregulated | Total |
|---|---|---|---|
| Control vs. LA | 98 | 162 | 260 |
| Control vs. AA | 512 | 1039 | 1551 |
| AA vs. LA | 546 | 153 | 699 |
| Biomarker | Control | LA | AA | p-Value |
|---|---|---|---|---|
| Mt_rRNA | 102,59.67 a ± 883.30 | 12,460.00 ab ± 1327.53 | 14,055.00 c ± 1911.033 | 0.048 |
| snoRNA | 549.67 a ± 32.50 | 624.33 a ± 18.56 | 775.67 b ± 111.07 | 0.017 |
| scaRNA | 45.00 ± 10.15 | 50.33 ± 8.51 | 47.33 ± 10.79 | 0.808 |
| lncRNA | 245,212.67 b ± 21,287.17 | 200,859.00 a ± 14,486.99 | 166,449.00 a ± 17,072.84 | 0.005 |
| snRNA | 193.33 ± 12.09 | 224.00 ± 46.57 | 254.67 ± 22.75 | 0.126 |
| miRNA | 722.67 ± 89.20 | 625.33 ± 35.16 | 622.33 ± 55.47 | 0.172 |
| Mt_tRNA | 1341.00 ± 154.70 | 1573.33 ± 197.06 | 1442.00 ± 228.50 | 0.403 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Abdelrahman, M.; Yang, Y.; Lv, H.; Yang, L. RNA Sequencing Reveals the Inhibitory Effect of High Levels of Arachidonic Acid and Linoleic Acid on C2C12 Differentiation and Myogenic Biomarkers. Nutrients 2024, 16, 706. https://doi.org/10.3390/nu16050706
Wang W, Abdelrahman M, Yang Y, Lv H, Yang L. RNA Sequencing Reveals the Inhibitory Effect of High Levels of Arachidonic Acid and Linoleic Acid on C2C12 Differentiation and Myogenic Biomarkers. Nutrients. 2024; 16(5):706. https://doi.org/10.3390/nu16050706
Chicago/Turabian StyleWang, Wei, Mohamed Abdelrahman, Ying Yang, Haimiao Lv, and Liguo Yang. 2024. "RNA Sequencing Reveals the Inhibitory Effect of High Levels of Arachidonic Acid and Linoleic Acid on C2C12 Differentiation and Myogenic Biomarkers" Nutrients 16, no. 5: 706. https://doi.org/10.3390/nu16050706
APA StyleWang, W., Abdelrahman, M., Yang, Y., Lv, H., & Yang, L. (2024). RNA Sequencing Reveals the Inhibitory Effect of High Levels of Arachidonic Acid and Linoleic Acid on C2C12 Differentiation and Myogenic Biomarkers. Nutrients, 16(5), 706. https://doi.org/10.3390/nu16050706






