Retrieval of Three-Dimensional Green Volume in Urban Green Space from Multi-Source Remote Sensing Data
Abstract
:1. Introduction
2. Materials and Methods
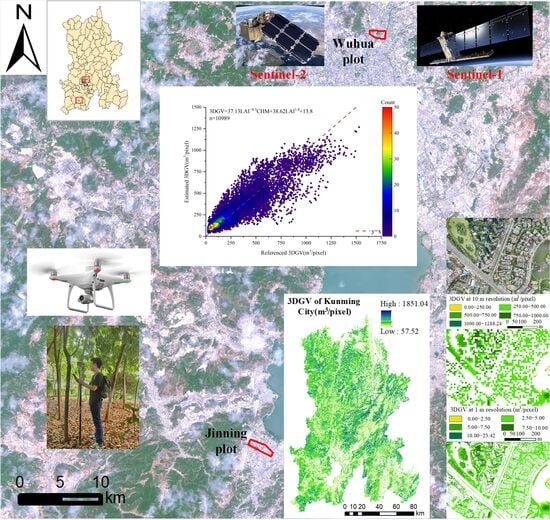
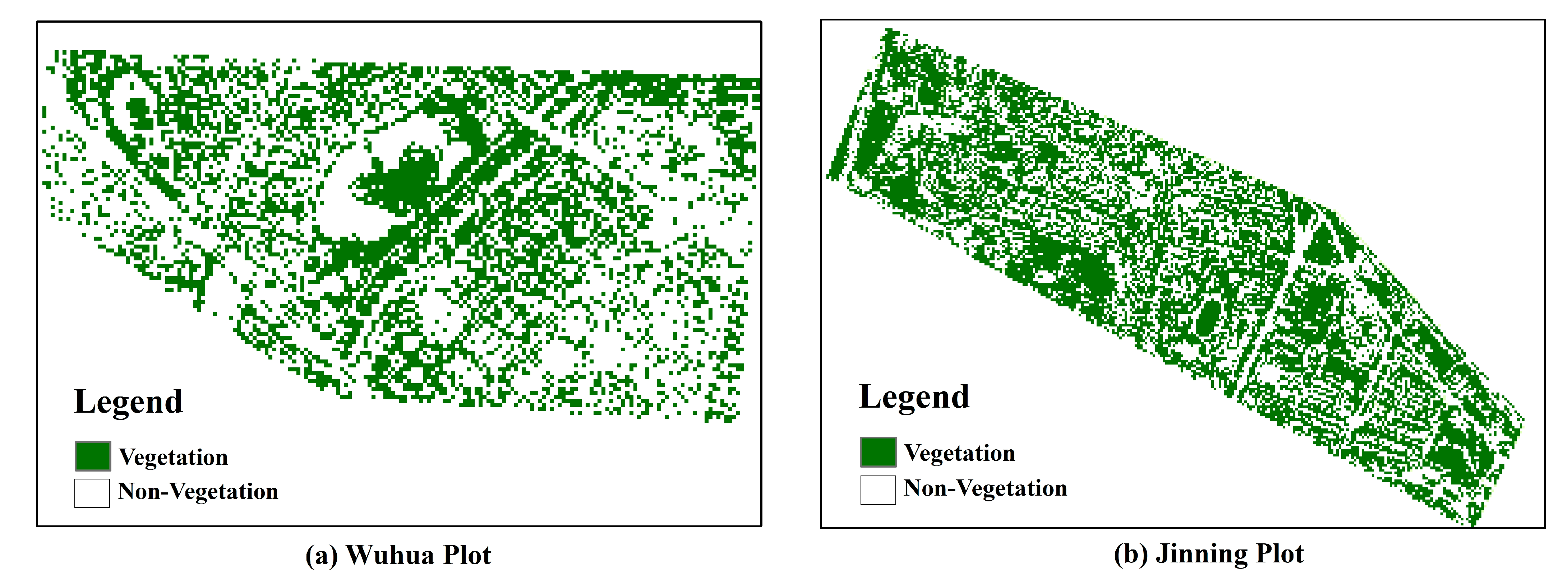
2.1. A Brief Description of Study Area and Study Sites
2.2. Data Acquisition and Processing
2.2.1. Sentinel-1 Images
2.2.2. Sentinel-2 Images
2.2.3. Acquisition and Preprocessing of UAV Images
2.2.4. Field Measurements
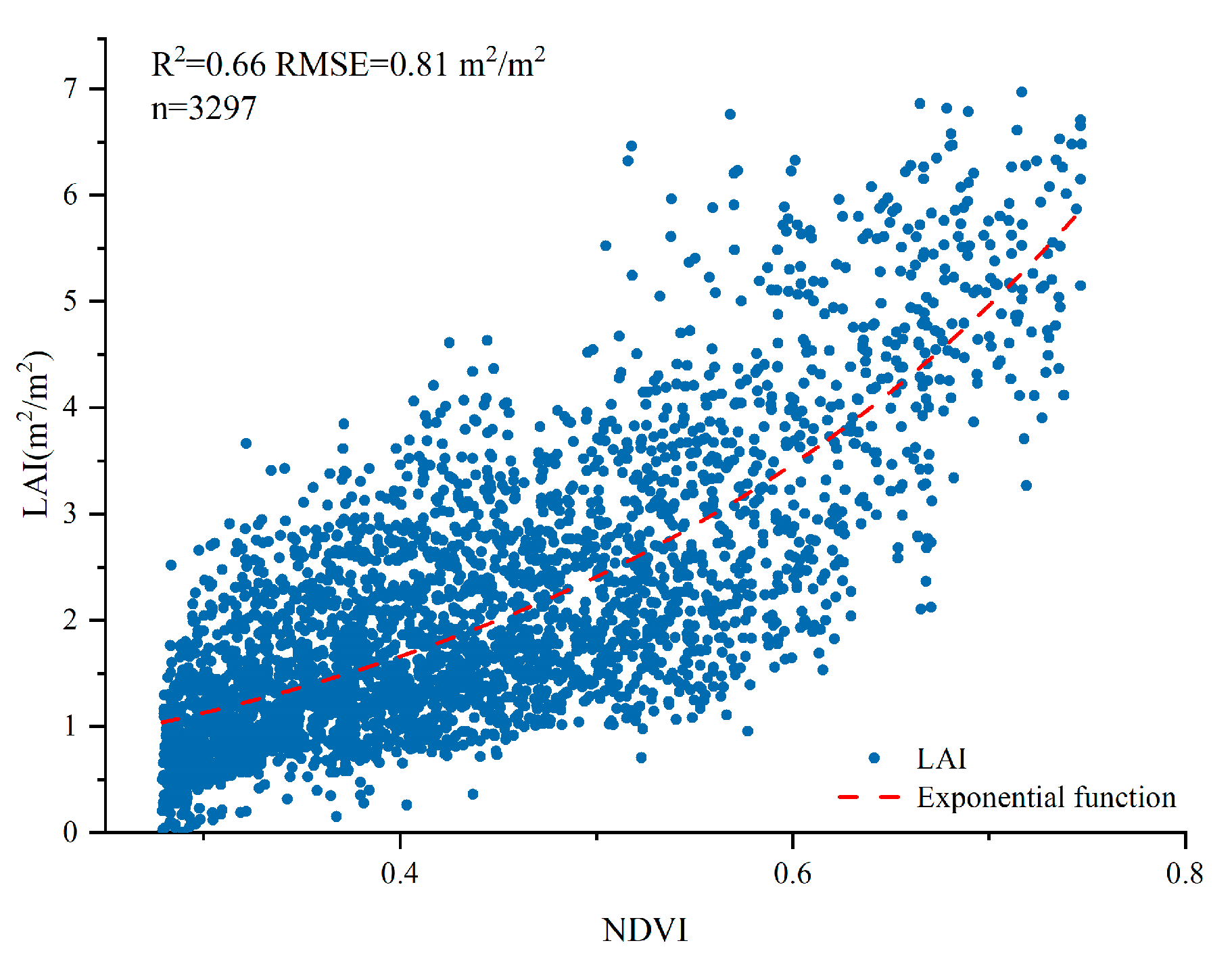
2.3. Calculating LAI Derived from Satellite Images
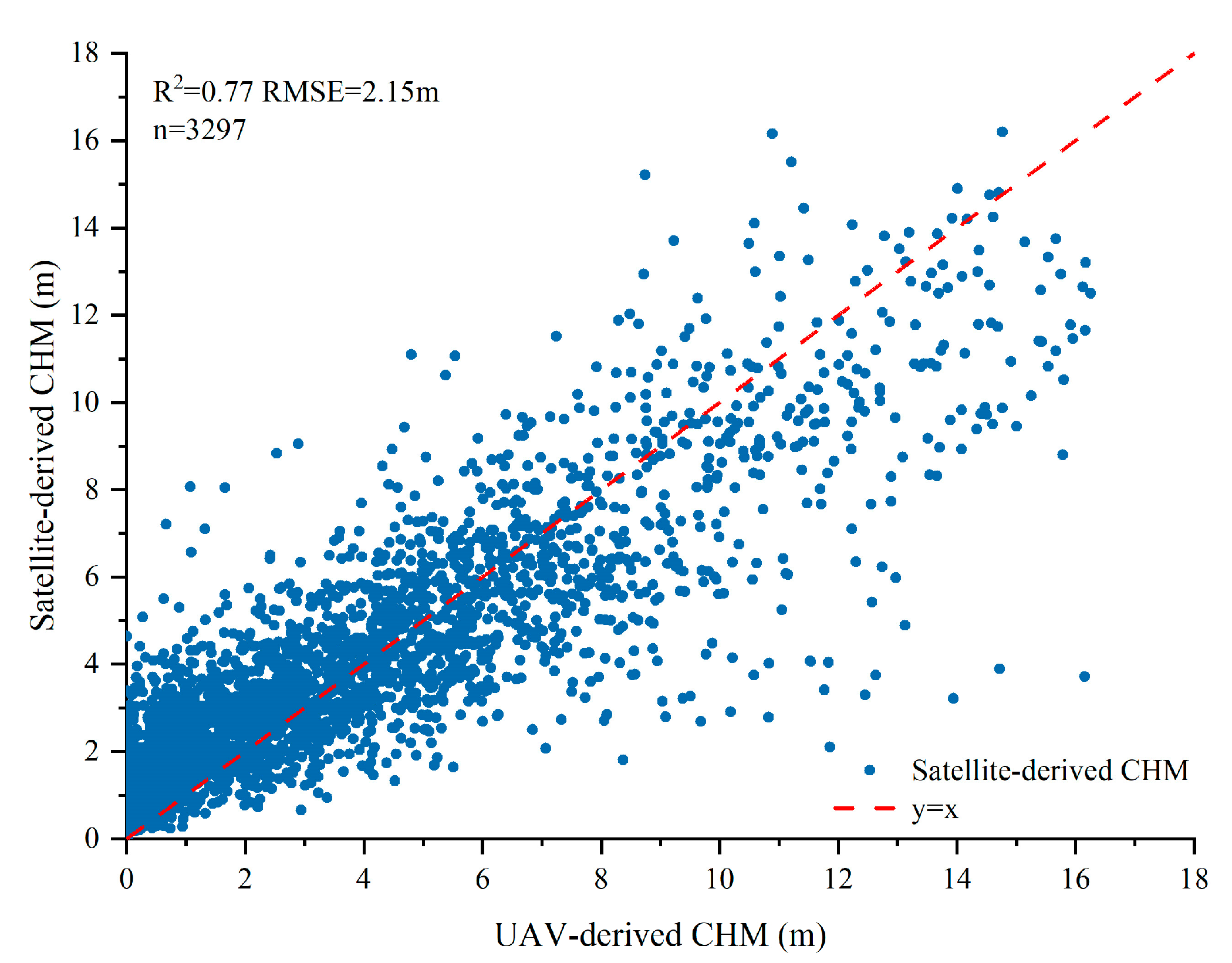
2.4. Calculating CHM Derived from Satellite Images
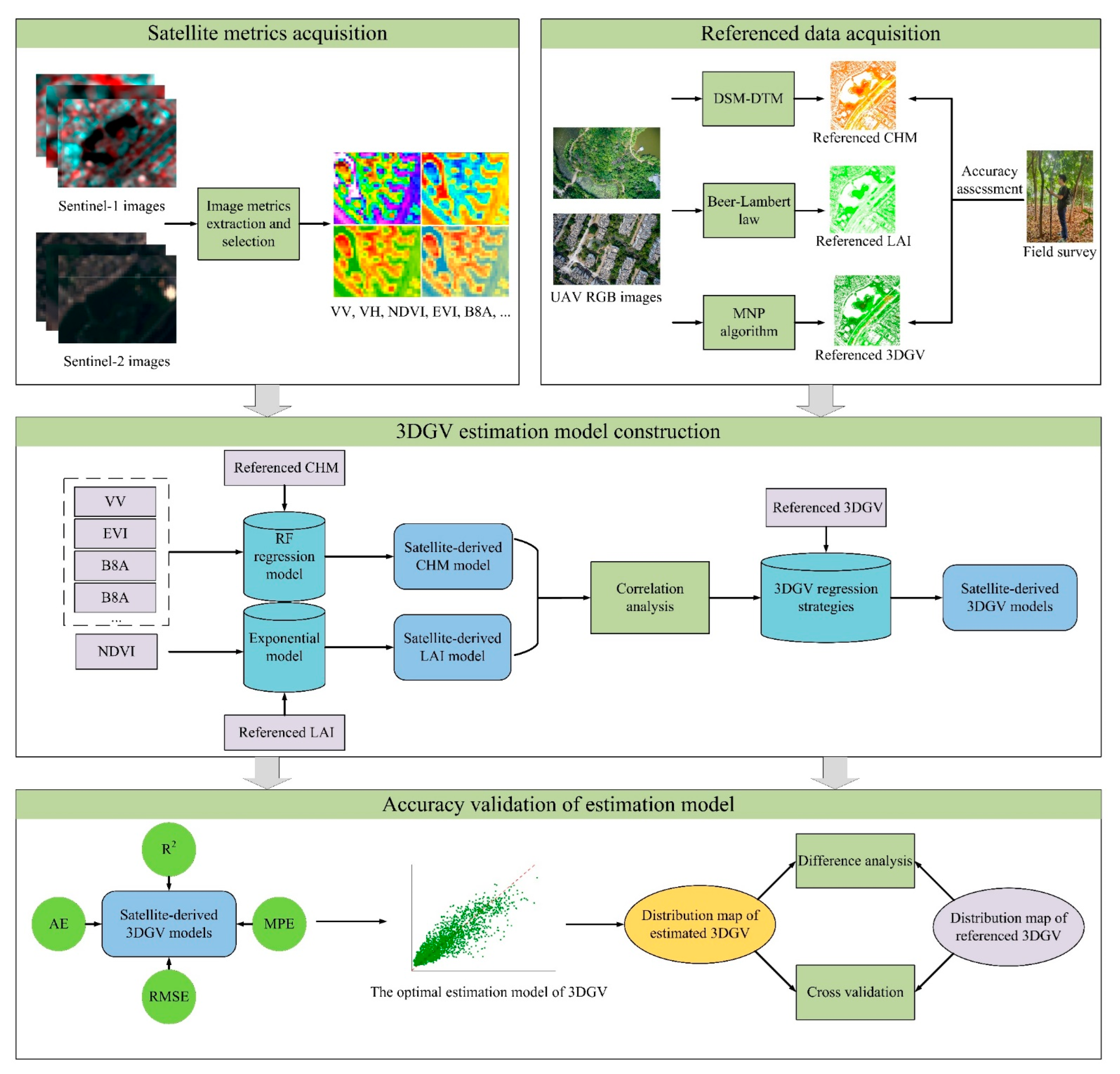
2.5. Construction of 3DGV Estimation Models
2.6. Accuracy Assessment of Estimation Models
3. Results
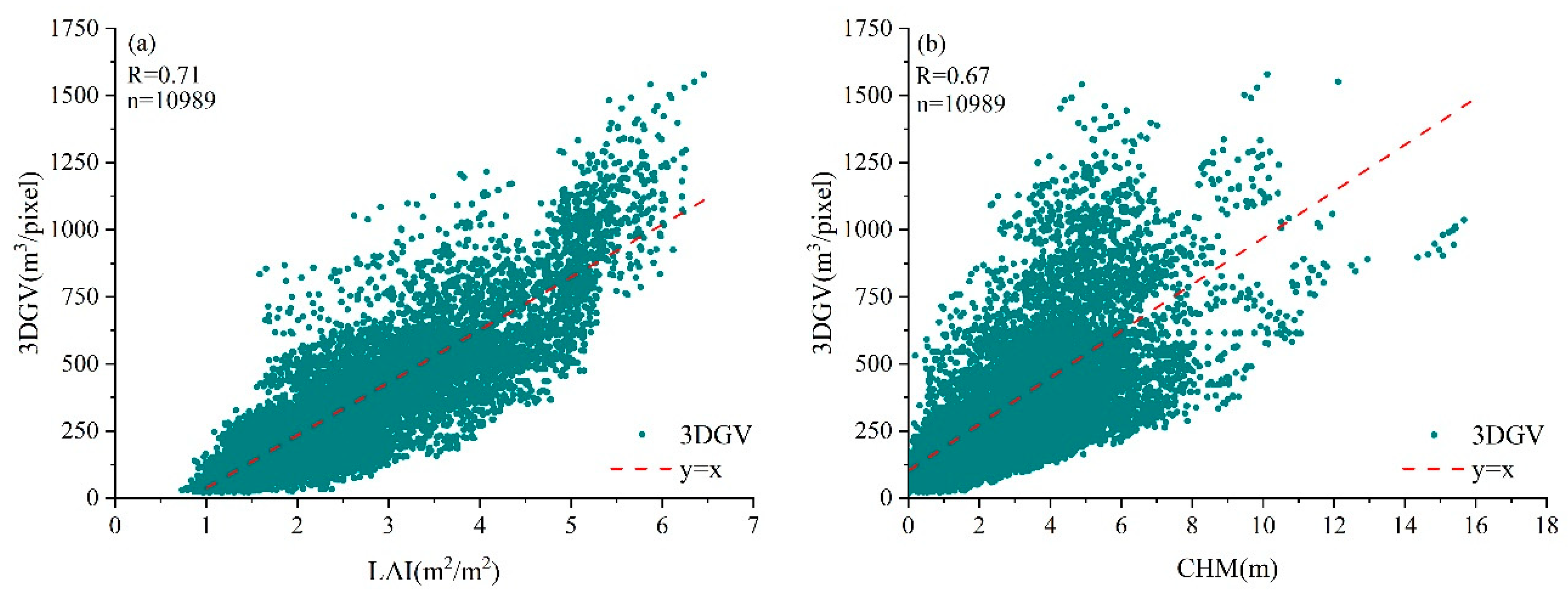
3.1. Univariate Estimation Models of 3DGV
3.2. Bivariate Estimation Models of 3DGV
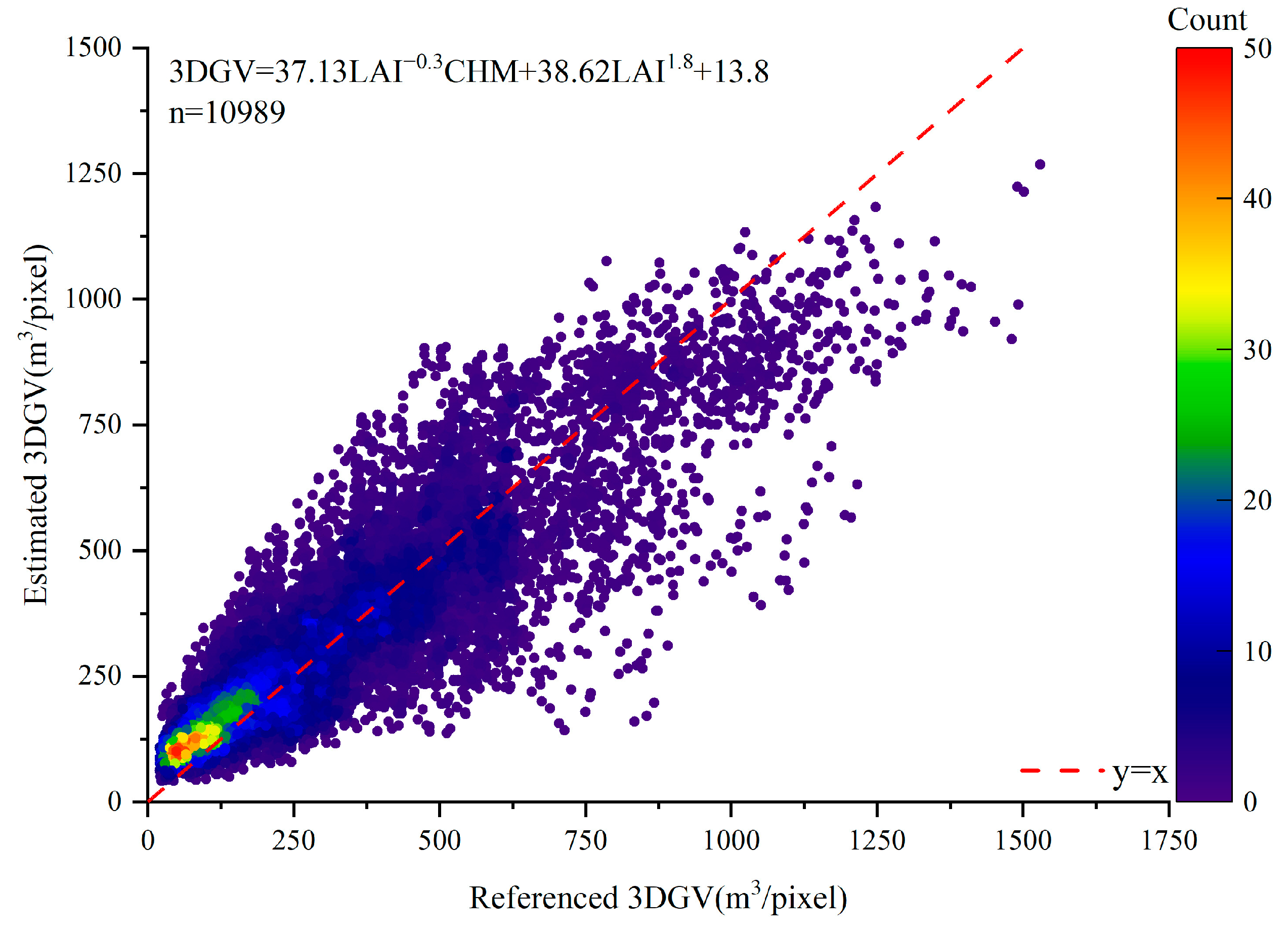
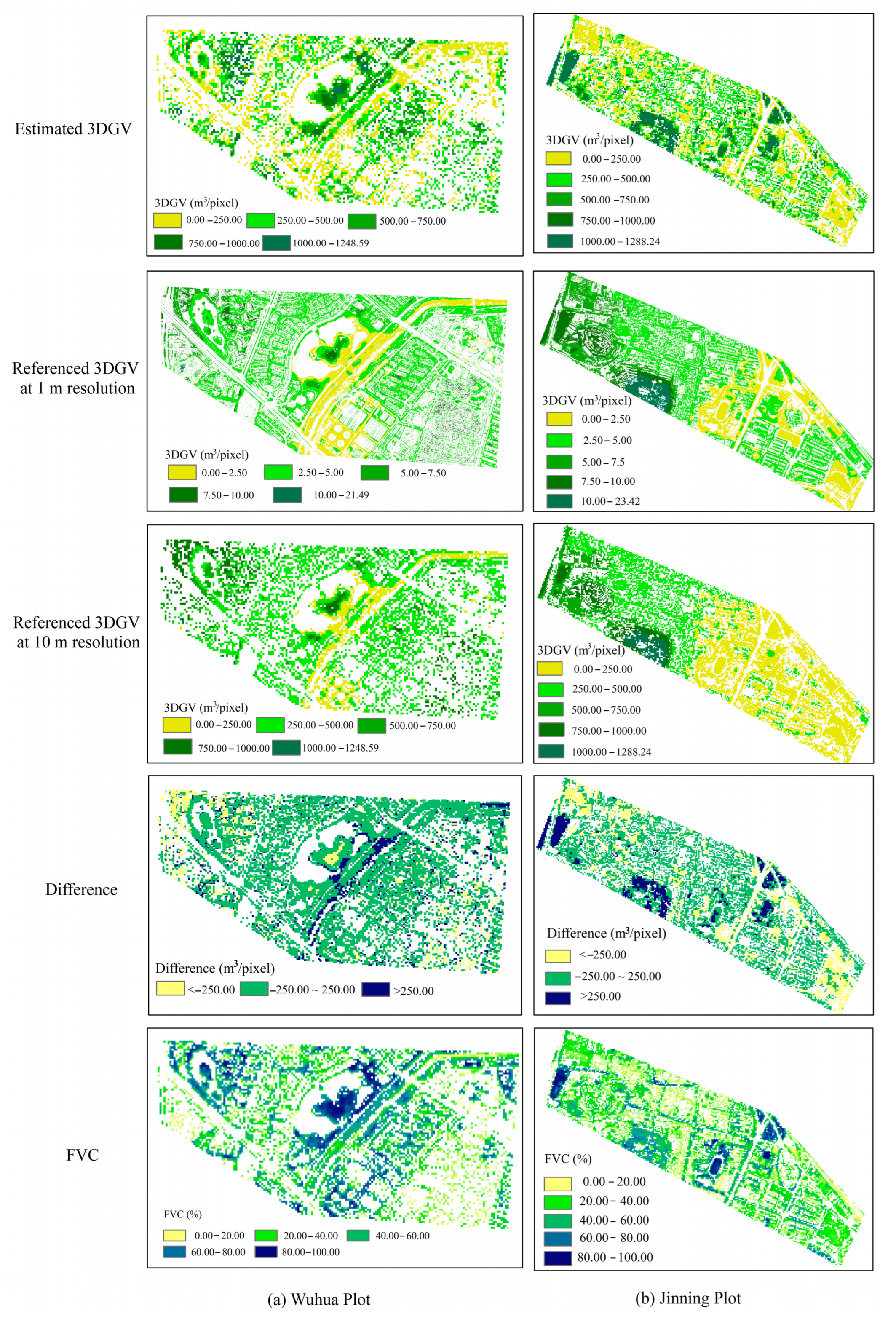
3.3. Validation of Estimated 3DGV
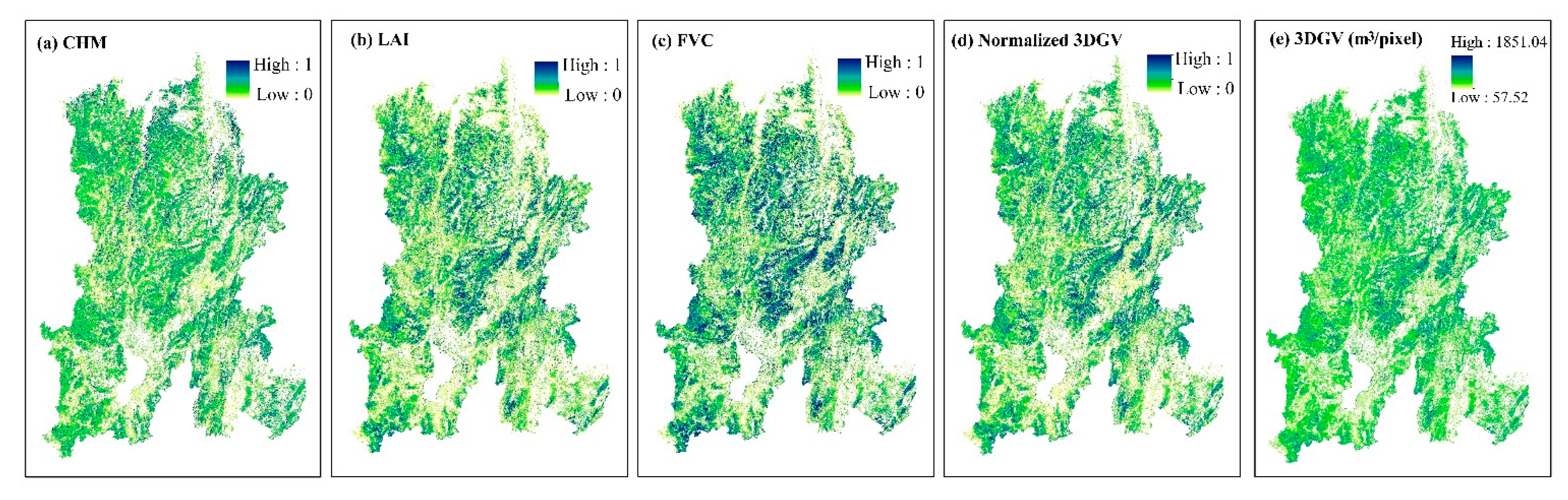
3.4. Spatial Pattern of 3DGV in Kunming City
4. Discussion
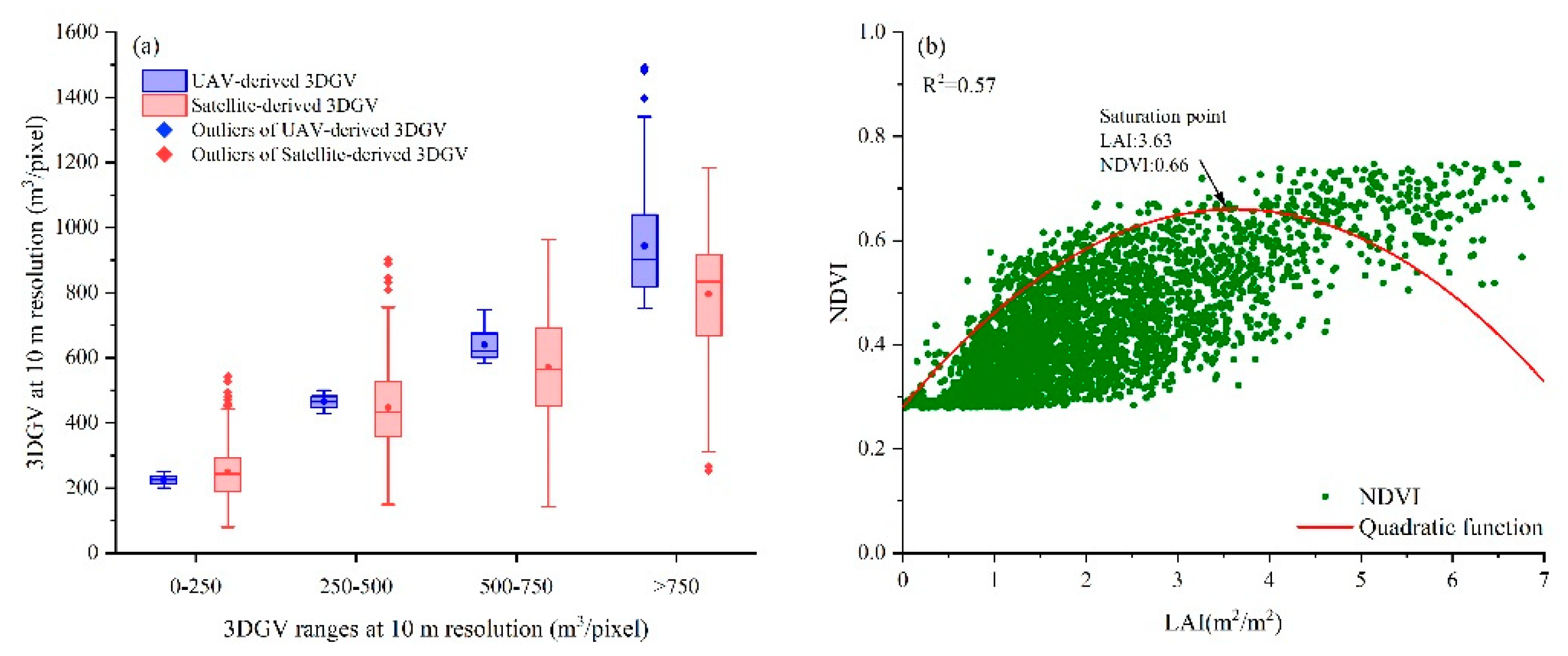
4.1. Analyzing the Effect of NDVI Saturation and Spatial Resolution of Sentinel Images
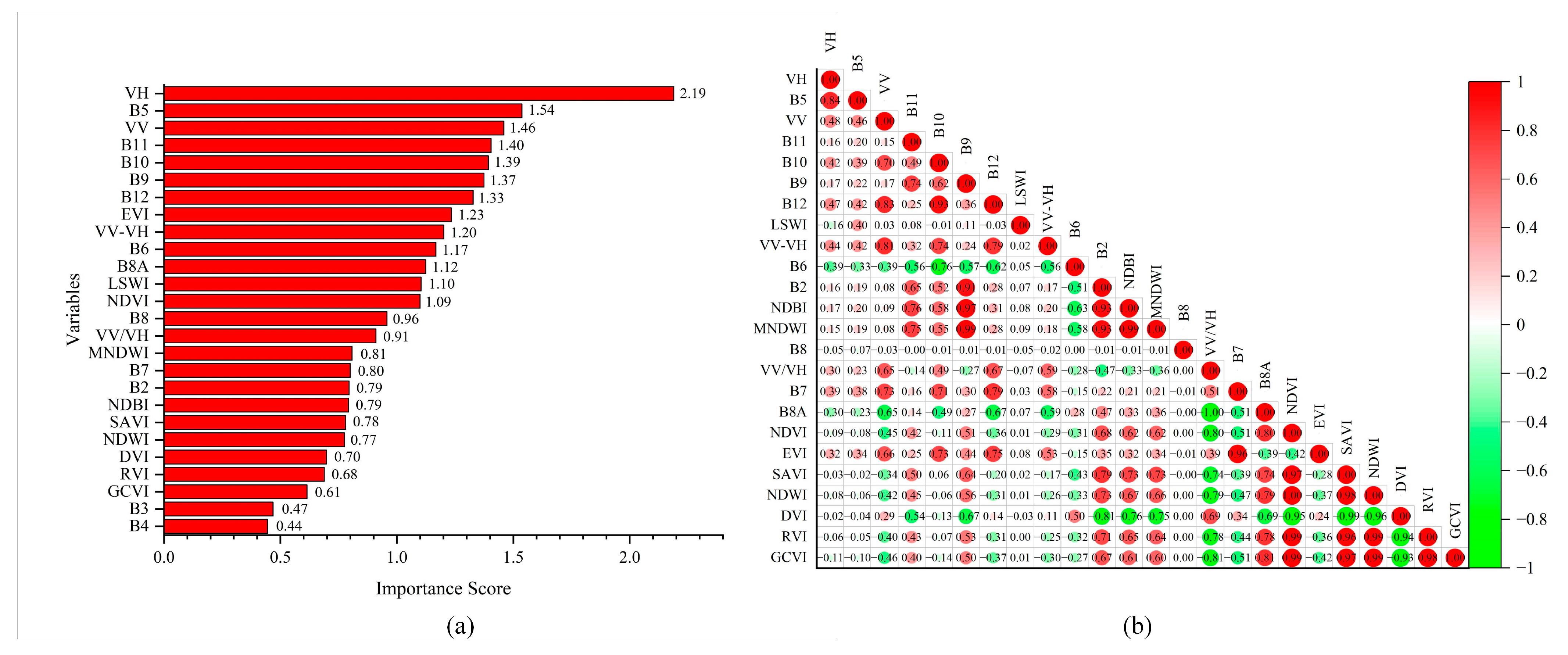
4.2. Predictor Variables Selection
4.3. Limitations and Strengths
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nath, T.K.; Han, S.S.Z.; Lechner, A.M. Urban green space and well-being in Kuala Lumpur, Malaysia. Urban For. Urban Green. 2018, 36, 34–41. [Google Scholar] [CrossRef]
- Dobbs, C.; Kendal, D.; Nitschke, C. The effects of land tenure and land use on the urban forest structure and composition of Melbourne. Urban For. Urban Green. 2013, 12, 417–425. [Google Scholar] [CrossRef]
- Wolch, J.R.; Byrne, J.; Newell, J.P. Urban green space, public health, and environmental justice: The challenge of making cities ‘just green enough’. Landsc. Urban Plan. 2014, 125, 234–244. [Google Scholar] [CrossRef]
- Wang, T.; Yang, X.; Hu, S.; Shi, H. Comparisons of methods measuring green quantity. China Acad. J. Electron. Publ. House 2010, 8, 36–38. [Google Scholar]
- Chen, Z. Research on the ecological benefits of urban landscaping in Beijing (2). China Gard. 1998, 14, 51–54. [Google Scholar]
- Zhou, J.H.; Sun, T.Z. Study on remote sensing model of three-dimensional green biomass and the estimation of environmental benefits of greenery. Remote Sens. Environ. China 1995, 3, 162–174. [Google Scholar]
- Song, Z.; Guo, X.; Ma, W. Study on green quantity of green space along road in Beijing plain area. Jilin For. Sci. Technol. 2008, 37, 11–15. [Google Scholar]
- Chen, F.; Zhou, Z.X.; Xiao, R.B.; Wang, P.C.; Li, H.F.; Guo, E.X. Estimation of ecosystem services of urban green-land in industrial areas: A case study on green-land in the workshop area of the Wuhan Iron and Steel Company. Acta Ecol. Sin. 2006, 26, 2230–2236. [Google Scholar]
- Shen, X.y.; Li, Z.d. Review of researches on the leaf area index of landscape plants. Jilin For. Sci. Technol. 2007, 36, 18–22. [Google Scholar]
- Zhou, J.H. Research on the green quantity group of urban living environment (5)—Research on greening 3D volume and its application. China Gard. 1998, 14, 61–63. [Google Scholar]
- Zhou, T.; Luo, H.; Guo, D. Remote sensing image based quantitative study on urban spatial 3D Green Quantity Virescence three dimension quantity. Acta Ecol. Sin. 2005, 25, 415–420. [Google Scholar]
- Zhou, Y.; Zhou, J. Fast method to detect and calculate LVV. Acta Ecol. Sin. Pap. 2006, 26, 4204–4211. [Google Scholar]
- Liu, C.; Li, L.; Zhao, G. Vertical Distribution of Tridimensional Green Biomass in Shenyang Urban Forests. J. Northeast. For. Univ. 2008, 36, 18. [Google Scholar]
- Zheng, G.; Moskal, L.M. Computational-Geometry-Based Retrieval of Effective Leaf Area Index Using Terrestrial Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3958–3969. [Google Scholar] [CrossRef]
- Ma, H.; Song, J.; Wang, J.; Xiao, Z.; Fu, Z. Improvement of spatially continuous forest LAI retrieval by integration of discrete airborne LiDAR and remote sensing multi-angle optical data. Agric. For. Meteorol. 2014, 189–190, 60–70. [Google Scholar] [CrossRef]
- Liu, Q.; Cai, E.; Zhang, J.; Song, Q.; Li, X.; Dou, B. A Modification of the Finite-length Averaging Method in Measuring Leaf Area Index in Field. Chin. Bull. Bot. 2018, 53, 671–685. [Google Scholar] [CrossRef]
- Li, F.; Li, M.; Feng, X.-g. High-Precision Method for Estimating the Three-Dimensional Green Quantity of an Urban Forest. J. Indian Soc. Remote Sens. 2021, 49, 1407–1417. [Google Scholar] [CrossRef]
- Hyyppä, E.; Kukko, A.; Kaijaluoto, R.; White, J.C.; Wulder, M.A.; Pyörälä, J.; Liang, X.; Yu, X.; Wang, Y.; Kaartinen, H. Accurate derivation of stem curve and volume using backpack mobile laser scanning. ISPRS J. Photogramm. Remote Sens. 2020, 161, 246–262. [Google Scholar] [CrossRef]
- Sun, Y.; GU, Z.; Li, D. Study on remote sensing retrieval of leaf area index based on unmanned aerial vehicle and satellite image. Sci. Surv. Mapp. 2021, 46, 106–112. [Google Scholar]
- Zhou, X.; Liao, H.; Cui, Y.; Wang, F. UAV remote sensing estimation of three-dimensional green volume in landscaping: A case study in the Qishang campus of Fuzhou university. J. Fuzhou Univ. 2020, 48, 699–705. [Google Scholar]
- Hong, Z.; Xu, W.; Liu, Y.; Wang, L.; Ou, G.; Lu, N.; Dai, Q. Estimation of the Three-Dimension Green Volume Based on UAV RGB Images: A Case Study in YueYaTan Park in Kunming, China. Forests 2023, 14, 752. [Google Scholar] [CrossRef]
- Zheng, S.; Meng, C.; Xue, J.; Wu, Y.; Liang, J.; Xin, L.; Zhang, L. UAV-based spatial pattern of three-dimensional green volume and its influencing factors in Lingang New City in Shanghai, China. Front. Earth Sci. 2021, 15, 543–552. [Google Scholar] [CrossRef]
- Schumacher, J.; Rattay, M.; Kirchhöfer, M.; Adler, P.; Kändler, G. Combination of Multi-Temporal Sentinel 2 Images and Aerial Image Based Canopy Height Models for Timber Volume Modelling. Forests 2019, 10, 746. [Google Scholar] [CrossRef]
- Silveira, E.M.O.; Radeloff, V.C.; Martinuzzi, S.; Pastur, G.J.M.; Bono, J.; Politi, N.; Lizarraga, L.; Rivera, L.O.; Ciuffoli, L.; Rosas, Y.M. Nationwide native forest structure maps for Argentina based on forest inventory data, SAR Sentinel-1 and vegetation metrics from Sentinel-2 imagery. Remote Sens. Environ. 2023, 285, 113391. [Google Scholar] [CrossRef]
- Kacic, P.; Thonfeld, F.; Gessner, U.; Kuenzer, C. Forest Structure Characterization in Germany: Novel Products and Analysis Based on GEDI, Sentinel-1 and Sentinel-2 Data. Remote Sens. 2023, 15, 1969. [Google Scholar] [CrossRef]
- Zhang, X.; Song, P. Estimating Urban Evapotranspiration at 10m Resolution Using Vegetation Information from Sentinel-2: A Case Study for the Beijing Sponge City. Remote Sens. 2021, 13, 2048. [Google Scholar] [CrossRef]
- Mannschatz, T.; Pflug, B.; Borg, E.; Feger, K.H.; Dietrich, P. Uncertainties of LAI estimation from satellite imaging due to atmospheric correction. Remote Sens. Environ. 2014, 153, 24–39. [Google Scholar] [CrossRef]
- Xie, Q.; Dash, J.; Huete, A.; Jiang, A.; Yin, G.; Ding, Y.; Peng, D.; Hall, C.C.; Brown, L.; Shi, Y.; et al. Retrieval of crop biophysical parameters from Sentinel-2 remote sensing imagery. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 187–195. [Google Scholar] [CrossRef]
- Abdi, A.M. Land cover and land use classification performance of machine learning algorithms in a boreal landscape using Sentinel-2 data. GIScience Remote Sens. 2020, 57, 1–20. [Google Scholar] [CrossRef]
- Meng, B.; Liang, T.; Yi, S.; Yin, J.; Cui, X.; Ge, J.; Hou, M.; Lv, Y.; Sun, Y. Modeling Alpine Grassland Above Ground Biomass Based on Remote Sensing Data and Machine Learning Algorithm: A Case Study in East of the Tibetan Plateau, China. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 2986–2995. [Google Scholar] [CrossRef]
- Kacic, P.; Hirner, A.; Da Ponte, E. Fusing Sentinel-1 and -2 to Model GEDI-Derived Vegetation Structure Characteristics in GEE for the Paraguayan Chaco. Remote Sens. 2021, 13, 5105. [Google Scholar] [CrossRef]
- Aryal, J.; Sitaula, C.; Aryal, S. NDVI Threshold-Based Urban Green Space Mapping from Sentinel-2A at the Local Governmental Area (LGA) Level of Victoria, Australia. Land 2022, 11, 351. [Google Scholar] [CrossRef]
- Hashim, H.; Abd Latif, Z.; Adnan, N.A. Urban vegetation classification with NDVI threshold value method with very high resolution (VHR) Pleiades imagery. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 42, 237–240. [Google Scholar] [CrossRef]
- Karimulla, S.; Ravi Raja, A. Tree Crown Delineation from High Resolution Satellite Images. Indian J. Sci. Technol. 2016, 9, S1. [Google Scholar] [CrossRef]
- Srinivas, C.; Prasad, M.; Sirisha, M. Remote sensing image segmentation using OTSU algorithm. Int. J. Comput. Appl. 2019, 975, 8887. [Google Scholar]
- Feng, Q.; Liu, J.; Gong, J. UAV Remote Sensing for Urban Vegetation Mapping Using Random Forest and Texture Analysis. Remote Sens. 2015, 7, 1074–1094. [Google Scholar] [CrossRef]
- Wang, X.; Wang, M.; Wang, S.; Wu, Y. Extraction of vegetation information from visible unmanned aerial vehicle images. Nongye Gongcheng Xuebao/Trans. Chin. Soc. Agric. Eng. 2015, 31, 152–159. [Google Scholar] [CrossRef]
- Chu, H.; Xiao, Q.; Bai, J. The Retrieval of Leaf Area Index based on Remote Sensing by Unmanned Aerial Vehicle. Remote Sens. Technol. Appl. 2017, 32, 141–147. [Google Scholar]
- Xu, W.; Feng, Z.; Su, Z.; Xu, H.; Jiao, Y.; Fan, J. Development and experiment of handheld digitalized and multi-functional forest measurement gun. Trans. Chin. Soc. Agric. Eng. 2013, 29, 90–99. [Google Scholar]
- Cañete-Salinas, P.; Zamudio, F.; Yáñez, M.; Gajardo, J.; Valdés, H.; Espinosa, C.; Venegas, J.; Retamal, L.; Ortega-Farias, S.; Acevedo-Opazo, C. Evaluation of models to determine LAI on poplar stands using spectral indices from Sentinel-2 satellite images. Ecol. Model. 2020, 428, 109058. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Veroustraete, F.; Muñoz-Marí, J.; Clevers, J.G.P.W.; Camps-Valls, G.; Moreno, J. Experimental Sentinel-2 LAI estimation using parametric, non-parametric and physical retrieval methods—A comparison. ISPRS J. Photogramm. Remote Sens. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, X.; Bajgain, R.; Starks, P.; Steiner, J.; Doughty, R.B.; Chang, Q. Estimating leaf area index and aboveground biomass of grazing pastures using Sentinel-1, Sentinel-2 and Landsat images. ISPRS J. Photogramm. Remote Sens. 2019, 154, 189–201. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, X.; Gao, X.; Gao, j. Estimating average tree height in Xixiaoshan Forest Farm, Northeast China based on Sentinel-1 with Sentinel-2A data. Chin. J. Appl. Ecol. 2021, 32, 2839–2846. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Shataee, S.; Kalbi, S.; Fallah, A.; Pelz, D. Forest attribute imputation using machine-learning methods and ASTER data: Comparison of k-NN, SVR and random forest regression algorithms. Int. J. Remote Sens. 2012, 33, 6254–6280. [Google Scholar] [CrossRef]
- Lyu, X.; Li, X.; Gong, J.; Li, S.; Dou, H.; Dang, D.; Xuan, X.; Wang, H. Remote-sensing inversion method for aboveground biomass of typical steppe in Inner Mongolia, China. Ecol. Indic. 2021, 120, 106883. [Google Scholar] [CrossRef]
- Zhang, R.P.; Zhou, J.H.; Guo, J.; Miao, Y.H.; Zhang, L.L. Inversion models of aboveground grassland biomass in Xinjiang based on multisource data. Front. Plant Sci. 2023, 14, 1152432. [Google Scholar] [CrossRef]
- Zeng, W.; Yang, X.; Chen, X. Comparison on Prediction Precision of One-variable and Two-variable Volume Modelson Tree-leveland Stand-level. Cent. South For. Inventory Plan. 2017, 36, 1–6. [Google Scholar]
- Lin, S.; Zhang, H.; Liu, S.; Gao, G.; Li, L.; Huang, H. Characterizing Post-Fire Forest Structure Recovery in the Great Xing’an Mountain Using GEDI and Time Series Landsat Data. Remote Sens. 2023, 15, 3107. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, W.; Hong, Z.; Wang, L.; Ou, G.; Lu, N.; Dai, Q. Integrating three-dimensional greenness into RSEI improved the scientificity of ecological environment quality assessment for forest. Ecol. Indic. 2023, 156, 111092. [Google Scholar] [CrossRef]
- Potapov, P.; Tyukavina, A.; Turubanova, S.; Talero, Y.; Hernandez-Serna, A.; Hansen, M.C.; Saah, D.; Tenneson, K.; Poortinga, A.; Aekakkararungroj, A.; et al. Annual continuous fields of woody vegetation structure in the Lower Mekong region from 2000–2017 Landsat time-series. Remote Sens. Environ. 2019, 232, 111278. [Google Scholar] [CrossRef]
- Brede, B.; Verrelst, J.; Gastellu-Etchegorry, J.P.; Clevers, J.; Goudzwaard, L.; den Ouden, J.; Verbesselt, J.; Herold, M. Assessment of Workflow Feature Selection on Forest LAI Prediction with Sentinel-2A MSI, Landsat 7 ETM+ and Landsat 8 OLI. Remote Sens. 2020, 12, 915. [Google Scholar] [CrossRef]
- Liu, Z.; Jin, G. Improving accuracy of optical methods in estimating leaf area index through empirical regression models in multiple forest types. Trees 2016, 30, 2101–2115. [Google Scholar] [CrossRef]
- Liu, X.; Su, Y.; Hu, T.; Yang, Q.; Liu, B.; Deng, Y.; Tang, H.; Tang, Z.; Fang, J.; Guo, Q. Neural network guided interpolation for mapping canopy height of China’s forests by integrating GEDI and ICESat-2 data. Remote Sens. Environ. 2022, 269, 112844. [Google Scholar] [CrossRef]
- Fang, H.; Baret, F.; Plummer, S.; Schaepman-Strub, G. An Overview of Global Leaf Area Index (LAI): Methods, Products, Validation, and Applications. Rev. Geophys. 2019, 57, 739–799. [Google Scholar] [CrossRef]
- Xu, C.; Hantson, S.; Holmgren, M.; van Nes, E.H.; Staal, A.; Scheffer, M. Remotely sensed canopy height reveals three pantropical ecosystem states. Ecology 2016, 97, 2518–2521. [Google Scholar] [CrossRef]
- Hill, A.; Breschan, J.; Mandallaz, D. Accuracy Assessment of Timber Volume Maps Using Forest Inventory Data and LiDAR Canopy Height Models. Forests 2014, 5, 2253–2275. [Google Scholar] [CrossRef]
- Tonolli, S.; Dalponte, M.; Neteler, M.; Rodeghiero, M.; Vescovo, L.; Gianelle, D. Fusion of airborne LiDAR and satellite multispectral data for the estimation of timber volume in the Southern Alps. Remote Sens. Environ. 2011, 115, 2486–2498. [Google Scholar] [CrossRef]
- Fang, G.; He, X.; Weng, Y.; Fang, L. Texture Features Derived from Sentinel-2 Vegetation Indices for Estimating and Mapping Forest Growing Stock Volume. Remote Sens. 2023, 15, 2821. [Google Scholar] [CrossRef]
- Li, X.; Tang, L.; Peng, W.; Chen, J. Estimation method of urban green space living vegetation volume based on backpack light detection and ranging. Chin. J. Appl. Ecol. 2021, 33, 2777–2784. [Google Scholar] [CrossRef]
- He, C.; Convertino, M.; Feng, Z.; Zhang, S. Using LiDAR data to measure the 3D green biomass of Beijing urban forest in China. PLoS ONE 2013, 8, e75920. [Google Scholar] [CrossRef] [PubMed]





| Tree Species | Geometrical Morphology | Calculation Formula | Description |
|---|---|---|---|
| Metasequoia glyptostroboides Hu and W. | cone | represents crown diameter and represents crown height. | |
| Salix babylonica L. | ovoid | ||
| Elaeis guineensis Jacq. | |||
| Osmanthus fragrans Makino. | sphere | ||
| Cinnamomum japonicum Sieb. | |||
| Ficus microcarpa L.f. | |||
| Elaeocarpus decipiens Linn. | flabellate | ||
| Cycas revoluta Thunb. |
| Variable | Formula | Explanation | Attribute |
|---|---|---|---|
| VH | Backscatter coefficient of VV (Vertical–Vertical) polarization modes | σ represents the backscatter coefficient after the projection angle correction, sigma represents the radar brightness value. α represents the projection angle. | |
| VV | Backscatter coefficient of VH (Vertical–Horizontal) polarization modes | ||
| VV/VH | |||
| LSWI | Land surface water index | B8 is NIR (Wavelength = 842 nm), B4 is red band (Wavelength = 665 nm), B3 is green band (Wavelength = 560 nm) | |
| EVI | Enhanced vegetation index | ||
| B2 | NIR (Wavelength = 705 nm) | ||
| B6 | NIR (Wavelength = 705 nm) | ||
| B8 | NIR (Wavelength = 842 nm) | ||
| B8A | NIR (Wavelength = 865 nm) | ||
| B11 | SWIR (Wavelength = 1610 nm) |
| Regression Model | Formula | R2 | RMSE (m3/Pixel) | AE (m3/Pixel) | MPE (%) | p-Value | |
|---|---|---|---|---|---|---|---|
| 3DGV models based on LAI | Linear model | 0.61 | 168.89 | 151.73 | 12.45 | <0.05 | |
| Exponential model | 0.67 | 146.17 | 129.94 | 11.43 | <0.05 | ||
| Power model | 0.68 | 144.92 | 126.81 | 11.07 | <0.05 | ||
| Logarithmic model | 0.36 | 313.26 | 276.14 | 24.08 | >0.05 | ||
| Polynomial model | 0.67 | 145.34 | 128.75 | 11.21 | <0.05 | ||
| 3DGV models based on CHM | Linear model | 0.59 | 180.68 | 163.25 | 13.37 | <0.05 | |
| Exponential model | 0.43 | 256.45 | 234.71 | 19.71 | >0.05 | ||
| Power model | 0.49 | 234.46 | 217.04 | 17.82 | >0.05 | ||
| Logarithmic model | 0.4 | 273.75 | 258.31 | 20.65 | >0.05 | ||
| Polynomial model | 0.51 | 234.19 | 206.58 | 17.16 | <0.05 |
| Regression Model | Formula | R2 | RMSE (m3/Pixel) | AE (m3/Pixel) | MPE (%) | p-Value | |
|---|---|---|---|---|---|---|---|
| Bivariate models | Linear model | 0.68 | 142.78 | 127.59 | 10.96 | <0.05 | |
| Exponential model | 0.76 | 126.17 | 109.94 | 8.94 | <0.05 | ||
| Power model | 0.77 | 124.92 | 106.81 | 8.83 | <0.05 | ||
| Logarithmic model | 0.53 | 227.94 | 199.83 | 16.45 | <0.05 | ||
| Polynomial model | 0.77 | 130.94 | 106.71 | 9.19 | <0.05 | ||
| Compound model | 0.78 | 122.36 | 103.98 | 8.71 | <0.05 |
| 3DGV | 0–250 m3/Pixel | 250–500 m3/Pixel | 500–750 m3/Pixel | >750 m3/Pixel | Total |
|---|---|---|---|---|---|
| 0–250 m3/pixel | 3936 | 852 | 163 | 0 | 4951 |
| 250–500 m3/pixel | 972 | 2992 | 110 | 65 | 4139 |
| 500–750 m3/pixel | 132 | 124 | 944 | 123 | 1323 |
| >750 m3/pixel | 17 | 85 | 89 | 385 | 576 |
| Total | 5057 | 4053 | 1306 | 573 | 10,989 |
| PA/% | 79.50 | 72.29 | 71.35 | 67.07 | |
| UA/% | 77.83 | 73.82 | 72.39 | 67.19 | |
| OA/% | 75.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Z.; Xu, W.; Liu, Y.; Wang, L.; Ou, G.; Lu, N.; Dai, Q. Retrieval of Three-Dimensional Green Volume in Urban Green Space from Multi-Source Remote Sensing Data. Remote Sens. 2023, 15, 5364. https://doi.org/10.3390/rs15225364
Hong Z, Xu W, Liu Y, Wang L, Ou G, Lu N, Dai Q. Retrieval of Three-Dimensional Green Volume in Urban Green Space from Multi-Source Remote Sensing Data. Remote Sensing. 2023; 15(22):5364. https://doi.org/10.3390/rs15225364
Chicago/Turabian StyleHong, Zehu, Weiheng Xu, Yun Liu, Leiguang Wang, Guanglong Ou, Ning Lu, and Qinling Dai. 2023. "Retrieval of Three-Dimensional Green Volume in Urban Green Space from Multi-Source Remote Sensing Data" Remote Sensing 15, no. 22: 5364. https://doi.org/10.3390/rs15225364
APA StyleHong, Z., Xu, W., Liu, Y., Wang, L., Ou, G., Lu, N., & Dai, Q. (2023). Retrieval of Three-Dimensional Green Volume in Urban Green Space from Multi-Source Remote Sensing Data. Remote Sensing, 15(22), 5364. https://doi.org/10.3390/rs15225364