Saccharomyces cerevisiae and Caffeine Implications on the Eukaryotic Cell
Abstract
1. Introduction
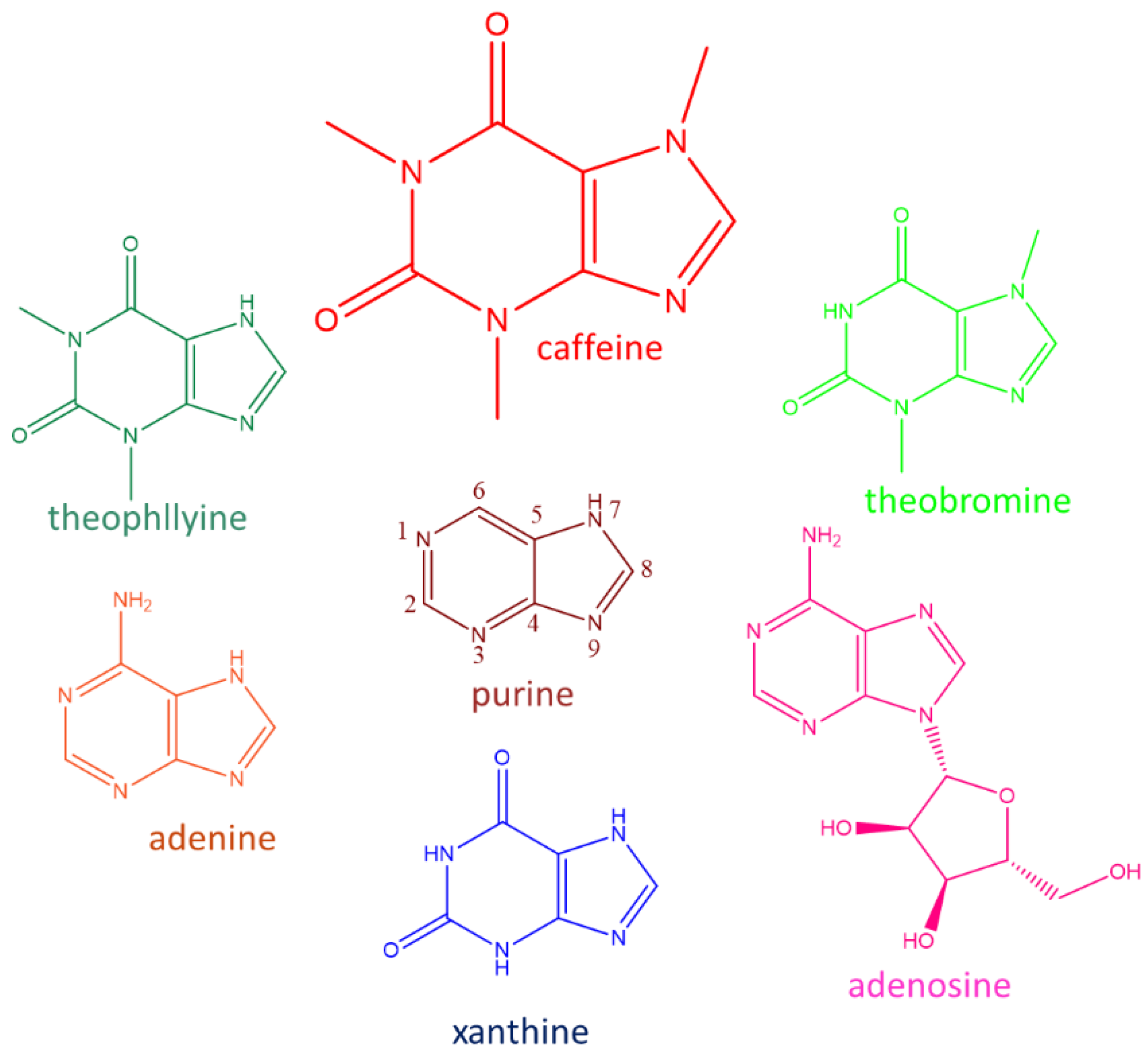
2. Caffeine: Transport and Toxicity in S. cerevisiae
3. Caffeine: Between Radio-Protector and Radio-Sensitizer
3.1. UV Irradiation
3.2. γ-Irradiation
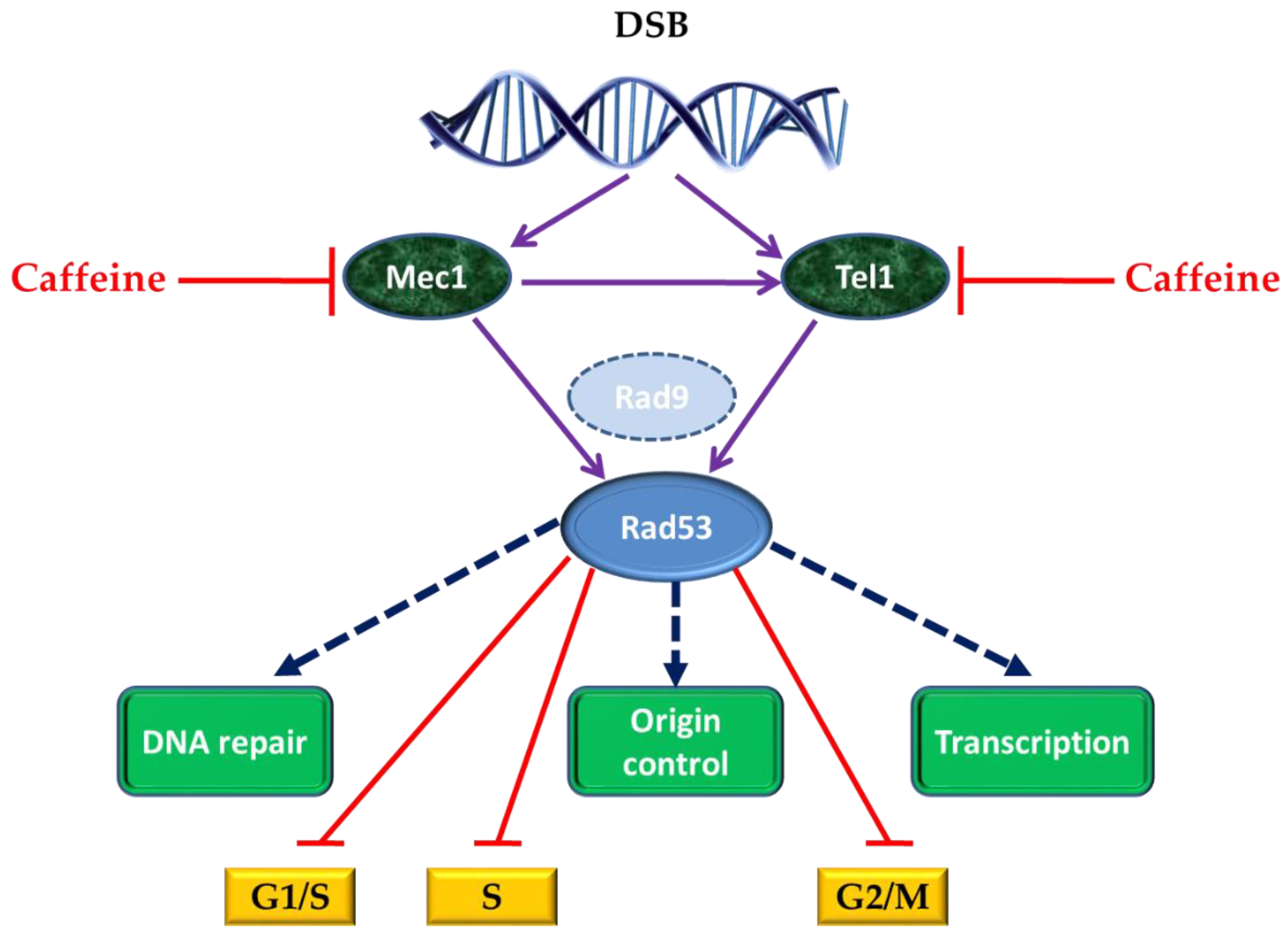
4. Caffeine and Cell Response to DNA Damage
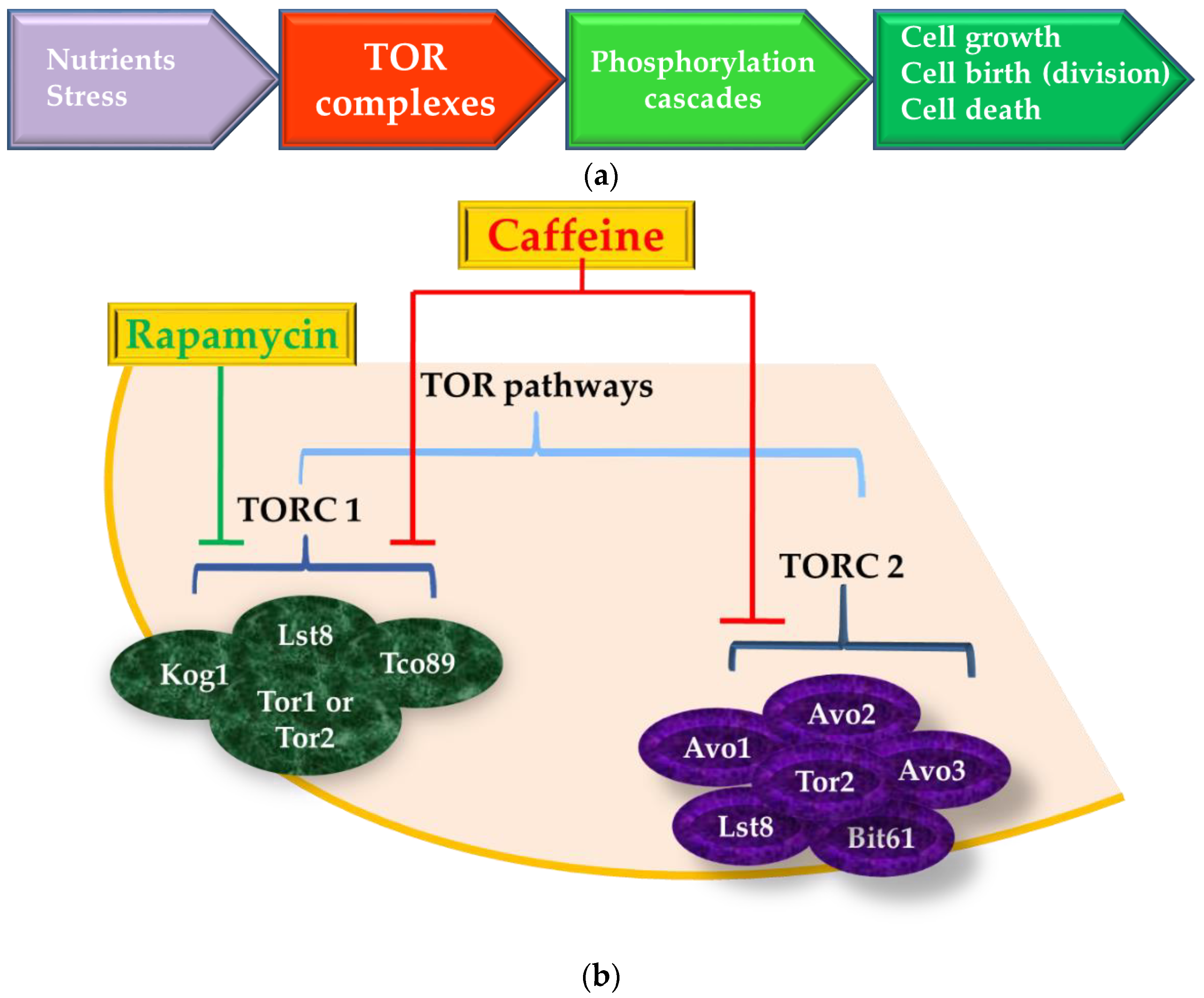
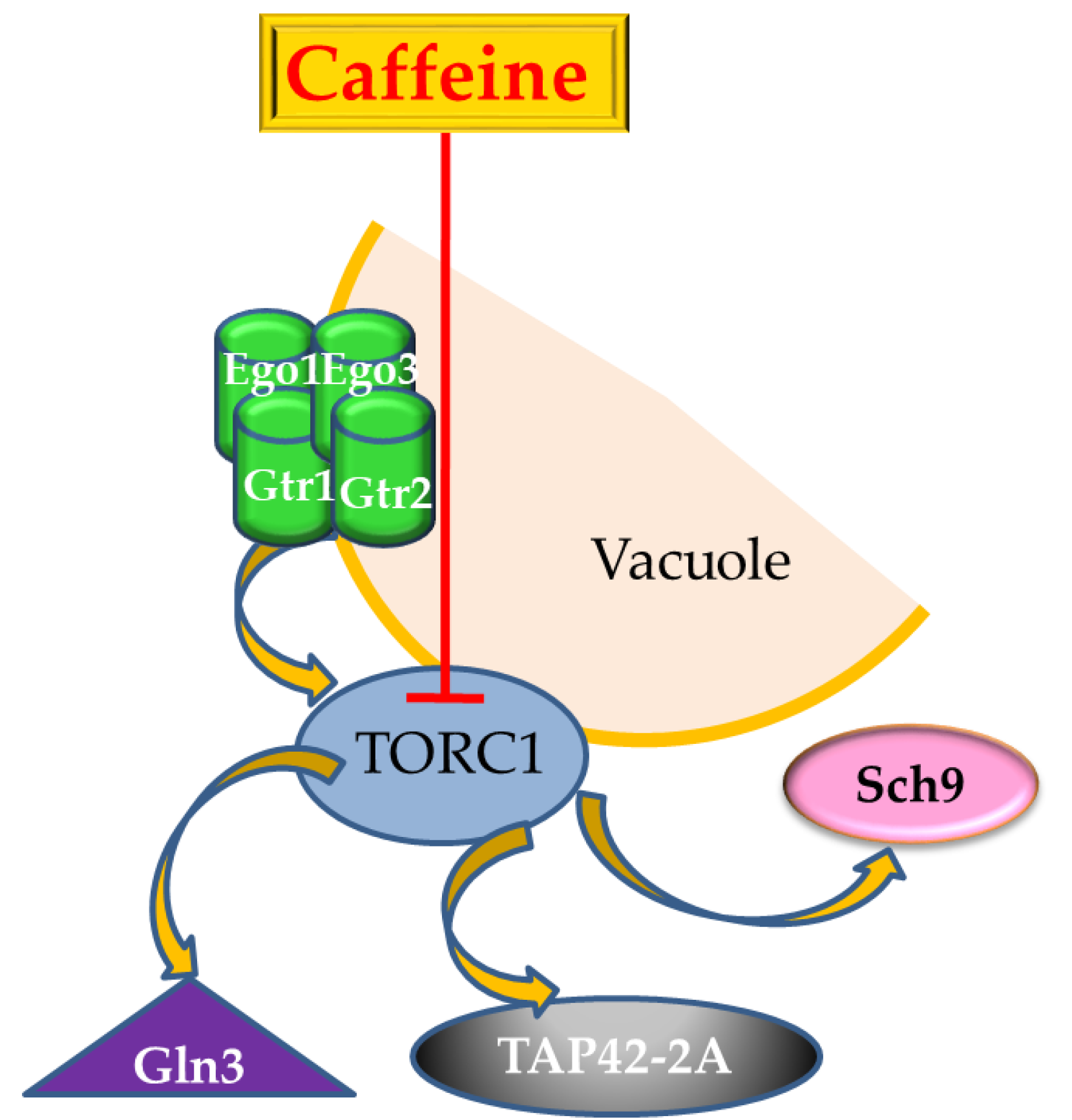
5. The Target-of-Rapamycin (TOR) Pathway is also the Target-of-Caffeine
6. Caffeine and the Yeast Cell Wall Integrity Pathway
7. Other Pathways Susceptible to Caffeine
8. Caffeine and Lifespan
9. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Cappelletti, S.; Piacentino, D.; Sani, G.; Aromatario, M. Caffeine: Cognitive and physical performance enhancer or psychoactive drug? Curr. Neuropharmacol. 2015, 13, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Dewick, P.M. Medicinal Natural Products: A Biosynthetic Approach, 3rd ed.; John Wiley & Sons: West Sussex, UK, 2009; pp. 413–416. [Google Scholar]
- Kaufmann, W.K.; Heffernan, T.P.; Beaulieu, L.M.; Doherty, S.; Frank, A.R.; Zhou, Y.; Bryant, M.F.; Zhou, T.; Luche, D.D.; Nikolaishvili-Feinberg, N.; et al. Caffeine and human DNA metabolism: The magic and the mystery. Mutat. Res. 2003, 532, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Karathia, H.; Vilaprinyo, E.; Sorribas, A.; Alves, R. Saccharomyces cerevisiae as a model organism: A comparative study. PLoS ONE 2011, 6, e16015. [Google Scholar] [CrossRef] [PubMed]
- Duina, A.A.; Miller, M.E.; Keeney, J.B. Budding yeast for budding geneticists: A primer on the Saccharomyces cerevisiae model system. Genetics 2014, 197, 33–48. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef]
- Castrillo, J.I.; Oliver, S. Yeast as a touchstone in post-genomic research: Strategies for integrative analysis in functional genomics. J. Biochem. Mol. Biol. 2004, 37, 93–106. [Google Scholar] [CrossRef]
- Matuo, R.; Sousa, F.G.; Soares, D.G.; Bonatto, D.; Saffi, J.; Escargueil, A.E.; Larsen, A.K.; Henriques, J.A. Saccharomyces cerevisiae as a model system to study the response to anticancer agents. Cancer Chemother. Pharmacol. 2012, 70, 491–502. [Google Scholar] [CrossRef]
- Dos Santos, S.C.; Sá-Correia, I. Yeast toxicogenomics: Lessons from a eukaryotic cell model and cell factory. Curr. Opin. Biotechnol. 2015, 33, 183–191. [Google Scholar] [CrossRef]
- Lian, J.; Mishra, S.; Zhao, H. Recent advances in metabolic engineering of Saccharomyces cerevisiae: New tools and their applications. Metab. Eng. 2018, 50, 85–108. [Google Scholar] [CrossRef]
- Nielsen, J. Yeast systems biology: Model organism and cell factory. Biotechnol. J. 2019, 14, e1800421. [Google Scholar] [CrossRef]
- Coronas-Serna, J.M.; Valenti, M.; Del Val, E.; Fernández-Acero, T.; Rodríguez-Escudero, I.; Mingo, J.; Luna, S.; Torices, L.; Pulido, R.; Molina, M.; et al. Modeling human disease in yeast: Recreating the PI3K-PTEN-Akt signaling pathway in Saccharomyces cerevisiae. Int. Microbiol. 2020, 23, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Kuranda, K.; Leberre, V.; Sokol, S.; Palamarczyk, G.; François, J. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 2006, 61, 1147–1166. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Rodriguez, C.; Chevallier, M.R.; Jund, R. The purine-cytosine permease gene of Saccharomyces cerevisiae: Primary structure and deduced protein sequence of the FCY2 gene product. Mol. Microbiol. 1990, 4, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Xiong, L. Characterization of a purine permease family gene OsPUP7 involved in growth and development control in rice. J. Integr. Plant. Biol. 2013, 55, 1119–1135. [Google Scholar] [CrossRef]
- Wagner, R.; Straub, M.L.; Souciet, J.L.; Potier, S.; de Montigny, J. New plasmid system to select for Saccharomyces cerevisiae purine-cytosine permease affinity mutants. J. Bacteriol. 2001, 183, 4386–4388. [Google Scholar] [CrossRef][Green Version]
- Tsujimoto, Y.; Shimizu, Y.; Otake, K.; Nakamura, T.; Okada, R.; Miyazaki, T.; Watanabe, K. Multidrug resistance transporters Snq2p and Pdr5p mediate caffeine efflux in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2015, 79, 1103–1110. [Google Scholar] [CrossRef]
- Sürmeli, Y.; Holyavkin, C.; Topaloğlu, A.; Arslan, M.; Kısakesen, H.İ.; Çakar, Z.P. Evolutionary engineering and molecular characterization of a caffeine-resistant Saccharomyces cerevisiae strain. World J. Microbiol. Biotechnol. 2019, 35, 183. [Google Scholar] [CrossRef]
- Wang, M.; Deng, W.W.; Zhang, Z.Z.; Yu, O. Engineering an ABC transporter for enhancing resistance to caffeine in Saccharomyces cerevisiae. J. Agric. Food Chem. 2016, 64, 7973–7978. [Google Scholar] [CrossRef]
- Kot, M.; Daniel, W.A. Caffeine as a marker substrate for testing cytochrome P450 activity in human and rat. Pharmacol. Rep. 2008, 60, 789–797. [Google Scholar]
- Cusinato, D.A.C.; Martinez, E.Z.; Cintra, M.T.C.; Filgueira, G.C.O.; Berretta, A.A.; Lanchote, V.L.; Coelho, E.B. Evaluation of potential herbal-drug interactions of a standardized propolis extract (EPP-AF®) using an in vivo cocktail approach. J. Ethnopharmacol. 2019, 245, 112174. [Google Scholar] [CrossRef]
- Aronsen, L.; Orvoll, E.; Lysaa, R.; Ravna, A.W.; Sager, G. Modulation of high affinity ATP-dependent cyclic nucleotide transporters by specific and non-specific cyclic nucleotide phosphodiesterase inhibitors. Eur. J. Pharmacol. 2014, 745, 249–253. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ding, R.; Shi, J.; Pabon, K.; Scotto, K.W. Xanthines down-regulate the drug transporter ABCG2 and reverse multidrug resistance. Mol. Pharmacol. 2012, 81, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Fuhr, U.; Jetter, A.; Kirchheiner, J. Appropriate phenotyping procedures for drug metabolizing enzymes and transporters in humans and their simultaneous use in the “cocktail” approach. Clin. Pharmacol. Ther. 2007, 81, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Bilodeau, P.S.; Winistorfer, S.C.; Kearney, W.R.; Robertson, A.D.; Piper, R.C. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 2003, 63, 237–243. [Google Scholar] [CrossRef]
- Samanta, M.P.; Liang, S. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. USA 2003, 100, 12579–12583. [Google Scholar] [CrossRef]
- Goldar, M.M.; Nishie, T.; Ishikura, Y.; Fukuda, T.; Takegawa, K.; Kawamukai, M. Functional conservation between fission yeast moc1/sds23 and its two orthologs, budding yeast SDS23 and SDS24, and phenotypic differences in their disruptants. Biosci. Biotechnol. Biochem. 2005, 69, 1422–1426. [Google Scholar] [CrossRef]
- Wiederkehr, A.; Meier, K.D.; Riezman, H. Identification and characterization of Saccharomyces cerevisiae mutants defective in fluid-phase endocytosis. Yeast 2001, 18, 759–773. [Google Scholar] [CrossRef]
- Hood-DeGrenier, J.K. Identification of phosphatase 2A-like Sit4-mediated signalling and ubiquitin-dependent protein sorting as modulators of caffeine sensitivity in S. cerevisiae. Yeast 2011, 28, 189–204. [Google Scholar] [CrossRef]
- Schmitt, M.; Schwanewilm, P.; Ludwig, J.; Lichtenberg-Fraté, H. Use of PMA1 as a housekeeping biomarker for assessment of toxicant-induced stress in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006, 72, 1515–1522. [Google Scholar] [CrossRef]
- Sayyed, K.; Le Vée, M.; Chamieh, H.; Fardel, O.; Abdel-Razzak, Z. Cigarette smoke condensate alters Saccharomyces cerevisiae efflux transporter mRNA and activity and increases caffeine toxicity. Toxicology 2018, 409, 129–136. [Google Scholar] [CrossRef]
- Candreva, E.C.; Keszenman, D.J.; Barrios, E.; Gelós, U.; Nunes, E. Mutagenicity induced by hyperthermia, hot mate infusion, and hot caffeine in Saccharomyces cerevisiae. Cancer Res. 1993, 53, 5750–5753. [Google Scholar] [PubMed]
- Dubeau, H.; Chung, Y.S. Effect of caffeine on ozone-sensitivity in Saccharomyces cerevisiae. Mol. Gen. Genet. 1984, 195, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R. Mutagenic and recombinogenic consequences of DNA-repair inhibition during treatment with 1,3-bis(2-chloroethyl)-1-nitrosourea in Saccharomyces cerevisiae. Mutat. Res. 1990, 241, 369–377. [Google Scholar] [CrossRef]
- Ferguson, L.R.; Turner, P.M.; Baguley, B.C. Induction of mitotic crossing-over by the topoisomerase II poison DACA (N-[2-dimethylamino)ethyl]acridine-4-carboxamide) in Saccharomyces cerevisiae. Mutat. Res. 1993, 289, 157–163. [Google Scholar] [CrossRef]
- Fominov, G.V.; Ter-Avanesian, M.D. Caffeine sensitivity of the yeast Saccharomyces cerevisiae MCD4 mutant is related to alteration of calcium homeostasis and degradation of misfolded proteins. Mol. Biol. (Mosk) 2005, 39, 464–476. [Google Scholar] [CrossRef]
- Courchesne, W.E.; Ozturk, S. Amiodarone induces a caffeine-inhibited, MID1-depedent rise in free cytoplasmic calcium in Saccharomyces cerevisiae. Mol. Microbiol. 2003, 47, 223–234. [Google Scholar] [CrossRef]
- Kihlman, B.A.; Sturelid, S.; Hartley-Asp, B.; Nilsson, K. The enhancement by caffeine of the frequencies of chromosomal aberrations induced in plant and animal cells by chemical and physical agents. Mutat. Res. 1974, 26, 105–122. [Google Scholar] [CrossRef]
- Nunes, E.; Brum, G.; Candreva, E.C.; Schenberg Frascino, A.C. Common repair pathways acting upon U.V.- and X-ray induced damage in diploid cells of Saccharomyces cerevisiae. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1984, 45, 593–606. [Google Scholar] [CrossRef]
- Hannan, M.A.; Nasim, A. Caffeine enhancement of radiation killing in different strains of Saccharomyces cerevisiae. Mol. Gen. Genet. 1977, 158, 111–116. [Google Scholar] [CrossRef]
- Siede, W.; Obermaier, S.; Eckardt, F. Influence of different inhibitors on the activity of the RAD54 dependent step of DNA repair in Saccharomyces cerevisiae. Radiat. Environ. Biophys. 1985, 24, 1–7. [Google Scholar] [CrossRef]
- Haynes, R.H. DNA repair and the genetic control of radiation sensitivity in yeast. In Molecular Mechanisms for Repair of DNA; Part B; Hanawalt, P., Setlow, R.B., Eds.; Academic Press: New York, NY, USA, 1975; pp. 529–540. [Google Scholar]
- Li, H.; Zeng, J.; Shen, K. PI3K/AKT/mTOR signaling pathway as a therapeutic target for ovarian cancer. Arch. Gynecol. Obstet. 2014, 290, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Zaborowska, D.; Zuk, J. The effect of DNA replication on mutation of the Saccharomyces cerevisiae CDC8 gene. Curr. Genet. 1990, 17, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Kori, M.; Aydin, B.; Arga, K. A comprehensive overview of signaling pathways and their crosstalk in human cancers. In The Most Recent Studies in Science and Art; Arapgirlioglu, H., Atik, A., Hızıroglu, S., Elliott, R.L., Tasxlıdere, E., Eds.; Gece Kitapligi: Ankara, Turkey, 2018; pp. 771–784. [Google Scholar]
- Nemavarkar, P.S.; Chourasia, B.K.; Pasupathy, K. Detection of gamma-irradiation induced DNA damage and radioprotection of compounds in yeast using comet assay. J. Radiat. Res. 2004, 45, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, P.J.; Pasupathy, K. Radioprotective action of caffeine: Use of Saccharomyces cerevisiae as a test system. Indian J. Exp. Biol. 2001, 39, 1254–1257. [Google Scholar]
- Anjaria, K.B.; Rao, B.S. Effect of caffeine on the genotoxic effects of gamma radiation and 4-NQO in diploid yeast. J. Environ. Pathol. Toxicol. Oncol. 2001, 20, 39–45. [Google Scholar] [CrossRef]
- Tsabar, M.; Eapen, V.V.; Mason, J.M.; Memisoglu, G.; Waterman, D.P.; Long, M.J.; Bishop, U.K.; Haber, S.N. Caffeine impairs resection during DNA break repair by reducing the levels of nucleases Sae2 and Dna2. Nucleic Acids Res. 2015, 43, 6889–6901. [Google Scholar] [CrossRef]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef]
- Hustedt, N.; Gasser, S.M.; Shimada, K. Replication checkpoint: Tuning and coordination of replication forks in s phase. Genes (Basel). 2013, 4, 388–434. [Google Scholar] [CrossRef]
- Cussiol, J.R.R.; Soares, B.L.; Oliveira, F.M.B. From yeast to humans: Understanding the biology of DNA Damage Response (DDR) kinases. Genet. Mol. Biol. 2019, 43, e20190071. [Google Scholar] [CrossRef]
- Heffernan, T.P.; Simpson, D.A.; Frank, A.R.; Heinloth, A.N.; Paules, R.S.; Cordeiro-Stone, M.; Kaufmann, W.K. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell Biol. 2002, 22, 8552–8561. [Google Scholar] [CrossRef]
- Barlow, J.H.; Rothstein, R. Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J. 2009, 28, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.C.; Haber, J.E. Surviving the breakup: The DNA damage checkpoint. Annu. Rev. Genet. 2006, 40, 209–235. [Google Scholar] [CrossRef] [PubMed]
- Sandell, L.L.; Zakian, V.A. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell 1993, 75, 729–739. [Google Scholar] [CrossRef]
- Lee, S.E.; Moore, J.K.; Holmes, A.; Umezu, K.; Kolodner, R.D.; Haber, J.E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 1998, 94, 399–409. [Google Scholar] [CrossRef]
- Vaze, M.B.; Pellicioli, A.; Lee, S.E.; Ira, G.; Liberi, G.; Arbel-Eden, A.; Foiani, M.; Haber, J.E. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 2002, 10, 373–385. [Google Scholar] [CrossRef]
- Barton, A.B.; Davies, C.J.; Hutchison, C.A., 3rd; Kaback, D.B. Cloning of chromosome I DNA from Saccharomycescerevisiae: Analysis of the FUN52 gene, whose product has homology to protein kinases. Gene 1992, 117, 137–140. [Google Scholar] [CrossRef]
- Schweitzer, B.; Philippsen, P. NPK1, a nonessential protein kinase gene in Saccharomyces cerevisiae with similarity to Aspergillus nidulans nimA. Mol. Gen. Genet. 1992, 234, 164–167. [Google Scholar] [CrossRef]
- Löffler, H.; Bochtler, T.; Fritz, B.; Tews, B.; Ho, A.D.; Lukas, J.; Bartek, J.; Krämer, A. DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle 2007, 6, 2541–2548. [Google Scholar] [CrossRef]
- Moura, D.J.; Castilhos, B.; Immich, B.F.; Cañedo, A.D.; Henriques, J.A.P.; Lenz, G.; Saffi, J. Kin3 protein, a NIMA-related kinase of Saccharomyces cerevisiae, is involved in DNA adduct damage response. Cell Cycle 2010, 9, 2220–2229. [Google Scholar] [CrossRef]
- Chakraverty, R.K.; Kearsey, J.M.; Oakley, T.J.; Grenon, M.; de la Torre Ruiz, M.A.; Lowndes, N.F.; Hickson, I.D. Topoisomerase III acts upstream of Rad53p in the S-phase DNA damage checkpoint. Mol. Cell Biol. 2001, 21, 7150–7162. [Google Scholar] [CrossRef]
- Mankouri, H.W.; Hickson, I.D. Top3 processes recombination intermediates and modulates checkpoint activity after DNA damage. Mol. Biol. Cell 2006, 17, 4473–4483. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crouch, R.J. Ribonuclease H: From discovery to 3D structure. New Biol. 1990, 2, 771–777. [Google Scholar] [PubMed]
- Hyjek, M.; Figiel, M.; Nowotny, M. RNases H: Structure and mechanism. DNA Repair 2019, 84, 102672. [Google Scholar] [CrossRef] [PubMed]
- Arudchandran, A.; Cerritelli, S.; Narimatsu, S.; Itaya, M.; Shin, D.Y.; Shimada, Y.; Crouch, R.J. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: Implications for roles of RNases H in DNA replication and repair. Genes Cells 2000, 5, 789–802. [Google Scholar] [CrossRef]
- Beauchamp, E.M.; Platanias, L.C. The evolution of the TOR pathway and its role in cancer. Oncogene 2013, 32, 3923–3932. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Costa, R.L.B.; Han, H.S.; Gradishar, W.J. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: A review. Breast. Cancer Res. Treat. 2018, 169, 397–406. [Google Scholar] [CrossRef]
- Janku, F.; Yap, T.A.; Meric-Bernstam, F. Targeting the PI3K pathway in cancer: Are we making headway? Nat. Rev. Clin. Oncol. 2018, 15, 273–291. [Google Scholar] [CrossRef]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Mohler, K.; Mann, R.; Kyle, A.; Reynolds, N.; Ibba, M. Aminoacyl-tRNA quality control is required for efficient activation of the TOR pathway regulator Gln3p. RNA Biol. 2018, 15, 594–603. [Google Scholar] [CrossRef]
- Corona Velazquez, A.F.; Jackson, W.T. So many roads: The multifaceted regulation of autophagy induction. Mol. Cell Biol. 2018, 38, e00303–e00318. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhou, C.; Kennedy, B.K. The yeast replicative aging model. Biochim. Biophys. Acta Mol. Basis. Dis. 2018, 1864, 2690–2696. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Awasthi, A.; Nain, V.; Issac, B.; Puria, R. Novel insights into TOR signalling in Saccharomyces cerevisiae through Torin2. Gene 2018, 669, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Luo, Y.; Huang, S. Updates of mTOR inhibitors. Anti-Cancer Agents Med. Chem. 2010, 10, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef]
- Reinke, A.; Chen, J.C.; Aronova, S.; Powers, T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J. Biol. Chem. 2006, 281, 31616–31626. [Google Scholar] [CrossRef]
- Dikicioglu, D.; Dereli, E.E.; Eraslan, S.; Oliver, S.G.; Kirdar, B. Saccharomyces cerevisiae adapted to grow in the presence of low-dose rapamycin exhibit altered amino acid metabolism. Cell Commun. Signal. 2018, 16, 85. [Google Scholar] [CrossRef]
- Rallis, C.; Codlin, S.; Bähler, J. TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell 2013, 12, 563–573. [Google Scholar] [CrossRef]
- Scott, P.H.; Lawrence, J.C., Jr. Attenuation of mammalian target of rapamycin activity by increased cAMP in 3T3-L1 adipocytes. J. Biol. Chem. 1998, 273, 34496–34501. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.B.; Carr, D.T.; Kiflezghi, M.G.; Zhao, Y.T.; Kim, D.B.; Thon, S.; Moore, M.D.; Li, M.A.K.; Kaeberlein, M. A system to identify inhibitors of mTOR signaling using high-resolution growth analysis in Saccharomyces cerevisiae. Geroscience 2017, 39, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.K.; Burgess, K.E.; Gray, J.V. Recovery from rapamycin: Drug-insensitive activity of yeast target of rapamycin complex 1 (TORC1) supports residual proliferation that dilutes rapamycin among progeny cells. J. Biol. Chem. 2014, 289, 26554–26565. [Google Scholar] [CrossRef] [PubMed]
- Wanke, V.; Cameroni, E.; Uotila, A.; Piccolis, M.; Urban, J.; Loewith, R.; De Virgilio, C. Caffeine extends yeast lifespan by targeting TORC1. Mol. MicroBiol. 2008, 69, 277–285. [Google Scholar] [CrossRef]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Heitman, J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 1995, 270, 27531–27537. [Google Scholar] [CrossRef]
- Kingsbury, J.M.; Cardenas, M.E. Vesicular trafficking systems impact TORC1-controlled transcriptional programs in Saccharomyces cerevisiae. G3 (Bethesda) 2016, 6, 641–652. [Google Scholar] [CrossRef][Green Version]
- Shamji, A.F.; Kuruvilla, F.G.; Schreiber, S.L. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 2000, 10, 1574–1581. [Google Scholar] [CrossRef]
- Komeili, A.; Wedaman, K.P.; O’Shea, E.K.; Powers, T. Mechanism of metabolic control: Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 2000, 151, 863–878. [Google Scholar] [CrossRef]
- Cooper, T.G. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol. Rev. 2002, 26, 223–238. [Google Scholar] [CrossRef]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed]
- Numamoto, M.; Tagami, S.; Ueda, Y.; Imabeppu, Y.; Sasano, Y.; Sugiyama, M.; Maekawa, H.; Harashima, S. Nuclear localization domains of GATA activator Gln3 are required for transcription of target genes through dephosphorylation in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2015, 120, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Narumiya, S.; Thumkeo, D. Rho signaling research: History, current status and future directions. FEBS Lett. 2018, 592, 1763–1776. [Google Scholar] [CrossRef] [PubMed]
- Lawson, C.D.; Ridley, A.J. Rho GTPase signaling complexes in cell migration and invasion. J. Cell Biol. 2018, 217, 447–457. [Google Scholar] [CrossRef]
- Guan, X.; Guan, X.; Dong, C.; Jiao, Z. Rho GTPases and related signaling complexes in cell migration and invasion. Exp. Cell Res. 2020, 388, 111824. [Google Scholar] [CrossRef]
- Delley, P.A.; Hall, M.N. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 1999, 147, 163–174. [Google Scholar] [CrossRef]
- Yan, G.; Lai, Y.; Jiang, Y. The TOR complex 1 is a direct target of Rho1 GTPase. Mol. Cell 2012, 45, 743–753. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, X.Q. Regulation of mTORC1 by small GTPases in response to nutrients. J. Nutr. 2020, 150, 1004–1011. [Google Scholar] [CrossRef]
- Schmitz, H.P.; Huppert, S.; Lorberg, A.; Heinisch, J.J. Rho5p downregulates the yeast cell integrity pathway. J. Cell Sci. 2002, 115, 3139–3148. [Google Scholar]
- Philip, B.; Levin, D.E. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell Biol. 2001, 21, 271–280. [Google Scholar] [CrossRef]
- Guo, S.; Shen, X.; Yan, G.; Ma, D.; Bai, X.; Li, S.; Jiang, Y. A MAP kinase dependent feedback mechanism controls Rho1 GTPase and actin distribution in yeast. PLoS ONE 2009, 4, e6089. [Google Scholar] [CrossRef]
- Yan, G.; Lai, Y.; Jiang, Y. TOR under stress: Targeting TORC1 by Rho1 GTPase. Cell Cycle 2012, 11, 3384–3388. [Google Scholar] [CrossRef] [PubMed]
- Hirasaki, M.; Horiguchi, M.; Numamoto, M.; Sugiyama, M.; Kaneko, Y.; Nogi, Y.; Harashima, S. Saccharomyces cerevisiae protein phosphatase Ppz1 and protein kinases Sat4 and Hal5 are involved in the control of subcellular localization of Gln3 by likely regulating its phosphorylation state. J. Biosci. Bioeng. 2011, 111, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Tzima, E. Role of small GTPases in endothelial cytoskeletal dynamics and the shear stress response. Circ. Res. 2006, 98, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, N.; Noguchi, E.; Nishimoto, T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 1999, 152, 853–867. [Google Scholar]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: A conserved switch for diverse cell functions. Nature 1990, 348, 125–132. [Google Scholar] [CrossRef]
- Wang, Y.; Kurihara, Y.; Sato, T.; Toh, H.; Kobayashi, H.; Sekiguchi, T. Gtr1p differentially associates with Gtr2p and Ego1p. Gene 2009, 437, 32–38. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, H.; Haggarty, S.J.; Spring, D.R.; Hwang, H.; Jin, F.; Snyder, M.; Schreiber, S.L. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. USA 2004, 101, 16594–16599. [Google Scholar] [CrossRef]
- Gao, M.; Kaiser, C.A. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 2006, 8, 657–667. [Google Scholar] [CrossRef]
- Dereli, E.E.; Arga, K.Y.; Dikicioglu, D.; Eraslan, S.; Erkol, E.; Celik, A.; Kirdar, B.; Di Camillo, B. Identification of novel components of target-of-rapamycin signaling pathway by network-based multi-omics integrative analysis. OMICS 2019, 23, 274–284. [Google Scholar] [CrossRef]
- González, A.; Ruiz, A.; Casamayor, A.; Ariño, J. Normal function of the yeast TOR pathway requires the type 2C protein phosphatase Ptc1. Mol. Cell Biol. 2009, 29, 2876–2888. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zaborske, J.M.; Wu, X.; Wek, R.C.; Pan, T. Selective control of amino acid metabolism by the GCN2 eIF2 kinase pathway in Saccharomyces cerevisiae. BMC Biochem. 2010, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- White, J.T.; Cato, T.; Deramchi, N.; Gabunilas, J.; Roy, K.R.; Wang, C.; Chanfreau, G.F.; Clarke, S.G. Protein methylation and translation: Role of lysine modification on the function of yeast elongation factor 1A. Biochemistry 2019, 58, 4997–5010. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Muñoz, G.A.; Kane, P. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 2008, 283, 20309–20319. [Google Scholar] [CrossRef] [PubMed]
- Banuelos, M.G.; Moreno, D.E.; Olson, D.K.; Nguyen, Q.; Ricarte, F.; Aguilera-Sandoval, C.R.; Gharakhanian, E. Genomic analysis of severe hypersensitivity to hygromycin B reveals linkage to vacuolar defects and new vacuolar gene functions in Saccharomyces cerevisiae. Curr. Genet. 2010, 56I, 121–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lum, P.Y.; Armour, C.D.; Stepaniants, S.B.; Cavet, G.; Wolf, M.K.; Butler, J.S.; Hinshaw, J.C.; Garnier, P.; Prestwich, G.D.; Leonardson, A.; et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 2004, 116, 121–137. [Google Scholar] [CrossRef]
- Moser, B.A.; Brondello, J.M.; Baber-Furnari, B.; Russell, P. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol. Cell Biol. 2000, 20, 4288–4294. [Google Scholar] [CrossRef][Green Version]
- Cid, V.J.; Durán, A.; del Rey, F.; Snyder, M.P.; Nombela, C.; Sánchez, M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 1995, 59, 345–386. [Google Scholar] [CrossRef]
- Vicente, M.F.; Basilio, A.; Cabello, A.; Peláez, F. Microbial natural products as a source of antifungals. Clin. Microbiol. Infect. 2003, 9, 15–32. [Google Scholar] [CrossRef]
- Levin, D.E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 262–291. [Google Scholar] [CrossRef]
- Truman, A.W.; Kim, K.Y.; Levin, D.E. Mechanism of Mpk1 mitogen-activated protein kinase binding to the Swi4 transcription factor and its regulation by a novel caffeine-induced phosphorylation. Mol. Cell Biol. 2009, 29, 6449–6461. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Vavassori, S.; Schweizer, L.M.; Schweizer, M. Impaired PRPP-synthesizing capacity compromises cell integrity signalling in Saccharomyces cerevisiae. Microbiology 2004, 150, 3327–3339. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ugbogu, E.A.; Wippler, S.; Euston, M.; Kouwenhoven, E.N.; de Brouwer, A.P.M.; Schweizer, L.M.; Schweizer, M. The contribution of the nonhomologous region of Prs1 to the maintenance of cell wall integrity and cell viability. FEMS Yeast Res. 2013, 13, 291–301. [Google Scholar] [CrossRef]
- Sipling, T.; Zhai, C.; Panaretou, B. Emw1p/YNL313cp is essential for maintenance of the cell wall in Saccharomyces cerevisiae. Microbiology 2011, 157, 1032–1041. [Google Scholar] [CrossRef]
- Castrejon, F.; Gomez, A.; Sanz, M.; Duran, A.; Roncero, C. The RIM101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p. Eukaryot. Cell 2006, 5, 507–517. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zolnierowicz, S.; Bollen, M. Protein phosphorylation and protein phosphatases. EMBO J. 2000, 19, 483–488. [Google Scholar] [CrossRef]
- Sakumoto, N.; Mukai, Y.; Uchida, K.; Kouchi, T.; Kuwajima, J.; Nakagawa, Y.; Sugioka, S.; Yamamoto, E.; Furuyama, T.; Mizubuchi, H.; et al. A series of protein phosphatase gene disruptants in Saccharomyces cerevisiae. Yeast 1999, 15, 1669–1679. [Google Scholar] [CrossRef]
- Yenush, L.; Merchan, S.; Holmes, J.; Serrano, R. pH-Responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol. Cell Biol. 2005, 25, 8683–8692. [Google Scholar] [CrossRef]
- Wharton, R.P.; Aggarwal, A.K. mRNA regulation by Puf domain proteins. Sci. STKE 2006, 2006, pe37. [Google Scholar] [CrossRef]
- Traven, A.; Lo, T.L.; Lithgow, T.; Heierhorst, J. The yeast PUF protein Puf5 has Pop2-independent roles in response to DNA replication stress. PLoS ONE 2010, 5, e10651. [Google Scholar] [CrossRef]
- Saiardi, A.; Caffrey, J.J.; Snyder, S.H.; Shears, S.B. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 2000, 275, 24686–24692. [Google Scholar] [CrossRef] [PubMed]
- Dubois, E.; Scherens, B.; Vierendeels, F.; Ho, M.M.; Messenguy, F.; Shears, S.B. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J. Biol. Chem. 2002, 277, 23755–23763. [Google Scholar] [CrossRef] [PubMed]
- Saiardi, A.; Bhandari, R.; Resnick, A.C.; Snowman, A.M.; Snyder, S.H. Phosphorylation of proteins by inositol pyrophosphates. Science 2004, 306, 2101–2105. [Google Scholar] [CrossRef] [PubMed]
- Saiardi, A.; Resnick, A.C.; Snowman, A.M.; Wendland, B.; Snyder, S.H. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. USA 2005, 102, 1911–1914. [Google Scholar] [CrossRef]
- Gallegos, S.; Pacheco, C.; Peters, C.; Opazo, C.M.; Aguayo, L.G. Features of alpha-synuclein that could explain the progression and irreversibility of Parkinson’s disease. Front. Neurosci. 2015, 9, 59. [Google Scholar] [CrossRef]
- Kardani, J.; Roy, I. Understanding caffeine’s role in attenuating the toxicity of α-synuclein aggregates: Implications for risk of Parkinson’s disease. ACS Chem. Neurosci. 2015, 6, 1613–1625. [Google Scholar] [CrossRef]
- Hong, C.T.; Chan, L.; Bai, C.-H. The Effect of caffeine on the risk and progression of Parkinson’s Disease: A meta-analysis. Nutrients 2020, 12, 1860. [Google Scholar] [CrossRef]
- Tsuzuki, J.; Newburgh, R.W. Inhibition of 5’-nucleotidase in rat brain by methylxanthines. J. Neurochem. 1975, 25, 895–896. [Google Scholar] [CrossRef]
- Liao, H.H.; Thorner, J. Adenosine 3’,5’-phosphate phosphodiesterase and pheromone response in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1981, 148, 919–925. [Google Scholar] [CrossRef]
- Tsuboi, M.; Yanagishima, N. Effect of cyclic AMP, theophylline and caffeine on the glucose repression of sporulation in Saccharomyces cerevisiae. Arch. MikroBiol. 1973, 93, 1–12. [Google Scholar] [CrossRef]
- Tortora, P.; Burlini, N.; Hanozet, G.M.; Guerritore, A. Effect of caffeine on glucose-induced inactivation of gluconeogenetic enzymes in Saccharomyces cerevisiae. A possible role of cyclic AMP. Eur. J. Biochem. 1982, 126, 617–622. [Google Scholar] [CrossRef]
- Sarinová, M.; Tichá, E.; Obernauerová, M.; Gbelská, Y. Impact of mitochondrial function on yeast susceptibility to antifungal compounds. Folia Microbiol. (Praha) 2007, 52, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.W., 3rd; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Baiges, I.; Arola, L. COCOA (Theobroma cacao) Polyphenol-rich extract increases the chronological lifespan of Saccharomyces cerevisiae. J. Frailty Aging 2016, 5, 186–190. [Google Scholar] [PubMed]
- Rockenfeller, P.; Madeo, F. Ageing and eating. Biochim. Biophys. Acta 2010, 1803, 499–506. [Google Scholar] [CrossRef]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef]
- Fabrizio, P.; Pozza, F.; Pletcher, S.D.; Gendron, C.M.; Longo, V.D. Regulation of longevity and stress resistance by Sch9 in yeast. Science 2001, 292, 288–290. [Google Scholar] [CrossRef]
- Bridi, J.C.; Barros, A.G.; Sampaio, L.R.; Ferreira, J.C.; Antunes Soares, F.; Romano-Silva, M.A. Lifespan extension induced by caffeine in Caenorhabditis elegans is partially dependent on adenosine signaling. Front. Aging Neurosci. 2015, 7, 220. [Google Scholar] [CrossRef]
- Sutphin, G.L.; Bishop, E.; Yanos, M.E.; Moller, R.M.; Kaeberlein, M. Caffeine extends life span, improves healthspan, and delays age-associated pathology in Caenorhabditis elegans. Longev. Healthspan 2012, 1, 9. [Google Scholar] [CrossRef]
- Loftfield, E.; Freedman, N.D.; Graubard, B.I.; Guertin, K.A.; Black, A.; Huang, W.-Y.; Shebl, F.M.; Mayne, S.T.; Sinha, R. Association of coffee consumption with overall and cause-specific mortality in a large US prospective cohort study. Am. J. Epidemiol. 2015, 182, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Fortes, C.; Forastiere, F.; Farchi, S.; Rapiti, E.; Pastori, G.; Perucci, C.A. Diet and overall survival in a cohort of very elderly people. Epidemiology 2000, 11, 440–445. [Google Scholar] [CrossRef]
- Paganini-Hill, A.; Kawas, C.H.; Corrada, M.M. Non-alcoholic beverage and caffeine consumption and mortality: The leisure world cohort study. Prev. Med. 2007, 44, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.M.; Lee, H.L.; Kwon, Y.Y.; Kang, M.S.; Lee, S.K.; Lee, C.K. Enhancement of mitochondrial function correlates with the extension of lifespan by caloric restriction and caloric restriction mimetics in yeast. Biochem. Biophys. Res. Commun. 2013, 441, 236–242. [Google Scholar] [CrossRef]
- Nagai, S. Stabilizing effect of caffeine on a respirationally unstable strain of yeast. Exp. Cell Res. 1962, 26, 253–259. [Google Scholar] [CrossRef]
- Pongpanich, M.; Patchsung, M.; Mutirangura, A. Pathologic replication-independent endogenous DNA double-strand breaks repair defect in chronological aging yeast. Front. Genet. 2018, 9, 501. [Google Scholar] [CrossRef] [PubMed]
- Thongsroy, J.; Patchsung, M.; Pongpanich, M.; Settayanon, S.; Mutirangura, A. Reduction in replication-independent endogenous DNA double-strand breaks promotes genomic instability during chronological aging in yeast. FASEB J. 2018, fj201800218RR. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009, 361, 1475–1485. [Google Scholar] [CrossRef]
- Thongsroy, J.; Matangkasombut, O.; Thongnak, A.; Rattanatanyong, P.; Jirawatnotai, S.; Mutirangura, A. Replication-independent endogenous DNA double-strand breaks in Saccharomyces cerevisiae model. PLoS ONE 2013, 8, e72706. [Google Scholar] [CrossRef]
- Kolb, H.; Kempf, K.; Martin, S. Health effects of eoffee: Mechanism unraveled? Nutrients 2020, 12, 1842. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruta, L.L.; Farcasanu, I.C. Saccharomyces cerevisiae and Caffeine Implications on the Eukaryotic Cell. Nutrients 2020, 12, 2440. https://doi.org/10.3390/nu12082440
Ruta LL, Farcasanu IC. Saccharomyces cerevisiae and Caffeine Implications on the Eukaryotic Cell. Nutrients. 2020; 12(8):2440. https://doi.org/10.3390/nu12082440
Chicago/Turabian StyleRuta, Lavinia Liliana, and Ileana Cornelia Farcasanu. 2020. "Saccharomyces cerevisiae and Caffeine Implications on the Eukaryotic Cell" Nutrients 12, no. 8: 2440. https://doi.org/10.3390/nu12082440
APA StyleRuta, L. L., & Farcasanu, I. C. (2020). Saccharomyces cerevisiae and Caffeine Implications on the Eukaryotic Cell. Nutrients, 12(8), 2440. https://doi.org/10.3390/nu12082440




