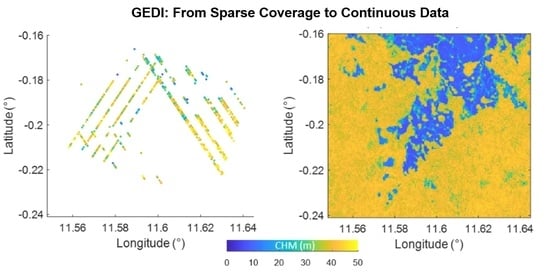
Tropical Forest Top Height by GEDI: From Sparse Coverage to Continuous Data
Abstract
1. Introduction
2. Method
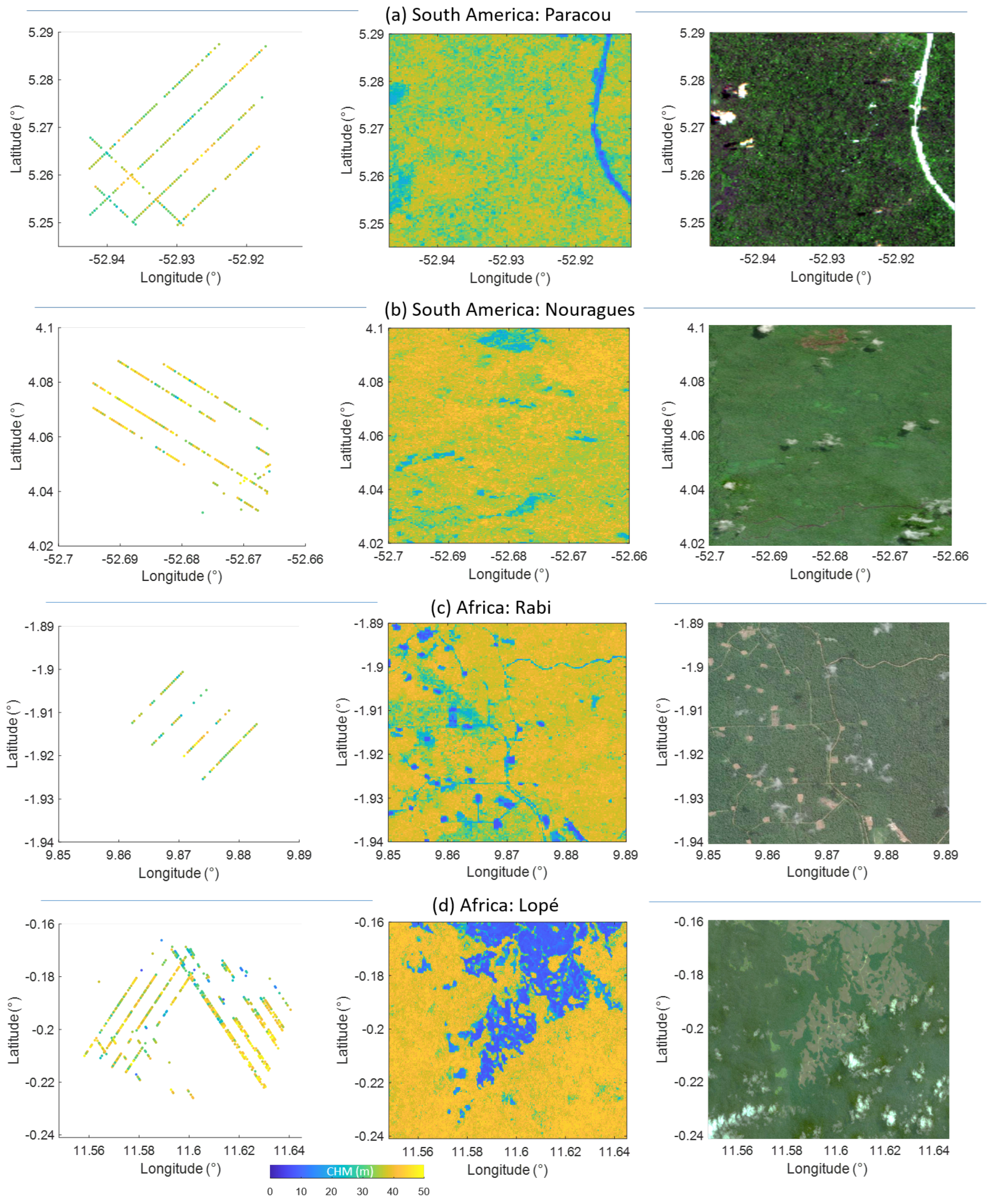
2.1. Study Sites
2.2. GEDI Processing
- Num-detected modes, which shows the detected modes in number.
- Canopy position: longitude, latitude, and elevation of the highest return (EHR).
- Ground position: longitude, latitude, and elevation of the lowest return (ELR).
- Relative height metrics RHn, for which n varies from 0% (lowest detectable return, ground position) to 100% (highest detectable return, canopy top). RHn is the height above the ground position and at a certain n% in the cumulative energy.
- Sensitivity. The shot sensitivity is the probability of a given canopy cover reaching the ground.
- num-detectedmodes = 0. These shot signals can be mostly noisy without any detected modes.
- Shots where the absolute difference between the ELM and the corresponding SRTM DEM is higher than 75 m ().
- Shots where RH98 < 3 m. These shots most likely correspond to bare soil or low vegetation.
2.3. Remote Sensing Image Processing
2.4. Random Forest
3. Results
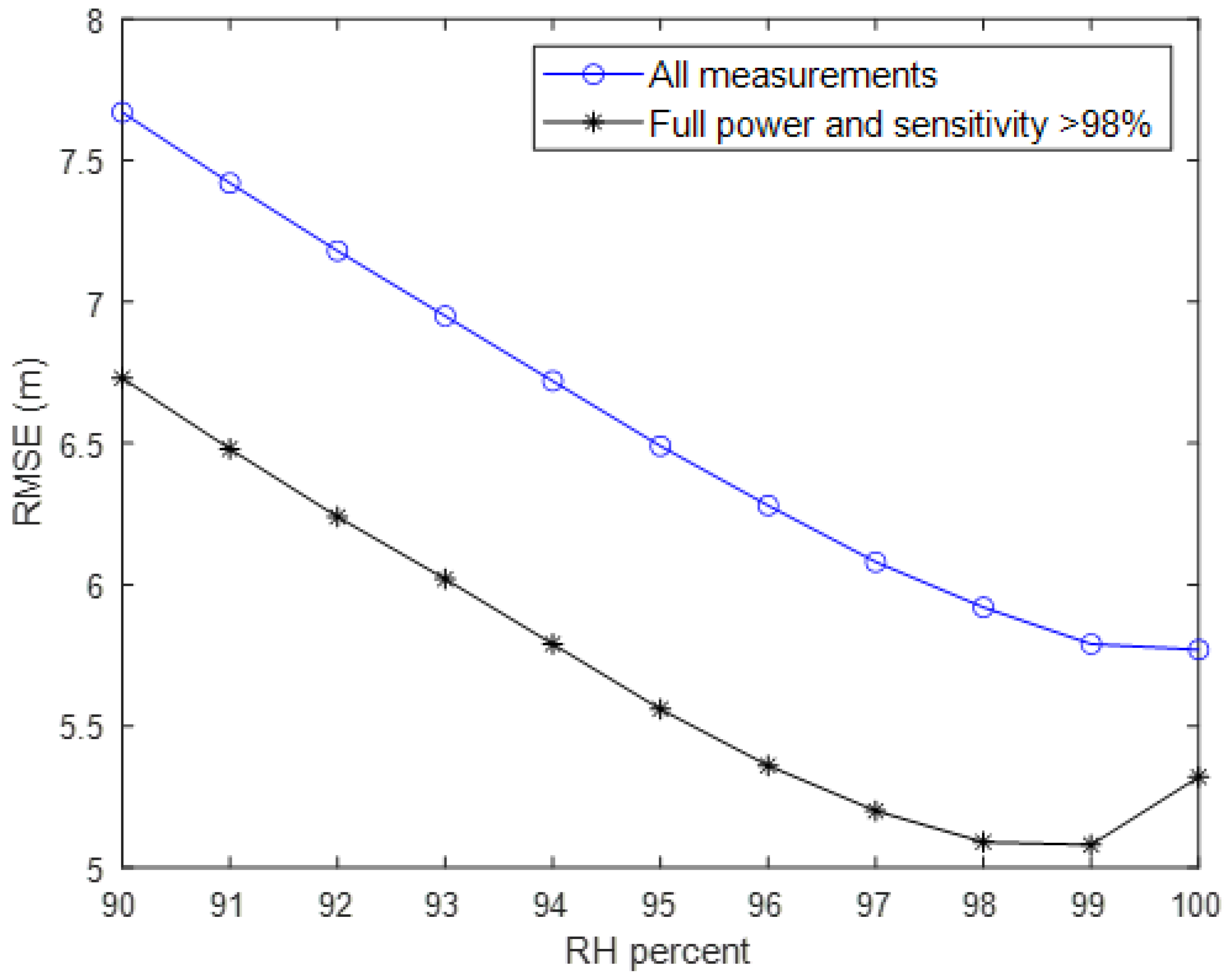
3.1. Select Suitable GEDI RH Metric
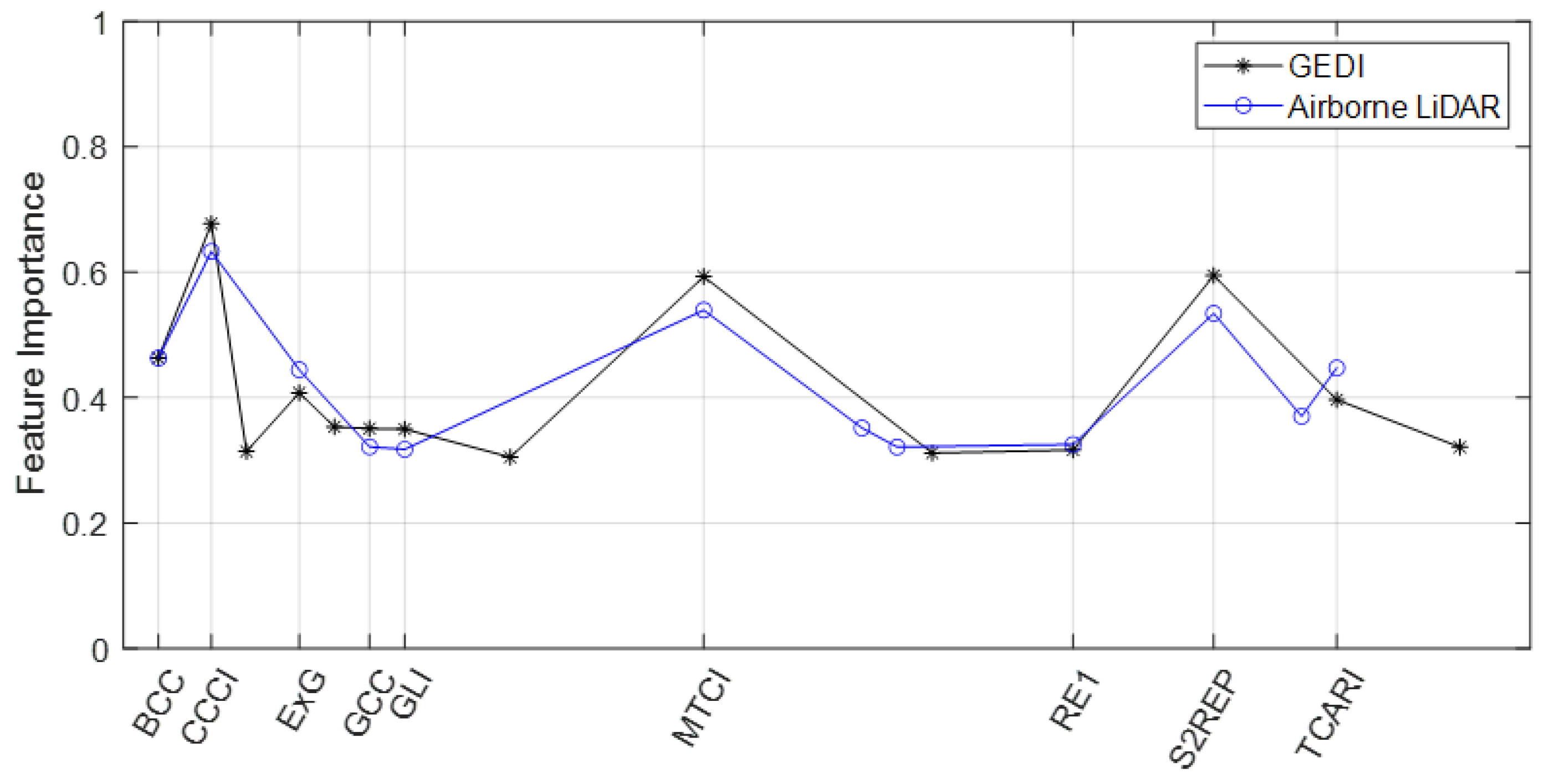
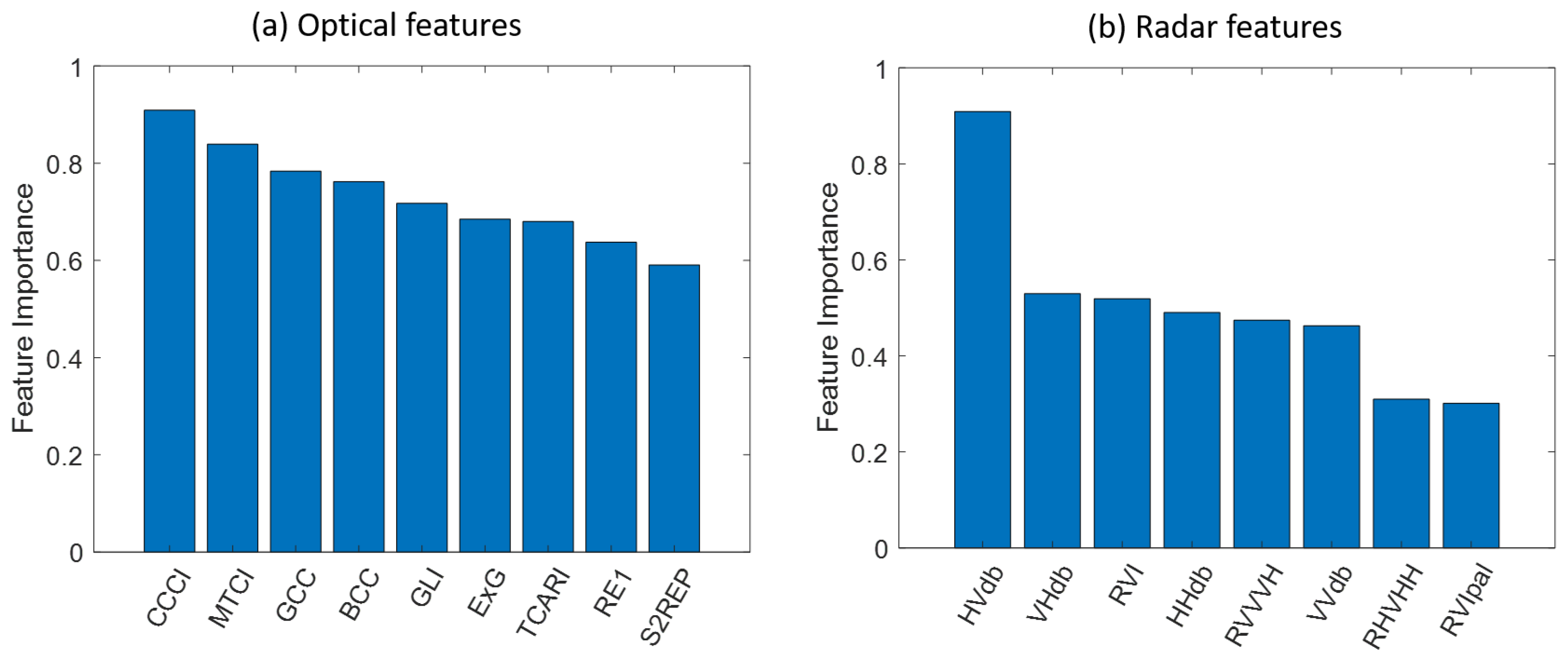
3.2. Feature Selection
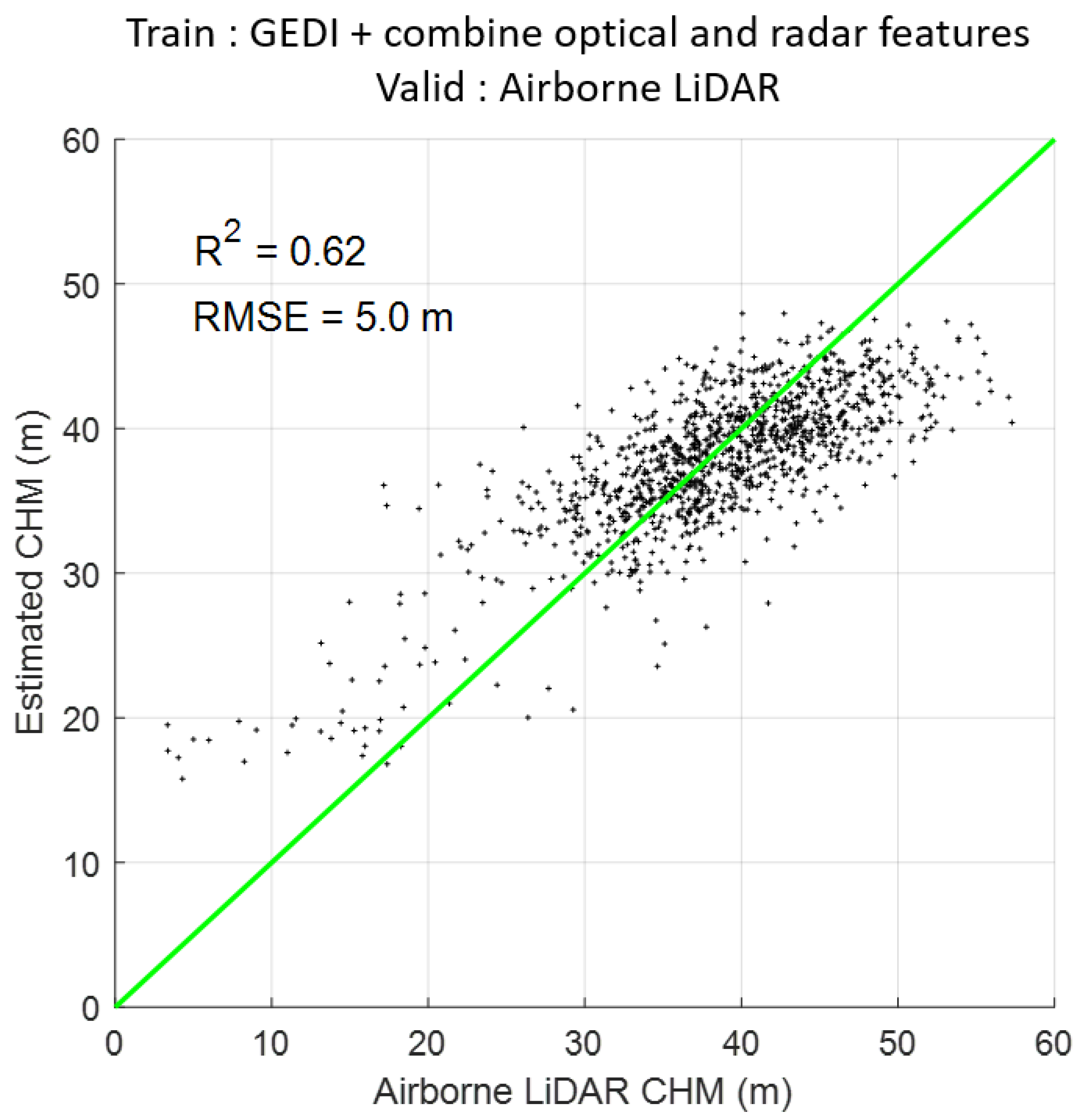
3.3. Combine Optical and Radar Features
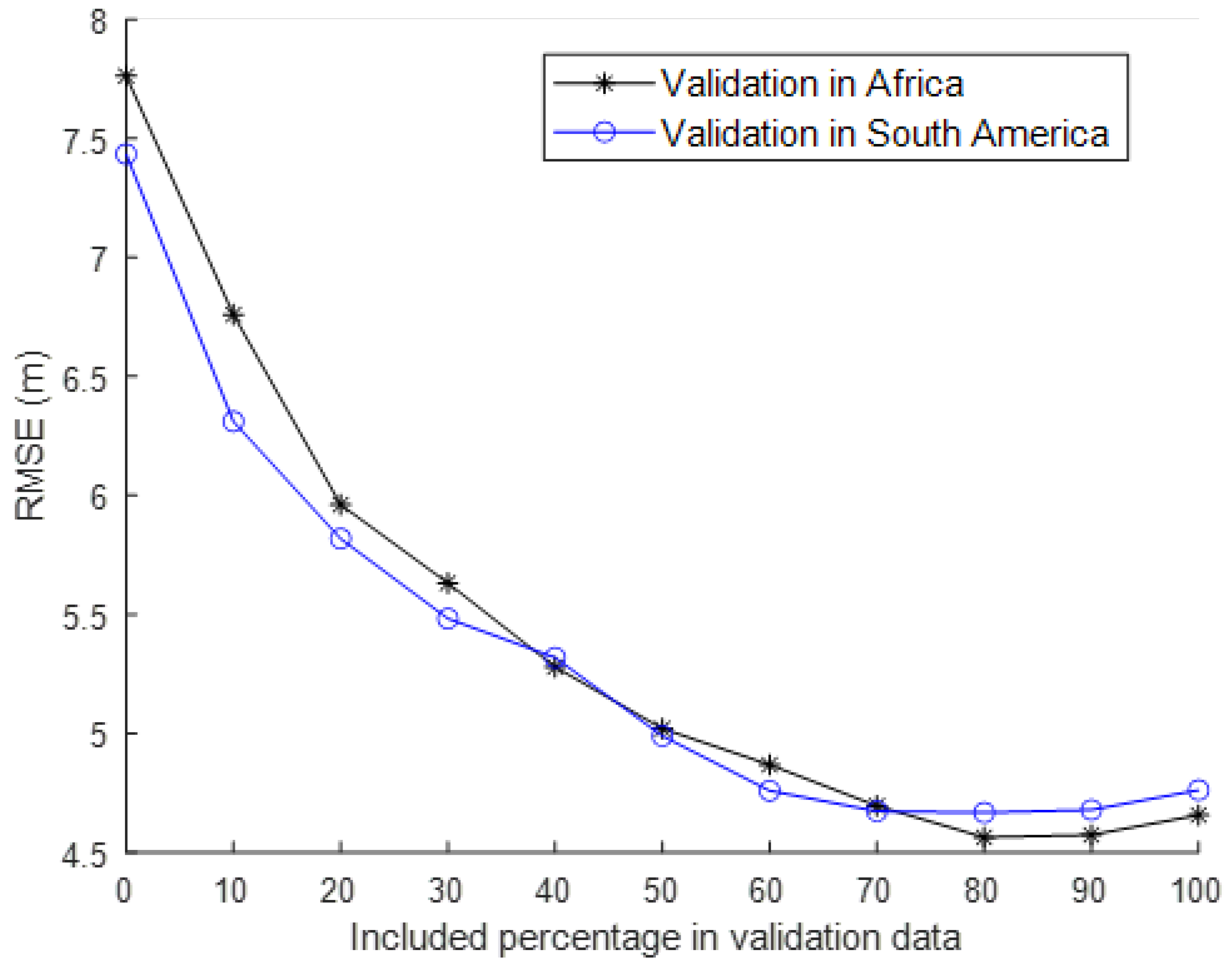
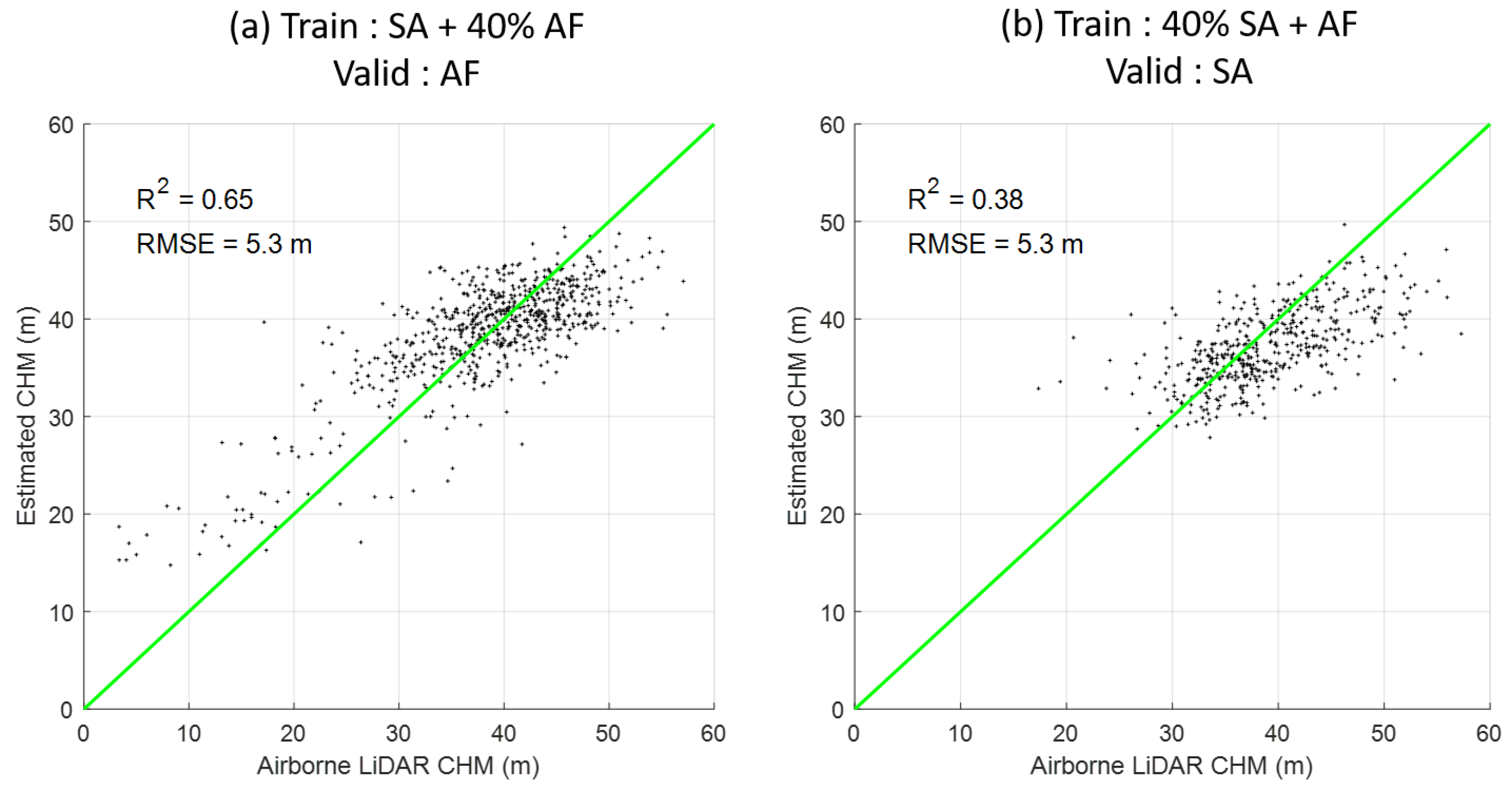
3.4. The Robustness of the Selected Features
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ho Tong Minh, D.; Tebaldini, S.; Rocca, F.; Le Toan, T.; Villard, L.; Dubois-Fernandez, P. Capabilities of BIOMASS Tomography for Investigating Tropical Forests. Geosci. Remote Sens. IEEE Trans. 2015, 53, 965–975. [Google Scholar] [CrossRef]
- Quegan, S.; Le Toan, T.; Chave, J.; Dall, J.; Exbrayat, J.-F.; Tong Minh, D.H.; Lomas, M.; D’Alessandro, M.M.; Paillou, P.; Papathanassiou, K.; et al. The European Space Agency BIOMASS mission: Measuring forest above-ground biomass from space. Remote Sens. Environ. 2019, 227, 44–60. [Google Scholar] [CrossRef]
- Duncanson, L.; Kellner, J.R.; Armston, J.; Dubayah, R.; Minor, D.M.; Hancock, S.; Healey, S.P.; Patterson, P.L.; Saarela, S.; Marselis, S.; et al. Aboveground biomass density models for NASA’s Global Ecosystem Dynamics Investigation (GEDI) lidar mission. Remote Sens. Environ. 2022, 270, 112845. [Google Scholar] [CrossRef]
- Dubayah, R.; Blair, J.B.; Goetz, S.; Fatoyinbo, L.; Hansen, M.; Healey, S.; Hofton, M.; Hurtt, G.; Kellner, J.; Luthcke, S.; et al. The Global Ecosystem Dynamics Investigation: High-resolution laser ranging of the Earth’s forests and topography. Sci. Remote Sens. 2020, 1, 100002. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-Resolution Global Maps of 21st-Century Forest Cover Change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Franklin, S.E.; Guo, X.; Cattet, M. Remote Sensing of Ecology, Biodiversity and Conservation: A Review from the Perspective of Remote Sensing Specialists. Sensors 2010, 10, 9647–9667. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cabello, F.; Montorio, R.; Alves, D.B. Remote sensing techniques to assess post-fire vegetation recovery. Curr. Opin. Environ. Sci. Health 2021, 21, 100251. [Google Scholar] [CrossRef]
- Torres, R.; Snoeij, P.; Geudtner, D.; Bibby, D.; Davidson, M.; Attema, E.; Potin, P.; Rommen, B.; Floury, N.; Brown, M.; et al. GMES Sentinel-1 mission. Remote Sens. Environ. 2012, 120, 9–24. [Google Scholar] [CrossRef]
- Shimada, M.; Itoh, T.; Motooka, T.; Watanabe, M.; Thapa, R. Generation of the first ’PALSAR-2. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016. [Google Scholar]
- Drusch, M.; Bello, U.D.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2 ESA Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Zhu, X.; Dong, Z.; Guo, W. Estimation of biomass in wheat using random forest regression algorithm and remote sensing data. Crop. J. 2016, 4, 212–219. [Google Scholar] [CrossRef]
- Ho Tong Minh, D.; Ndikumana, E.; Vieilledent, G.; McKey, D.; Baghdadi, N. Potential value of combining ALOS PALSAR and Landsat-derived tree cover data for forest biomass retrieval in Madagascar. Remote Sens. Environ. 2018, 213, 206–214. [Google Scholar] [CrossRef]
- Labrière, N.; Tao, S.; Chave, J.; Scipal, K.; Toan, T.L.; Abernethy, K.; Alonso, A.; Barbier, N.; Bissiengou, P.; Casal, T.; et al. In Situ Reference Datasets From the TropiSAR and AfriSAR Campaigns in Support of Upcoming Spaceborne Biomass Missions. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3617–3627. [Google Scholar] [CrossRef]
- Potapov, P.; Li, X.; Hernandez-Serna, A.; Tyukavina, A.; Hansen, M.C.; Kommareddy, A.; Pickens, A.; Turubanova, S.; Tang, H.; Silva, C.E.; et al. Mapping global forest canopy height through integration of GEDI and Landsat data. Remote Sens. Environ. 2021, 253, 112165. [Google Scholar] [CrossRef]
- Kotsiantis, S.B.; Zaharakis, I.D.; Pintelas, P.E. Machine learning: A review of classification and combining techniques. Artif. Intell. Rev. 2006, 26, 159–190. [Google Scholar] [CrossRef]
- Ienco, D.; Interdonato, R.; Gaetano, R.; Ho Tong Minh, D. Combining Sentinel-1 and Sentinel-2 Satellite Image Time Series for land cover mapping via a multi-source deep learning architecture. ISPRS J. Photogramm. Remote Sens. 2019, 158, 11–22. [Google Scholar] [CrossRef]
- Ndikumana, E.; Ho Tong Minh, D.; Dang Nguyen, H.T.; Baghdadi, N.; Courault, D.; Hossard, L.; El Moussawi, I. Estimation of Rice Height and Biomass Using Multitemporal SAR Sentinel-1 for Camargue, Southern France. Remote Sens. 2018, 10, 1394. [Google Scholar] [CrossRef]
- Zhang, L.; Du, B. Deep Learning for Remote Sensing Data: A Technical Tutorial on the State of the Art. IEEE Geosci. Remote Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.C.; Vincent, P. Representation Learning: A Review and New Perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]
- Ho Tong Minh, D.; Ienco, D.; Gaetano, R.; Lalande, N.; Ndikumana, E.; Osman, F.; Maurel, P. Deep Recurrent Neural Networks for Winter Vegetation Quality Mapping via Multitemporal SAR Sentinel-1. IEEE Geosci. Remote Sens. Lett. 2018, 15, 464–468. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Dubois-Fernandez, P.C.; Le Toan, T.; Daniel, S.; Oriot, H.; Chave, J.; Blanc, L.; Villard, L.; Davidson, M.W.J.; Petit, M. The TropiSAR Airborne Campaign in French Guiana: Objectives, Description, and Observed Temporal Behavior of the Backscatter Signal. Geosci. Remote Sens. IEEE Trans. 2012, 8, 3228–3241. [Google Scholar] [CrossRef]
- Pardini, M.; Tello, M.; Cazcarra-Bes, V.; Papathanassiou, K.P.; Hajnsek, I. L- and P-Band 3-D SAR Reflectivity Profiles Versus Lidar Waveforms: The AfriSAR Case. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 3386–3401. [Google Scholar] [CrossRef]
- Fayad, I.; Baghdadi, N.; Lahssini, K. An Assessment of the GEDI Lasers Capabilities in Detecting Canopy Tops and Their Penetration in a Densely Vegetated, Tropical Area. Remote Sens. 2022, 14, 2969. [Google Scholar] [CrossRef]
- Ngo, Y.N.; Huang, Y.; Ho Tong Minh, D.; Ferro-Famil, L.; Fayad, I.; Baghdadi, N. Tropical forest vertical structure characterization: From GEDI to P-band SAR tomography. IEEE Geosci. Remote Sens. Lett. 2022, 19, 7004705. [Google Scholar] [CrossRef]
- Gillespie, A.R.; Kahle, A.B.; Walker, R.E. Color enhancement of highly correlated images. II. Channel ratio and “chromaticity” transformation techniques. Remote Sens. Environ. 1987, 22, 343–365. [Google Scholar] [CrossRef]
- Hancock, D.W.; Dougherty, C.T. Relationships between Blue- and Red-based Vegetation Indices and Leaf Area and Yield of Alfalfa. Crop Sci. 2007, 47, 2547–2556. [Google Scholar] [CrossRef]
- Li, F.; Miao, Y.; Feng, G.; Yuan, F.; Yue, S.; Gao, X.; Liu, Y.; Liu, B.; Ustin, S.L.; Chen, X. Improving estimation of summer maize nitrogen status with red edge-based spectral vegetation indices. Field Crop. Res. 2014, 157, 111–123. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Vincini, M.; Frazzi, E.; D’Alessio, P. A broad-band leaf chlorophyll vegetation index at the canopy scale. Precis. Agric. 2008, 9, 303–319. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Huete, A.; Liu, H.; Batchily, K.; van Leeuwen, W. A comparison of vegetation indices over a global set of TM images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Yang, P.; van der Tol, C.; Campbell, P.K.; Middleton, E.M. Fluorescence Correction Vegetation Index (FCVI): A physically based reflectance index to separate physiological and non-physiological information in far-red sun-induced chlorophyll fluorescence. Remote Sens. Environ. 2020, 240, 111676. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Wang, F.-M.; Huang, J.-F.; Tang, Y.-L.; Wang, X.-Z. New Vegetation Index and Its Application in Estimating Leaf Area Index of Rice. Rice Sci. 2007, 14, 195–203. [Google Scholar] [CrossRef]
- Pinty, B.; Verstraete, M.G. GEMI: A non-linear index to monitor global vegetation from satellites. Vegetatio 1992, 101, 15–20. [Google Scholar] [CrossRef]
- Louhaichi, M.; Borman, M.M.; Johnson, D.E. Spatially Located Platform and Aerial Photography for Documentation of Grazing Impacts on Wheat. Geocarto Int. 2001, 16, 65–70. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Crist, E.P.; Cicone, R.C. A Physically-Based Transformation of Thematic Mapper Data—The TM Tasseled Cap. IEEE Trans. Geosci. Remote Sens. 1984, GE-22, 256–263. [Google Scholar] [CrossRef]
- Ceccato, P.; Gobron, N.; Flasse, S.; Pinty, B.; Tarantola, S. Designing a spectral index to estimate vegetation water content from remote sensing data: Part 1: Theoretical approach. Remote Sens. Environ. 2002, 82, 188–197. [Google Scholar] [CrossRef]
- Crippen, R.E. Calculating the vegetation index faster. Remote Sens. Environ. 1990, 34, 71–73. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. Isprs J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef]
- Jurgens, C. The modified normalized difference vegetation index (mNDVI) a new index to determine frost damages in agriculture based on Landsat TM data. Int. J. Remote Sens. 1997, 18, 3583–3594. [Google Scholar] [CrossRef]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A.R. A review of vegetation indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.; Kerr, Y.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of Vegetation Indices and a Modified Simple Ratio for Boreal Applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Wilson, E.H.; Sader, S.A. Detection of forest harvest type using multiple dates of Landsat TM imagery. Remote Sens. Environ. 2002, 80, 385–396. [Google Scholar] [CrossRef]
- Yang, W.; Kobayashi, H.; Wang, C.; Shen, M.; Chen, J.; Matsushita, B.; Tang, Y.; Kim, Y.; Bret-Harte, M.S.; Zona, D.; et al. A semi-analytical snow-free vegetation index for improving estimation of plant phenology in tundra and grassland ecosystems. Remote Sens. Environ. 2019, 228, 31–44. [Google Scholar] [CrossRef]
- Sulik, J.J.; Long, D.S. Spectral considerations for modeling yield of canola. Remote Sens. Environ. 2016, 184, 161–174. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Badgley, G.; Field, C.B.; Berry, J.A. Canopy near-infrared reflectance and terrestrial photosynthesis. Sci. Adv. 2017, 3, e1602244. [Google Scholar] [CrossRef] [PubMed]
- Goel, N.S.; Qin, W. Influences of canopy architecture on relationships between various vegetation indices and LAI and Fpar: A computer simulation. Remote Sens. Rev. 1994, 10, 309–347. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Zheng, Q.; Huang, W.; Cui, X.; Shi, Y.; Liu, L. New Spectral Index for Detecting Wheat Yellow Rust Using Sentinel-2 Multispectral Imagery. Sensors 2018, 18, 868. [Google Scholar] [CrossRef]
- Kim, Y.; van Zyl, J.J. A Time-Series Approach to Estimate Soil Moisture Using Polarimetric Radar Data. IEEE Trans. Geosci. Remote Sens. 2009, 47, 2519–2527. [Google Scholar] [CrossRef]
- Huete, A. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Pasqualotto, N.; Delegido, J.; Van Wittenberghe, S.; Rinaldi, M.; Moreno, J. Multi-Crop Green LAI Estimation with a New Simple Sentinel-2 LAI Index (SeLI). Sensors 2019, 19, 904. [Google Scholar] [CrossRef]
- Birth, G.S.; McVey, G.R. Measuring the Color of Growing Turf with a Reflectance Spectrophotometer. Agron. J. 1968, 60, 640–643. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Haboudane, D.; Tremblay, N.; Miller, J.R.; Vigneault, P. Remote Estimation of Crop Chlorophyll Content Using Spectral Indices Derived From Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2008, 46, 423–437. [Google Scholar] [CrossRef]
- Bannari, A.; Asalhi, H.; Teillet, P. Transformed difference vegetation index (TDVI) for vegetation cover mapping. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Toronto, ON, Canada, 24–28 June 2002; Volume 5, pp. 3053–3055. [Google Scholar] [CrossRef]
- Blanco, V.; Blaya-Ros, P.J.; Castillo, C.; Soto-Vallés, F.; Torres-Sánchez, R.; Domingo, R. Potential of UAS-Based Remote Sensing for Estimating Tree Water Status and Yield in Sweet Cherry Trees. Remote Sens. 2020, 12, 2359. [Google Scholar] [CrossRef]
- Xing, N.; Huang, W.; Xie, Q.; Shi, Y.; Ye, H.; Dong, Y.; Wu, M.; Sun, G.; Jiao, Q. A Transformed Triangular Vegetation Index for Estimating Winter Wheat Leaf Area Index. Remote Sens. 2020, 12, 16. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Milenković, M.; Reiche, J.; Armston, J.; Neuenschwander, A.; De Keersmaecker, W.; Herold, M.; Verbesselt, J. Assessing Amazon rainforest regrowth with GEDI and ICESat-2 data. Sci. Remote Sens. 2022, 5, 100051. [Google Scholar] [CrossRef]
- Lang, N.; Jetz, W.; Schindler, K.; Wegner, J.D. A high-resolution canopy height model of the Earth. arXiv 2022, arXiv:2204.08322. [Google Scholar]
- Ho Tong Minh, D.; Le Toan, T.; Rocca, F.; Tebaldini, S.; d’Alessandro, M.M.; Villard, L. Relating P-band Synthetic Aperture Radar Tomography to Tropical Forest Biomass. IEEE Trans. Geosci. Remote Sens. 2014, 52, 967–979. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Mermoz, S.; Réjou-Méchain, M.; Villard, L.; Toan, T.L.; Rossi, V.; Gourlet-Fleury, S. Decrease of L-band SAR backscatter with biomass of dense forests. Remote Sens. Environ. 2015, 159, 307–317. [Google Scholar] [CrossRef]
- Dietterich, T.G. Ensemble Methods in Machine Learning. In Proceedings of the Multiple Classifier Systems, First International Workshop, MCS 2000, Cagliari, Italy, 21–23 June 2000; Springer: Berlin/Heidelberg, Germany, 2000; pp. 1–15. [Google Scholar]
- Zawadzki, J.; Cieszewski, C.J.; Zasada, M.; Lowe, R.C. Applying geostatistics for investigations of forest ecosystems using remote sensing imagery. Silva Fenn. 2005, 39, 599. [Google Scholar] [CrossRef]
- Lang, N.; Kalischek, N.; Armston, J.; Schindler, K.; Dubayah, R.; Wegner, J.D. Global canopy height regression and uncertainty estimation from GEDI LIDAR waveforms with deep ensembles. Remote Sens. Environ. 2022, 268, 112760. [Google Scholar] [CrossRef]



| ID | Indices | Description | Formulation |
|---|---|---|---|
| (1) | Green | Green | G |
| (2) | BCC [26] | Blue Chromatic Coordinate | B/(R + G + B) |
| (3) | Blue | Blue | B |
| (4) | BNDVI [27] | Blue Normalized Difference Vegetation Index | (N − B)/(N + B) |
| (5) | CCCI [28] | Canopy Chlorophyll Content Index | ((N − RE1)/(N + RE1))/(( N − R)/(N + R)) |
| (6) | CLGREEN [29] | Chlorophyll Index Green | (N/G) − 1.0 |
| (7) | CVI [30] | Chlorophyll Vegetation Index | (N/G) × (R/G) |
| (8) | DVI [31] | Difference Vegetation Index | N − R |
| (9) | EVI [32] | Enhanced Vegetation Index | g × (N − R)/(N + C1 × R − C2 × B + L) |
| (10) | ExG [33] | Excess Green Index | 2 × G − (R + B) |
| (11) | FCVI [34] | Fluorescence Correction Vegetation Index | N − ((R + G + B)/3.0) |
| (12) | GARI [35] | Green Atmospherically Resistant Vegetation Index | (N − (G − (B − R)))/(N − (G + (B − R))) |
| (13) | GBNDVI [36] | Green-Blue Normalized Difference Vegetation Index | (N − (G + B))/(N + (G + B)) |
| (14) | GCC [26] | Green Chromatic Coordinate | G/(R + G + B) |
| (15) | GEMI [37] | Global Environment Monitoring Index | ((2.0 × ((N ** 2.0) − (R ** 2.0)) + 1.5 × N + 0.5 × R)/ (N + R + 0.5)) × (1.0 − 0.25 × ((2.0 × ((N ** 2.0) − (R ** 2)) + 1.5 × N + 0.5 × R)/(N + R + 0.5))) − ((R − 0.125)/(1 − R)) |
| (16) | GLI [38] | Green Leaf Index | (2.0 × G − R − B)/(2.0 × G + R + B) |
| (17) | GNDVI [35] | Green Normalized Difference Vegetation Index | (N − G)/(N + G) |
| (18) | GRNDVI [36] | Green-Red Normalized Difference Vegetation Index | (N − (G + R))/(N + (G + R)) |
| (19) | GRVI [39] | Green Ratio Vegetation Index | N/G |
| (20) | GSAVI [40] | Green Soil-Adjusted Vegetation Index | (N − G)/((N + G + 0.5) × (1 + 0.5)) |
| (21) | GVI [40] | Green Vegetation Index | (−0.290 × G − 0.562 × R + 0.600 × RE1 + 0.491 × N) |
| (22) | GVMI [41] | Global Vegetation Moisture Index | ((N + 0.1) − (S2 + 0.02))/((N + 0.1) + (S2 + 0.02)) |
| (23) | lHHdb | PALSAR-2 HH | HH |
| (24) | HVdb | PALSAR-2 HV | HV |
| (25) | IPVI [42] | Infrared Percentage Vegetation Index | ((N/(N + R))/2) × ((N − R)/(N + R) + 1) |
| (26) | IRECI [43] | Inverted Red-Edge Chlorophyll Index | (RE3 − R)/(RE1/RE2) |
| (27) | MCARI [43] | Modified Chlorophyll Absorption in Reflectance Index | ((RE1 − R) − 0.2 × (RE1 − G)) × (RE1/R) |
| (28) | MNDVI [44] | Modified Normalized Difference Vegetation Index | (N − S2)/(N + S2) |
| (29) | MNSI [45] | Misra Non Such Index | −0.404 × G − 0.039 × R − 0.505 × RE1 + 0.762 × N |
| (30) | MSAVI [46] | Modified Soil-Adjusted Vegetation Index | 0.5 × (2.0 × N + 1 − (((2 × N + 1) ** 2) − 8 × (N − R)) ** 0.5) |
| (31) | MSBI [45] | Misra Soil Brightness Index | 0.406 × G + 0.600 × R + 0.645 × RE1 + 0.243 × N |
| (32) | MSR [47] | Modified Simple Ratio | (N/R − 1)/((N/R + 1) ** 0.5) |
| (33) | MTCI [43] | MERIS Terrestrial Chlorophyll Index | (RE2 − RE1)/(RE1 − R) |
| (34) | MTVI2 [48] | Modified Triangular Vegetation Index 2 | (1.5 × (1.2 × (N − G) − 2.5 × (R − G)))/((((2.0 × N + 1) ** 2) − (6.0 × N − 5 × (R ** 0.5)) − 0.5) ** 0.5) |
| (35) | MYVI [45] | Misra Yellow Vegetation Index | −0.723 × G − 0.597 × R + 0.206 × RE1 − 0.278 × N |
| (36) | NBR [49] | Normalized Blue Red | (N − S2)/(N + S2) |
| (37) | NDGI [50] | Normalized Difference Greenness Index | (G − R)/(G + R) |
| (38) | NDMI [49] | Normalized Difference Moisture Index | (N − S1)/(N + S1) |
| (39) | NDVI [42] | Normalized Difference Vegetation Index | (N − R)/(N + R) |
| (40) | NDWI [49] | Normalized Difference Water Index | (N − S2)/(N + S2) |
| (41) | NDYI [51] | Normalized Difference Yellowness Index | (G − B)/(G + B) |
| (42) | NGRDI [52] | Normalized Green Red Difference Index | (G − B)/(G + B) |
| (43) | NIR | NIR | N |
| (44) | NRS1 | NIR/SWIR1 | N/S1 |
| (45) | NIRv [53] | Near-Infrared Reflectance of Vegetation | ((N − R)/(N + R)) × N |
| (46) | NLI [54] | Non-Linear Vegetation Index | ((N ** 2) − R)/((N ** 2) + R) |
| (47) | OSAVI [55] | Optimized Soil-Adjusted Vegetation Index | (1.16) × (N − R)/(N + R + 0.16) |
| (48) | PNDVI [11] | Pan NDVI | (N − (B + G + R))/(N + (B + G + R)) |
| (49) | PSRI [56] | Plant Senescence Reflectance Index | (R − B)/RE2 |
| (50) | RHVHH | PALSAR-2 HV/HH | HV/HH |
| (51) | RVVVH | Sentinel-1 VV/VH | VV/VH |
| (52) | RCC [26] | Red Chromatic Coordinate | R/(R + G + B) |
| (53) | RDVI [57] | Renormalized Difference Vegetation Index | (N − R)/((N + R) ** 0.5) |
| (54) | RE1 | Red Edge 1 | RE1 |
| (55) | RE2 | Red Edge 2 | RE2 |
| (56) | RE3 | Red Edge 3 | RE3 |
| (57) | RE4 | Red Edge 4 | RE4 |
| (58) | Red | Red | R |
| (59) | REDSI [58] | Red-Edge Disease Stress Index | ((705.0 − 665.0) × (RE3 − R) − (783.0 − 665.0) × (RE1 − R))/ (2.0 × R) |
| (60) | RVI [59] | Radar Vegetation Index Sentinel-1 | (4 × VHdb)/(VVdb + VHdb) |
| (61) | RVIpal [59] | Radar Vegetation Index PALSAR-2 | (4 × HVdb)/(HHdb + HVdb) |
| (62) | S2REP [43] | Sentinel-2 Red-Edge Position | 705.0 + 35.0 × ((((RE3 + R)/2.0) − RE1)/(RE2 − RE1)) |
| (63) | SAVI [60] | Soil-Adjusted Vegetation Index | (1.0 + L) × (N − R)/(N + R + L) |
| (64) | SeLI [61] | Sentinel-2 LAI Green Index | (RE4 − RE1)/(RE4 + RE1) |
| (65) | SR [62] | Simple Ratio | N/R |
| (66) | SWIR1 | SWIR1 | S1 |
| (67) | S1RS2 | SWIR1/SWIR2 | S1/S2 |
| (68) | SWIR2 | SWIR2 | S2 |
| (69) | TCARI [63] | Transformed Chlorophyll Absorption in Reflectance Index | 3 × ((RE1 − R) − 0.2 × (RE1 − G) × (RE1/R)) |
| (70) | TCI [64] | Triangular Chlorophyll Index | 1.2 × (RE1 − G) − 1.5 × (R − G) × (RE1/R) ** 0.5 |
| (71) | TDVI [65] | Trasformed NDVI | 1.5 × ((N)/((N ** 2 + R + 0.5) ** 0.5)) |
| (72) | TRRVI [66] | Transformed Red Range Vegetation Index | ((RE2 − R)/(RE2 + R))/(((N − R)/(N + R)) + 1.0) |
| (73) | TTVI [67] | Transformed Triangular Vegetation Index | 0.5 × ((865.0 − 740.0) × (RE3 − RE2) − (RE4 − RE2) × (783.0 − 740)) |
| (74) | VARI [39] | Visible Atmospherically Resistant Index | (RE1 − 1.7 × R + 0.7 × B)/(RE1 + 1.3 × R − 1.3 × B) |
| (75) | VHdb | Sentinel-1 VH | VH |
| (76) | VIG [39] | Vegetation Index Green | (G − R)/(G + R) |
| (77) | VVdb | Sentinel-1 VV | VV |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngo, Y.-N.; Ho Tong Minh, D.; Baghdadi, N.; Fayad, I. Tropical Forest Top Height by GEDI: From Sparse Coverage to Continuous Data. Remote Sens. 2023, 15, 975. https://doi.org/10.3390/rs15040975
Ngo Y-N, Ho Tong Minh D, Baghdadi N, Fayad I. Tropical Forest Top Height by GEDI: From Sparse Coverage to Continuous Data. Remote Sensing. 2023; 15(4):975. https://doi.org/10.3390/rs15040975
Chicago/Turabian StyleNgo, Yen-Nhi, Dinh Ho Tong Minh, Nicolas Baghdadi, and Ibrahim Fayad. 2023. "Tropical Forest Top Height by GEDI: From Sparse Coverage to Continuous Data" Remote Sensing 15, no. 4: 975. https://doi.org/10.3390/rs15040975
APA StyleNgo, Y.-N., Ho Tong Minh, D., Baghdadi, N., & Fayad, I. (2023). Tropical Forest Top Height by GEDI: From Sparse Coverage to Continuous Data. Remote Sensing, 15(4), 975. https://doi.org/10.3390/rs15040975