Interplay between Cruciferous Vegetables and the Gut Microbiome: A Multi-Omic Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Ex Vivo Fecal Incubation Model
2.2. Microbial Sequencing
2.3. Microbiome Data Management and Quantification of ASVs
2.4. Diversity Analysis and Visualization
2.5. Beta-Diversity Analysis
2.6. Differential Abundance Analysis
2.7. Metabolomic Analysis
2.8. Statistical Methods Summary
2.9. Statistical Analysis of Metabolomics Data
2.10. Metabolomics Data Annotation
2.11. Chemical Similarity Enrichment Analysis
2.12. Multi-Block PLS-DA
2.13. Microbial Metabolite Identification
2.14. Data and Code Availability
3. Results
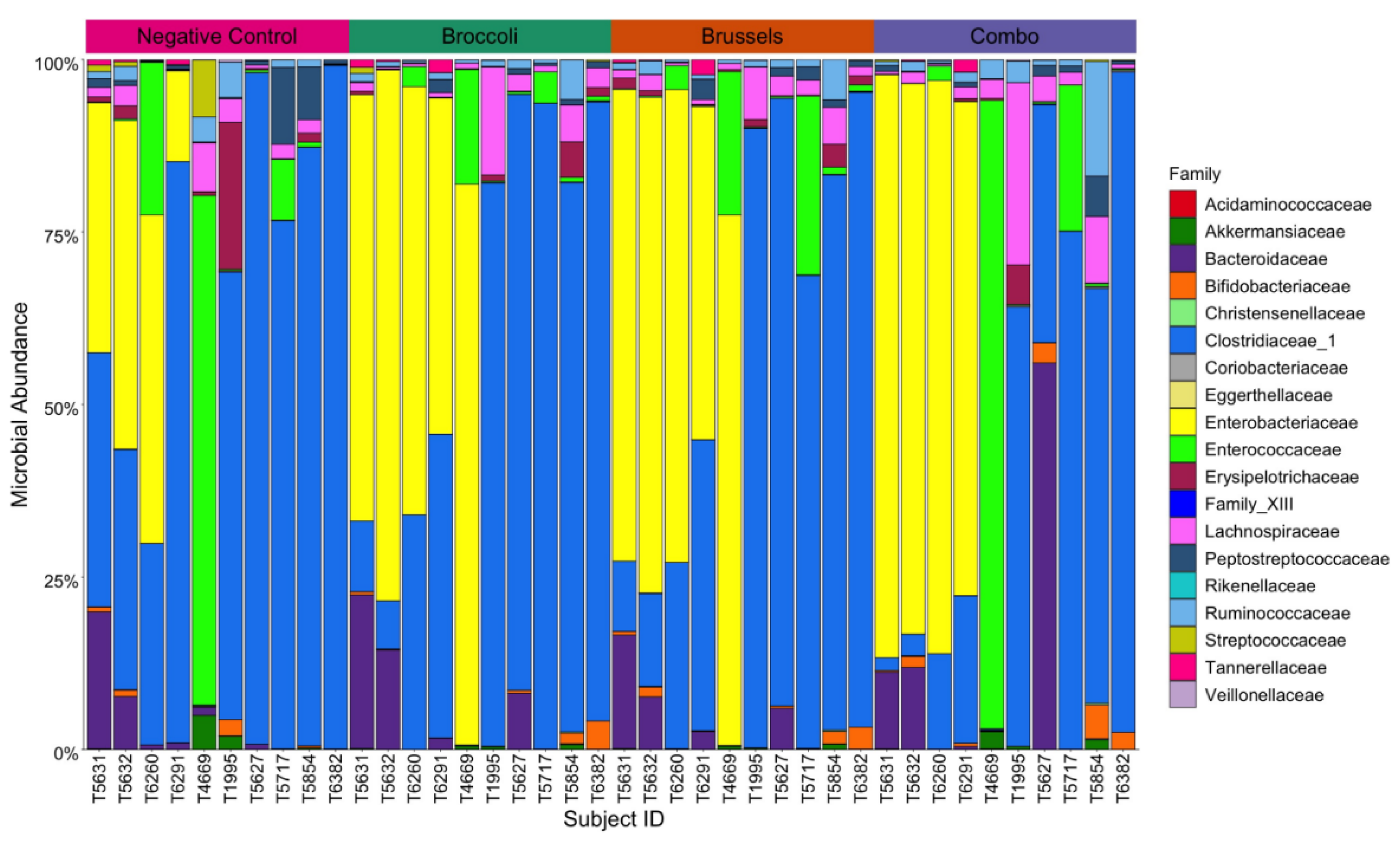
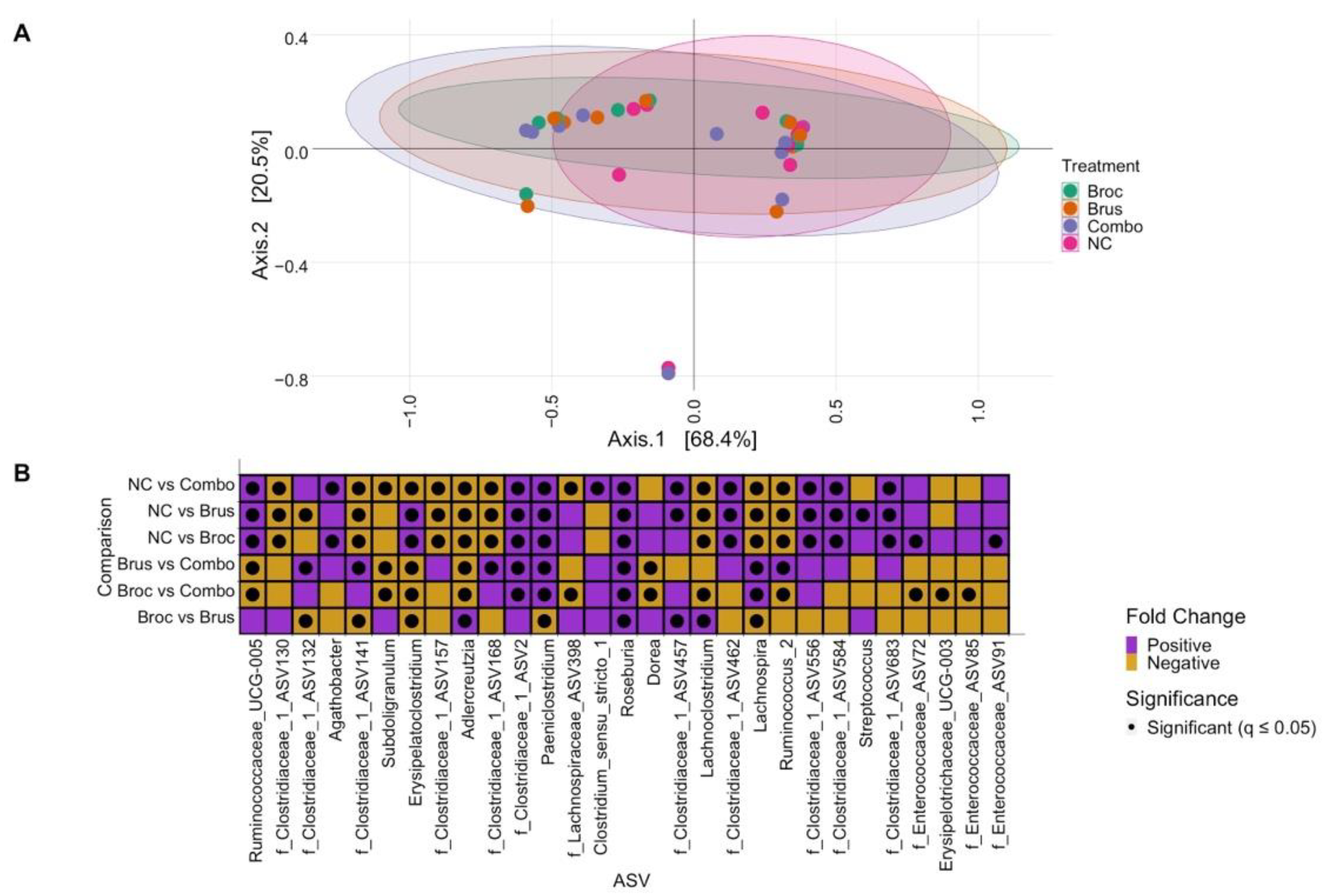
3.1. Impact of Cruciferous Vegetables on Microbiome Composition
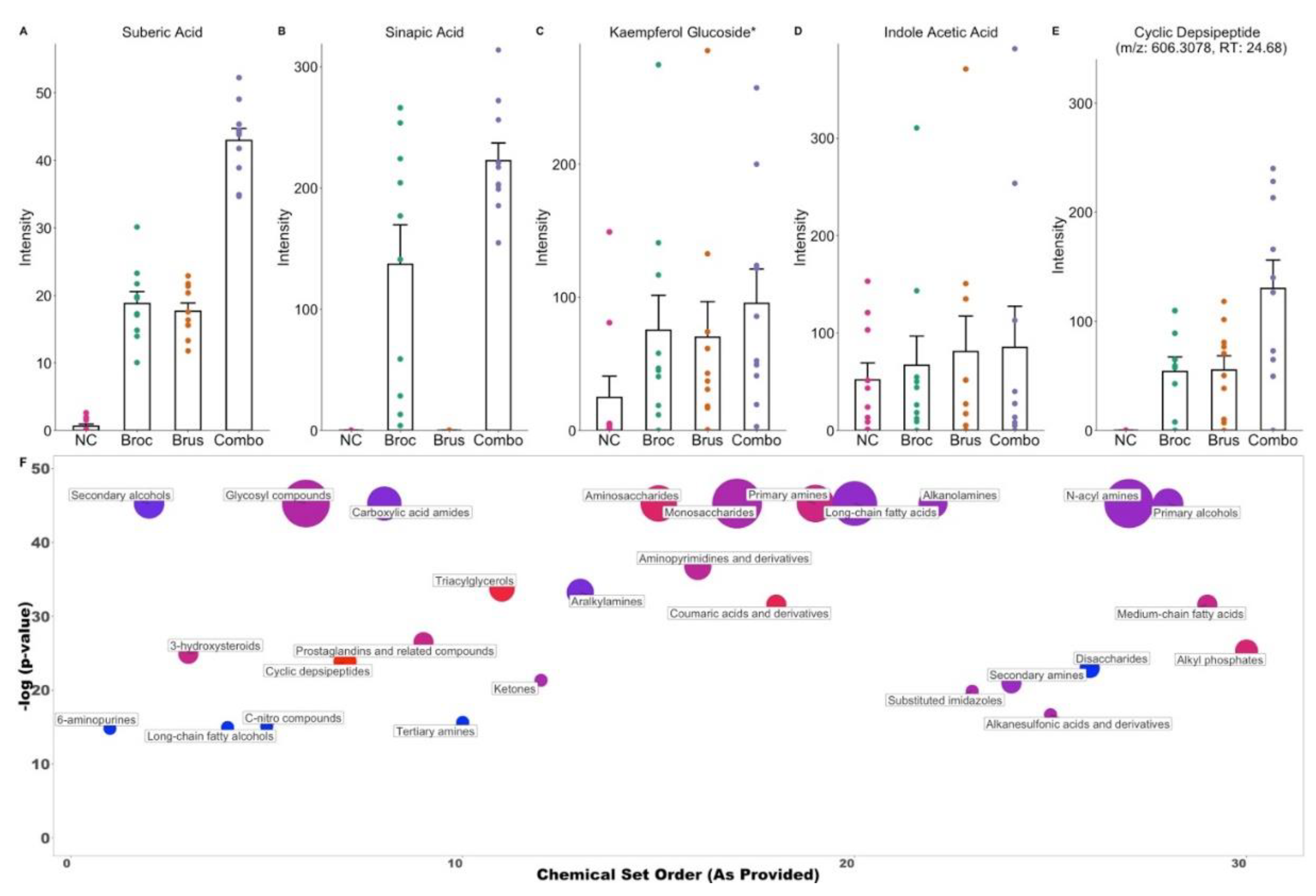
3.2. Cruciferous Vegetable Consumption Alters the Digestive Metabolome
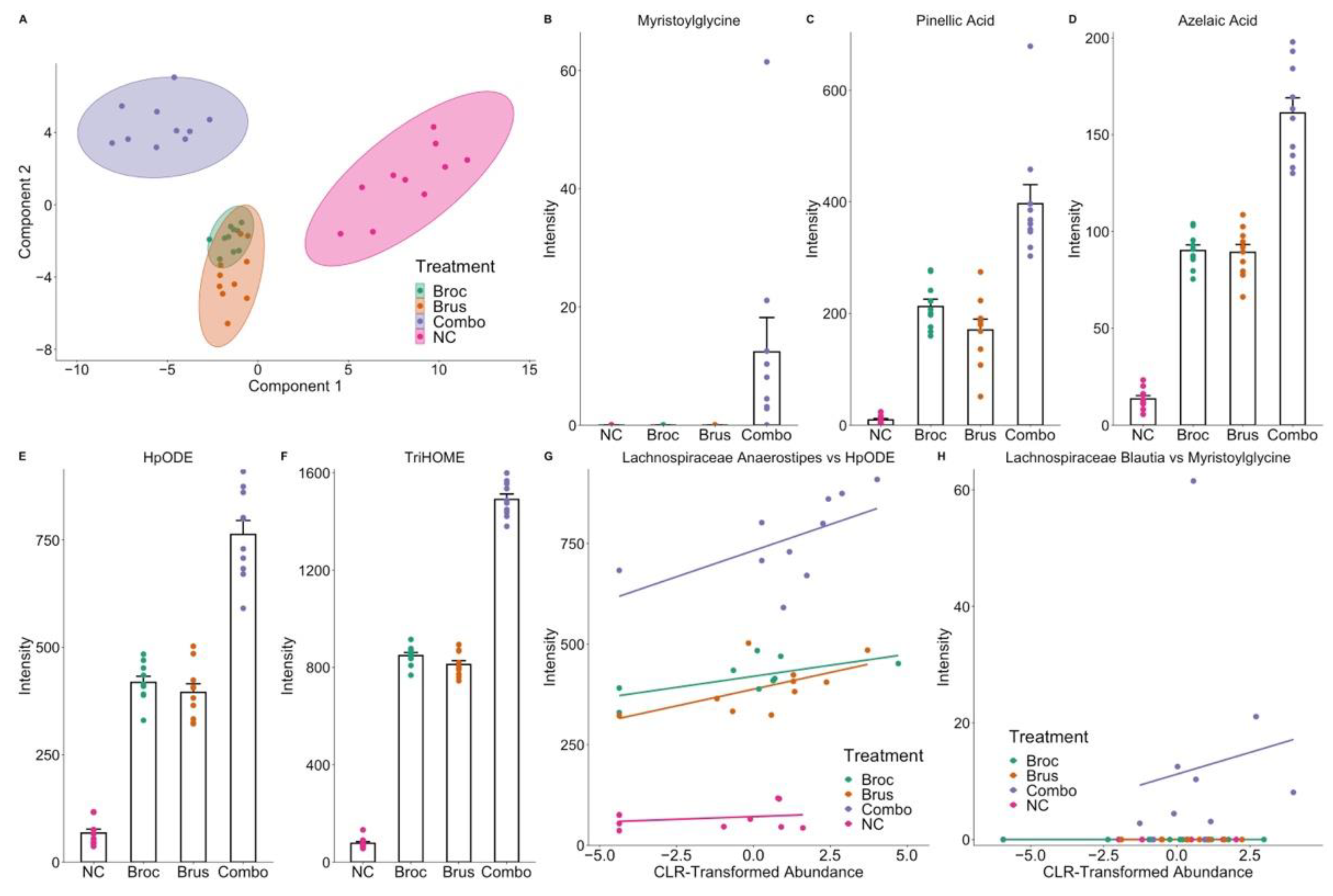
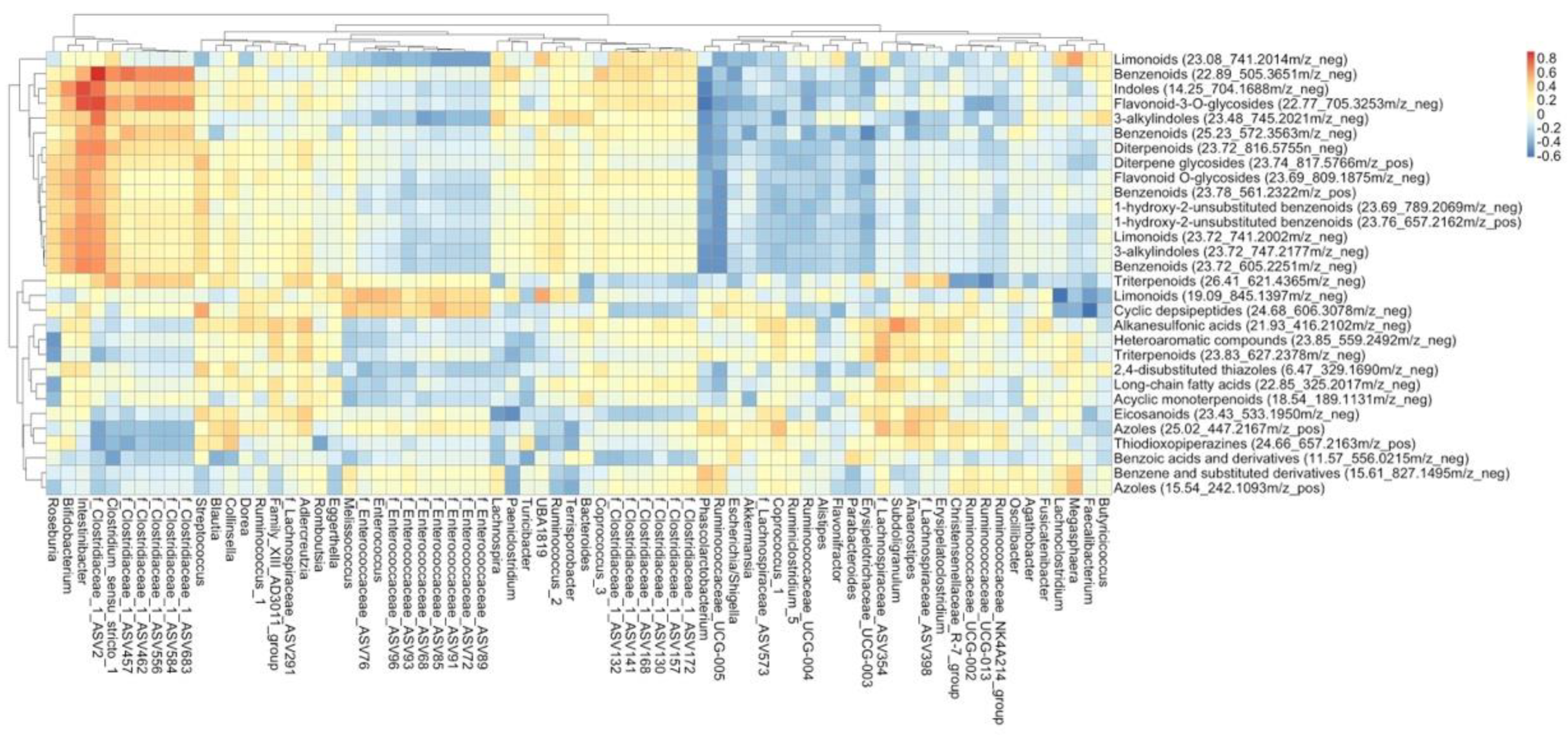
3.3. Interplay between Cruciferous Vegetables and the Gut Microbiome
3.4. Identification of Microbial Metabolites of Cruciferous Vegetables
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aune, D.; Giovannucci, E.; Boffetta, P.; Fadnes, L.T.; Keum, N.; Norat, T.; Greenwood, D.C.; Riboli, E.; Vatten, L.J.; Tonstad, S. Fruit and vegetable intake and the risk of cardiovascular disease, total cancer and all-cause mortality—A systematic review and dose-response meta-analysis of prospective studies. Int. J. Epidemiol. 2017, 46, 1029–1056. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.K.; Gallicchio, L.; Boyd, K.; Shiels, M.; Hammond, E.; Tao, X.G.; Chen, L.; Robinson, K.A.; Caulfield, L.E.; Herman, J.G.; et al. Cruciferous vegetable consumption and lung cancer risk: A systematic review. Cancer Epidemiol. Biomark. Prev. 2009, 18, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Turati, F.; Rossi, M.; Pelucchi, C.; Levi, F.; Vecchia, C.L. Fruit and vegetables and cancer risk: A review of southern European studies. Br. J. Nutr. 2015, 113, S102–S110. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Mao, Q.; Lin, Y.; Zhou, F.; Xie, L. The Association of cruciferous vegetables intake and risk of bladder cancer: A meta-analysis. World J. Urol. 2013, 31, 127–133. [Google Scholar] [CrossRef]
- Yao, B.; Yan, Y.; Ye, X.; Fang, H.; Xu, H.; Liu, Y.; Li, S.; Zhao, Y. Intake of fruit and vegetables and risk of bladder cancer: A dose-response meta-analysis of observational studies. Cancer Causes Control 2014, 25, 1645–1658. [Google Scholar] [CrossRef]
- Vieira, A.R.; Vingeliene, S.; Chan, D.S.M.; Aune, D.; Abar, L.; Navarro Rosenblatt, D.; Greenwood, D.C.; Norat, T. Fruits, vegetables, and bladder cancer risk: A systematic review and meta-analysis. Cancer Med. 2015, 4, 136–146. [Google Scholar] [CrossRef]
- Xu, C.; Zeng, X.-T.; Liu, T.-Z.; Zhang, C.; Yang, Z.-H.; Li, S.; Chen, X.-Y. Fruits and vegetables intake and risk of bladder cancer: A PRISMA-compliant systematic review and dose-response meta-analysis of prospective cohort studies. Medicine 2015, 94, e759. [Google Scholar] [CrossRef]
- Liu, X.; Lv, K. Cruciferous vegetables intake is inversely associated with risk of breast cancer: A meta-analysis. Breast 2013, 22, 309–313. [Google Scholar] [CrossRef]
- Wu, Q.J.; Yang, Y.; Vogtmann, E.; Wang, J.; Han, L.H.; Li, H.L.; Xiang, Y.B. Cruciferous vegetables intake and the risk of colorectal cancer: A meta-analysis of observational studies. Ann. Oncol. 2013, 24, 1079–1087. [Google Scholar] [CrossRef]
- Tse, G.; Eslick, G.D. Cruciferous vegetables and risk of colorectal neoplasms: A systematic review and meta-analysis. Nutr. Cancer 2014, 66, 128–139. [Google Scholar] [CrossRef]
- Bandera, E.V.; Kushi, L.H.; Moore, D.F.; Gifkins, D.M.; McCullough, M.L. Fruits and vegetables and endometrial cancer risk: A systematic literature review and meta-analysis. Nutr. Cancer 2007, 58, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Li, X.; Yu, T. Cruciferous vegetables consumption and the risk of ovarian cancer: A meta-analysis of observational studies. Diagn. Pathol. 2014, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hu, Y.; Hu, Y.; Zheng, S. Intake of cruciferous vegetables is associated with reduced risk of ovarian cancer: A meta-analysis. Asia Pac. J. Clin. Nutr. 2015, 24, 101–109. [Google Scholar] [CrossRef]
- Li, L.; Luo, Y.; Lu, M.; Xu, X.; Lin, H.; Zheng, Z. Cruciferous vegetable consumption and the risk of pancreatic cancer: A meta-analysis. World J. Surg. Oncol. 2015, 13, 44. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Mao, Q.; Cao, M.; Xie, L. Cruciferous vegetables intake and risk of prostate cancer: A meta-analysis. Int. J. Urol. 2012, 19, 134–141. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, L. Cruciferous vegetables intake is associated with lower risk of renal cell carcinoma: Evidence from a meta-analysis of observational studies. PLoS ONE 2013, 8, e75732. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Mao, Q.; Wang, X.; Zhou, F.; Luo, J.; Wang, C.; Lin, Y.; Zheng, X.; Xie, L. Cruciferous vegetables consumption and risk of renal cell carcinoma: A meta-analysis. Nutr. Cancer 2013, 65, 668–676. [Google Scholar] [CrossRef]
- Clarke, J.D.; Dashwood, R.H.; Ho, E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008, 269, 291–304. [Google Scholar] [CrossRef]
- Atwell, L.L.; Beaver, L.M.; Shannon, J.; Williams, D.E.; Dashwood, R.H.; Ho, E. Epigenetic regulation by sulforaphane: Opportunities for breast and prostate cancer chemoprevention. Curr. Pharm. Rep. 2015, 1, 102–111. [Google Scholar] [CrossRef]
- Watson, G.W.; Beaver, L.M.; Williams, D.E.; Dashwood, R.H.; Ho, E. Phytochemicals from cruciferous vegetables, epigenetics, and prostate cancer prevention. AAPS J. 2013, 15, 951–961. [Google Scholar] [CrossRef]
- Verkerk, R.; Schreiner, M.; Krumbein, A.; Ciska, E.; Holst, B.; Rowland, I.; Schrijver, R.D.; Hansen, M.; Gerhäuser, C.; Mithen, R.; et al. Glucosinolates in brassica vegetables: The influence of the food supply chain on intake, bioavailability and human health. Mol. Nutr. Food Res. 2009, 53, S219. [Google Scholar] [CrossRef] [PubMed]
- Bouranis, J.A.; Beaver, L.M.; Choi, J.; Wong, C.P.; Jiang, D.; Sharpton, T.J.; Stevens, J.F.; Ho, E. Composition of the gut microbiome influences production of sulforaphane-nitrile and iberin-nitrile from glucosinolates in broccoli sprouts. Nutrients 2021, 13, 3013. [Google Scholar] [CrossRef]
- Guadamuro, L.; Dohrmann, A.B.; Tebbe, C.C.; Mayo, B.; Delgado, S. Bacterial communities and metabolic activity of faecal cultures from equol producer and non-producer menopausal women under treatment with soy isoflavones. BMC Microbiol. 2017, 17, 93. [Google Scholar] [CrossRef] [PubMed]
- Setchell, K.D.R.; Brown, N.M.; Lydeking-Olsen, E. The clinical importance of the metabolite equol—A clue to the effectiveness of soy and its isoflavones. J. Nutr. 2002, 132, 3577–3584. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; González-Sarrías, A.; García-Villalba, R.; Núñez-Sánchez, M.A.; Selma, M.V.; García-Conesa, M.T.; Espín, J.C. Urolithins, the rescue of “old” metabolites to understand a “new” concept: Metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol. Nutr. Food Res. 2017, 61, 1500901. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Hoeflinger, J.L.; Neme, B.P.; Jeffery, E.H.; Miller, M.J. Dietary broccoli alters rat cecal microbiota to improve glucoraphanin hydrolysis to bioactive isothiocyanates. Nutrients 2017, 9, 262. [Google Scholar] [CrossRef]
- Liou, C.S.; Sirk, S.J.; Diaz, C.A.C.; Klein, A.P.; Fischer, C.R.; Higginbottom, S.K.; Erez, A.; Donia, M.S.; Sonnenburg, J.L.; Sattely, E.S. A metabolic pathway for activation of dietary glucosinolates by a human gut symbiont. Cell 2020, 180, 717–728.e19. [Google Scholar] [CrossRef]
- Wu, Y.; Shen, Y.; Zhu, Y.; Mupunga, J.; Zou, L.; Liu, C.; Liu, S.; Mao, J. Broccoli ingestion increases the glucosinolate hydrolysis activity of microbiota in the mouse gut. Int. J. Food Sci. Nutr. 2019, 70, 585–594. [Google Scholar] [CrossRef]
- Luang-In, V.; Narbad, A.; Nueno-Palop, C.; Mithen, R.; Bennett, M.; Rossiter, J.T. The metabolism of methylsulfinylalkyl- and methylthioalkyl-glucosinolates by a selection of human gut bacteria. Mol. Nutr. Food Res. 2014, 58, 875–883. [Google Scholar] [CrossRef]
- Luang-In, V.; Albaser, A.A.; Nueno-Palop, C.; Bennett, M.H.; Narbad, A.; Rossiter, J.T. Glucosinolate and desulfo-glucosinolate metabolism by a selection of human gut bacteria. Curr. Microbiol. 2016, 73, 442–451. [Google Scholar] [CrossRef]
- Kellingray, L.; Tapp, H.S.; Saha, S.; Doleman, J.F.; Narbad, A.; Mithen, R.F. Consumption of a diet rich in brassica vegetables is associated with a reduced abundance of sulphate-reducing bacteria: A randomised crossover study. Mol. Nutr. Food Res. 2017, 61, 1600992. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, J.L.; Liu, X.; Charron, C.S.; Novotny, J.A.; Jeffery, E.H.; Seifried, H.E.; Ross, S.A.; Miller, M.J.; Swanson, K.S.; Holscher, H.D. Broccoli consumption affects the human gastrointestinal microbiota. J. Nutr. Biochem. 2019, 63, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hullar, M.A.J.; Schwarz, Y.; Lampe, J.W. Human gut bacterial communities are altered by addition of cruciferous vegetables to a controlled fruit- and vegetable-free diet. J. Nutr. 2009, 139, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hullar, M.A.J.; Beresford, S.A.A.; Lampe, J.W. Variation of glucoraphanin metabolism in vivo and ex vivo by human gut bacteria. Br. J. Nutr. 2011, 106, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Zandani, G.; Kaftori-Sandler, N.; Sela, N.; Nyska, A.; Madar, Z. Dietary broccoli improves markers associated with glucose and lipid metabolism through modulation of gut microbiota in mice. Nutrition 2021, 90, 111240. [Google Scholar] [CrossRef]
- Eve, A.A.; Liu, X.; Wang, Y.; Miller, M.J.; Jeffery, E.H.; Madak-Erdogan, Z. Biomarkers of broccoli consumption: Implications for glutathione metabolism and liver health. Nutrients 2020, 12, 2514. [Google Scholar] [CrossRef]
- Jun, S.-R.; Cheema, A.; Bose, C.; Boerma, M.; Palade, P.T.; Carvalho, E.; Awasthi, S.; Singh, S.P. Multi-omic analysis reveals different effects of sulforaphane on the microbiome and metabolome in old compared to young mice. Microorganisms 2020, 8, 1500. [Google Scholar] [CrossRef]
- He, C.; Huang, L.; Lei, P.; Liu, X.; Li, B.; Shan, Y. Sulforaphane normalizes intestinal flora and enhances gut barrier in mice with BBN-induced bladder cancer. Mol. Nutr. Food Res. 2018, 62, e1800427. [Google Scholar] [CrossRef]
- Thomaz, F.S.; Altemani, F.; Panchal, S.K.; Worrall, S.; Dekker Nitert, M. The influence of wasabi on the gut microbiota of high-carbohydrate, high-fat diet-induced hypertensive wistar rats. J. Hum. Hypertens. 2021, 35, 170–180. [Google Scholar] [CrossRef]
- Xu, X.; Dai, M.; Lao, F.; Chen, F.; Hu, X.; Liu, Y.; Wu, J. Effect of glucoraphanin from broccoli seeds on lipid levels and gut microbiota in high-fat diet-fed mice. J. Funct. Foods 2020, 68, 103858. [Google Scholar] [CrossRef]
- Housley, L.; Magana, A.A.; Hsu, A.; Beaver, L.M.; Wong, C.P.; Stevens, J.F.; Choi, J.; Jiang, Y.; Bella, D.; Williams, D.E.; et al. Untargeted metabolomic screen reveals changes in human plasma metabolite profiles following consumption of fresh broccoli sprouts. Mol. Nutr. Food Res. 2018, 62, 1700665. [Google Scholar] [CrossRef] [PubMed]
- Charron, C.S.; Vinyard, B.T.; Ross, S.A.; Seifried, H.E.; Jeffery, E.H.; Novotny, J.A. Absorption and metabolism of isothiocyanates formed from broccoli glucosinolates: Effects of BMI and daily consumption in a randomised clinical trial. Br. J. Nutr. 2018, 120, 1370–1379. [Google Scholar] [CrossRef]
- Charron, C.S.; Vinyard, B.T.; Jeffery, E.H.; Ross, S.A.; Seifried, H.E.; Novotny, J.A. BMI is associated with increased plasma and urine appearance of glucosinolate metabolites after consumption of cooked broccoli. Br. J. Nutr. 2020, 7, 575092. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Riedl, K.; Bella, D.; Schwartz, S.J.; Stevens, J.F.; Ho, E. Comparison of isothiocyanate metabolite levels and histone deacetylase activity in human subjects consuming broccoli sprouts or broccoli supplement. J. Agric. Food Chem. 2011, 59, 10955–10963. [Google Scholar] [CrossRef]
- Atwell, L.L.; Hsu, A.; Wong, C.P.; Stevens, J.F.; Bella, D.; Yu, T.-W.; Pereira, C.B.; Löhr, C.V.; Christensen, J.M.; Dashwood, R.H.; et al. Absorption and chemopreventive targets of sulforaphane in humans following consumption of broccoli sprouts or a myrosinase-treated broccoli sprout extract. Mol. Nutr. Food Res. 2015, 59, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Hsu, A.; Riedl, K.; Bella, D.; Schwartz, S.J.; Stevens, J.F.; Ho, E. Bioavailability and inter-conversion of sulforaphane and erucin in human subjects consuming broccoli sprouts or broccoli supplement in a cross-over study design. Pharm. Res. 2011, 64, 456–463. [Google Scholar] [CrossRef]
- Atwell, L.L.; Zhang, Z.; Mori, M.; Farris, P.; Vetto, J.T.; Naik, A.M.; Oh, K.Y.; Thuillier, P.; Ho, E.; Shannon, J. Sulforaphane bioavailability and chemopreventive activity in women scheduled for breast biopsy. Cancer Prev. Res. 2015, 8, 1184–1191. [Google Scholar] [CrossRef]
- Fahey, J.W.; Wehage, S.L.; Holtzclaw, W.D.; Kensler, T.W.; Egner, P.A.; Shapiro, T.A.; Talalay, P. Protection of humans by plant glucosinolates: Efficiency of conversion of glucosinolates to isothiocyanates by the gastrointestinal microflora. Cancer Prev. Res. 2012, 5, 603–611. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The human metabolome database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Guijas, C.; Benton, H.P.; Warth, B.; Siuzdak, G. METLIN MS2 molecular standards database: A broad chemical and biological resource. Nat. Methods 2020, 17, 953–954. [Google Scholar] [CrossRef] [PubMed]
- Aura, A.-M.; Härkönen, H.; Fabritius, M.; Poutanen, K. Development of an in vitro enzymic digestion method for removal of starch and protein and assessment of its performance using rye and wheat breads. J. Cereal Sci. 1999, 29, 139–152. [Google Scholar] [CrossRef]
- Gil-Izquierdo, A.; Zafrilla, P.; Tomás-Barberán, F.A. An in vitro method to simulate phenolic compound release from the food matrix in the gastrointestinal tract. Eur. Food Res. Technol. 2002, 214, 155–159. [Google Scholar] [CrossRef]
- Vallejo, F.; Gil-Izquierdo, A.; Pérez-Vicente, A.; García-Viguera, C. In vitro gastrointestinal digestion study of broccoli inflorescence phenolic compounds, glucosinolates, and vitamin C. J. Agric. Food Chem. 2004, 52, 135–138. [Google Scholar] [CrossRef]
- Sarvan, I.; Kramer, E.; Bouwmeester, H.; Dekker, M.; Verkerk, R. Sulforaphane formation and bioaccessibility are more affected by steaming time than meal composition during in vitro digestion of broccoli. Food Chem. 2017, 214, 580–586. [Google Scholar] [CrossRef]
- Rychlik, J.; Olejnik, A.; Olkowicz, M.; Kowalska, K.; Juzwa, W.; Myszka, K.; Dembczyński, R.; Moyer, M.P.; Grajek, W. Antioxidant capacity of broccoli sprouts subjected to gastrointestinal digestion. J. Sci. Food Agric. 2015, 95, 1892–1902. [Google Scholar] [CrossRef]
- Center for Food Safety and Applied Nutrition. Guidance for Industry: Compliance with and Recommendations for Implementation of the Standards for the Growing, Harvesting, Packing, and Holding of Produce for Human Consumption for Sprout Operations; Center for Food Safety and Applied Nutrition: College Park, MD, USA, 2017. [Google Scholar]
- Henson, W.Y. U.S. EPA, Pesticides, Label, ECR CALCIUM HYPOCHLORITE T, 3/17/2011. 2011. Available online: https://www3.epa.gov/pesticides/chem_search/ppls/086460-00004-20110317.pdf (accessed on 29 June 2021).
- Fahey, J.W.; Zalcmann, A.T.; Talalay, P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001, 56, 5–51. [Google Scholar] [CrossRef]
- National Advisory Committee on Microbiological Criteria for Foods. Microbiological safety evaluations and recommendations on sprouted seeds. Int. J. Food Microbiol. 1999, 52, 123–153. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Illumina 16S Sample Preparation Guide; Illumina: San Diego, CA, USA, 2013.
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soci. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; Hara, B.; Simpson, G.; Solymos, P.; Henry, M.; Stevens, H.; Maintainer, H.; Oksanen@oulu, J. The vegan package. Commun. Ecol. Package 2009, 10, 631–637. [Google Scholar]
- Wickham, H. Ggplot2. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- García-Jaramillo, M.; Beaver, L.M.; Truong, L.; Axton, E.R.; Keller, R.M.; Prater, M.C.; Magnusson, K.R.; Tanguay, R.L.; Stevens, J.F.; Hord, N.G. Nitrate and nitrite exposure leads to mild anxiogenic-like behavior and alters brain metabolomic profile in zebrafish. PLoS ONE 2020, 15, e0240070. [Google Scholar] [CrossRef]
- Kirkwood, J.S.; Lebold, K.M.; Miranda, C.L.; Wright, C.L.; Miller, G.W.; Tanguay, R.L.; Barton, C.L.; Traber, M.G.; Stevens, J.F. Vitamin C deficiency activates the purine nucleotide cycle in zebrafish. J. Biol. Chem. 2012, 287, 3833–3841. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. LmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Alcazar Magana, A.; Wright, K.; Vaswani, A.; Caruso, M.; Reed, R.L.; Bailey, C.F.; Nguyen, T.; Gray, N.E.; Soumyanath, A.; Quinn, J.; et al. Integration of mass spectral fingerprinting analysis with precursor ion (MS1) quantification for the characterisation of botanical extracts: Application to extracts of Centella asiatica (L.) urban. Phytochem. Anal. 2020, 31, 722–738. [Google Scholar] [CrossRef] [PubMed]
- Keller, R.M.; Beaver, L.M.; Reardon, P.N.; Prater, M.C.; Truong, L.; Robinson, M.M.; Tanguay, R.L.; Stevens, J.F.; Hord, N.G. Nitrate-induced improvements in exercise performance are coincident with exuberant changes in metabolic genes and the metabolome in zebrafish (Danio rerio) skeletal muscle. J. Appl. Physiol. 2021, 131, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
- Hoffmann, M.A.; Nothias, L.-F.; Ludwig, M.; Fleischauer, M.; Gentry, E.C.; Witting, M.; Dorrestein, P.C.; Dührkop, K.; Böcker, S. High-confidence structural annotation of metabolites absent from spectral libraries. Nat. Biotechnol. 2022, 40, 411–421. [Google Scholar] [CrossRef]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2021, 39, 462–471. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Böcker, S. Fragmentation trees reloaded. In Proceedings of the Research in Computational Molecular Biology; Przytycka, T.M., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 65–79. [Google Scholar]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [PubMed]
- Böcker, S.; Letzel, M.C.; Lipták, Z.; Pervukhin, A. SIRIUS: Decomposing isotope patterns for metabolite identification. Bioinformatics 2009, 25, 218–224. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fiehn, O. Chemical similarity enrichment analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Shannon, C.P.; Gautier, B.; Rohart, F.; Vacher, M.; Tebbutt, S.J.; Lê Cao, K.-A. DIABLO: An integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 2019, 35, 3055–3062. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Santiago, M.; Fernández-Peralbo, M.A.; Priego-Capote, F.; Luque de Castro, M.D. MSCombine: A Tool for merging untargeted metabolomic data from high-resolution mass spectrometry in the positive and negative ionization modes. Metabolomics 2016, 12, 43. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. MixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Liquet, B.; Cao, K.-A.L.; Hocini, H.; Thiébaut, R. A novel approach for biomarker selection and the integration of repeated measures experiments from two assays. BMC Bioinform. 2012, 13, 325. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. PROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Kahalehili, H.M.; Newman, N.K.; Pennington, J.M.; Kolluri, S.K.; Kerkvliet, N.I.; Shulzhenko, N.; Morgun, A.; Ehrlich, A.K. Dietary indole-3-carbinol activates AhR in the gut, alters Th17-microbe interactions, and exacerbates insulitis in NOD mice. Front. Immunol. 2021, 11, 606441. [Google Scholar] [CrossRef]
- Zhang, L.S.; Davies, S.S. Microbial metabolism of dietary components to bioactive metabolites: Opportunities for new therapeutic interventions. Genome Med. 2016, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.E. Indoles derived from glucobrassicin: Cancer chemoprevention by indole-3-carbinol and 3,3′-diindolylmethane. Front. Nutr. 2021, 8, 734334. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, B.; Wang, B.; Zhang, X. Volatile components of ethanolic extract from broccolini leaves. Nat. Prod. Res. 2012, 26, 680–683. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Gao, Y.; Chen, H.; Yin, Y.; Zhang, W. Indole-3-acetic acid alleviates nonalcoholic fatty liver disease in mice via attenuation of hepatic lipogenesis, and oxidative and inflammatory stress. Nutrients 2019, 11, 2062. [Google Scholar] [CrossRef]
- Yadav, V.R.; Prasad, S.; Sung, B.; Kannappan, R.; Aggarwal, B.B. Targeting inflammatory pathways by triterpenoids for prevention and treatment of cancer. Toxins 2010, 2, 2428–2466. [Google Scholar] [CrossRef]
- Bishayee, A.; Ahmed, S.; Brankov, N.; Perloff, M. Triterpenoids as potential agents for the chemoprevention and therapy of breast cancer. Front. Biosci. 2011, 16, 980–996. [Google Scholar] [CrossRef]
- Carbonero, F.; Benefiel, A.; Alizadeh-Ghamsari, A.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouranis, J.A.; Beaver, L.M.; Jiang, D.; Choi, J.; Wong, C.P.; Davis, E.W.; Williams, D.E.; Sharpton, T.J.; Stevens, J.F.; Ho, E. Interplay between Cruciferous Vegetables and the Gut Microbiome: A Multi-Omic Approach. Nutrients 2023, 15, 42. https://doi.org/10.3390/nu15010042
Bouranis JA, Beaver LM, Jiang D, Choi J, Wong CP, Davis EW, Williams DE, Sharpton TJ, Stevens JF, Ho E. Interplay between Cruciferous Vegetables and the Gut Microbiome: A Multi-Omic Approach. Nutrients. 2023; 15(1):42. https://doi.org/10.3390/nu15010042
Chicago/Turabian StyleBouranis, John A., Laura M. Beaver, Duo Jiang, Jaewoo Choi, Carmen P. Wong, Edward W. Davis, David E. Williams, Thomas J. Sharpton, Jan F. Stevens, and Emily Ho. 2023. "Interplay between Cruciferous Vegetables and the Gut Microbiome: A Multi-Omic Approach" Nutrients 15, no. 1: 42. https://doi.org/10.3390/nu15010042
APA StyleBouranis, J. A., Beaver, L. M., Jiang, D., Choi, J., Wong, C. P., Davis, E. W., Williams, D. E., Sharpton, T. J., Stevens, J. F., & Ho, E. (2023). Interplay between Cruciferous Vegetables and the Gut Microbiome: A Multi-Omic Approach. Nutrients, 15(1), 42. https://doi.org/10.3390/nu15010042






