Mechanisms of Feedback Regulation of Vitamin A Metabolism
Abstract
1. Introduction
2. Bioactive Vitamin A Metabolites
3. Transcriptional Regulation Mediated by RAR-RXR
4. Vitamin A Supplementation
5. Vitamin A Absorption
6. Vitamin A Storage
7. Vitamin A Delivery to Target Tissues
8. Conversion of Retinol to RA
9. Cellular Fate of RA and RA Breakdown
10. Homeostasis in Vitamin A Metabolism
- upregulation of genes responsible for sequestering RA precursors such as Crbp1 and Lrat.
- upregulation of genes responsible for opposing RA formation (Dhrs3) and the degradation of RA (Cyp26a1)
- downregulation of genes involved in the synthesis of RA (Rdh10, Raldh2)
- downregulation of genes involved in the uptake of carotenoids (Srb1) and conversion of β-carotene to retinaldehyde (Bco1)
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKR | aldo-keto reductase |
| CRBP | cellular retinol binding protein |
| CRABP | cellular retinoic acid binding protein |
| CYP | cytochrome P450 |
| DHRS | dehydrogenase/reductase (SDR family) member |
| LRAT | lecithin:retinol acyltransferase |
| NAD | nicotinamide adenine dinucleotide |
| NADP | nicotinamide adenine dinucleotide phosphate |
| NHR | nuclear hormone receptor |
| RA | all-trans-retinoic acid |
| RAR | retinoic acid receptor |
| RXR | retinoid X receptor |
| RALDH | retinaldehyde dehydrogenase |
| RDH | retinol dehydrogenase |
| SDR | short-chain dehydrogenases reductase |
| TTR | transthyretin |
References
- Levin, E.R.; Hammes, S.R. Nuclear receptors outside the nucleus: Extranuclear signalling by steroid receptors. Nat. Rev. Mol. Cell. Biol. 2016, 17, 783–797. [Google Scholar] [CrossRef]
- Frigo, D.E.; Bondesson, M.; Williams, C. Nuclear receptors: From molecular mechanisms to therapeutics. Essays Biochem. 2021, 65, 847–856. [Google Scholar] [CrossRef]
- Weikum, E.R.; Liu, X.; Ortlund, E.A. The nuclear receptor superfamily: A structural perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef]
- Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell 1999, 97, 161–163.
- Cannon, W.B. Organization for Physiological Homeostasis. Physiol. Rev. 1929, 9, 399–431. [Google Scholar] [CrossRef]
- Grune, T.; Lietz, G.; Palou, A.; Ross, A.C.; Stahl, W.; Tang, G.; Thurnham, D.; Yin, S.A.; Biesalski, H.K. Beta-carotene is an important vitamin A source for humans. J. Nutr. 2010, 140, 2268S–2285S. [Google Scholar] [CrossRef]
- Sporn, M.B.; Dunlop, N.M.; Newton, D.L.; Smith, J.M. Prevention of chemical carcinogenesis by vitamin A and its synthetic analogs (retinoids). Fed. Proc. 1976, 35, 1332–1338. [Google Scholar]
- Dowling, J.E.; Wald, G. The Biological Function of Vitamin a Acid. Proc. Natl. Acad. Sci. USA 1960, 46, 587–608. [Google Scholar] [CrossRef]
- Alexander, S.P.H.; Cidlowski, J.A.; Kelly, E.; Mathie, A.; Peters, J.A.; Veale, E.L.; Armstrong, J.F.; Faccenda, E.; Harding, S.D.; Pawson, A.J.; et al. The Concise Guide to Pharmacology 2019/20: Nuclear hormone receptors. Br. J. Pharmacol. 2019, 176 (Suppl. 1), S229–S246. [Google Scholar] [CrossRef]
- Heyman, R.A.; Mangelsdorf, D.J.; Dyck, J.A.; Stein, R.B.; Eichele, G.; Evans, R.M.; Thaller, C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell 1992, 68, 397–406. [Google Scholar] [CrossRef]
- Zhang, X.K.; Lehmann, J.; Hoffmann, B.; Dawson, M.I.; Cameron, J.; Graupner, G.; Hermann, T.; Tran, P.; Pfahl, M. Homodimer formation of retinoid X receptor induced by 9-cis retinoic acid. Nature 1992, 358, 587–591. [Google Scholar] [CrossRef]
- Kliewer, S.A.; Umesono, K.; Noonan, D.J.; Heyman, R.A.; Evans, R.M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 1992, 358, 771–774. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Borgmeyer, U.; Heyman, R.A.; Zhou, J.Y.; Ong, E.S.; Oro, A.E.; Kakizuka, A.; Evans, R.M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992, 6, 329–344. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Evans, R.M. The RXR heterodimers and orphan receptors. Cell 1995, 83, 841–850. [Google Scholar] [CrossRef]
- Levin, A.A.; Sturzenbecker, L.J.; Kazmer, S.; Bosakowski, T.; Huselton, C.; Allenby, G.; Speck, J.; Kratzeisen, C.; Rosenberger, M.; Lovey, A.; et al. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature 1992, 355, 359–361. [Google Scholar] [CrossRef]
- Liu, L.; Derguini, F.; Gudas, L.J. Metabolism and regulation of gene expression by 4-oxoretinol versus all-trans retinoic acid in normal human mammary epithelial cells. J. Cell Physiol. 2009, 220, 771–779. [Google Scholar] [CrossRef]
- Idres, N.; Marill, J.; Flexor, M.A.; Chabot, G.G. Activation of retinoic acid receptor-dependent transcription by all-trans-retinoic acid metabolites and isomers. J. Biol. Chem. 2002, 277, 31491–31498. [Google Scholar] [CrossRef]
- Buck, J.; Derguini, F.; Levi, E.; Nakanishi, K.; Hammerling, U. Intracellular signaling by 14-hydroxy-4,14-retro-retinol. Science 1991, 254, 1654–1656. [Google Scholar] [CrossRef]
- Buck, J.; Grun, F.; Derguini, F.; Chen, Y.; Kimura, S.; Noy, N.; Hammerling, U. Anhydroretinol: A naturally occurring inhibitor of lymphocyte physiology. J. Exp. Med. 1993, 178, 675–680. [Google Scholar] [CrossRef]
- Pijnappel, W.W.; Hendriks, H.F.; Folkers, G.E.; van den Brink, C.E.; Dekker, E.J.; Edelenbosch, C.; van der Saag, P.T.; Durston, A.J. The retinoid ligand 4-oxo-retinoic acid is a highly active modulator of positional specification. Nature 1993, 366, 340–344. [Google Scholar] [CrossRef]
- Derguini, F.; Nakanishi, K.; Hammerling, U.; Buck, J. Intracellular signaling activity of synthetic (14R)-, (14S)-, and (14RS)-14-hydroxy-4,14-retro-retinol. Biochemistry 1994, 33, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Derguini, F.; Nakanishi, K.; Hammerling, U.; Chua, R.; Eppinger, T.; Levi, E.; Buck, J. 13,14-Dihydroxy-retinol, a new bioactive retinol metabolite. J. Biol. Chem. 1995, 270, 18875–18880. [Google Scholar] [CrossRef]
- Achkar, C.C.; Derguini, F.; Blumberg, B.; Langston, A.; Levin, A.A.; Speck, J.; Evans, R.M.; Bolado, J., Jr.; Nakanishi, K.; Buck, J.; et al. 4-Oxoretinol, a new natural ligand and transactivator of the retinoic acid receptors. Proc. Natl. Acad. Sci. USA 1996, 93, 4879–4884. [Google Scholar] [CrossRef]
- Blumberg, B.; Bolado, J., Jr.; Derguini, F.; Craig, A.G.; Moreno, T.A.; Chakravarti, D.; Heyman, R.A.; Buck, J.; Evans, R.M. Novel retinoic acid receptor ligands in Xenopus embryos. Proc. Natl. Acad. Sci. USA 1996, 93, 4873–4878. [Google Scholar] [CrossRef]
- Lane, M.A.; Chen, A.C.; Roman, S.D.; Derguini, F.; Gudas, L.J. Removal of LIF (leukemia inhibitory factor) results in increased vitamin A (retinol) metabolism to 4-oxoretinol in embryonic stem cells. Proc. Natl. Acad. Sci. USA 1999, 96, 13524–13529. [Google Scholar] [CrossRef]
- Moise, A.R.; Kuksa, V.; Imanishi, Y.; Palczewski, K. Identification of all-trans-retinol:all-trans-13,14-dihydroretinol saturase. J. Biol. Chem. 2004, 279, 50230–50242. [Google Scholar] [CrossRef]
- Moise, A.R.; Kuksa, V.; Blaner, W.S.; Baehr, W.; Palczewski, K. Metabolism and transactivation activity of 13,14-dihydroretinoic acid. J. Biol. Chem. 2005, 280, 27815–27825. [Google Scholar] [CrossRef]
- Moise, A.R.; Dominguez, M.; Alvarez, S.; Alvarez, R.; Schupp, M.; Cristancho, A.G.; Kiser, P.D.; de Lera, A.R.; Lazar, M.A.; Palczewski, K. Stereospecificity of retinol saturase: Absolute configuration, synthesis, and biological evaluation of dihydroretinoids. J. Am. Chem. Soc. 2008, 130, 1154–1155. [Google Scholar] [CrossRef]
- Moise, A.R.; Isken, A.; Dominguez, M.; de Lera, A.R.; von Lintig, J.; Palczewski, K. Specificity of zebrafish retinol saturase: Formation of all-trans-13,14-dihydroretinol and all-trans-7,8- dihydroretinol. Biochemistry 2007, 46, 1811–1820. [Google Scholar] [CrossRef]
- Moise, A.R.; Alvarez, S.; Dominguez, M.; Alvarez, R.; Golczak, M.; Lobo, G.P.; von Lintig, J.; de Lera, A.R.; Palczewski, K. Activation of retinoic acid receptors by dihydroretinoids. Mol. Pharmacol. 2009, 76, 1228–1237. [Google Scholar] [CrossRef]
- Moise, A.R.; Lobo, G.P.; Erokwu, B.; Wilson, D.L.; Peck, D.; Alvarez, S.; Dominguez, M.; Alvarez, R.; Flask, C.A.; de Lera, A.R.; et al. Increased adiposity in the retinol saturase-knockout mouse. FASEB J. 2010, 24, 1261–1270. [Google Scholar] [CrossRef]
- Shirley, M.A.; Bennani, Y.L.; Boehm, M.F.; Breau, A.P.; Pathirana, C.; Ulm, E.H. Oxidative and reductive metabolism of 9-cis-retinoic acid in the rat. Identification of 13,14-dihydro-9-cis-retinoic acid and its taurine conjugate. Drug Metab. Dispos. 1996, 24, 293–302. [Google Scholar] [PubMed]
- Schmidt, C.K.; Volland, J.; Hamscher, G.; Nau, H. Characterization of a new endogenous vitamin A metabolite. Biochim. Biophys. Acta 2002, 1583, 237–251. [Google Scholar] [CrossRef]
- Schmidt, C.K.; Hoegberg, P.; Fletcher, N.; Nilsson, C.B.; Trossvik, C.; Hakansson, H.; Nau, H. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters the endogenous metabolism of all-trans-retinoic acid in the rat. Arch. Toxicol. 2003, 77, 371–383. [Google Scholar] [PubMed]
- Hoegberg, P.; Schmidt, C.K.; Fletcher, N.; Nilsson, C.B.; Trossvik, C.; Gerlienke Schuur, A.; Brouwer, A.; Nau, H.; Ghyselinck, N.B.; Chambon, P.; et al. Retinoid status and responsiveness to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking retinoid binding protein or retinoid receptor forms. Chem. Biol. Interact. 2005, 156, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Schuchardt, J.P.; Wahlstrom, D.; Ruegg, J.; Giese, N.; Stefan, M.; Hopf, H.; Pongratz, I.; Hakansson, H.; Eichele, G.; Pettersson, K.; et al. The endogenous retinoid metabolite S-4-oxo-9-cis-13,14-dihydro-retinoic acid activates retinoic acid receptor signalling both in vitro and in vivo. FEBS J. 2009, 276, 3043–3059. [Google Scholar] [CrossRef]
- Ruhl, R.; Krzyzosiak, A.; Niewiadomska-Cimicka, A.; Rochel, N.; Szeles, L.; Vaz, B.; Wietrzych-Schindler, M.; Alvarez, S.; Szklenar, M.; Nagy, L.; et al. 9-cis-13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice. PLoS Genet. 2015, 11, e1005213. [Google Scholar] [CrossRef]
- Krezel, W.; Ruhl, R.; de Lera, A.R. Alternative retinoid X receptor (RXR) ligands. Mol. Cell Endocrinol. 2019, 491, 110436. [Google Scholar] [CrossRef]
- Weber, P.; Flores, R.E.; Kiefer, M.F.; Schupp, M. Retinol Saturase: More than the Name Suggests. Trends Pharmacol. Sci. 2020, 41, 418–427. [Google Scholar] [CrossRef]
- Sarang, Z.; Saghy, T.; Budai, Z.; Ujlaky-Nagy, L.; Bedekovics, J.; Beke, L.; Mehes, G.; Nagy, G.; Ruhl, R.; Moise, A.R.; et al. Retinol Saturase Knock-Out Mice are Characterized by Impaired Clearance of Apoptotic Cells and Develop Mild Autoimmunity. Biomolecules 2019, 9, 737. [Google Scholar] [CrossRef]
- Pang, X.Y.; Wang, S.; Jurczak, M.J.; Shulman, G.I.; Moise, A.R. Retinol saturase modulates lipid metabolism and the production of reactive oxygen species. Arch. Biochem. Biophys. 2017, 633, 93–102. [Google Scholar] [CrossRef]
- Heidenreich, S.; Witte, N.; Weber, P.; Goehring, I.; Tolkachov, A.; von Loeffelholz, C.; Docke, S.; Bauer, M.; Stockmann, M.; Pfeiffer, A.F.H.; et al. Retinol saturase coordinates liver metabolism by regulating ChREBP activity. Nat. Commun. 2017, 8, 384. [Google Scholar] [CrossRef]
- Schupp, M.; Lefterova, M.I.; Janke, J.; Leitner, K.; Cristancho, A.G.; Mullican, S.E.; Qatanani, M.; Szwergold, N.; Steger, D.J.; Curtin, J.C.; et al. Retinol saturase promotes adipogenesis and is downregulated in obesity. Proc. Natl. Acad. Sci. USA 2009, 106, 1105–1110. [Google Scholar] [CrossRef]
- Al Tanoury, Z.; Piskunov, A.; Rochette-Egly, C. Vitamin A and retinoid signaling: Genomic and nongenomic effects. J. Lipid Res. 2013, 54, 1761–1775. [Google Scholar] [CrossRef]
- Bruck, N.; Vitoux, D.; Ferry, C.; Duong, V.; Bauer, A.; de The, H.; Rochette-Egly, C. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. EMBO J. 2009, 28, 34–47. [Google Scholar] [CrossRef]
- Piskunov, A.; Rochette-Egly, C. A retinoic acid receptor RARalpha pool present in membrane lipid rafts forms complexes with G protein alphaQ to activate p38MAPK. Oncogene 2012, 31, 3333–3345. [Google Scholar] [CrossRef]
- Masia, S.; Alvarez, S.; de Lera, A.R.; Barettino, D. Rapid, nongenomic actions of retinoic acid on phosphatidylinositol-3-kinase signaling pathway mediated by the retinoic acid receptor. Mol. Endocrinol. 2007, 21, 2391–2402. [Google Scholar] [CrossRef]
- Chen, N.; Onisko, B.; Napoli, J.L. The nuclear transcription factor RARalpha associates with neuronal RNA granules and suppresses translation. J. Biol. Chem. 2008, 283, 20841–20847. [Google Scholar] [CrossRef]
- Shabrova, E.; Hoyos, B.; Vinogradov, V.; Kim, Y.K.; Wassef, L.; Leitges, M.; Quadro, L.; Hammerling, U. Retinol as a cofactor for PKCdelta-mediated impairment of insulin sensitivity in a mouse model of diet-induced obesity. FASEB J. 2016, 30, 1339–1355. [Google Scholar] [CrossRef]
- Park, S.W.; Nhieu, J.; Persaud, S.D.; Miller, M.C.; Xia, Y.; Lin, Y.W.; Lin, Y.L.; Kagechika, H.; Mayo, K.H.; Wei, L.N. A new regulatory mechanism for Raf kinase activation, retinoic acid-bound Crabp1. Sci. Rep. 2019, 9, 10929. [Google Scholar] [CrossRef]
- Kane, M.A.; Folias, A.E.; Pingitore, A.; Perri, M.; Krois, C.R.; Ryu, J.Y.; Cione, E.; Napoli, J.L. CrbpI modulates glucose homeostasis and pancreas 9-cis-retinoic acid concentrations. Mol. Cell. Biol. 2011, 31, 3277–3285. [Google Scholar] [CrossRef]
- Kane, M.A.; Folias, A.E.; Pingitore, A.; Perri, M.; Obrochta, K.M.; Krois, C.R.; Cione, E.; Ryu, J.Y.; Napoli, J.L. Identification of 9-cis-retinoic acid as a pancreas-specific autacoid that attenuates glucose-stimulated insulin secretion. Proc. Natl. Acad. Sci. USA 2010, 107, 21884–21889. [Google Scholar] [CrossRef]
- Niederreither, K.; Abu-Abed, S.; Schuhbaur, B.; Petkovich, M.; Chambon, P.; Dolle, P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat. Genet. 2002, 31, 84–88. [Google Scholar] [CrossRef]
- Calleja, C.; Messaddeq, N.; Chapellier, B.; Yang, H.; Krezel, W.; Li, M.; Metzger, D.; Mascrez, B.; Ohta, K.; Kagechika, H.; et al. Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev. 2006, 20, 1525–1538. [Google Scholar] [CrossRef]
- Ghyselinck, N.B.; Duester, G. Retinoic acid signaling pathways. Development 2019, 146, dev167502. [Google Scholar] [CrossRef]
- Shannon, S.R.; Moise, A.R.; Trainor, P.A. New insights and changing paradigms in the regulation of vitamin A metabolism in development. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e264. [Google Scholar] [CrossRef]
- Stefanovic, S.; Zaffran, S. Mechanisms of retinoic acid signaling during cardiogenesis. Mech. Dev. 2017, 143, 9–19. [Google Scholar] [CrossRef]
- Sirbu, I.O.; Chis, A.R.; Moise, A.R. Role of carotenoids and retinoids during heart development. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158636. [Google Scholar] [CrossRef]
- Billings, S.E.; Pierzchalski, K.; Butler Tjaden, N.E.; Pang, X.Y.; Trainor, P.A.; Kane, M.A.; Moise, A.R. The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J. 2013, 27, 4877–4889. [Google Scholar] [CrossRef]
- Wang, S.; Huang, W.; Castillo, H.A.; Kane, M.A.; Xavier-Neto, J.; Trainor, P.A.; Moise, A.R. Alterations in retinoic acid signaling affect the development of the mouse coronary vasculature. Dev. Dyn. 2018, 247, 976–991. [Google Scholar] [CrossRef]
- Napoli, J.L. Post-natal all-trans-retinoic acid biosynthesis. Methods Enzymol. 2020, 637, 27–54. [Google Scholar] [PubMed]
- Wu, L.; Belyaeva, O.V.; Adams, M.K.; Klyuyeva, A.V.; Lee, S.A.; Goggans, K.R.; Kesterson, R.A.; Popov, K.M.; Kedishvili, N.Y. Mice lacking the epidermal retinol dehydrogenases SDR16C5 and SDR16C6 display accelerated hair growth and enlarged meibomian glands. J. Biol. Chem. 2019, 294, 17060–17074. [Google Scholar] [CrossRef] [PubMed]
- Asson-Batres, M.A.; Ryzhov, S.; Tikhomirov, O.; Duarte, C.W.; Congdon, C.B.; Lessard, C.R.; McFarland, S.; Rochette-Egly, C.; Tran, T.L.; Galindo, C.L.; et al. Effects of vitamin A deficiency in the postnatal mouse heart: Role of hepatic retinoid stores. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H1773–H1789. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Vuckovic, M.G.; Smullin, C.P.; Kim, M.; Lo, C.P.; Devericks, E.; Yoo, H.S.; Tintcheva, M.; Deng, Y.; Napoli, J.L. Modest Decreases in Endogenous All-trans-Retinoic Acid Produced by a Mouse Rdh10 Heterozygote Provoke Major Abnormalities in Adipogenesis and Lipid Metabolism. Diabetes 2018, 67, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Dolle, P.; Ghyselinck, N.B.; Duester, G. Endogenous retinoic acid signaling is required for maintenance and regeneration of cornea. Exp. Eye Res. 2017, 154, 190–195. [Google Scholar] [CrossRef]
- Tong, M.H.; Yang, Q.E.; Davis, J.C.; Griswold, M.D. Retinol dehydrogenase 10 is indispensible for spermatogenesis in juvenile males. Proc. Natl. Acad. Sci. USA 2013, 110, 543–548. [Google Scholar] [CrossRef]
- Mendoza-Parra, M.A.; Malysheva, V.; Mohamed Saleem, M.A.; Lieb, M.; Godel, A.; Gronemeyer, H. Reconstructed cell fate-regulatory programs in stem cells reveal hierarchies and key factors of neurogenesis. Genome Res. 2016, 26, 1505–1519. [Google Scholar] [CrossRef]
- Moutier, E.; Ye, T.; Choukrallah, M.A.; Urban, S.; Osz, J.; Chatagnon, A.; Delacroix, L.; Langer, D.; Rochel, N.; Moras, D.; et al. Retinoic acid receptors recognize the mouse genome through binding elements with diverse spacing and topology. J. Biol. Chem. 2012, 287, 26328–26341. [Google Scholar] [CrossRef]
- Delacroix, L.; Moutier, E.; Altobelli, G.; Legras, S.; Poch, O.; Choukrallah, M.A.; Bertin, I.; Jost, B.; Davidson, I. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol. Cell. Biol. 2010, 30, 231–244. [Google Scholar] [CrossRef]
- Paschaki, M.; Schneider, C.; Rhinn, M.; Thibault-Carpentier, C.; Dembele, D.; Niederreither, K.; Dolle, P. Transcriptomic analysis of murine embryos lacking endogenous retinoic acid signaling. PLoS ONE 2013, 8, e62274. [Google Scholar] [CrossRef]
- Rochette-Egly, C. Retinoic Acid-Regulated Target Genes During Development: Integrative Genomics Analysis. Subcell. Biochem. 2020, 95, 57–85. [Google Scholar] [PubMed]
- de The, H.; Vivanco-Ruiz, M.M.; Tiollais, P.; Stunnenberg, H.; Dejean, A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature 1990, 343, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Penvose, A.; Keenan, J.L.; Bray, D.; Ramlall, V.; Siggers, T. Comprehensive study of nuclear receptor DNA binding provides a revised framework for understanding receptor specificity. Nat. Commun. 2019, 10, 2514. [Google Scholar] [CrossRef]
- Kato, S.; Sasaki, H.; Suzawa, M.; Masushige, S.; Tora, L.; Chambon, P.; Gronemeyer, H. Widely spaced, directly repeated PuGGTCA elements act as promiscuous enhancers for different classes of nuclear receptors. Mol. Cell. Biol. 1995, 15, 5858–5867. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.; Mullan, H.E.; Krumlauf, R. Long-range regulation by shared retinoic acid response elements modulates dynamic expression of posterior Hoxb genes in CNS development. Dev. Biol. 2014, 388, 134–144. [Google Scholar] [CrossRef][Green Version]
- Kurokawa, R.; Soderstrom, M.; Horlein, A.; Halachmi, S.; Brown, M.; Rosenfeld, M.G.; Glass, C.K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature 1995, 377, 451–454. [Google Scholar] [CrossRef]
- Chen, J.D.; Evans, R.M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 1995, 377, 454–457. [Google Scholar] [CrossRef]
- Nagy, L.; Kao, H.Y.; Chakravarti, D.; Lin, R.J.; Hassig, C.A.; Ayer, D.E.; Schreiber, S.L.; Evans, R.M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 1997, 89, 373–380. [Google Scholar] [CrossRef]
- Germain, P.; Iyer, J.; Zechel, C.; Gronemeyer, H. Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature 2002, 415, 187–192. [Google Scholar] [CrossRef]
- Epping, M.T.; Wang, L.; Edel, M.J.; Carlee, L.; Hernandez, M.; Bernards, R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 2005, 122, 835–847. [Google Scholar] [CrossRef]
- Kumar, S.; Duester, G. Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development 2014, 141, 2972–2977. [Google Scholar] [CrossRef] [PubMed]
- Gudas, L.J.; Wagner, J.A. Retinoids regulate stem cell differentiation. J. Cell Physiol. 2011, 226, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Mark, M.; Ghyselinck, N.B.; Chambon, P. Function of retinoic acid receptors during embryonic development. Nucl. Recept. Signal. 2009, 7, e002. [Google Scholar] [CrossRef] [PubMed]
- Wendling, O.; Ghyselinck, N.B.; Chambon, P.; Mark, M. Roles of retinoic acid receptors in early embryonic morphogenesis and hindbrain patterning. Development 2001, 128, 2031–2038. [Google Scholar] [CrossRef]
- Suzuki, M.; Wang, T.; Garretto, D.; Isasi, C.R.; Cardoso, W.V.; Greally, J.M.; Quadro, L. Disproportionate Vitamin A Deficiency in Women of Specific Ethnicities Linked to Differences in Allele Frequencies of Vitamin A-Related Polymorphisms. Nutrients 2021, 13, 1743. [Google Scholar] [CrossRef]
- Genaro Pde, S.; Martini, L.A. Vitamin A supplementation and risk of skeletal fracture. Nutr. Rev. 2004, 62, 65–67. [Google Scholar]
- Sheftel, J.; van Stuijvenberg, M.E.; Dhansay, M.A.; Suri, D.J.; Grahn, M.; Keuler, N.S.; Binkley, N.C.; Tanumihardjo, S.A. Chronic and acute hypervitaminosis A are associated with suboptimal anthropometric measurements in a cohort of South African preschool children. Am. J. Clin. Nutr. 2022, nqab422. [Google Scholar] [CrossRef]
- Rothman, K.J.; Moore, L.L.; Singer, M.R.; Nguyen, U.S.; Mannino, S.; Milunsky, A. Teratogenicity of high vitamin A intake. N. Engl. J. Med. 1995, 333, 1369–1373. [Google Scholar] [CrossRef]
- Goodman, G.E.; Thornquist, M.D.; Balmes, J.; Cullen, M.R.; Meyskens, F.L., Jr.; Omenn, G.S.; Valanis, B.; Williams, J.H., Jr. The Beta-Carotene and Retinol Efficacy Trial: Incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping beta-carotene and retinol supplements. J. Natl. Cancer Inst. 2004, 96, 1743–1750. [Google Scholar] [CrossRef]
- Hemila, H. The effect of beta-carotene on the mortality of male smokers is modified by smoking and by vitamins C and E: Evidence against a uniform effect of nutrient. J. Nutr. Sci. 2020, 9, e11. [Google Scholar] [CrossRef]
- Isoherranen, N.; Zhong, G. Biochemical and physiological importance of the CYP26 retinoic acid hydroxylases. Pharmacol. Ther. 2019, 204, 107400. [Google Scholar] [CrossRef]
- Belyaeva, O.V.; Adams, M.K.; Popov, K.M.; Kedishvili, N.Y. Generation of Retinaldehyde for Retinoic Acid Biosynthesis. Biomolecules 2019, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Widjaja-Adhi, M.A.K.; Golczak, M. The molecular aspects of absorption and metabolism of carotenoids and retinoids in vertebrates. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158571. [Google Scholar] [CrossRef] [PubMed]
- Napoli, J.L.; Yoo, H.S. Retinoid metabolism and functions mediated by retinoid binding-proteins. Methods Enzymol. 2020, 637, 55–75. [Google Scholar] [PubMed]
- von Lintig, J.; Moon, J.; Lee, J.; Ramkumar, S. Carotenoid metabolism at the intestinal barrier. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158580. [Google Scholar] [CrossRef]
- Saeed, A.; Hoekstra, M.; Hoeke, M.O.; Heegsma, J.; Faber, K.N. The interrelationship between bile acid and vitamin A homeostasis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 496–512. [Google Scholar] [CrossRef]
- Hoeke, M.O.; Plass, J.R.; Heegsma, J.; Geuken, M.; van Rijsbergen, D.; Baller, J.F.; Kuipers, F.; Moshage, H.; Jansen, P.L.; Faber, K.N. Low retinol levels differentially modulate bile salt-induced expression of human and mouse hepatic bile salt transporters. Hepatology 2009, 49, 151–159. [Google Scholar] [CrossRef]
- Ruiz, A.; Winston, A.; Lim, Y.H.; Gilbert, B.A.; Rando, R.R.; Bok, D. Molecular and biochemical characterization of lecithin retinol acyltransferase. J. Biol. Chem. 1999, 274, 3834–3841. [Google Scholar] [CrossRef]
- Batten, M.L.; Imanishi, Y.; Maeda, T.; Tu, D.C.; Moise, A.R.; Bronson, D.; Possin, D.; Van Gelder, R.N.; Baehr, W.; Palczewski, K. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem. 2004, 279, 10422–10432. [Google Scholar] [CrossRef]
- Napoli, J.L. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim. Biophys. Acta 2012, 1821, 152–167. [Google Scholar] [CrossRef]
- Wongsiriroj, N.; Piantedosi, R.; Palczewski, K.; Goldberg, I.J.; Johnston, T.P.; Li, E.; Blaner, W.S. The molecular basis of retinoid absorption: A genetic dissection. J. Biol. Chem. 2008, 283, 13510–13519. [Google Scholar] [CrossRef] [PubMed]
- Orland, M.D.; Anwar, K.; Cromley, D.; Chu, C.H.; Chen, L.; Billheimer, J.T.; Hussain, M.M.; Cheng, D. Acyl coenzyme A dependent retinol esterification by acyl coenzyme A: Diacylglycerol acyltransferase 1. Biochim. Biophys. Acta 2005, 1737, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.L.; Monetti, M.; Burri, B.J.; Farese, R.V., Jr. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J. Lipid Res. 2005, 46, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Ables, G.P.; Yang, K.J.; Vogel, S.; Hernandez-Ono, A.; Yu, S.; Yuen, J.J.; Birtles, S.; Buckett, L.K.; Turnbull, A.V.; Goldberg, I.J.; et al. Intestinal DGAT1 deficiency reduces postprandial triglyceride and retinyl ester excursions by inhibiting chylomicron secretion and delaying gastric emptying. J. Lipid Res. 2012, 53, 2364–2379. [Google Scholar] [CrossRef]
- Blaner, W.S.; Brun, P.J.; Calderon, R.M.; Golczak, M. Retinol-binding protein 2 (RBP2): Biology and pathobiology. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 197–218. [Google Scholar] [CrossRef] [PubMed]
- Xueping, E.; Zhang, L.; Lu, J.; Tso, P.; Blaner, W.S.; Levin, M.S.; Li, E. Increased neonatal mortality in mice lacking cellular retinol-binding protein II. J. Biol. Chem. 2002, 277, 36617–36623. [Google Scholar]
- Cai, K.; Gudas, L.J. Retinoic acid receptors and GATA transcription factors activate the transcription of the human lecithin:retinol acyltransferase gene. Int. J. Biochem. Cell Biol. 2009, 41, 546–553. [Google Scholar] [CrossRef]
- Wei, L.N.; Blaner, W.S.; Goodman, D.S.; Nguyen-Huu, M.C. Regulation of the cellular retinoid-binding proteins and their messenger ribonucleic acids during P19 embryonal carcinoma cell differentiation induced by retinoic acid. Mol. Endocrinol. 1989, 3, 454–463. [Google Scholar] [CrossRef]
- Zolfaghari, R.; Ross, A.C. An essential set of basic DNA response elements is required for receptor-dependent transcription of the lecithin:retinol acyltransferase (Lrat) gene. Arch. Biochem. Biophys. 2009, 489, 1–9. [Google Scholar] [CrossRef]
- Hodges, J.K.; Tan, L.; Green, M.H.; Ross, A.C. Vitamin A and retinoic acid combined have a more potent effect compared to vitamin A alone on the uptake of retinol into extrahepatic tissues of neonatal rats raised under vitamin A-marginal conditions. Curr. Dev. Nutr. 2017, 1, e000265. [Google Scholar] [CrossRef][Green Version]
- Ross, A.C.; Li, N.Q.; Wu, L. The components of VARA, a nutrient-metabolite combination of vitamin A and retinoic acid, act efficiently together and separately to increase retinyl esters in the lungs of neonatal rats. J. Nutr. 2006, 136, 2803–2807. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van Bennekum, A.; Werder, M.; Thuahnai, S.T.; Han, C.H.; Duong, P.; Williams, D.L.; Wettstein, P.; Schulthess, G.; Phillips, M.C.; Hauser, H. Class B scavenger receptor-mediated intestinal absorption of dietary beta-carotene and cholesterol. Biochemistry 2005, 44, 4517–4525. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.H. Mechanisms involved in the intestinal absorption of dietary vitamin A and provitamin A carotenoids. Biochim. Biophys. Acta 2012, 1821, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Clugston, R.D.; Huang, L.S.; Blaner, W.S. Chronic alcohol consumption has a biphasic effect on hepatic retinoid loss. FASEB J. 2015, 29, 3654–3667. [Google Scholar] [CrossRef] [PubMed]
- Widjaja-Adhi, M.A.; Lobo, G.P.; Golczak, M.; Von Lintig, J. A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Hum. Mol. Genet. 2015, 24, 3206–3219. [Google Scholar] [CrossRef]
- Kiefer, C.; Sumser, E.; Wernet, M.F.; Von Lintig, J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. USA 2002, 99, 10581–10586. [Google Scholar] [CrossRef]
- Schweigert, F.J.; Trupschuch, A.; Hantschel, C. Modulation of absorption of beta-carotene and tissue accumulation of beta-carotene and vitamin A by different surfactants in rats. Ann. Nutr. Metab. 2002, 46, 200–204. [Google Scholar] [CrossRef]
- Voolstra, O.; Kiefer, C.; Hoehne, M.; Welsch, R.; Vogt, K.; von Lintig, J. The Drosophila class B scavenger receptor NinaD-I is a cell surface receptor mediating carotenoid transport for visual chromophore synthesis. Biochemistry 2006, 45, 13429–13437. [Google Scholar] [CrossRef]
- Toomey, M.B.; Lopes, R.J.; Araujo, P.M.; Johnson, J.D.; Gazda, M.A.; Afonso, S.; Mota, P.G.; Koch, R.E.; Hill, G.E.; Corbo, J.C.; et al. High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl. Acad. Sci. USA 2017, 114, 5219–5224. [Google Scholar] [CrossRef]
- Wang, X.D.; Tang, G.W.; Fox, J.G.; Krinsky, N.I.; Russell, R.M. Enzymatic conversion of beta-carotene into beta-apo-carotenals and retinoids by human, monkey, ferret, and rat tissues. Arch. Biochem. Biophys. 1991, 285, 8–16. [Google Scholar] [CrossRef]
- Kiefer, C.; Hessel, S.; Lampert, J.M.; Vogt, K.; Lederer, M.O.; Breithaupt, D.E.; von Lintig, J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 2001, 276, 14110–14116. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.E.; Ramkumar, S.; Sun, W.; Colon Ortiz, C.; Kiser, P.D.; Golczak, M.; von Lintig, J. The Biochemical Basis of Vitamin A Production from the Asymmetric Carotenoid beta-Cryptoxanthin. ACS Chem. Biol. 2018, 13, 2121–2129. [Google Scholar] [CrossRef]
- Bandara, S.; Thomas, L.D.; Ramkumar, S.; Khadka, N.; Kiser, P.D.; Golczak, M.; von Lintig, J. The Structural and Biochemical Basis of Apocarotenoid Processing by beta-Carotene Oxygenase-2. ACS Chem. Biol. 2021, 16, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.D.; Bandara, S.; Parmar, V.M.; Srinivasagan, R.; Khadka, N.; Golczak, M.; Kiser, P.D.; von Lintig, J. The human mitochondrial enzyme BCO2 exhibits catalytic activity toward carotenoids and apocarotenoids. J. Biol. Chem. 2020, 295, 15553–15565. [Google Scholar] [CrossRef] [PubMed]
- Wassef, L.; Shete, V.; Costabile, B.; Rodas, R.; Quadro, L. High Preformed Vitamin A Intake during Pregnancy Prevents Embryonic Accumulation of Intact beta-Carotene from the Maternal Circulation in Mice. J. Nutr. 2015, 145, 1408–1414. [Google Scholar] [CrossRef] [PubMed]
- Quadro, L.; Giordano, E.; Costabile, B.K.; Nargis, T.; Iqbal, J.; Kim, Y.; Wassef, L.; Hussain, M.M. Interplay between beta-carotene and lipoprotein metabolism at the maternal-fetal barrier. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158591. [Google Scholar] [CrossRef]
- Ferrucci, L.; Perry, J.R.; Matteini, A.; Perola, M.; Tanaka, T.; Silander, K.; Rice, N.; Melzer, D.; Murray, A.; Cluett, C.; et al. Common variation in the beta-carotene 15,15′-monooxygenase 1 gene affects circulating levels of carotenoids: A genome-wide association study. Am. J. Hum. Genet. 2009, 84, 123–133. [Google Scholar] [CrossRef]
- Lobo, G.P.; Hessel, S.; Eichinger, A.; Noy, N.; Moise, A.R.; Wyss, A.; Palczewski, K.; von Lintig, J. ISX is a retinoic acid-sensitive gatekeeper that controls intestinal beta,beta-carotene absorption and vitamin A production. FASEB J. 2010, 24, 1656–1666. [Google Scholar] [CrossRef]
- Lobo, G.P.; Amengual, J.; Baus, D.; Shivdasani, R.A.; Taylor, D.; von Lintig, J. Genetics and diet regulate vitamin A production via the homeobox transcription factor ISX. J. Biol. Chem. 2013, 288, 9017–9027. [Google Scholar] [CrossRef]
- Seino, Y.; Miki, T.; Kiyonari, H.; Abe, T.; Fujimoto, W.; Kimura, K.; Takeuchi, A.; Takahashi, Y.; Oiso, Y.; Iwanaga, T.; et al. Isx participates in the maintenance of vitamin A metabolism by regulation of beta-carotene 15,15′-monooxygenase (Bcmo1) expression. J. Biol. Chem. 2008, 283, 4905–4911. [Google Scholar] [CrossRef]
- Widjaja-Adhi, M.A.K.; Palczewski, G.; Dale, K.; Knauss, E.A.; Kelly, M.E.; Golczak, M.; Levine, A.D.; von Lintig, J. Transcription factor ISX mediates the cross talk between diet and immunity. Proc. Natl. Acad. Sci. USA 2017, 114, 11530–11535. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, S.; Moon, J.; Golczak, M.; von Lintig, J. LRAT coordinates the negative-feedback regulation of intestinal retinoid biosynthesis from beta-carotene. J. Lipid Res. 2021, 62, 100055. [Google Scholar] [CrossRef] [PubMed]
- Blomhoff, R.; Holte, K.; Naess, L.; Berg, T. Newly administered [3H]retinol is transferred from hepatocytes to stellate cells in liver for storage. Exp. Cell Res. 1984, 150, 186–193. [Google Scholar] [CrossRef]
- Senoo, H.; Stang, E.; Nilsson, A.; Kindberg, G.M.; Berg, T.; Roos, N.; Norum, K.R.; Blomhoff, R. Internalization of retinol-binding protein in parenchymal and stellate cells of rat liver. J. Lipid Res. 1990, 31, 1229–1239. [Google Scholar] [CrossRef]
- Ong, D.E.; Chytil, F. Specificity of cellular retinol-binding protein for compounds with vitamin A activity. Nature 1975, 255, 74–75. [Google Scholar] [CrossRef]
- Ong, D.E.; Chytil, F. Changes in levels of cellular retinol- and retinoic-acid-binding proteins of liver and lung during perinatal development of rat. Proc. Natl. Acad. Sci. USA 1976, 73, 3976–3978. [Google Scholar] [CrossRef]
- Ghyselinck, N.B.; Bavik, C.; Sapin, V.; Mark, M.; Bonnier, D.; Hindelang, C.; Dierich, A.; Nilsson, C.B.; Hakansson, H.; Sauvant, P.; et al. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 1999, 18, 4903–4914. [Google Scholar] [CrossRef]
- Pierzchalski, K.; Yu, J.; Norman, V.; Kane, M.A. CrbpI regulates mammary retinoic acid homeostasis and the mammary microenvironment. FASEB J. 2013, 27, 1904–1916. [Google Scholar] [CrossRef]
- Wake, K. “Sternzellen” in the liver: Perisinusoidal cells with special reference to storage of vitamin A. Am. J. Anat. 1971, 132, 429–462. [Google Scholar] [CrossRef]
- Okabe, T.; Yorifuji, H.; Yamada, E.; Takaku, F. Isolation and characterization of vitamin-A-storing lung cells. Exp. Cell Res. 1984, 154, 125–135. [Google Scholar] [CrossRef]
- Apte, M.V.; Haber, P.S.; Applegate, T.L.; Norton, I.D.; McCaughan, G.W.; Korsten, M.A.; Pirola, R.C.; Wilson, J.S. Periacinar stellate shaped cells in rat pancreas: Identification, isolation, and culture. Gut 1998, 43, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Orban, T.; Palczewska, G.; Palczewski, K. Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. J. Biol. Chem. 2011, 286, 17248–17258. [Google Scholar] [CrossRef]
- Imanishi, Y.; Gerke, V.; Palczewski, K. Retinosomes: New insights into intracellular managing of hydrophobic substances in lipid bodies. J. Cell Biol. 2004, 166, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, A.; He, Y.G.; Andersson, S. Cell type-specific expression of beta-carotene 9′,10′-monooxygenase in human tissues. J. Histochem. Cytochem. 2005, 53, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Shmarakov, I.; Fleshman, M.K.; D’Ambrosio, D.N.; Piantedosi, R.; Riedl, K.M.; Schwartz, S.J.; Curley, R.W., Jr.; von Lintig, J.; Rubin, L.P.; Harrison, E.H.; et al. Hepatic stellate cells are an important cellular site for beta-carotene conversion to retinoid. Arch. Biochem. Biophys. 2010, 504, 3–10. [Google Scholar] [CrossRef]
- Raghuvanshi, S.; Reed, V.; Blaner, W.S.; Harrison, E.H. Cellular localization of beta-carotene 15,15′ oxygenase-1 (BCO1) and beta-carotene 9′,10′ oxygenase-2 (BCO2) in rat liver and intestine. Arch. Biochem. Biophys. 2015, 572, 19–27. [Google Scholar] [CrossRef]
- Wagner, C.; Hois, V.; Eggeling, A.; Pusch, L.M.; Pajed, L.; Starlinger, P.; Claudel, T.; Trauner, M.; Zimmermann, R.; Taschler, U.; et al. KIAA1363 affects retinyl ester turnover in cultured murine and human hepatic stellate cells. J. Lipid Res. 2022, 63, 100173. [Google Scholar] [CrossRef]
- Wagner, C.; Hois, V.; Pajed, L.; Pusch, L.M.; Wolinski, H.; Trauner, M.; Zimmermann, R.; Taschler, U.; Lass, A. Lysosomal acid lipase is the major acid retinyl ester hydrolase in cultured human hepatic stellate cells but not essential for retinyl ester degradation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158730. [Google Scholar] [CrossRef]
- Taschler, U.; Schreiber, R.; Chitraju, C.; Grabner, G.F.; Romauch, M.; Wolinski, H.; Haemmerle, G.; Breinbauer, R.; Zechner, R.; Lass, A.; et al. Adipose triglyceride lipase is involved in the mobilization of triglyceride and retinoid stores of hepatic stellate cells. Biochim. Biophys. Acta 2015, 1851, 937–945. [Google Scholar] [CrossRef]
- Grumet, L.; Taschler, U.; Lass, A. Hepatic Retinyl Ester Hydrolases and the Mobilization of Retinyl Ester Stores. Nutrients 2016, 9, 13. [Google Scholar] [CrossRef]
- Grumet, L.; Eichmann, T.O.; Taschler, U.; Zierler, K.A.; Leopold, C.; Moustafa, T.; Radovic, B.; Romauch, M.; Yan, C.; Du, H.; et al. Lysosomal Acid Lipase Hydrolyzes Retinyl Ester and Affects Retinoid Turnover. J. Biol. Chem. 2016, 291, 17977–17987. [Google Scholar] [CrossRef] [PubMed]
- Tuohetahuntila, M.; Molenaar, M.R.; Spee, B.; Brouwers, J.F.; Houweling, M.; Vaandrager, A.B.; Helms, J.B. ATGL and DGAT1 are involved in the turnover of newly synthesized triacylglycerols in hepatic stellate cells. J. Lipid Res. 2016, 57, 1162–1174. [Google Scholar] [CrossRef] [PubMed]
- Peterson, P.A. Studies on the interaction between prealbumin, retinol-binding protein, and vitamin A. J. Biol. Chem. 1971, 246, 44–49. [Google Scholar] [CrossRef]
- van Jaarsveld, P.P.; Edelhoch, H.; Goodman, D.S.; Robbins, J. The interaction of human plasma retinol-binding protein and prealbumin. J. Biol. Chem. 1973, 248, 4698–4705. [Google Scholar] [CrossRef]
- Muto, Y.; Smith, J.E.; Milch, P.O.; Goodman, D.S. Regulation of retinol-binding protein metabolism by vitamin A status in the rat. J. Biol. Chem. 1972, 247, 2542–2550. [Google Scholar] [CrossRef]
- Wei, S.; Episkopou, V.; Piantedosi, R.; Maeda, S.; Shimada, K.; Gottesman, M.E.; Blaner, W.S. Studies on the metabolism of retinol and retinol-binding protein in transthyretin-deficient mice produced by homologous recombination. J. Biol. Chem. 1995, 270, 866–870. [Google Scholar] [CrossRef]
- Bellovino, D.; Morimoto, T.; Tosetti, F.; Gaetani, S. Retinol binding protein and transthyretin are secreted as a complex formed in the endoplasmic reticulum in HepG2 human hepatocarcinoma cells. Exp. Cell Res. 1996, 222, 77–83. [Google Scholar] [CrossRef]
- O’Byrne, S.M.; Wongsiriroj, N.; Libien, J.; Vogel, S.; Goldberg, I.J.; Baehr, W.; Palczewski, K.; Blaner, W.S. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT). J. Biol. Chem. 2005, 280, 35647–35657. [Google Scholar] [CrossRef]
- Thompson, S.J.; Sargsyan, A.; Lee, S.A.; Yuen, J.J.; Cai, J.; Smalling, R.; Ghyselinck, N.; Mark, M.; Blaner, W.S.; Graham, T.E. Hepatocytes Are the Principal Source of Circulating RBP4 in Mice. Diabetes 2017, 66, 58–63. [Google Scholar] [CrossRef]
- Du, M.; Phelps, E.; Balangue, M.J.; Dockins, A.; Moiseyev, G.; Shin, Y.; Kane, S.; Otalora, L.; Ma, J.-X.; Farjo, R.; et al. Transgenic Mice Over-Expressing RBP4 Have RBP4-Dependent and Light-Independent Retinal Degeneration. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4375–4383. [Google Scholar] [CrossRef]
- Quadro, L.; Blaner, W.S.; Hamberger, L.; Van Gelder, R.N.; Vogel, S.; Piantedosi, R.; Gouras, P.; Colantuoni, V.; Gottesman, M.E. Muscle expression of human retinol-binding protein (RBP). Suppression of the visual defect of RBP knockout mice. J. Biol. Chem. 2002, 277, 30191–30197. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Suzuki, T.; Shen, J.; Wakana, S.; Araki, K.; Yamamura, K.I.; Lei, L.; Li, Z. Rescue of retinal morphology and function in a humanized mouse at the mouse retinol-binding protein locus. Lab. Investig. 2017, 97, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, T.; Ross, A.C. Regulation of hepatic lecithin: Retinol acyltransferase activity by retinoic acid. Arch. Biochem. Biophys. 1993, 301, 221–227. [Google Scholar] [CrossRef]
- Saeed, A.; Yang, J.; Heegsma, J.; Groen, A.K.; van Mil, S.W.C.; Paulusma, C.C.; Zhou, L.; Wang, B.; Faber, K.N. Farnesoid X receptor and bile acids regulate vitamin A storage. Sci. Rep. 2019, 9, 19493. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; He, Y.; Liu, H.X.; Tsuei, J.; Jiang, X.; Yang, L.; Wang, Z.T.; Wan, Y.J. All-trans retinoic acid regulates hepatic bile acid homeostasis. Biochem. Pharmacol. 2014, 91, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.Y.; He, H.; Nguyen, T.; Mennone, A.; Boyer, J.L. Retinoic acid represses CYP7A1 expression in human hepatocytes and HepG2 cells by FXR/RXR-dependent and independent mechanisms. J. Lipid Res. 2010, 51, 2265–2274. [Google Scholar] [CrossRef]
- Li, B.; Cai, S.Y.; Boyer, J.L. The role of the retinoid receptor, RAR/RXR heterodimer, in liver physiology. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166085. [Google Scholar] [CrossRef]
- Leo, M.A.; Lieber, C.S. Hepatic vitamin A depletion in alcoholic liver injury. N. Engl. J. Med. 1982, 307, 597–601. [Google Scholar] [CrossRef]
- Trasino, S.E.; Tang, X.H.; Jessurun, J.; Gudas, L.J. A retinoic acid receptor beta2 agonist reduces hepatic stellate cell activation in nonalcoholic fatty liver disease. J. Mol. Med. 2016, 94, 1143–1151. [Google Scholar] [CrossRef]
- Wang, S.; Yu, J.; Kane, M.A.; Moise, A.R. Modulation of retinoid signaling: Therapeutic opportunities in organ fibrosis and repair. Pharmacol. Ther. 2020, 205, 107415. [Google Scholar] [CrossRef]
- Butler, J.M.; Supharattanasitthi, W.; Yang, Y.C.; Paraoan, L. RNA-seq analysis of ageing human retinal pigment epithelium: Unexpected up-regulation of visual cycle gene transcription. J. Cell. Mol. Med. 2021, 25, 5572–5585. [Google Scholar] [CrossRef] [PubMed]
- Kiser, P.D.; Palczewski, K. Pathways and disease-causing alterations in visual chromophore production for vertebrate vision. J. Biol. Chem. 2021, 296, 100072. [Google Scholar] [CrossRef] [PubMed]
- Bouillet, P.; Sapin, V.; Chazaud, C.; Messaddeq, N.; Decimo, D.; Dolle, P.; Chambon, P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech. Dev. 1997, 63, 173–186. [Google Scholar] [CrossRef]
- Kawaguchi, R.; Yu, J.; Honda, J.; Hu, J.; Whitelegge, J.; Ping, P.; Wiita, P.; Bok, D.; Sun, H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science 2007, 315, 820–825. [Google Scholar] [CrossRef]
- Chen, Y.; Clarke, O.B.; Kim, J.; Stowe, S.; Kim, Y.K.; Assur, Z.; Cavalier, M.; Godoy-Ruiz, R.; von Alpen, D.C.; Manzini, C.; et al. Structure of the STRA6 receptor for retinol uptake. Science 2016, 353, aad8266. [Google Scholar] [CrossRef]
- Kawaguchi, R.; Zhong, M.; Kassai, M.; Ter-Stepanian, M.; Sun, H. STRA6-catalyzed vitamin A influx, efflux, and exchange. J. Membr. Biol. 2012, 245, 731–745. [Google Scholar] [CrossRef]
- Amengual, J.; Zhang, N.; Kemerer, M.; Maeda, T.; Palczewski, K.; Von Lintig, J. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum. Mol. Genet. 2014, 23, 5402–5417. [Google Scholar] [CrossRef]
- Kelly, M.; von Lintig, J. STRA6: Role in cellular retinol uptake and efflux. Hepatobiliary Surg. Nutr. 2015, 4, 229–242. [Google Scholar]
- Kelly, M.; Widjaja-Adhi, M.A.; Palczewski, G.; von Lintig, J. Transport of vitamin A across blood-tissue barriers is facilitated by STRA6. FASEB J. 2016, 30, 2985–2995. [Google Scholar] [CrossRef]
- Alapatt, P.; Guo, F.; Komanetsky, S.M.; Wang, S.; Cai, J.; Sargsyan, A.; Rodriguez Diaz, E.; Bacon, B.T.; Aryal, P.; Graham, T.E. Liver retinol transporter and receptor for serum retinol-binding protein (RBP4). J. Biol. Chem. 2013, 288, 1250–1265. [Google Scholar] [CrossRef]
- Shi, Y.; Obert, E.; Rahman, B.; Rohrer, B.; Lobo, G.P. The Retinol Binding Protein Receptor 2 (Rbpr2) is required for Photoreceptor Outer Segment Morphogenesis and Visual Function in Zebrafish. Sci. Rep. 2017, 7, 16207. [Google Scholar] [CrossRef] [PubMed]
- Skarnes, W.C.; Rosen, B.; West, A.P.; Koutsourakis, M.; Bushell, W.; Iyer, V.; Mujica, A.O.; Thomas, M.; Harrow, J.; Cox, T.; et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011, 474, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Naylor, H.M.; Newcomer, M.E. The structure of human retinol-binding protein (RBP) with its carrier protein transthyretin reveals an interaction with the carboxy terminus of RBP. Biochemistry 1999, 38, 2647–2653. [Google Scholar] [CrossRef]
- van Bennekum, A.M.; Wei, S.; Gamble, M.V.; Vogel, S.; Piantedosi, R.; Gottesman, M.; Episkopou, V.; Blaner, W.S. Biochemical basis for depressed serum retinol levels in transthyretin-deficient mice. J. Biol. Chem. 2001, 276, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Motani, A.; Wang, Z.; Conn, M.; Siegler, K.; Zhang, Y.; Liu, Q.; Johnstone, S.; Xu, H.; Thibault, S.; Wang, Y.; et al. Identification and characterization of a non-retinoid ligand for retinol-binding protein 4 which lowers serum retinol-binding protein 4 levels in vivo. J. Biol. Chem. 2009, 284, 7673–7680. [Google Scholar] [CrossRef] [PubMed]
- Raila, J.; Willnow, T.E.; Schweigert, F.J. Megalin-mediated reuptake of retinol in the kidneys of mice is essential for vitamin A homeostasis. J. Nutr. 2005, 135, 2512–2516. [Google Scholar] [CrossRef]
- Marino, M.; Andrews, D.; Brown, D.; McCluskey, C.R. Transcytosis of retinol-binding protein across renal proximal tubule cells after megalin (gp 330)-mediated endocytosis. J. Am. Soc. Nephrol. 2001, 12, 637–648. [Google Scholar] [CrossRef]
- Christensen, E.I.; Moskaug, J.O.; Vorum, H.; Jacobsen, C.; Gundersen, T.E.; Nykjaer, A.; Blomhoff, R.; Willnow, T.E.; Moestrup, S.K. Evidence for an essential role of megalin in transepithelial transport of retinol. J. Am. Soc. Nephrol. 1999, 10, 685–695. [Google Scholar] [CrossRef]
- Biesalski, H.K.; Frank, J.; Beck, S.C.; Heinrich, F.; Illek, B.; Reifen, R.; Gollnick, H.; Seeliger, M.W.; Wissinger, B.; Zrenner, E. Biochemical but not clinical vitamin A deficiency results from mutations in the gene for retinol binding protein. Am. J. Clin. Nutr. 1999, 69, 931–936. [Google Scholar] [CrossRef]
- Quadro, L.; Hamberger, L.; Gottesman, M.E.; Colantuoni, V.; Ramakrishnan, R.; Blaner, W.S. Transplacental delivery of retinoid: The role of retinol-binding protein and lipoprotein retinyl ester. Am. J. Physiol. Endocrinol. Metab. 2004, 286, E844–E851. [Google Scholar] [CrossRef]
- Wassef, L.; Quadro, L. Uptake of dietary retinoids at the maternal-fetal barrier: In vivo evidence for the role of lipoprotein lipase and alternative pathways. J. Biol. Chem. 2011, 286, 32198–32207. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wongsiriroj, N.; Blaner, W.S. The multifaceted nature of retinoid transport and metabolism. Hepatobiliary Surg. Nutr. 2014, 3, 126–139. [Google Scholar] [PubMed]
- Blaner, W.S.; Obunike, J.C.; Kurlandsky, S.B.; al-Haideri, M.; Piantedosi, R.; Deckelbaum, R.J.; Goldberg, I.J. Lipoprotein lipase hydrolysis of retinyl ester. Possible implications for retinoid uptake by cells. J. Biol. Chem. 1994, 269, 16559–16565. [Google Scholar] [CrossRef]
- Quadro, L.; Hamberger, L.; Gottesman, M.E.; Wang, F.; Colantuoni, V.; Blaner, W.S.; Mendelsohn, C.L. Pathways of vitamin A delivery to the embryo: Insights from a new tunable model of embryonic vitamin A deficiency. Endocrinology 2005, 146, 4479–4490. [Google Scholar] [CrossRef]
- Quadro, L.; Blaner, W.S.; Salchow, D.J.; Vogel, S.; Piantedosi, R.; Gouras, P.; Freeman, S.; Cosma, M.P.; Colantuoni, V.; Gottesman, M.E. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 1999, 18, 4633–4644. [Google Scholar] [CrossRef]
- Quadro, L.; Spiegler, E.K. Maternal-Fetal Transfer of Vitamin A and Its Impact on Mammalian Embryonic Development. Subcell. Biochem. 2020, 95, 27–55. [Google Scholar]
- Panariello, L.; Quadro, L.; Trematerra, S.; Colantuoni, V. Identification of a novel retinoic acid response element in the promoter region of the retinol-binding protein gene. J. Biol. Chem. 1996, 271, 25524–25532. [Google Scholar] [CrossRef]
- Jessen, K.A.; Satre, M.A. Mouse retinol binding protein gene: Cloning, expression and regulation by retinoic acid. Mol. Cell. Biochem. 2000, 211, 85–94. [Google Scholar] [CrossRef]
- Soprano, D.R.; Wyatt, M.L.; Dixon, J.L.; Soprano, K.J.; Goodman, D.S. Retinol-binding protein synthesis and secretion by the rat visceral yolk sac. Effect of retinol status. J. Biol. Chem. 1988, 263, 2934–2938. [Google Scholar] [CrossRef]
- Smith, J.E.; Milch, P.O.; Muto, Y.; Goodman, D.S. The plasma transport and metabolism of retinoic acid in the rat. Biochem. J. 1973, 132, 821–827. [Google Scholar] [CrossRef]
- Melhus, H.; Laurent, B.; Rask, L.; Peterson, P.A. Ligand-dependent secretion of rat retinol-binding protein expressed in HeLa cells. J. Biol. Chem. 1992, 267, 12036–12041. [Google Scholar] [CrossRef]
- Chazaud, C.; Bouillet, P.; Oulad-Abdelghani, M.; Dolle, P. Restricted expression of a novel retinoic acid responsive gene during limb bud dorsoventral patterning and endochondral ossification. Dev. Genet. 1996, 19, 66–73. [Google Scholar] [CrossRef]
- Laursen, K.B.; Kashyap, V.; Scandura, J.; Gudas, L.J. An alternative retinoic acid-responsive Stra6 promoter regulated in response to retinol deficiency. J. Biol. Chem. 2015, 290, 4356–4366. [Google Scholar] [CrossRef] [PubMed]
- Young, B.D.; Varney, K.M.; Wilder, P.T.; Costabile, B.K.; Pozharski, E.; Cook, M.E.; Godoy-Ruiz, R.; Clarke, O.B.; Mancia, F.; Weber, D.J. Physiologically Relevant Free Ca(2+) Ion Concentrations Regulate STRA6-Calmodulin Complex Formation via the BP2 Region of STRA6. J. Mol. Biol. 2021, 433, 167272. [Google Scholar] [CrossRef]
- Liu, W.; Yu, W.R.; Carling, T.; Juhlin, C.; Rastad, J.; Ridefelt, P.; Akerstrom, G.; Hellman, P. Regulation of gp330/megalin expression by vitamins A and D. Eur. J. Clin. Investig. 1998, 28, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Belyaeva, O.V.; Kedishvili, N.Y. Human pancreas protein 2 (PAN2) has a retinal reductase activity and is ubiquitously expressed in human tissues. FEBS Lett. 2002, 531, 489–493. [Google Scholar] [CrossRef]
- Kedishvili, N.Y. Multifunctional nature of human retinol dehydrogenases. Curr. Org. Chem. 2002, 6, 1247–1257. [Google Scholar] [CrossRef]
- Haeseleer, F.; Palczewski, K. Short-chain dehydrogenases/reductases in retina. Methods Enzymol. 2000, 316, 372–383. [Google Scholar]
- Haeseleer, F.; Huang, J.; Lebioda, L.; Saari, J.C.; Palczewski, K. Molecular characterization of a novel short-chain dehydrogenase/reductase that reduces all-trans-retinal. J. Biol. Chem. 1998, 273, 21790–21799. [Google Scholar] [CrossRef]
- Haeseleer, F.; Jang, G.F.; Imanishi, Y.; Driessen, C.; Matsumura, M.; Nelson, P.S.; Palczewski, K. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J. Biol. Chem. 2002, 277, 45537–45546. [Google Scholar] [CrossRef]
- Wu, B.X.; Chen, Y.; Chen, Y.; Fan, J.; Rohrer, B.; Crouch, R.K.; Ma, J.X. Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3365–3372. [Google Scholar]
- Molotkov, A.; Fan, X.; Duester, G. Excessive vitamin A toxicity in mice genetically deficient in either alcohol dehydrogenase Adh1 or Adh3. Eur. J. Biochem. 2002, 269, 2607–2612. [Google Scholar] [CrossRef] [PubMed]
- Pares, X.; Farres, J.; Kedishvili, N.; Duester, G. Medium- and short-chain dehydrogenase/reductase gene and protein families: Medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism. Cell. Mol. Life Sci. 2008, 65, 3936–3949. [Google Scholar] [CrossRef] [PubMed]
- Sandell, L.L.; Sanderson, B.W.; Moiseyev, G.; Johnson, T.; Mushegian, A.; Young, K.; Rey, J.P.; Ma, J.X.; Staehling-Hampton, K.; Trainor, P.A. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007, 21, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.K.; Belyaeva, O.V.; Wu, L.; Kedishvili, N.Y. The retinaldehyde reductase activity of DHRS3 is reciprocally activated by retinol dehydrogenase 10 to control retinoid homeostasis. J. Biol. Chem. 2014, 289, 14868–14880. [Google Scholar] [CrossRef]
- Kurosaka, H.; Wang, Q.; Sandell, L.; Yamashiro, T.; Trainor, P.A. Rdh10 loss-of-function and perturbed retinoid signaling underlies the etiology of choanal atresia. Hum. Mol. Genet. 2017, 26, 1268–1279. [Google Scholar] [CrossRef]
- Sandell, L.L.; Lynn, M.L.; Inman, K.E.; McDowell, W.; Trainor, P.A. RDH10 oxidation of Vitamin A is a critical control step in synthesis of retinoic acid during mouse embryogenesis. PLoS ONE 2012, 7, e30698. [Google Scholar] [CrossRef]
- Farjo, K.M.; Moiseyev, G.; Nikolaeva, O.; Sandell, L.L.; Trainor, P.A.; Ma, J.X. RDH10 is the primary enzyme responsible for the first step of embryonic Vitamin A metabolism and retinoic acid synthesis. Dev. Biol. 2011, 357, 347–355. [Google Scholar] [CrossRef]
- Cunningham, T.J.; Chatzi, C.; Sandell, L.L.; Trainor, P.A.; Duester, G. Rdh10 mutants deficient in limb field retinoic acid signaling exhibit normal limb patterning but display interdigital webbing. Dev. Dyn. 2011, 240, 1142–1150. [Google Scholar] [CrossRef]
- Wang, S.; Yu, J.; Jones, J.W.; Pierzchalski, K.; Kane, M.A.; Trainor, P.A.; Xavier-Neto, J.; Moise, A.R. Retinoic acid signaling promotes the cytoskeletal rearrangement of embryonic epicardial cells. FASEB J. 2018, 32, 3765–3781. [Google Scholar] [CrossRef]
- Xiao, Y.; Hill, M.C.; Zhang, M.; Martin, T.J.; Morikawa, Y.; Wang, S.; Moise, A.R.; Wythe, J.D.; Martin, J.F. Hippo Signaling Plays an Essential Role in Cell State Transitions during Cardiac Fibroblast Development. Dev. Cell 2018, 45, 153–169.e156. [Google Scholar] [CrossRef] [PubMed]
- Rhinn, M.; Schuhbaur, B.; Niederreither, K.; Dolle, P. Involvement of retinol dehydrogenase 10 in embryonic patterning and rescue of its loss of function by maternal retinaldehyde treatment. Proc. Natl. Acad. Sci. USA 2011, 108, 16687–16692. [Google Scholar] [CrossRef] [PubMed]
- Kam, R.K.; Shi, W.; Chan, S.O.; Chen, Y.; Xu, G.; Lau, C.B.; Fung, K.P.; Chan, W.Y.; Zhao, H. Dhrs3 protein attenuates retinoic acid signaling and is required for early embryonic patterning. J. Biol. Chem. 2013, 288, 31477–31487. [Google Scholar] [CrossRef]
- Feng, L.; Hernandez, R.E.; Waxman, J.S.; Yelon, D.; Moens, C.B. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev. Biol. 2010, 338, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.K.; Lee, S.A.; Belyaeva, O.V.; Wu, L.; Kedishvili, N.Y. Characterization of human short chain dehydrogenase/reductase SDR16C family members related to retinol dehydrogenase 10. Chem. Biol. Interact. 2017, 276, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Belyaeva, O.V.; Chang, C.; Berlett, M.C.; Kedishvili, N.Y. Evolutionary origins of retinoid active short-chain dehydrogenases/reductases of SDR16C family. Chem. Biol. Interact. 2015, 234, 135–143. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, P.; Krois, C.R.; Kane, M.A.; Napoli, J.L. Altered vitamin A homeostasis and increased size and adiposity in the rdh1-null mouse. FASEB J. 2007, 21, 2886–2896. [Google Scholar] [CrossRef]
- Kiser, P.D.; Golczak, M.; Maeda, A.; Palczewski, K. Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochim. Biophys. Acta 2012, 1821, 137–151. [Google Scholar] [CrossRef]
- Zolfaghari, R.; Chen, Q.; Ross, A.C. DHRS3, a retinal reductase, is differentially regulated by retinoic acid and lipopolysaccharide-induced inflammation in THP-1 cells and rat liver. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G578–G588. [Google Scholar] [CrossRef]
- Cerignoli, F.; Guo, X.; Cardinali, B.; Rinaldi, C.; Casaletto, J.; Frati, L.; Screpanti, I.; Gudas, L.J.; Gulino, A.; Thiele, C.J.; et al. retSDR1, a short-chain retinol dehydrogenase/reductase, is retinoic acid-inducible and frequently deleted in human neuroblastoma cell lines. Cancer Res. 2002, 62, 1196–1204. [Google Scholar]
- Persson, B.; Kallberg, Y.; Bray, J.E.; Bruford, E.; Dellaporta, S.L.; Favia, A.D.; Duarte, R.G.; Jornvall, H.; Kavanagh, K.L.; Kedishvili, N.; et al. The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem. Biol. Interact. 2009, 178, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Belyaeva, O.V.; Adams, M.K.; Wu, L.; Kedishvili, N.Y. The antagonistically bifunctional retinoid oxidoreductase complex is required for maintenance of all-trans-retinoic acid homeostasis. J. Biol. Chem. 2017, 292, 5884–5897. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.K.; Belyaeva, O.V.; Wu, L.; Chaple, I.F.; Dunigan-Russell, K.; Popov, K.M.; Kedishvili, N.Y. Characterization of subunit interactions in the hetero-oligomeric retinoid oxidoreductase complex. Biochem. J. 2021, 478, 3597–3611. [Google Scholar] [CrossRef] [PubMed]
- Niederreither, K.; Subbarayan, V.; Dolle, P.; Chambon, P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 1999, 21, 444–448. [Google Scholar] [CrossRef]
- Teletin, M.; Vernet, N.; Ghyselinck, N.B.; Mark, M. Roles of Retinoic Acid in Germ Cell Differentiation. Curr. Top Dev. Biol. 2017, 125, 191–225. [Google Scholar]
- Gyongyosi, A.; Szatmari, I.; Pap, A.; Dezso, B.; Pos, Z.; Szeles, L.; Varga, T.; Nagy, L. RDH10, RALDH2, and CRABP2 are required components of PPARgamma-directed ATRA synthesis and signaling in human dendritic cells. J. Lipid Res. 2013, 54, 2458–2474. [Google Scholar] [CrossRef]
- Nallamshetty, S.; Wang, H.; Rhee, E.J.; Kiefer, F.W.; Brown, J.D.; Lotinun, S.; Le, P.; Baron, R.; Rosen, C.J.; Plutzky, J. Deficiency of retinaldehyde dehydrogenase 1 induces BMP2 and increases bone mass in vivo. PLoS ONE 2013, 8, e71307. [Google Scholar]
- Ziouzenkova, O.; Orasanu, G.; Sharlach, M.; Akiyama, T.E.; Berger, J.P.; Viereck, J.; Hamilton, J.A.; Tang, G.; Dolnikowski, G.G.; Vogel, S.; et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat. Med. 2007, 13, 695–702. [Google Scholar] [CrossRef]
- Teletin, M.; Vernet, N.; Yu, J.; Klopfenstein, M.; Jones, J.W.; Feret, B.; Kane, M.A.; Ghyselinck, N.B.; Mark, M. Two functionally redundant sources of retinoic acid secure spermatogonia differentiation in the seminiferous epithelium. Development 2019, 146, dev170225. [Google Scholar] [CrossRef]
- Yasmeen, R.; Reichert, B.; Deiuliis, J.; Yang, F.; Lynch, A.; Meyers, J.; Sharlach, M.; Shin, S.; Volz, K.S.; Green, K.B.; et al. Autocrine function of aldehyde dehydrogenase 1 as a determinant of diet- and sex-specific differences in visceral adiposity. Diabetes 2013, 62, 124–136. [Google Scholar] [CrossRef]
- Fan, X.; Molotkov, A.; Manabe, S.; Donmoyer, C.M.; Deltour, L.; Foglio, M.H.; Cuenca, A.E.; Blaner, W.S.; Lipton, S.A.; Duester, G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell. Biol. 2003, 23, 4637–4648. [Google Scholar] [CrossRef] [PubMed]
- Dupe, V.; Matt, N.; Garnier, J.M.; Chambon, P.; Mark, M.; Ghyselinck, N.B. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl. Acad. Sci. USA 2003, 100, 14036–14041. [Google Scholar] [CrossRef] [PubMed]
- Elizondo, G.; Corchero, J.; Sterneck, E.; Gonzalez, F.J. Feedback inhibition of the retinaldehyde dehydrogenase gene ALDH1 by retinoic acid through retinoic acid receptor alpha and CCAAT/enhancer-binding protein beta. J. Biol. Chem. 2000, 275, 39747–39753. [Google Scholar] [CrossRef] [PubMed]
- Elizondo, G.; Medina-Diaz, I.M.; Cruz, R.; Gonzalez, F.J.; Vega, L. Retinoic acid modulates retinaldehyde dehydrogenase 1 gene expression through the induction of GADD153-C/EBPbeta interaction. Biochem. Pharmacol. 2009, 77, 248–257. [Google Scholar] [CrossRef]
- Zhong, G.; Seaman, C.J.; Paragas, E.M.; Xi, H.; Herpoldt, K.L.; King, N.P.; Jones, J.P.; Isoherranen, N. Aldehyde Oxidase Contributes to All-Trans-Retinoic Acid Biosynthesis in Human Liver. Drug Metab. Dispos. 2021, 49, 202–211. [Google Scholar] [CrossRef]
- Terao, M.; Kurosaki, M.; Barzago, M.M.; Fratelli, M.; Bagnati, R.; Bastone, A.; Giudice, C.; Scanziani, E.; Mancuso, A.; Tiveron, C.; et al. Role of the molybdoflavoenzyme aldehyde oxidase homolog 2 in the biosynthesis of retinoic acid: Generation and characterization of a knockout mouse. Mol. Cell. Biol. 2009, 29, 357–377. [Google Scholar] [CrossRef]
- Chambers, D.; Wilson, L.; Maden, M.; Lumsden, A. RALDH-independent generation of retinoic acid during vertebrate embryogenesis by CYP1B1. Development 2007, 134, 1369–1383. [Google Scholar] [CrossRef]
- Maguire, M.; Larsen, M.C.; Vezina, C.M.; Quadro, L.; Kim, Y.K.; Tanumihardjo, S.A.; Jefcoate, C.R. Cyp1b1 directs Srebp-mediated cholesterol and retinoid synthesis in perinatal liver; Association with retinoic acid activity during fetal development. PLoS ONE 2020, 15, e0228436. [Google Scholar] [CrossRef]
- Maguire, M.; Larsen, M.C.; Foong, Y.H.; Tanumihardjo, S.; Jefcoate, C.R. Cyp1b1 deletion and retinol deficiency coordinately suppress mouse liver lipogenic genes and hepcidin expression during post-natal development. Mol. Cell Endocrinol. 2017, 454, 50–68. [Google Scholar] [CrossRef]
- Sirbu, I.O.; Gresh, L.; Barra, J.; Duester, G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development 2005, 132, 2611–2622. [Google Scholar] [CrossRef]
- Schilling, T.F.; Sosnik, J.; Nie, Q. Visualizing retinoic acid morphogen gradients. Methods Cell Biol. 2016, 133, 139–163. [Google Scholar]
- Schilling, T.F.; Nie, Q.; Lander, A.D. Dynamics and precision in retinoic acid morphogen gradients. Curr. Opin. Genet. Dev. 2012, 22, 562–569. [Google Scholar] [CrossRef]
- Delva, L.; Bastie, J.N.; Rochette-Egly, C.; Kraiba, R.; Balitrand, N.; Despouy, G.; Chambon, P.; Chomienne, C. Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol. Cell. Biol. 1999, 19, 7158–7167. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.H.; Peng, C.C.; Lutz, J.D.; Yeung, C.K.; Zelter, A.; Isoherranen, N. Direct protein-protein interactions and substrate channeling between cellular retinoic acid binding proteins and CYP26B1. FEBS Lett. 2016, 590, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Ortiz, D.; Zelter, A.; Nath, A.; Isoherranen, N. CYP26C1 Is a Hydroxylase of Multiple Active Retinoids and Interacts with Cellular Retinoic Acid Binding Proteins. Mol. Pharmacol. 2018, 93, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, I.; Wei, L.N. All-trans Retinoic Acid as a Versatile Cytosolic Signal Modulator Mediated by CRABP1. Int. J. Mol. Sci. 2019, 20, 3610. [Google Scholar] [CrossRef] [PubMed]
- Durand, B.; Saunders, M.; Leroy, P.; Leid, M.; Chambon, P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell 1992, 71, 73–85. [Google Scholar] [CrossRef]
- Li, Z.; Shen, J.; Wu, W.K.; Wang, X.; Liang, J.; Qiu, G.; Liu, J. Vitamin A deficiency induces congenital spinal deformities in rats. PLoS ONE 2012, 7, e46565. [Google Scholar] [CrossRef]
- Leroy, P.; Nakshatri, H.; Chambon, P. Mouse retinoic acid receptor alpha 2 isoform is transcribed from a promoter that contains a retinoic acid response element. Proc. Natl. Acad. Sci. USA 1991, 88, 10138–10142. [Google Scholar] [CrossRef]
- Davis, K.D.; Lazar, M.A. Induction of retinoic acid receptor-beta by retinoic acid is cell specific. Endocrinology 1993, 132, 1469–1474. [Google Scholar] [CrossRef]
- Hoffmann, B.; Lehmann, J.M.; Zhang, X.K.; Hermann, T.; Husmann, M.; Graupner, G.; Pfahl, M. A retinoic acid receptor-specific element controls the retinoic acid receptor-beta promoter. Mol. Endocrinol. 1990, 4, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.M.; Zhang, X.K.; Pfahl, M. RAR gamma 2 expression is regulated through a retinoic acid response element embedded in Sp1 sites. Mol. Cell. Biol. 1992, 12, 2976–2985. [Google Scholar]
- Sucov, H.M.; Murakami, K.K.; Evans, R.M. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc. Natl. Acad. Sci. USA 1990, 87, 5392–5396. [Google Scholar] [CrossRef] [PubMed]
- Kopf, E.; Plassat, J.L.; Vivat, V.; de The, H.; Chambon, P.; Rochette-Egly, C. Dimerization with retinoid X receptors and phosphorylation modulate the retinoic acid-induced degradation of retinoic acid receptors alpha and gamma through the ubiquitin-proteasome pathway. J. Biol. Chem. 2000, 275, 33280–33288. [Google Scholar] [CrossRef] [PubMed]
- Ferry, C.; Gianni, M.; Lalevee, S.; Bruck, N.; Plassat, J.L.; Raska, I., Jr.; Garattini, E.; Rochette-Egly, C. SUG-1 plays proteolytic and non-proteolytic roles in the control of retinoic acid target genes via its interaction with SRC-3. J. Biol. Chem. 2009, 284, 8127–8135. [Google Scholar] [CrossRef] [PubMed]
- Carrier, M.; Lutzing, R.; Gaouar, S.; Rochette-Egly, C. TRIM24 mediates the interaction of the retinoic acid receptor alpha with the proteasome. FEBS Lett. 2018, 592, 1426–1433. [Google Scholar] [CrossRef]
- Cheng, X.; Pei, P.; Yu, J.; Zhang, Q.; Li, D.; Xie, X.; Wu, J.; Wang, S.; Zhang, T. F-box protein FBXO30 mediates retinoic acid receptor gamma ubiquitination and regulates BMP signaling in neural tube defects. Cell Death Dis. 2019, 10, 551. [Google Scholar] [CrossRef]
- Ross, A.C.; Zolfaghari, R. Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu. Rev. Nutr. 2011, 31, 65–87. [Google Scholar] [CrossRef]
- Pennimpede, T.; Cameron, D.A.; MacLean, G.A.; Li, H.; Abu-Abed, S.; Petkovich, M. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res. Part A Clin. Mol. Teratol. 2010, 88, 883–894. [Google Scholar] [CrossRef]
- Dubey, A.; Saint-Jeannet, J.P. Anterior patterning genes induced by Zic1 are sensitive to retinoic acid and its metabolite, 4-oxo-RA. Dev. Dyn. 2021, 251, 498–512. [Google Scholar] [CrossRef]
- Baron, J.M.; Heise, R.; Blaner, W.S.; Neis, M.; Joussen, S.; Dreuw, A.; Marquardt, Y.; Saurat, J.H.; Merk, H.F.; Bickers, D.R.; et al. Retinoic acid and its 4-oxo metabolites are functionally active in human skin cells in vitro. J. Investig. Dermatol. 2005, 125, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Zolfaghari, R.; Ross, A.C. Liver-specific cytochrome P450 CYP2C22 is a direct target of retinoic acid and a retinoic acid-metabolizing enzyme in rat liver. J. Lipid Res. 2010, 51, 1781–1792. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, J.E.; Zelter, A.; Isoherranen, N. The relative importance of CYP26A1 in hepatic clearance of all-trans retinoic acid. Biochem. Pharmacol. 2010, 80, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Topletz, A.R.; Zhong, G.; Isoherranen, N. Scaling in vitro activity of CYP3A7 suggests human fetal livers do not clear retinoic acid entering from maternal circulation. Sci. Rep. 2019, 9, 4620. [Google Scholar] [CrossRef]
- Kramlinger, V.M.; Nagy, L.D.; Fujiwara, R.; Johnson, K.M.; Phan, T.T.; Xiao, Y.; Enright, J.M.; Toomey, M.B.; Corbo, J.C.; Guengerich, F.P. Human cytochrome P450 27C1 catalyzes 3,4-desaturation of retinoids. FEBS Lett. 2016, 590, 1304–1312. [Google Scholar] [CrossRef] [PubMed]
- Rollman, O.; Wood, E.J.; Olsson, M.J.; Cunliffe, W.J. Biosynthesis of 3,4-didehydroretinol from retinol by human skin keratinocytes in culture. Biochem. J. 1993, 293 Pt 3, 675–682. [Google Scholar] [CrossRef]
- Johnson, K.M.; Phan, T.T.N.; Albertolle, M.E.; Guengerich, F.P. Human mitochondrial cytochrome P450 27C1 is localized in skin and preferentially desaturates trans-retinol to 3,4-dehydroretinol. J. Biol. Chem. 2017, 292, 13672–13687. [Google Scholar] [CrossRef]
- Enright, J.M.; Toomey, M.B.; Sato, S.Y.; Temple, S.E.; Allen, J.R.; Fujiwara, R.; Kramlinger, V.M.; Nagy, L.D.; Johnson, K.M.; Xiao, Y.; et al. Cyp27c1 Red-Shifts the Spectral Sensitivity of Photoreceptors by Converting Vitamin A1 into A2. Curr. Biol. 2015, 25, 3048–3057. [Google Scholar] [CrossRef]
- Maclean, G.; Dolle, P.; Petkovich, M. Genetic disruption of CYP26B1 severely affects development of neural crest derived head structures, but does not compromise hindbrain patterning. Dev. Dyn. 2009, 238, 732–745. [Google Scholar] [CrossRef]
- Abu-Abed, S.; Dolle, P.; Metzger, D.; Wood, C.; MacLean, G.; Chambon, P.; Petkovich, M. Developing with lethal RA levels: Genetic ablation of Rarg can restore the viability of mice lacking Cyp26a1. Development 2003, 130, 1449–1459. [Google Scholar] [CrossRef]
- Dranse, H.J.; Sampaio, A.V.; Petkovich, M.; Underhill, T.M. Genetic deletion of Cyp26b1 negatively impacts limb skeletogenesis by inhibiting chondrogenesis. J. Cell Sci. 2011, 124, 2723–2734. [Google Scholar] [CrossRef] [PubMed]
- Abu-Abed, S. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001, 15, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, K.; Zhao, X.; Uehara, M.; Yamashita, K.; Nishijima, M.; Nishino, J.; Saijoh, Y.; Sakai, Y.; Hamada, H. Regulation of Retinoic Acid Distribution Is Required for Proximodistal Patterning and Outgrowth of the Developing Mouse Limb. Dev. Cell 2004, 6, 411–422. [Google Scholar] [CrossRef]
- Topletz, A.R.; Tripathy, S.; Foti, R.S.; Shimshoni, J.A.; Nelson, W.L.; Isoherranen, N. Induction of CYP26A1 by metabolites of retinoic acid: Evidence that CYP26A1 is an important enzyme in the elimination of active retinoids. Mol. Pharmacol. 2015, 87, 430–441. [Google Scholar] [CrossRef] [PubMed]
- White, J.A.; Beckett-Jones, B.; Guo, Y.D.; Dilworth, F.J.; Bonasoro, J.; Jones, G.; Petkovich, M. cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J. Biol. Chem. 1997, 272, 18538–18541. [Google Scholar] [CrossRef]
- White, J.A.; Guo, Y.D.; Baetz, K.; Beckett-Jones, B.; Bonasoro, J.; Hsu, K.E.; Dilworth, F.J.; Jones, G.; Petkovich, M. Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. J. Biol. Chem. 1996, 271, 29922–29927. [Google Scholar] [CrossRef]
- White, J.A.; Ramshaw, H.; Taimi, M.; Stangle, W.; Zhang, A.; Everingham, S.; Creighton, S.; Tam, S.P.; Jones, G.; Petkovich, M. Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proc. Natl. Acad. Sci. USA 2000, 97, 6403–6408. [Google Scholar] [CrossRef]
- Loudig, O.; Maclean, G.A.; Dore, N.L.; Luu, L.; Petkovich, M. Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem. J. 2005, 392, 241–248. [Google Scholar] [CrossRef]
- Zhang, Y.; Zolfaghari, R.; Ross, A.C. Multiple retinoic acid response elements cooperate to enhance the inducibility of CYP26A1 gene expression in liver. Gene 2010, 464, 32–43. [Google Scholar] [CrossRef][Green Version]
- Zolfaghari, R.; Ross, A.C. Hepatocyte nuclear factor 4alpha (HNF4alpha) in coordination with retinoic acid receptors increases all-trans-retinoic acid-dependent CYP26A1 gene expression in HepG2 human hepatocytes. J. Cell Biochem. 2014, 115, 1740–1751. [Google Scholar] [CrossRef]
- Loudig, O.; Babichuk, C.; White, J.; Abu-Abed, S.; Mueller, C.; Petkovich, M. Cytochrome P450RAI(CYP26) promoter: A distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol. Endocrinol. 2000, 14, 1483–1497. [Google Scholar] [CrossRef]
- Ribes, V.; Fraulob, V.; Petkovich, M.; Dolle, P. The oxidizing enzyme CYP26a1 tightly regulates the availability of retinoic acid in the gastrulating mouse embryo to ensure proper head development and vasculogenesis. Dev. Dyn. 2007, 236, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Zile, M.H.; Schnoes, H.K.; DeLuca, H.F. Characterization of retinoyl beta-glucuronide as a minor metabolite of retinoic acid in bile. Proc. Natl. Acad. Sci. USA 1980, 77, 3230–3233. [Google Scholar] [CrossRef] [PubMed]
- Goswami, B.C.; Reida, A.K.; Ivanoff, K.D.; Barua, A.B.; Olson, J.A. Intestinal absorption and metabolism of retinoyl beta-glucuronide in humans, and of 15-[14C]-retinoyl beta-glucuronide in rats of different vitamin A status. J. Nutr. Biochem. 2003, 14, 703–709. [Google Scholar] [CrossRef]
- Barua, A.B.; Olson, J.A. Retinoyl beta-glucuronide: An endogenous compound of human blood. Am. J. Clin. Nutr. 1986, 43, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Samokyszyn, V.M.; Gall, W.E.; Zawada, G.; Freyaldenhoven, M.A.; Chen, G.; Mackenzie, P.I.; Tephly, T.R.; Radominska-Pandya, A. 4-hydroxyretinoic acid, a novel substrate for human liver microsomal UDP-glucuronosyltransferase(s) and recombinant UGT2B7. J. Biol. Chem. 2000, 275, 6908–6914. [Google Scholar] [CrossRef]
- Barua, A.B.; Batres, R.O.; Olson, J.A. Characterization of retinyl beta-glucuronide in human blood. Am. J. Clin. Nutr. 1989, 50, 370–374. [Google Scholar] [CrossRef]
- Khoo, K.C.; Reik, D.; Colburn, W.A. Pharmacokinetics of isotretinoin following a single oral dose. J. Clin. Pharmacol. 1982, 22, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Reboul, E.; Abou, L.; Mikail, C.; Ghiringhelli, O.; Andre, M.; Portugal, H.; Jourdheuil-Rahmani, D.; Amiot, M.J.; Lairon, D.; Borel, P. Lutein transport by Caco-2 TC-7 cells occurs partly by a facilitated process involving the scavenger receptor class B type I (SR-BI). Biochem. J. 2005, 387, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Reboul, E.; Klein, A.; Bietrix, F.; Gleize, B.; Malezet-Desmoulins, C.; Schneider, M.; Margotat, A.; Lagrost, L.; Collet, X.; Borel, P. Scavenger receptor class B type I (SR-BI) is involved in vitamin E transport across the enterocyte. J. Biol. Chem. 2006, 281, 4739–4745. [Google Scholar] [CrossRef]
- Goncalves, A.; Margier, M.; Roi, S.; Collet, X.; Niot, I.; Goupy, P.; Caris-Veyrat, C.; Reboul, E. Intestinal scavenger receptors are involved in vitamin K1 absorption. J. Biol. Chem. 2014, 289, 30743–30752. [Google Scholar] [CrossRef]
- Lalevée, S.; Anno, Y.N.; Chatagnon, A.; Samarut, E.; Poch, O.; Laudet, V.; Benoit, G.; Lecompte, O.; Rochette-Egly, C. Genome-wide in Silico Identification of New Conserved and Functional Retinoic Acid Receptor Response Elements (Direct Repeats Separated by 5 bp). J. Biol. Chem. 2011, 286, 33322–33334. [Google Scholar] [CrossRef] [PubMed]
- Gould, A.; Itasaki, N.; Krumlauf, R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron 1998, 21, 39–51. [Google Scholar] [CrossRef]
- Korkmaz, G.; Lopes, R.; Ugalde, A.P.; Nevedomskaya, E.; Han, R.; Myacheva, K.; Zwart, W.; Elkon, R.; Agami, R. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.H.; Hennighausen, L.; Shin, H.Y. Dissecting Tissue-Specific Super-Enhancers by Integrating Genome-Wide Analyses and CRISPR/Cas9 Genome Editing. J. Mammary Gland Biol. Neoplasia 2019, 24, 47–59. [Google Scholar] [CrossRef]
- Daniel, B.; Nagy, G.; Hah, N.; Horvath, A.; Czimmerer, Z.; Poliska, S.; Gyuris, T.; Keirsse, J.; Gysemans, C.; Van Ginderachter, J.A.; et al. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev. 2014, 28, 1562–1577. [Google Scholar] [CrossRef] [PubMed]
- Parihar, M.; Bendelac-Kapon, L.; Gur, M.; Abbou, T.; Belorkar, A.; Achanta, S.; Kinberg, K.; Vadigepalli, R.; Fainsod, A. Retinoic Acid Fluctuation Activates an Uneven, Direction-Dependent Network-Wide Robustness Response in Early Embryogenesis. Front. Cell Dev. Biol. 2021, 9, 747969. [Google Scholar] [CrossRef]
- Lee, L.M.; Leung, C.Y.; Tang, W.W.; Choi, H.L.; Leung, Y.C.; McCaffery, P.J.; Wang, C.C.; Woolf, A.S.; Shum, A.S. A paradoxical teratogenic mechanism for retinoic acid. Proc. Natl. Acad. Sci. USA 2012, 109, 13668–13673. [Google Scholar] [CrossRef]
- D’Aniello, E.; Rydeen, A.B.; Anderson, J.L.; Mandal, A.; Waxman, J.S. Depletion of retinoic acid receptors initiates a novel positive feedback mechanism that promotes teratogenic increases in retinoic acid. PLoS Genet. 2013, 9, e1003689. [Google Scholar] [CrossRef]
- D’Aniello, E.; Waxman, J.S. Input overload: Contributions of retinoic acid signaling feedback mechanisms to heart development and teratogenesis. Dev. Dyn. 2015, 244, 513–523. [Google Scholar] [CrossRef]
- Rydeen, A.; Voisin, N.; D’Aniello, E.; Ravisankar, P.; Devignes, C.S.; Waxman, J.S. Excessive feedback of Cyp26a1 promotes cell non-autonomous loss of retinoic acid signaling. Dev. Biol. 2015, 405, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Shoemaker, A.; Simsiman, R.; Denning, M.; Zachman, R.D. Expression of retinoic acid nuclear receptors and tissue transglutaminase is altered in various tissues of rats fed a vitamin A-deficient diet. J. Nutr. 1992, 122, 2144–2152. [Google Scholar] [CrossRef] [PubMed]
- Kostetskii, I.; Linask, K.K.; Zile, M.H. Vitamin A deficiency and the expression of retinoic acid receptors during early cardiogenesis in quail embryo. Rouxs Arch. Dev. Biol. 1996, 205, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Vega, V.A.; Anzulovich, A.C.; Varas, S.M.; Bonomi, M.R.; Gimenez, M.S.; Oliveros, L.B. Effect of nutritional vitamin A deficiency on lipid metabolism in the rat heart: Its relation to PPAR gene expression. Nutrition 2009, 25, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Zolfaghari, R.; Ross, A.C. Lecithin:retinol acyltransferase expression is regulated by dietary vitamin A and exogenous retinoic acid in the lung of adult rats. J. Nutr. 2002, 132, 1160–1164. [Google Scholar] [CrossRef]
- Ross, A.C.; Zolfaghari, R. Regulation of hepatic retinol metabolism: Perspectives from studies on vitamin A status. J. Nutr. 2004, 134, 269S–275S. [Google Scholar] [CrossRef]
- Liang, X.; Chen, M.; Wang, D.; Wen, J.; Chen, J. Vitamin A deficiency indicating as low expression of LRAT may be a novel biomarker of primary hypertension. Clin. Exp. Hypertens. 2021, 43, 151–163. [Google Scholar] [CrossRef]
- Amengual, J.; Golczak, M.; Palczewski, K.; von Lintig, J. Lecithin:Retinol Acyltransferase Is Critical for Cellular Uptake of Vitamin A from Serum Retinol-binding Protein*. J. Biol. Chem. 2012, 287, 24216–24227. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, Y.; Moiseyev, G.; Takahashi, Y.; Mott, R.; Ma, J.X. Comparison of ocular pathologies in vitamin A-deficient mice and RPE65 gene knockout mice. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5507–5514. [Google Scholar] [CrossRef]
- Kurlandsky, S.B.; Duell, E.A.; Kang, S.; Voorhees, J.J.; Fisher, G.J. Auto-regulation of retinoic acid biosynthesis through regulation of retinol esterification in human keratinocytes. J. Biol. Chem. 1996, 271, 15346–15352. [Google Scholar] [CrossRef]
- Ross, A.C.; Foulke, D.T.; Matsuura, T.; Tresini, M.; Breen, J.J.; Gurr, J.A. Hepatic lecithin: Retinol acyltransferase activity is induced in vivo by retinoic acid, but not by triiodothyronine, in vitamin A-deficient, hypothyroid rats. J. Nutr. Biochem. 1997, 8, 456–460. [Google Scholar] [CrossRef]
- Chertow, B.S.; Blaner, W.S.; Baranetsky, N.G.; Sivitz, W.I.; Cordle, M.B.; Thompson, D.; Meda, P. Effects of vitamin A deficiency and repletion on rat insulin secretion in vivo and in vitro from isolated islets. J. Clin. Investig. 1987, 79, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Perozzi, G.; Mengheri, E.; Colantuoni, V.; Gaetani, S. Vitamin A intake and in vivo expression of the genes involved in retinol transport. Eur. J. Biochem. 1991, 196, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Rajan, N.; Blaner, W.S.; Soprano, D.R.; Suhara, A.; Goodman, D.S. Cellular retinol-binding protein messenger RNA levels in normal and retinoid-deficient rats. J. Lipid Res. 1990, 31, 821–829. [Google Scholar] [CrossRef]
- Smith, W.C.; Nakshatri, H.; Leroy, P.; Rees, J.; Chambon, P. A retinoic acid response element is present in the mouse cellular retinol binding protein I (mCRBPI) promoter. EMBO J. 1991, 10, 2223–2230. [Google Scholar] [CrossRef]
- Husmann, M.; Hoffmann, B.; Stump, D.G.; Chytil, F.; Pfahl, M. A retinoic acid response element from the rat CRBPI promoter is activated by an RAR/RXR heterodimer. Biochem. Biophys. Res. Commun. 1992, 187, 1558–1564. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Umesono, K.; Kliewer, S.A.; Borgmeyer, U.; Ong, E.S.; Evans, R.M. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell 1991, 66, 555–561. [Google Scholar] [CrossRef]
- Nakshatri, H.; Chambon, P. The directly repeated RG(G/T)TCA motifs of the rat and mouse cellular retinol-binding protein II genes are promiscuous binding sites for RAR, RXR, HNF-4, and ARP-1 homo- and heterodimers. J. Biol. Chem. 1994, 269, 890–902. [Google Scholar] [CrossRef]
- Zhang, L.; Xueping, E.; Luker, K.E.; Shao, J.S.; Levin, M.S.; Suh, E.; Li, E. Analysis of human cellular retinol-binding protein II promoter during enterocyte differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 282, G1079–G1087. [Google Scholar] [CrossRef][Green Version]
- Reijntjes, S.; Zile, M.H.; Maden, M. The expression of Stra6 and Rdh10 in the avian embryo and their contribution to the generation of retinoid signatures. Int. J. Dev. Biol. 2010, 54, 1267–1275. [Google Scholar] [CrossRef]
- Strate, I.; Min, T.H.; Iliev, D.; Pera, E.M. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development 2009, 136, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Wassef, L.; Spiegler, E.; Quadro, L. Embryonic phenotype, beta-carotene and retinoid metabolism upon maternal supplementation of beta-carotene in a mouse model of severe vitamin A deficiency. Arch. Biochem. Biophys. 2013, 539, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Sperkova, Z.; Napoli, J.L. Cellular expression of retinal dehydrogenase types 1 and 2: Effects of vitamin A status on testis mRNA. J. Cell Physiol. 2001, 186, 220–232. [Google Scholar] [CrossRef]
- Trasino, S.E.; Benoit, Y.D.; Gudas, L.J. Vitamin A deficiency causes hyperglycemia and loss of pancreatic beta-cell mass. J. Biol. Chem. 2015, 290, 1456–1473. [Google Scholar] [CrossRef] [PubMed]
- Harari, A.; Melnikov, N.; Kandel Kfir, M.; Kamari, Y.; Mahler, L.; Ben-Amotz, A.; Harats, D.; Cohen, H.; Shaish, A. Dietary beta-Carotene Rescues Vitamin A Deficiency and Inhibits Atherogenesis in Apolipoprotein E-Deficient Mice. Nutrients 2020, 12, 1625. [Google Scholar] [CrossRef] [PubMed]
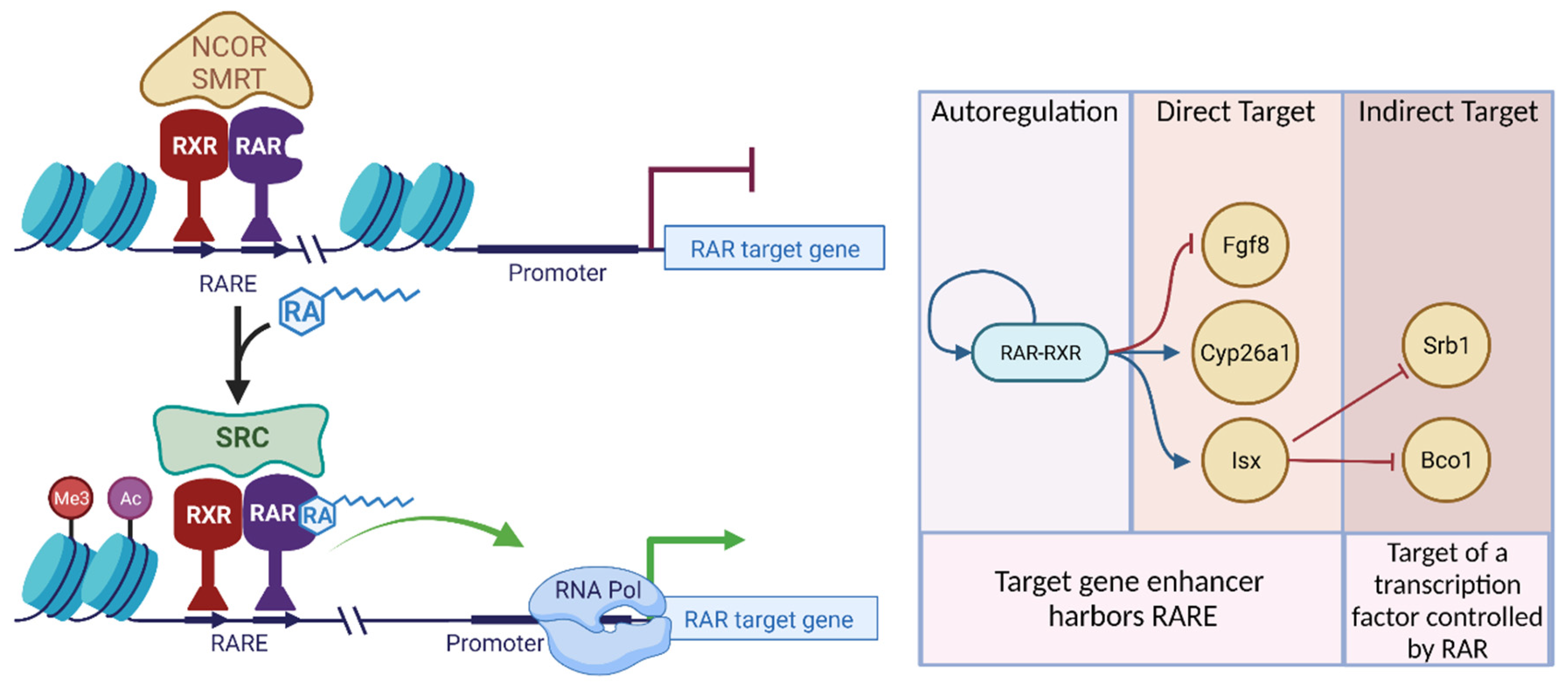
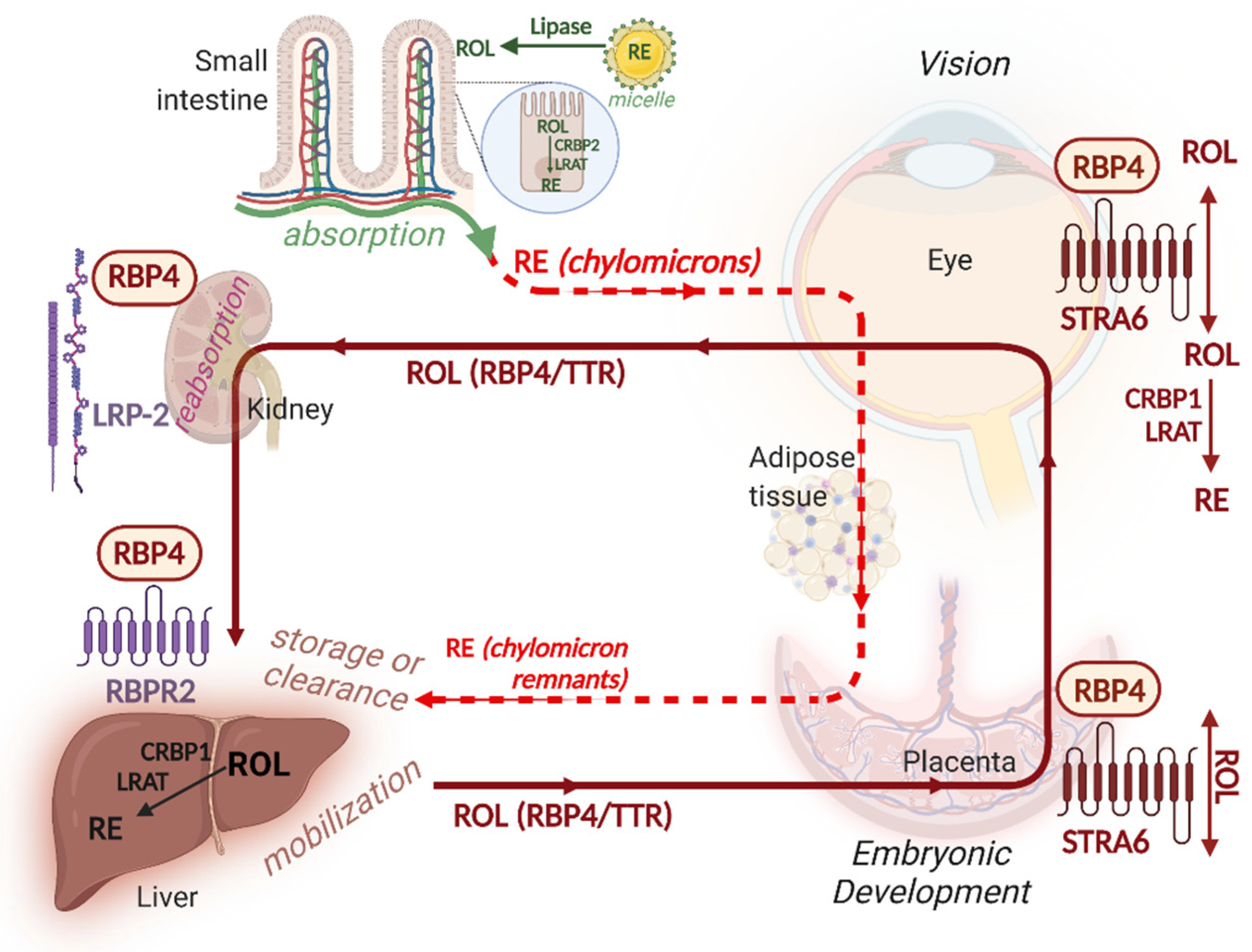
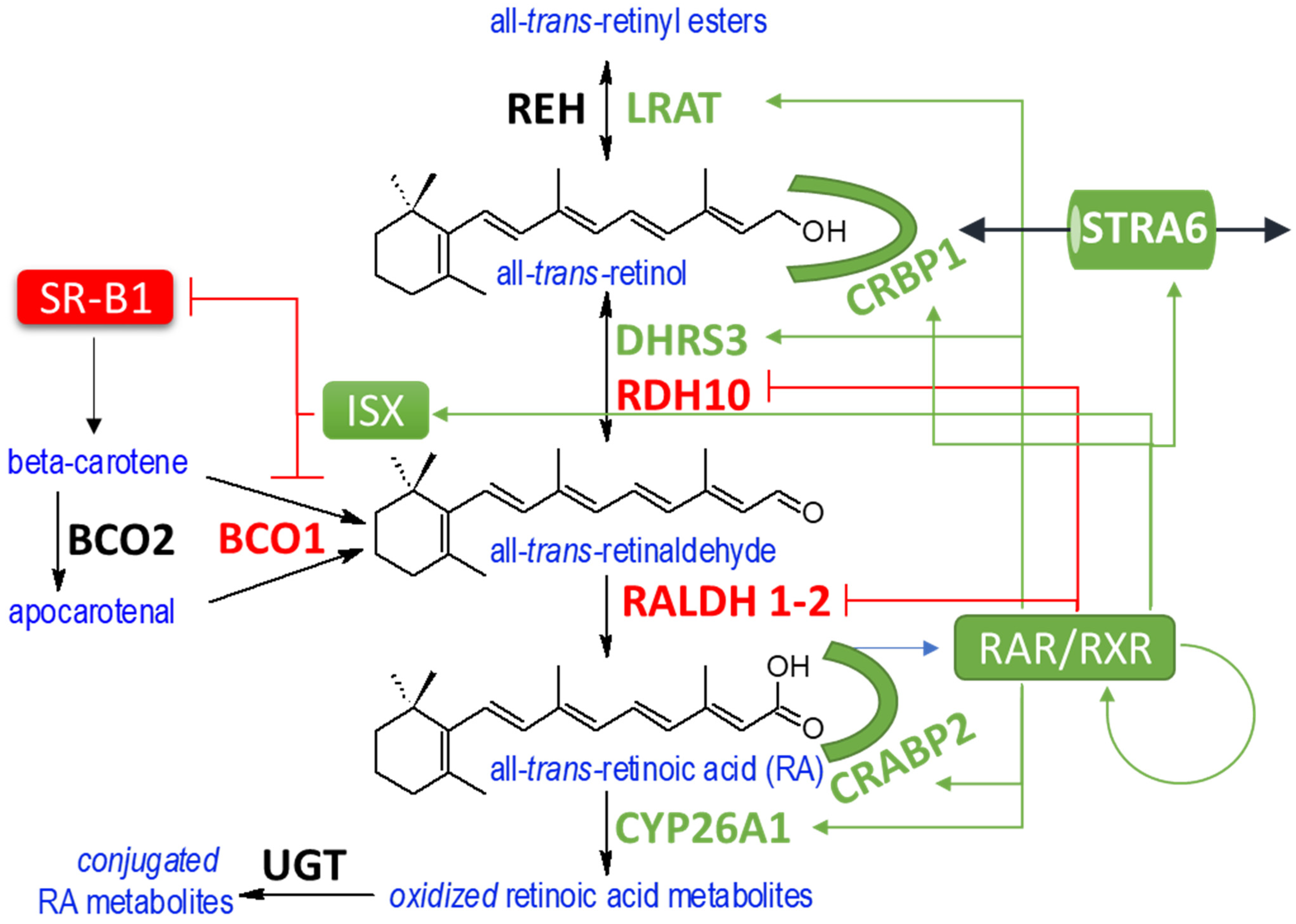
| Role in Vitamin A Metabolism | Gene Name | Acronym | Effect of VAD on Gene Expression | Effect of RA on Gene Expression |
|---|---|---|---|---|
| Signaling | Retinoic acid receptors | RARα RARβ RARγ | Downregulated in some tissues of VAD rats and quail [312,313] | Directly upregulated in response to RA via conserved RARE [72,258,259,260,261,262] |
| Retinoid X receptors | RXRα RXRβ RXRγ | Downregulated of Rxra and Rxrb in hearts of VAD rats, corrected with VA supplementation. [314] | Not clear if Rxr genes are RAR-targets | |
| Conversion of provitamin A carotenoids to retinol | B-carotene-15,15-dioxygenase 1 | BCO1 | Upregulated in VAD mice [130]. | Expression is suppressed by RA via RAR-mediated induction of the transcription factor ISX [115,128,129,130,131,132] |
| Storage | Lecithin retinal acyltransferase | LRAT | Protein and transcript levels of LRAT decrease in the many tissues of VAD animals [315,316,317,318,319]. There is evidence that the magnitude and direction of response is tissue-specific. | Indirectly upregulated in response to RA, suggested by fact that upregulation pf LRAT and LRAT activity by RA is blocked by the translation inhibitor, cycloheximide [163,316,320,321]. No functional RARE sites have been identified. A genomic region of the Lrat promoter confers RA-inducibility and contains binding sites for SP1 [109] and GATA transcription factors [107] |
| Retinol Binding Proteins | Cellular retinol-binding proteins | CRBP1 | Decreased expression of Rbp1 in VAD rats [322,323,324] | Upregulated by RAR via a direct mechanism unaffected by cycloheximide and including a functional RARE [108,325,326] |
| CRBP2 | Upregulated in the intestine of VAD rats [324] | Not clear if regulated in response to RA. Promoter appears to harbor a poorly conserved response element for RXR or HNF-4 [327,328] and whose physiological relevance is currently, unclear [105,329]. | ||
| Retinol binding protein | RBP4 | VAD causes reduced secretion of RBP4 from liver cells [155,200,201] | Expression induced in response to RA [197,198] but has not been clearly demonstrated to be via direct mechanism or to harbor a functional RARE. | |
| RBP4 Receptors | Stimulated by retinoic acid 6 | STRA6 | VAD causes expansion of domains of expression of STRA6 in quail embryos [330]. Alternatively spliced Stra6 mouse isoforms are differentially regulated by VAD [203]. | Directly induced by RA via a functional RARE [173,174,202,203] |
| Retinol binding protein receptor 2 | RBPR2 | Expression is inversely correlated with liver retinol stores [180]. | Expression is downregulated by RA or retinol treatment [180]. | |
| RA synthetic enzymes | Retinol dehydrogenase 10 | RDH10 | Expression of Rdh10 is upregulated in genetic models of RA-deficiency [217] | Rdh10 is negatively regulated by RA [224,331]. Β-carotene supplementation leads to downregulation of Rdh10 [332]. Rdh10 is downregulated in genetic models of RA-excess [59] |
| Retinaldehyde dehydrogenases 1-2 | RALDH1-2 | VAD causes upregulation of Raldh1 and downregulation of Raldh2 in rat testes [333], | Raldh1 and Raldh2 are downregulated in genetic models of RA-excess in mouse [59]. Suppression of Raldh1 expression by RA is via direct RAR binding [243,244] | |
| Enzymes which prevent RA formation or reduce RA levels | Short-chain dehydrogenase reductase family member 3 | DHRS3 | Expression is decreased in the liver and hearts of VAD rats [63,229]. | Directly upregulated by RA, though a functional RARE has not been identified [229,230]. |
| Cytochrome P450 26 A1 | CYP26A1 | CYP26A1 is downregulated in liver and pancreatic tissues of VAD mice [334,335]. | Directly upregulated via an identified RARE [288,289]. HNF4A cooperates with RAR in the regulation of CYP26A1 [284,290] | |
| Cytochrome P450 enzymes family 2 C22 | CYP2C22 | Directly upregulated by RA [272] | ||
| RA binding proteins | Cellular retinoic acid-binding proteins | CRABP1 CRABP2 | Crabp1 and Crabp2 are downregulated in liver and pancreatic tissues of VAD mice [334] | Crabp1 is indirectly upregulated by RA [108]. Crabp2 is directly upregulated by RA via an identified RARE [257]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Connor, C.; Varshosaz, P.; Moise, A.R. Mechanisms of Feedback Regulation of Vitamin A Metabolism. Nutrients 2022, 14, 1312. https://doi.org/10.3390/nu14061312
O’Connor C, Varshosaz P, Moise AR. Mechanisms of Feedback Regulation of Vitamin A Metabolism. Nutrients. 2022; 14(6):1312. https://doi.org/10.3390/nu14061312
Chicago/Turabian StyleO’Connor, Catherine, Parisa Varshosaz, and Alexander R. Moise. 2022. "Mechanisms of Feedback Regulation of Vitamin A Metabolism" Nutrients 14, no. 6: 1312. https://doi.org/10.3390/nu14061312
APA StyleO’Connor, C., Varshosaz, P., & Moise, A. R. (2022). Mechanisms of Feedback Regulation of Vitamin A Metabolism. Nutrients, 14(6), 1312. https://doi.org/10.3390/nu14061312






